Abstract
Phage-mediated lysis has been an essential laboratory tool for rapidly identifying Bacillus anthracis for more than 40 years, relying on the γ phage derivative of a Bacillus cereus prophage called W. The complete genomic sequences of the temperate W phage, referred to as Wβ, and its lytic variant γ were determined and found to encode 53 open reading frames each, spanning 40,864 bp and 37,373 bp, respectively. Direct comparison of the genomes showed that γ evolved through mutations at key loci controlling host recognition, lysogenic growth, and possibly host phenotypic modification. Included are a cluster of point mutations at the gp14 tail fiber locus of γ, encoding a protein that, when fused to green fluorescent protein, binds specifically to B. anthracis. A large 2,003-bp deletion was also identified at the γ lysogeny module, explaining its shift from a temperate to a lytic lifestyle. Finally, evidence of recombination was observed at a dicistronic Wβ locus, encoding putative bacterial cell surface-modifying proteins, replaced in γ with a locus, likely obtained from a B. anthracis prophage, encoding demonstrable fosfomycin resistance. Reverse transcriptase PCR analysis confirmed strong induction at the dicistronic Wβ locus and at four other phage loci in B. anthracis and/or B. cereus lysogens. In all, this study represents the first genomic and functional description of two historically important phages and is part of a broader investigation into contributions of phage to the B. anthracis life cycle. Initial findings suggest that lysogeny of B. anthracis promotes ecological adaptation, rather than virulence, as with other gram-positive pathogens.
In 1898, Nikolay Gamaleya, a famous Russian microbiologist, reported on a transmissible lytic agent specifically active against “the malignant anthrax bacteria” Bacillus anthracis (7, 27). This observation of bacteriolysis described the activity of a bacterial virus, or bacteriophage, and predated the acknowledged discoveries of phages in 1915 and 1917, respectively, by Frederick Twort (64) and Felix d'Herelle (22). More-detailed descriptions of bacteriophages infecting B. anthracis were reported in 1931 by Cowles (18) and in 1951 by McCloy (46). Specifically, McCloy identified an “atypical” Bacillus cereus soil strain (now designated ATCC 11950) with an inducible prophage, called W, capable of infecting many B. anthracis isolates. A lytic variant of W, called γ, was ultimately isolated by Brown and Cherry in 1955 (11) which more efficiently infected encapsulated bacilli and became a tool for rapid clinical diagnosis of B. anthracis (1). To this day, the γ phage lysis assay remains a standard protocol for B. anthracis identification used by the Centers for Disease Control and Prevention, as well as veterinary and public health laboratories throughout the United States. As a result of the widespread use of γ and the genetic instability inherent to most viral forms, there are now several very similar, yet genetically distinct, γ phage types, including isolates LSU (γL), USAMRIID (γU), and Cherry (γC) [(represented by GenBank accession numbers DQ222853, DQ222855, and DQ222851, respectively, and described elsewhere by Fouts et al. [26]), as well as isolate d'Herelle (this study). The differences that define these phages have not been characterized previously.
More-recent reports of B. anthracis phages present a morphologically and genetically diverse group, including members of the families Siphoviridae, Podoviridae, Myoviridae, and Tectiviridae, with various degrees of infective specificity for different B. anthracis isolates and certain highly related B. cereus strains (2, 34, 49, 51, 66). This variation is not surprising in light of our increased recognition of the staggering diversity of bacteriophage types in the environment, with an estimated ≥1031 phages in the biosphere composed of roughly 108 distinct species (16, 54). With this understanding has come a “new age of phage research,” focusing on topics ranging from genomics and ecology to the impact of phages on bacterial community structure and the global cycling of organic matter (16, 68). Of particular interest is the role of phage in generating prokaryotic genetic variability, including a process called lysogenic conversion, whereby bacterial virulence and/or fitness traits are spread horizontally by phage vectors through bacterial populations (12). Well-studied examples of this include the acquisition of phage-encoded toxins, enzymes, mitogens, and colonization factors by emerging or reemerging human pathogens. Presumably, this phenomenon also drives ecological functioning and evolutionary change in any environmental niche, as evidenced by a recent genomic analysis of cyanophage showing the presence of phage genes involved in host photosynthetic activity and carbon-phosphate utilization pathways that likely contribute to cyanobacterial adaptations to low-nutrient oceanic environments (60). Considering the ubiquity of phage in the biosphere and the enormous possibilities for DNA transfer (for example, an estimated 2 ×1016 phage-mediated DNA transfer events occur per second in the ocean [14] and perhaps in the soil), phage-mediated gene flow is likely a dynamic process throughout the biosphere, shaping microbial genomes and driving bacterial adaptation and speciation.
B. anthracis is a single, genetically homogeneous group within the otherwise very heterogeneous B. cereus lineage of organisms that predominantly includes B. cereus and Bacillus thuringiensis strains (37, 40, 50). The emergence of B. anthracis as an obligate animal pathogen from this lineage (otherwise including soil saprophytes and insect symbionts and pathogens) is clearly associated with its acquisition of two large virulence plasmids encoding anti-host toxins and a capsular structure (48); however, it is also notable that B. anthracis is distinguished from its closest relatives by, among other things, the presence of integrated viral forms or prophage (24, 50). Contributions of these prophages, or indeed any viral form, to the B. anthracis phenotype are as yet unknown.
As part of the overall goal of discerning the influence of phage-mediated processes on B. anthracis, we initiated study of two historically important phages, Wβ and γ (isolate d'Herelle, herein designated simply γ). First, we report on the basic ultrastructural characteristics and nucleotide sequences of these phages. In a comparative analysis of the two genomes, we found that the γ variant likely evolved from a parental Wβ phage in a process involving small and large deletions, point mutations, and homologous recombination with the B. anthracis genome. Additionally, functional assays were used to identify two phage proteins required for antibiotic resistance and host cell surface binding. Finally, we identified several phage loci including genes encoding a putative lipoprotein, sigma factor, and cell surface carbohydrate-modifying enzyme, expressed in Wβ lysogens that may act in a process of lysogenic conversion. The work presented here is the first detailed genomic and functional analysis of any phage infecting B. anthracis.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
B. anthracis ΔSterne is a pXO1− derivative of the Sterne strain (36) that produces no capsule or toxins. B. cereus strains RSVF1 (ATCC 4342), ATCC 14579, ATCC 10987, and ATCC 11950 were obtained from the American Type Culture Collection (Manassas, VA). Strains CDC 13100, CDC 13140, and CDC 32805 are atypical B. cereus isolates obtained from the Centers for Disease Control and Prevention. B. thuringiensis strains BGSC 4AA1, BGSC 4AK1, BGSC 4AY1, BGSC 4AG1, BGSC 4BR1, and BGSC 4BU1 and B. cereus strains UM101, DP-B5184, and DP-B5185 were obtained from Richard Calendar. All other B. cereus and B. thuringiensis strains used in this work were exactly as described in a previous phage host range study (55). ΔSterne::φWβ and RSVF1::φWβ are stable lysogens obtained by infection with Wβ as follows. Five milliliters of a mid-log-phase brain heart infusion (BHI) broth culture was infected at a multiplicity of ∼1, incubated for 24 h at 30°C with agitation, and plated on BHI agar, and individual colonies were screened by PCR with Wβ-specific primers to identify lysogens. The Bacillus strains were usually grown on BHI broth and agar plates. For sporulation, Leighton-Doi (LD) liquid sporulation medium was used.
Escherichia coli strain XL1-Blue (Stratagene) was used to propagate pDG148 (59) and pDG148::gp41, and transformants were selected on Luria broth (LB) agar supplemented with 100 μg of ampicillin per ml. Multicopy plasmid pDG148 is an E. coli-Bacillus subtilis shuttle vector (also replicating in B. cereus) with an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible Pspac promoter. For introduction into B. cereus RSVF1, electrocompetent cells were prepared as previously described (9) and transformants were isolated on BHI agar with 5 μg of kanamycin per ml.
Bacteriophage isolation and propagation.
Phage γ was obtained from Hans Ackermann at the Felix d'Herelle Reference Centre for Bacterial Viruses (Laval University, Quebec, Canada). Phage Wβ was induced from its prophage state by plating B. cereus strain ATCC 11950 on BHI agar plates containing 20 μg of fosfomycin (Sigma-Aldrich) per ml. Ring-shaped colonies formed in the presence of fosfomycin were picked at central positions with micropipette tips and streaked onto a soft BHI agar overlay plate (0.75% agar) containing a fresh RSVF1 lawn. After incubation for 24 h at 30°C, the agar overlay was recovered in 3 ml of 5 mM CaCl2 and pelleted by centrifugation, and the phage-bearing supernatant was recovered.
The γ and Wβ phages were propagated on B. cereus RSVF1 in BHI broth. Briefly, a phage preparation was added to an equivalent volume of mid-log-phase bacteria and incubated at 30°C with agitation for 24 h. Phage supernatants were recovered, and this process was repeated until a sufficiently high-titer phage stock was obtained (>109/ml). All phage preparations were filter sterilized prior to use.
A large-plaque Wβ variant was identified by infecting B. anthracis at a multiplicity of infection of 1 and visually observing subsequent plaque fields on BHI soft-agar overlays. After 16 h at 30°C, rare 2-mm round plaques were observed amid predominantly 1-mm plaques. The large plaques were picked, subcultured on B. anthracis overlay plates, and used as the template in PCRs with primers flanking wp14. The resulting product was sequenced and translated to identify the Sp14 tail fiber.
Bacteriophage host range analysis.
Only high-titer phage stocks were used to determine host range. For each strain, 0.1-ml aliquots of an overnight BHI broth culture were added separately to 0.1 ml of undiluted phage and each of three 100-fold serial dilutions, in four sterile, 10-ml, round-bottom polypropylene tubes. After incubation at 37°C for 15 min, 3 ml of soft BHI agar was added to each tube, gently mixed by inversion, and poured over the surface of a prewarmed BHI agar plate. Plates were incubated for 24 h at 30°C, and plaques were enumerated to determine the number of PFU per milliliter (PFU). Averages were determined on the basis of the results of three different infections.
Fluorescence studies.
Mid-log-phase samples in 5 ml of BHI broth were washed and resuspended in 1 ml of 0.1 M phosphate-buffered saline (PBS) containing 3% formalin. Cells were then incubated for 15 min at 24°C, washed three times, and resuspended in 1 ml of PBS. A 200-μl aliquot of the green fluorescent protein (GFP)-Gp14 extract was added, and the mixture was incubated for 1 min at room temperature, after which the unbound protein was removed by washing with PBS. Samples were mounted on slides with ProLong Gold antifade reagent (Molecular Probes) and visualized at ×1,000 magnification under UV irradiation (the excitation and emission wavelengths were 480 and 535 nm, respectively) with an Eclipse E400 microscope (Nikon). Each fluorescence image represents a 1-s exposure time.
The microplate fluorometric assay was performed with a SpectraMax M5 (Molecular Devices) microplate reader with a fluorescence intensity detection modality. For this, overnight cultures were diluted 1:100 in 30 ml of BHI broth and incubated for 3 h at 30°C with shaking at 150 rpm. Cells were harvested by centrifugation, washed, and resuspended in 2 ml of PBS (∼109 bacteria per ml). A 400-μl aliquot of cells was then added to 400 μl of the GFP-Gp14 extract, mixed gently, and incubated at room temperature for 2 min. For the bacterial combinations, 400 μl of each strain was first combined in a single tube (with or without B. anthracis), washed, resuspended in 400 μl of PBS, and treated as described above with 400 μl of GFP-Gp14 extract. After labeling, cells were washed twice and resuspended in 400 μl of PBS. Two-hundred-microliter aliquots of each sample were added to black 96-well Costar plates, and fluorescence intensity was measured as relative light units in a top-read based on six samplings per well with excitation and emission wavelengths of 485 nm and 538 nm, respectively.
Electron microscopy.
Bacteriophages were prepared from 30-ml high-titer stocks by centrifugation in a Beckman model L5-65 ultracentrifuge (25,000 rpm, 2 h, 4°C, type 35 rotor) and very gentle suspension in 100 μl of 5 mM CaCl2. At the Rockefeller University Bio-Imaging Resource Center, samples were added to carbon- and Formvar-coated grids (400 mesh) and placed in a drop of 2% phosphotungstic acid, pH 7.0, for 2 min at 25°C. Excess fluid was removed, and the specimen was examined on a JEOL CX 100 II transmission electron microscope.
DNA manipulation.
Restriction enzymes, T4 DNA ligase, and Antarctic phosphatase were purchased from New England Biolabs. Phage DNA was prepared with the Lambda Maxi Kit (QIAGEN). DNA fragments and plasmids were purified with NucleoSpin extraction and plasmid kits (BD Biosciences), respectively. For construction of the pDG148::gp41 derivative, gp41 was PCR amplified with primers Fos1 (5′-CCAAAGCTTAGGTGGTTTGTTTTTCTGTATCAAACTTG-3′) and Fos2 (5′-ACCATGCATGCGAAAGCCCTAGCTTTCTTATTA-3′). Fos1 contains the gp41 start codon, an exogenous ribosome binding site (underlined), and a 5′ HindIII site. Fos2 contains the gp41 stop codon and a 5′ SphI site. The PCR product was digested with HindIII-SphI and ligated into the HindIII-SphI sites of plasmid pDG148 (treated with Antarctic phosphatase). For construction of the gfp-gp14 and gfp-wp14 fusions, the tail fiber open reading frames (ORFs) were PCR amplified with primers Tail1 (5′-ACAGATATCTTGGGGAAACTTAGTTTTACTT-3′) and Tail2 (5′-CCCAAGCTTTCATCTATATCTCTCCCTATAACTGA-3′), digested with EcoRV-HindIII (primer-encoded sites), and ligated into the phosphatase-treated SmaI-HindIII sites of plasmid pBAD24::gfpmut2 (56). Here, ligation places the 5′ ends of gp14 and wp14 downstream of, and in frame with, the 3′ end of gfpmut2. gfpmut2 encodes a red-shifted GFP variant (17). Constructions were confirmed by DNA sequence analysis at the Rockefeller University DNA Sequencing Resource Center.
Protein expression.
E. coli strains XL1-Blue/pBAD24::gfp-gp14 and XL1-Blue/pBAD24::gfp-wp14 were grown overnight in 20 ml of LB with 100 μg of ampicillin per ml, diluted 1:100 into 1 liter of LB with ampicillin, and grown for 3 h at 37°C with shaking at 250 rpm. To induce expression from the PBAD promoter of pBAD24 (29), l-arabinose (Sigma-Aldrich) was added to a final concentration of 0.2%, and the cultures were shifted to 30°C for 4 h of shaking at 250 rpm and then to 4°C overnight with shaking at 80 rpm. Cultures were pelleted by centrifugation, washed twice, resuspended in 50 ml of PBS, and lysed with chloroform added to a final concentration of 15%. For lysis, the cultures were gently rocked at 24°C for 1 h and centrifuged (10 min, 3,220 × g, 4°C in an Eppendorf model 5810 R tabletop centrifuge) and the soluble protein-bearing supernatants were recovered and sterilely filtered. Subsequent sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis revealed that while GFP-Gp14 was produced in a soluble form, GFP-Wp14 was not. Numerous attempts to purify a soluble GFP-Wp14 fusion were unsuccessful.
DNA sequence analysis and bioinformatics.
Genomic DNA sequencing was performed to 10-fold redundancy at Genome Therapeutics Corp. (Waltham, MA) and at the Rockefeller University DNA Sequencing Resource Center. The complete genomes were assembled with SeqMan II (DNASTAR, Inc.) sequence analysis software, and genes were identified with GeneMark.hmm for prokaryotes (42), with FramePlot 2.3.2 (35), and by visual inspection guided by BLAST (3) results. Gene numbers are preceded by a wp or gp designation for Wβ and γ phage loci, respectively. ORFs were searched against the protein database with the BLAST and PSI-BLAST (4) algorithms. For predictions of protein sequence structure, the following programs were used: PSORT-B (28), SOSUI (33), and COILS (43). Clustal alignments of sequences were performed with the BCM Search Launcher (57) and BOXSHADE 3.21. To determine the nucleotide sequences of cos sites, we sequenced the ends of γ and Wβ DNAs directly with multiple different primers.
Reverse transcriptase (RT) PCR.
RNA was isolated from strains grown for 16 h at 30°C in 5 ml of BHI broth with shaking at 150 rpm or from 5-h cultures established from overnight cultures diluted 1:100 into either 5 ml of BHI broth or 5 ml of LD medium and grown at 30°C with shaking at 150 rpm. Samples were prepared from 100-μl culture aliquots with the RNeasy Mini Kit (QIAGEN), followed by treatment with 4 U of RNase-free DNase I (Ambion Inc.) and inactivation according to the manufacturer's protocol. RNA was ethanol precipitated, resuspended in distilled H2O, and quantified spectrophotometrically. Quality was assessed both by agarose gel electrophoresis and by PCR analysis of 1-μg RNA samples with primer sets specific for phage and host loci (which yielded no PCR products, showing the absence of contaminating DNA). cDNA was synthesized from 500 ng of RNA with the SuperScript First Strand Synthesis System for RT-PCR (Invitrogen). Subsequent PCRs were carried out for 30 cycles with primer pairs specific for Wβ phage loci 2, 8, 9, 10, 14, 16, 17, 18, 25, 27, 29, 35, 37, 41, 45, 47, 48, 50, and 52. Despite the use of several distinct primer pairs, reactions specific for wp22 and wp40 always yielded background chromosomal signals and had to be dropped from consideration. The vrrA and BA4073 primer sets were included as internal controls. Negative controls included a no-template control and a no-RT control, both of which did not yield PCR products. Products were separated by agarose gel electrophoresis and stained with ethidium bromide.
Fosfomycin resistance assay.
The pDG148 derivatives were grown overnight at 30°C with shaking at 150 rpm in BHI broth supplemented with 10 μg of kanamycin per ml. Cultures were then diluted 1:100 into BHI broth containing 1 mM IPTG and kanamycin and grown for 6 h at 30°C with agitation. Serial dilutions of each culture were plated on BHI agar plates supplemented with a range of fosfomycin concentrations and incubated 24 h at 30°C. Resulting colonies were enumerated to evaluate survival. For analysis of strains lacking pDG148, IPTG and kanamycin were omitted from all incubations.
The MICs of fosfomycin for RSVF1, RSVF1/pDG148, and RSVF1/pDG148::gp41 were determined by a microplate dilution method (39) with BHI broth and an inoculum of 107 stationary-phase bacteria. The MIC was defined as the lowest concentration of fosfomycin inhibiting visible bacterial growth in a 96-well plate after 18 h of incubation at 30°C. For this, sets of 10 twofold serial dilutions of fosfomycin were established in 100 μl of BHI broth per well (starting with 1 mg of fosfomycin per ml); bacteria from overnight BHI broth cultures (grown with kanamycin and IPTG, except in the case of RSVF1) were added to each well in 100-μl aliquots.
Nucleotide sequence accession numbers.
The phage genomes described in this report have been submitted to the GenBank database and assigned accession numbers DQ289556 (for γd'Herelle) and DQ289555 (for Wβ).
RESULTS
Host range and ultrastructure of the γ phage.
The analysis of the γ phage host range in Table 1 demonstrated specificity for B. anthracis and, despite extensive laboratory passage on B. anthracis, for B. cereus strain RSVF1. A particularly close genetic relationship exists between B. anthracis and RSVF1, and RSVF1 is an “atypical” B. cereus strain that manifests phenotypes of both B. anthracis and B. cereus (30, 55). Three other atypical B. cereus strains, CDC 13100, CDC 13140, and CDC 32805, were also infected, confirming specificity for a range of such strains, as well as B. anthracis. Infection of a large panel of more-typical B. cereus and B. thuringiensis strains did not occur. As expected for a lytic phage, γ formed clear plaques (discrete clearing zones within infected bacterial lawns, indicative of total phage-mediated lysis) on all sensitive strains. The inability of γ to infect either ATCC 11950 or ΔSterne::φWβ is a likely example of superinfection immunity, a process whereby an integrated prophage (W, in this case) prevents infection with related phage.
TABLE 1.
Phage specificities within the B. cereus lineage
| Organism or strain | γ titerc | Wβ titerc |
|---|---|---|
| B. anthracis strains | ||
| ΔSterne | 1.0 × 109 | 2.5 × 109 |
| ΔSterne::φWβ | <10 | <10 |
| Atypical B. cereus strainsa | ||
| RSVF1 | 3.0 × 108 | 3.3 × 109 |
| CDC 13100 | 1.4 × 107 | 1.5 × 106 |
| CDC 13140 | 5.0 × 106 | 5.4 × 106 |
| CDC 32805 | 4.5 × 107 | 3.7 × 107 |
| ATCC 11950 | <10 | <10 |
| B. cereusb | 0 | 0 |
| B. thuringiensisb | 0 | 0 |
Atypical B. cereus strains are those that express certain phenotypes characteristic of B. anthracis, like sensitivity to the γ phage.
Multiple strain types were analyzed. The B. cereus strains used include ATCC designations 14579, 10987, 27348, 15816, 13472, 25261, 43881, and 14893, as well as RTS134, RTS100, NRL579, UM101, DP-B5184, DP-B5185, and HER1414. The B. thuringiensis strains used include ATCC 33679, ATCC 35866, and BGSC designations 4AA1, 4AK1, 4AY1, 4AG1, 4BR1, and 4BU1. No plaque formation was ever observed upon infection of these organisms.
Titers are expressed in PFU per milliliter.
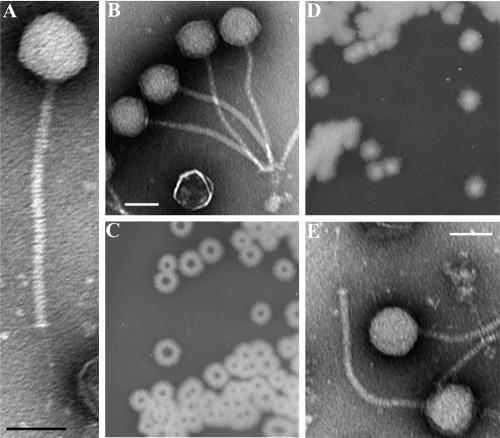
Morphologically, γ (Fig. 1A) is similar to the Siphoviridae family of tailed phages (double-stranded DNA viruses of the order Caudovirales) consisting of an icosahedral head (∼56 nm in diameter) and a long noncontractile tail (∼200 nm long) connected distally to a small plate and a fibrous tail extension (∼63 nm long). This structure is similar to that reported for older γ phage isolates (2, 67); thus, gross morphological changes have not occurred over time. Interestingly, phage particles usually demonstrated a bouquet-like aggregation (Fig. 1B) created by tail fiber adherence to bacterial debris and/or interbaseplate affinity.
FIG. 1.
B. anthracis phage morphologies and isolation. (A and B) Transmission electron micrographs of γ phage virions showing its isometric head and long noncontractile tail. (C and D) Single colonies of B. cereus ATCC 11950 grown for 16 h on BHI agar plates with and without 20 μg ml−1 fosfomycin, respectively. The central holes in panel C are enriched for the Wβ phage. (E) A single intact Wβ phage virion. All scale bars represent 50 nm.
Isolation and analysis of the W phage from B. cereus ATCC 11950.
We also sought the parental lysogenic phage of γ, called W, encoded in ATCC 11950. Initially, we could not induce W from its lysogenic state by standard methods (data not shown); however, we fortuitously did so in a study of fosfomycin resistance in B. cereus. Fosfomycin is a broad-spectrum bactericidal antibiotic produced in the soil by Streptomyces spp. (31). Unlike B. anthracis and other B. cereus strains (data not shown), ATCC 11950 produced donut-shaped bacterial colonies on agar plates in the presence of fosfomycin (Fig. 1C) but not in its absence (Fig. 1D). The central clearing zones on fosfomycin plates were enriched for infectious W particles. This effect was not observed with sublethal levels of other antibiotics, including penicillin, streptomycin, tetracycline, and ciprofloxacin (data not shown).
The purified W phage was morphologically identical to γ (Fig. 1E) and had a host range restricted to B. anthracis and the atypical B. cereus strains (Table 1). As expected for a temperate phage, W produced turbid plaques. The inability of an induced W phage isolate to infect lysogenic strain ATCC 11950 has been previously observed by McCloy (45), who originally isolated two phage types from this strain, a rare α form and a dominant β form; the β phage, unlike α, could not infect ATCC 11950. We were therefore working with the major phenotypic form of this phage, Wβ.
Genomic analysis of γ and Wβ phages.
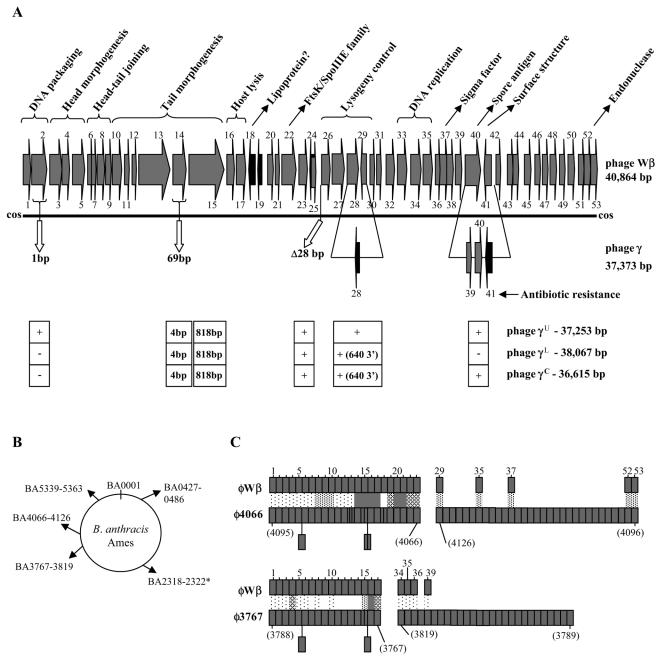
The genomic sequences of both γ and Wβ were determined and shown, by pairwise comparison, to be 100% identical, with five exceptions (Fig. 2A; Tables S1 and S2 in the supplemental material). The G+C contents of γ and Wβ were 35.1% and 35.3%, respectively, similar to that of the B. anthracis genome (36.4%). The γ phage encoded 53 ORFs over 37,373 nucleotides, while the Wβ phage had 53 ORFs within its 40,864-bp genome. Each genome was flanked by the same complementary 9-bp single-stranded cohesive ends (cos sites). Though very similar in gene order to that of the λ supergroup of the family Siphoviridae, the overall sequences of γ and Wβ were distinct from all others in the database. Even highly related γ isolates γL, γU, and γC were distinguished from our γ isolate and Wβ at several distinct loci (Fig. 2A). In the B. anthracis Ames genome, we discerned three largely intact prophages (BA3767 to BA3819, BA4066 to BA4126, and BA0427 to BA0486) and one defective prophage (BA5339 to BA5363) with notable similarities to γ and Wβ sequences (Fig. 2B). These prophages correspond to the B. anthracis loci referred to as LambdaBa1 to LambdaBa4 (50).
FIG. 2.
Genometric analysis of the γ and Wβ phages. (A) Schematic representation of the entire Wβ phage genome. Numerical gene designations, directions of transcription, and assumed functions are indicated. ORFs 24 and 25 overlap in opposite orientations. The sites of five inserts, deletions, and point mutations that distinguish the γd'Herelle phage (referred to as γ) are shown below the solid line with γ gene number designations. Below the γ phage representation, genetic differences that define the other γ phage isolates, γL, γU, and γC and Cherry, are indicated in the boxed areas below the appropriate genetic positions. The presence or absence of alterations identical to those in d'Herelle is indicated by a plus or a minus sign below gp2, the 28-bp deletion between gp25 and gp26, and the Fosr island (gp39 to gp41). The 2,003-bp deletion in the γ lysogeny region (forming gp28) is observed in each isolate, although the 3′ end extends an additional 640 bp in the cases of isolates γL and γC. A set of only four point mutations at orf14 (identical in isolates γL, γU, and γC and distinct from d'Herelle) is shown. The orf15 locus, identical in Wβ and d'Herelle, is distinguished by a set of 818 base differences, identical in isolates γL, γU and γC. Other than the mutations described here, the Wβ sequence is 100% identical to that of the γL, γU and γC, and the d'Herelle phages. (B) Genomic loci of the B. anthracis Ames strain that bear similarity to γ and Wβ phage sequences. GenBank sequences BA3767 to BA3819, BA4066 to BA4126, BA5339 to BA5363, and BA0427 to BA0486 correspond to known Ames prophages (LambdaBa01 to LambdaBa04, respectively). The asterisk denotes five loci with products >40% identical to five contiguous γ and Wβ loci. (C) Pairwise comparison of Wβ loci with homologous sequences of B. anthracis prophages φ4066 and φ3767. Wβ gene numbers are indicated at the top, while prophage gene designations are indicated below (in parentheses) in numerical order. The prophages are represented not as their linear integrated forms but rather as their inverted left and right halves (gene numbers are indicated below in parentheses) in order to maintain gene order with Wβ. Light stippling represents genes encoding proteins with 18 to 30% identity, medium is 31 to 40%, heavy is 41 to 75%, and filled is 76 to 100%. Since Wβ encodes fewer genes, some φ4066 and φ3767 loci are inserted either as small rectangles within the gene order or as rectangles extending below each sequence. For φ3767, the following genes are absent from the GenBank sequence and are not included in the gene order shown: BA3772, BA3773, BA3790, BA3794, and BA3798.
Lambdoid phage genomes are genetic mosaics arising from mutations and rampant recombination events that include the horizontal transfer of gene fragments or functional gene modules among phages (12). The resulting genomes are “pastings” of conserved modules from different phages, encoding basic functions like capsid building, host lysis, lysogeny, and replication. Interspersed are distinguishing loci, either conserved among related phages or unique, which may encode lysogen conversion factors. The architecture of the γ and Wβ genomes is consistent with this model. The DNA packaging, structural (head and tail components), and host lysis proteins (ORFs 1 to 17) are well conserved among elements of phages φ3626 of Clostridium perfringens and φ105 of B. subtilis and prophages φ4066 and φ3767 of B. anthracis. The modules specifying lysogeny (ORFs wp26 to wp30, including resolvase and genetic switch functions) and phage DNA replication (including ORFs wp33 to wp35) are most similar to corresponding elements of B. cereus, B. thuringiensis, Clostridium thermocellum, Lactococcus lactis, and lactobacillus phages. Aside from the other γ isolates (γL, γU, and γC), the sequence and gene order of γ and Wβ are most similar overall to those of B. anthracis prophages φ4066 and φ3767 (and, to a lesser extent, prophages of the closely related species B. cereus and B. thuringiensis)—10 genes are unique to this group, while the products of 23 genes, including those for host recognition, are primarily related to each other on the basis of BLASTP scores. Alignment of the Wβ genome with all discernible φ4066 and φ3767 genes (Fig. 2C) shows the extent of this similarity but also illustrates the diversity that still distinguishes them.
Recently, a highly conserved programmed translational −1 frameshift was found to be common among the tail assembly genes of most double-stranded DNA phage (70). Here, a short “slippery” sequence between overlapping genes controls the production of two proteins, exemplified by the products of phage λ genes G and T, essential for the progression of tail subunit polymerization. Analysis of the γ and Wβ sequences did identify two putative orthologs of G and T, orf11 and orf12, which are of the appropriate size and, like G an T, are encoded between a major tail protein (orf10) and a tape measure protein (orf13). Unlike, G and T, however, the orf11 and orf12 loci do not overlap and appear to lack a conventional slippery sequence. Despite this, a nonconventional slippery sequence, providing either a −2 or a +1 frameshift, could fuse the orf11 and orf12 products and is worthy of further investigation.
Expression analysis of Wβ prophage loci.
A previous study of S. thermophilus prophage gene transcription in the lysogenic state showed that only those few products required to maintain lysogeny and with putative lysogenic conversion functions are expressed (65). We similarly analyzed phage gene expression in Wβ lysogens of both B. anthracis (ΔSterne::φWβ) and B. cereus (RSVF1::φWβ). Total RNAs prepared from BHI broth (both growing and in late stationary phase) and sporulating LD medium cultures were analyzed by RT-PCR with primer pairs specific to 19 Wβ loci distributed throughout the entire integrated genome. We observed that six loci were expressed in the lysogenic state (Fig. 3A and B). From within the highly conserved gene modules encoding obvious phage functions, only the wp29 locus of the lysogeny module (encoding a putative cI-like repressor protein) was expressed (Fig. 3B). All other expressed loci were detected outside of conserved phage gene modules.
FIG. 3.
RT-PCR analysis of Wβ phage gene expression during the lysogenic state. RNA prepared from cultures grown under three different conditions was reverse transcribed with random primers. Subsequent PCR was performed with primers specific for the indicated Wβ phage genes. (A) Analysis with RNA from overnight BHI broth cultures of two different B. anthracis (ΔSterne::φWβ) lysogens (+) and the parental nonlysogen (−) and from the B. cereus (RSVF1::φWβ) lysogen (+) and nonlysogen (−). (B) Analysis of RNA isolated from only B. anthracis and B. cereus Wβ lysogens grown for 5 h in either liquid sporulation medium (+) or BHI medium (−).
Conspicuous within the γ and Wβ genomes are a set of seven genes, wp18 to wp25, between the lysis and lysogeny modules, that are unique to B. anthracis, B. cereus, and B. thuringiensis phages and encode no obvious phage functions. Comparative genomic studies of temperate members of the family Siphoviridae from low G+C content gram-positive bacteria have shown this to be a region of high variability among different phages, encoding one to six genes (21); the presence of prominent virulence factors at this position within prophages of pathogenic Streptococcus pyogenes and Staphylococcus aureus suggests that this region serves lysogen conversion functions. While no obvious virulence factor(s) is encoded within this region of Wβ or γ, it does include two putative surface proteins (Wp18 and Wp25), a transcriptional repressor (Wp19), and a prominent cell division protein (Wp22). RT-PCR analysis showed that the first and last genes of this locus, wp18 and wp25, were transcribed in both the B. anthracis and B. cereus lysogens (Fig. 3A and B). While wp18 was expressed under all growth conditions, wp25 was primarily expressed in the early stationary-phase sporulation and BHI broth cultures (Fig. 3B) but not in the overnight BHI broth cultures (Fig. 3A). A possible relationship between a section of the wp18-to-wp25 locus and a nonprophage region of B. anthracis was identified in BLASTP analyses with Wβ sequences. Here, a set of five contiguous B. anthracis loci were identified (BA2318 to BA2322; Fig. 2B), encoding proteins with 46 to 80% identity to products of the similarly organized wp19 to wp23 span of loci. While homologs of Wp19 to -23 are also encoded within B. anthracis prophage φ4066, only at BA2318 to BA2322 is there a stretch of encoded proteins with such extensive sequence similarity. This conserved sequence and gene order suggests a common evolutionary origin.
For the Siphoviridae family members of low G+C content gram-positive bacteria, the region downstream of the phage replication module is often marked by numerous insertions and deletions and is a recombination hot spot (41). The equivalent region in both Wβ and γ is populated by small loci encoding proteins either with no obvious homologs or with homology only to closely related B. anthracis, B. cereus, and B. thuringiensis phage and/or nonprophage genes. Prominent in this group is orf37 (wp37 in Wβ and gp36 in γ), encoding a putative bacterial RNA polymerase sigma factor that may act to regulate host gene transcription. RT-PCR analysis indicated that wp37 was expressed predominantly during sporulation (Fig. 3B), a process well known to involve a cascade of multiple distinct sigma factors. Additionally, we observed that the products of both wp41 and wp45 were transcriptionally active during growth and/or sporulation and may thus impact host phenotypic expression (Fig. 3A and B). Wp41 is likely of bacterial, not phage, origin and is discussed below. Wp45 is weakly similar to RNA polymerase sigma subunits, and its gene is primarily transcribed only during sporulation in B. anthracis. The poor expression of wp45 in RSVF1::φWβ, relative to ΔSterne::φWβ, was the only reproducible difference between the B. anthracis and B. cereus lysogens in the RT-PCR studies.
Deletions in the lysogeny control region.
A genetic basis for the difference in the γ and Wβ lifestyles (lytic versus lysogenic, respectively) became clearer in a direct comparison of their genomes. There were two polymorphisms at the γ lysogeny control region (wp26 to wp30), a 28-bp deletion in an intergenic region immediately upstream of the module and a 2,003-bp deletion removing wp28 and the 5′ end of wp29, yielding a small partial ORF in γ, gp28. The wp29 locus is similar in sequence and genetic position to functional homologs of the cI repressor of phage λ, which acts to repress lytic proliferation and promote lysogeny. In the λ system, most cI mutants are lytic variants (6). Therefore, the conversion of γ to an obligately lytic phage through loss of its repressor is likely. Sequence analysis of highly related γ isolates γL, γU, and γC shows that γU arose from a 2,003-bp deletion identical to that observed in our d'Herelle isolate, while γL and γC arose from identical 2,643-bp deletions exactly encompassing the above 2,003-bp deletion and an additional 640-bp 3′ stretch. The deletion in isolates γL and γC removes those genes corresponding to wp28 to wp31 in the lysogeny control region. Comparative genomic studies of Streptococcus thermophilus phages show that lytic phages do arise from temperate phages by deletion and rearrangement events in the lysogeny module (41). Interestingly, clear-plaque lytic variants of Wβ occur at a frequency of ∼2% and were often attributed by PCR analysis with wp28-to-wp29-specific primers, to deletions akin to that observed in γ (data not shown). A pool of lytic variants seems to represent a significant apparent subpopulation of Wβ.
Point mutations in the tail fiber locus orf14.
The γ phage was originally distinguished from Wβ by its ability to penetrate the surface capsular structure expressed by vegetative B. anthracis under certain conditions (11). This altered tropism may be related to our observation of 69 missense mutations in the tail fiber gene gp14, compared to wp14, resulting in 24 amino acid changes (Fig. 4A). Phage tail fibers mediate recognition and binding to target cell receptors during adsorption, and mutational events in tail fibers can modify infective specificity. The incidence of so many distinct alterations in gp14 suggests that this is a hot spot for change, as has been observed with phage loci specifying host tropism (23, 41). Gp14 is a more basic protein overall compared to Wp14 (with pI values of 6.66 and 7.74, respectively), which is consistent with a form that more efficiently penetrates the negatively charged capsule of B. anthracis to reach subjacent binding receptors.
FIG. 4.
Tail fiber sequence and binding analysis. (A) Sequence alignment of the Gp14 and Wp14 tail fibers of γ and Wβ, respectively, with Sp14 (a plaque size variant of Wβ), Cp14 (of the B. anthracis Cherry phage, accession number YP_338146), and BA4079 (of the B. anthracis φ4066 prophage, accession number NP_846318). Conserved residues are indicated by dark boxes, while conservative changes are indicated by gray boxes. (B) Fluorescence micrograph of GFP-Gp14 binding to the surface of B. anthracis ΔSterne. (C) GFP-Gp14 binding to the surface of B. cereus RSVF1. The contrast in this image was manipulated with Adobe Photoshop 7.0 in order to better illustrate binding. The actual polar/septal fluorescence signal was more akin to that observed in the background of panel D. (D) Rare whole-cell binding of GFP-Gp14 binding to B. cereus RSVF1. (E) Phase-contrast image of B. cereus ATCC 10987 after treatment with GFP-Gp14. (F) Fluorescence image corresponding to the field shown in panel E. (G) Fluorescence-based method for distinguishing B. anthracis from B. cereus and B. thuringiensis in pure and mixed samples. The indicated bacterial organisms were treated with the GFP-Gp14 fusion, washed, and then examined to quantify retained surface fluorescence. The results represent averages of three independent experiments. Bc, B. cereus; Ba, B. anthracis; Bt, B. thuringiensis.
In agreement with a major role for the Orf14 tail fibers in the infective process, we found that mutations in at least one large-plaque mutant of Wβ mapped to the wp14 locus. Sequence analysis indicated that a shift to a larger plaque size was associated with a two-residue change at the C-terminal end of this tail fiber, referred to as Sp14 (Fig. 4A).
To determine if Gp14 and Wp14 can mediate host recognition, we sought to perform cell wall binding studies with GFP-Gp14 and GFP-Wp14 fusions. We were unable to express GFP-Wp14 in a soluble form despite using numerous expression strategies; however, we did isolate a soluble and fluorescent GFP-Gp14 fusion suitable for surface labeling analyses. By fluorescence microscopy, we showed that the Gp14 moiety specifically directed GFP over the entire surface of B. anthracis (Fig. 4B) and to polar and septal positions of RSVF1 (Fig. 4C). No labeling of γ phage-resistant B. cereus ATCC 10987 (Fig. 4E and F) or B. thuringiensis HD1 (data not shown) was observed. A quantitative method for detecting surface binding was also developed whereby labeled bacteria were washed and analyzed with a microplate-based fluorometric assay (Fig. 4G). Here, B. anthracis labeled with GFP-Gp14 yielded a strong signal while γ phage-sensitive strains RSVF1 and CDC 13100 were closer to the background levels observed with γ-resistant B. cereus ATCC 10987 and 14579 and B. thuringiensis HD1. The ability here to discern B. anthracis from atypical B. cereus strains was likely attributable to the differential binding (whole cell versus polar/septal) displayed by these bacterial types above. The polar/septal binding to atypical B. cereus may not have been sufficiently above the background to be distinguished from unlabeled bacteria in this assay.
The altered spatial distribution of Gp14 binding to B. anthracis and atypical B. cereus was unexpected and indicated that binding epitopes on B. anthracis and RSVF1 are differentially arrayed. For RSVF1, we did observe that <0.1% of the visualized population, regardless of growth stage, displayed a B. anthracis-like whole-cell binding pattern with GFP-Gp14 (Fig. 4D). A reverse situation, in which B. anthracis displayed polar/septal labeling, was not observed.
Evidence of homologous recombination with the B. anthracis genome.
A major genetic difference distinguishing γ from Wβ arose from substitution of a 2,823-bp segment encompassing wp40 and wp41 with a 1,360-bp locus encoding three distinct genes, gp39 to wp41. In a BLAST analyses, we observed that the 1,360-bp γ island was 100% identical at the DNA level to an otherwise poorly conserved region of the φ4066 prophage of B. anthracis Ames (Fig. 5A) and may have been acquired by homologous recombination from this prophage during the evolution of γ. The ability of bacteriophages to acquire prophage genes from their host via recombination, thus creating chimeric forms, is well documented. Three findings support this acquisition from B. anthracis: (i) the only Gp40 homolog in protein database is the corresponding φ4066 product, (ii) the product of gp41 is most closely related to that of BA4109 from prophage φ4066 (Fig. 5B), and (iii) 21-bp and 230-bp regions of homologous DNA (100% and 88.4% identity, respectively) flank the 2,823-bp region of Wβ and the BA4111-to-BA4109 locus in φ4066 and could support recombination (Fig. 5A). This flanking homology suggests that transfer of the 1,360-bp island into an infecting γ-like phage (and the reciprocal transfer of wp40 to wp41 into the B. anthracis chromosome) may not be an isolated event. Additionally, this is indirect evidence that gene flux exists between infecting bacteriophage and B. anthracis.
FIG. 5.
Acquisition of the gp41 locus and the role for its product in fosfomycin resistance. (A) Pairwise comparison of the gp39-to-gp41 γ phage locus to the corresponding BA4111-to-BA4009 region of the B. anthracis φ4066 prophage. Bold values between aligned phage sequences indicate the percent identity of the respective protein products. The 5′ 21-bp and 3′ 230-bp regions of DNA sequence homology (100% and 88.4% identity, respectively) shared by φ4066 and γ (and Wβ as well), and presumed to support recombination, are indicated as the cross-hatched regions extending above the γ phage sequence. The cross-hatching between genomes indicates the region of 100% DNA identity observed between γ and φ4066 but absent in Wβ. Arrows indicate the direction of gene transcription. Flanking loci are not homologous. (B) Phylogenetic representation of sequence relationships between fosfomycin resistance proteins from a variety of bacterial species. Most are encoded in the host chromosome, with the exception of E. coli Fosr (transposon Tn2921 of plasmid pSU961), B. anthracis BA4109 (φ4066 encoded), Staphylococcus epidermidis (plasmid pIP1842), and Pseudomonas aeruginosa (class I integron). The consensus tree was obtained by a bootstrap analysis of an alignment (1,000 replicates) obtained with the MacVector program (Accelrys Inc.). Nodes that are well supported by the phylogenetic analysis are indicated by the percentages of replicates for which those nodes were recovered. The root was arbitrarily set. (C) Fosfomycin resistance conferred by Gp41. Bacterial cultures were plated in the presence of increasing concentrations of fosfomycin, and resultant colonies were enumerated. RSVF1 (open squares), RSVF1/pDG148 (cross-hatches), and RSVF1/pDG148::gp41 (open circles) are shown. (D) Inherent fosfomycin resistance in B. anthracis and other soil organisms. The following strains were plated in the presence of increasing concentrations of fosfomycin, and the resulting colonies were enumerated: B. cereus RSVF1 (open squares), B. cereus ATCC 14579 (closed circles), B. cereus ATCC 13472 (closed squares), B. thuringiensis HD1 (open triangles), and B. anthracis ΔSterne (closed diamonds). Values represent the averages of four independent experiments.
Antibiotic resistance encoded within a γ-specific gene module.
Acquisition and transfer of the 1,360-bp γ island is significant considering that one of its products, Gp41, is highly related to a family of fosfomycin resistance (Fosr) proteins of prokaryotic genomes and plasmids (5). In particular, Gp41 is >60% identical to proteins of a variety of Bacillus species, including the FosB metallothiol transferase of B. subtilis (15); homology includes well-conserved fosfomycin and cofactor binding motifs required to form inactivated fosfomycin adducts. Gp41 is, in fact, highly related to Fosr proteins of a wide range of both gram-positive and -negative microorganisms (Fig. 5B), though it is most similar to BA4109 from B. anthracis prophage φ4066. Phage-encoded antibiotic resistance, like that encoded in γ and φ4066, has not been previously reported. To confirm the ability of Gp41 to confer Fosr, it was expressed from an IPTG-inducible expression vector, pDG148, in an otherwise Foss RSVF1 background. The resulting Gp41 expression led to high levels of resistance to Fos and growth over a range of concentrations that were largely inhibitory to RSVF1 (Fig. 5C). Additionally, the MICs of fosfomycin for RSVF1/pDG148 and RSVF1/pDG148::gp41 were determined by an agar dilution method to be 62.5 and 500 μg/ml, respectively.
During the course of this work, we noted that B. anthracis exhibited very high levels of intrinsic Fosr (Fig. 5D) not observed in B. cereus and B. thuringiensis strains. The reason for this variation is unclear, as BLASTP analyses showed Gp41 homologs distributed throughout the B. cereus lineage of organisms. While it is notable that a Fosr protein is phage encoded with B. anthracis, homologous Fosr proteins are encoded on mobile insertion cassettes in both B. cereus and B. thuringiensis (20). Nevertheless, the elevated resistance to fosfomycin observed in B. anthracis, combined with the phage-mediated mechanism for Fosr gene movement implied here, could reflect an adaptive mechanism for B. anthracis in the environment.
Putative products of a Wβ-specific gene module.
The 2,823-bp Wβ phage island encodes two proteins, Wp40 and Wp41, with possible lysogen conversion functions. On the basis of its sequence, Wp40 may be a collagen-like glycoprotein (Table S2 in the supplemental material), similar to the BclA proteins of B. anthracis, B. cereus, and B. thuringiensis, which assemble as extended hair-like filaments on the outer spore surface or exosporium (61). While the function of these appendages is unknown, BclA is the immunodominant protein of the B. anthracis exosporium (58) and may act as a permeability barrier around the spore (10).
BclA of B. anthracis forms trimeric, collagen-like filaments from monomers that have a 38-residue N-terminal domain, a 266-residue collagen-like region with 76 XXG repeat elements, and a 134-residue C-terminal domain that stabilizes the trimeric structure via interactions between beta strands (10). The number of XXG repeats varies in different naturally occurring B. anthracis isolates, ranging from 17 to 91, and the length of this region determines the length of a hair-like nap on the spore surface. The corresponding structure of Wp40 consists of a 19-residue N-terminal anchor, a 289-residue central domain containing 95 XXG repeats, and a beta strand-rich 161-residue C-terminal domain. The proteins most similar to Wp40, detected by a BLASTP search, were the putative spore surface antigens BC4769 and BCE4869 of B. cereus strains 14579 and 10987, respectively (Fig. 6A), although neither is phage encoded. Additionally, the collagen-like domains of BC4769 and BCE4869 code for only 42 and 31 XXG repeats, respectively. Whether Wp40 contributes to spore surface structure, like BclA, remains unknown, although it is notable both that Wp40 has more XXG repeats than previously described spore antigens and that immediately upstream of wp40 is a putative σK sporulation-specific promoter similar to that identified upstream of bclA (61). All attempts to analyze wp40 gene expression by RT-PCR were unsuccessful owing to primer cross-reactivity with a host gene(s).
FIG. 6.
Alignments of the wp40 and wp41 products of Wβ with homologous protein sequences. (A) Alignment of the putative spore surface antigen Wp40 with the collagen triple-helix repeat proteins of B. cereus strain ATCC 14579 (BA4769; accession number NP_834473) and B. cereus strain ATCC10987 (BCE4869; accession number NP_981162). (B) Alignment of the putative mannose 6-phosphate isomerase Wp41 with similar proteins from a variety of bacterial organisms, including Clostridium tetani E88 (CTC01899; accession number NP_782477), B. thuringiensis ATCC 35646 (RBTH_04512; accession number ZP_00741184), B. subtilis 168 (BSU26560; accession number NP_390533), Lactobacillus plantarum (pLP9000_05; accession number NP_631996), and Methanosarcina barkeri (MbarDRAFT_2481; accession number ZP_00543223). Black-shaded residues are identical in the alignment, while gray-shaded residues denote conservative changes.
The second protein encoded within the 2,823-bp Wβ phage island, Wp41, was also of interest as it encoded a putative member the ManC family of bacterial phosphomannose isomerases that catalyze the reversible isomerization of d-mannose 6-phosphate and d-fructose 6-phosphate (38). Bacterial phosphomannose isomerases are important for mannose catabolism and the production of GDP-d-mannose, a nucleotide sugar necessary for mannosylation of, among other things, exopolysaccharides that decorate some bacterial surfaces. The ManC sequences are strongly conserved across a broad range of bacterial species. Accordingly, we observed Wp41 homologs in multiple distinct organisms, including gram-positive and -negative bacteria, as well as archaebacteria (Fig. 6B and data not shown). While no Wp41 homologs appear to be phage encoded, they have been identified in a plasmid of Lactobacillus plantarum (52), and a potentially mobile genetic element in Haemophilus influenzae (8). Contributions of Wp41 to nutrient acquisition or surface structure determination could be significant considering that N-acetylmannosamine is a major component of the B. anthracis surface polysaccharide structure (25) and that B. anthracis has no ManC homolog, lacks mannose catabolic pathways, has a reduced number of phosphotransferases, and has a reduced capacity for sugar utilization (50). Expression analysis of Wp41 does support a role for this protein in vegetative cells (Fig. 3B); however, its strong expression during sporulation also supports an additional function, perhaps in the glycosylation of Wp40-like spore surface protein.
DISCUSSION
Our experiments show that the γ lytic phage evolved from the temperate Wβ phage in a process involving distinct DNA recombination events and the accumulation of both point mutations and small and large deletions. These changes include (i) a point mutation in wp2 causing an A569S shift in Gp2, (ii) a 28-bp deletion between ORFs 25 and 26, (iii) 69 point mutations in the gp14 locus, (iv) a 2,003-bp deletion in the γ lysogeny module removing most of the cI homolog and an upstream locus (ORFs 28 and 29 from Wβ), and (v) a 2,823-bp locus encoding wp40 and wp41 that was replaced by a 1,360-bp island (Fosr module) in a likely recombination process with a B. anthracis prophage. These findings largely agree with the results of other comparative genomic analyses, particularly of S. thermophilus lytic and temperate phages (13, 41), which show similar accumulations of major and minor modifications and suggest that such changes are driving factors in the diversification of bacterial DNA viruses.
A trait-driven model is presented (Fig. 7) that summarizes the genetic changes defining the pool of obviously related phages infecting B. anthracis discussed in this work. Included are the Wβ and γd'Herelle forms, as well as recently reported γ isolates γL, γU, and γC. While each is nearly identical to the others over most of its sequence, these forms are nonetheless distinct on the basis of a series of major and minor genetic alterations described in the model. The 28-bp deletion identified upstream of the γd'Herelle lysogeny module is identical in all γ variants, distinguishing the γ grouping from β. Within γ, an identical 2,003-bp deletion in the lysogeny modules of γd'Herelle and γU subdivides these phages from γL and γC, which share a 2,643-bp deletion. In γL and γC, this 2,643-bp deletion exactly encompasses both the 2,003-bp deletion (with an identical 5′ end) and an additional 640 bp of 3′ sequence, removing a Cro repressor homolog and an additional gene. Presumably, the larger deletion also favors lytic growth. While the mutation described in orf2 (the A569S shift) subdivides the γ group in a manner similar to that of the lysogeny module deletions, the grouping collapses upon integration of the Fosr island acquisition and tail fiber mutation data. The 1,360-bp Fosr islands were acquired by each of the γd'Herelle, γU, and γC forms (with identical insert sites), though not by γL, which retains the 2,823-bp “spore antigen” island of Wβ. Additionally, the 24 residue changes that define Gp14 of γd'Herelle are clearly distinct from the single amino acid change common to the Orf14 tail fibers of γL, γU, and γC. Unlike γd'Herelle, the γL, γU, and γC isolates accumulated tail fiber mutations primarily at the orf15 locus, with each isolate bearing an identical array of changes that accounts for an 818-base difference compared to the γd'Herelle and Wβ sequences. The relationship between the Orf14 and Orf15 putative tail fiber proteins in the process of tail fiber synthesis or binding remains unknown.
FIG. 7.
Graded relatedness that defines the Wβ-and-γ group of phages. The flow chart is based on genetic differences described in this work and is intended only to illustrate the mutations and recombination events that currently distinguish each phage. Other than those differences reported here, all phage sequences are identical. The series of alterations is presented in an arbitrary manner and is not intended to imply any true phylogeny. The γ variants listed are γd'H (isolate d'Herelle described in this study), γU (isolate USAMRIID), γL (isolate LSU), and γC (Cherry isolate). aa, amino acids.
On the basis of the series of genetic differences that define each γ isolate, the phylogeny of this family is difficult to reproduce. Whether the acquisition of Fosr occurred once in a common ancestor of γd'Herelle, γU, and γC (thus distinguishing them from γL) or if it occurred independently after the distinct lysogeny module deletions and/or other alterations is impossible to determine on the basis of the current genomic sequences. It is possible that future sequence analyses of γ variants from other sources will enable a more accurate view of γ phage evolution and, indeed, afford insights into phage evolution distinct from that provided by other lambdoid phages and the dairy phages. At the conclusion of this work, another B. anthracis phage, called Fah, was reported in the literature (47). While no Fah sequence was reported in GenBank, the genome description is most similar to that of Wβ, differing primarily with respect to the absence of ORFs wp38 to wp40. This is consistent with our prediction of this particular region as a mutational hot spot.
The induced form of prophage W from ATCC 11950 is apparently well suited to genetic change, as evidenced both by the natural occurrence of variants (including the dominant β form and the α and γ variants) and by the recombination and numerous mutations that now distinguish the γ variants. This is not unexpected, considering the coevolutionary arms race that exists between bacteria and phage (69), in which phage may respond to ecological pressure by expanding certain preexisting subpopulations and modifying these forms over time. Here, the evolution of γ is a response to pressure imposed by its use as a diagnostic phage in a laboratory environment. Diversification of the γ lysogeny region and tail fiber has likely occurred because of intentional selection for improved infectivity and, despite the artificial nature of the selective force, does reflect genetic modifications that occur naturally to alter phenotypes of host resource consumption and phage adsorption. The driving force behind the acquisition of the Fosr island is less clear, as no advantage is obvious for this trait in the laboratory. Rather, this may reflect the inherent instability of the 2,823-bp Wβ island during passage through B. anthracis, owing to the presence of homologous sequences within a host prophage. The exchange of novel gene modules (or “morons”) among diverse lambdoid phage and/or prophage via homologous recombination is, in fact, considered a major force driving phage evolution (12, 32). Here, conserved linker sequences flanking the units of exchange drive module transfer in a recombination process mediated by either a host enzyme (like RecA) or a phage enzyme (like lambda red). A well-studied example concerns the highly conserved sopE locus of several distinct Salmonella sp. prophages which, like the Fosr island, is surrounded by different sequences but is immediately bordered by regions with considerable sequence homology (12). It is likely that Fosr was acquired by homologous recombination from a B. anthracis prophage and that phage populations dominated by Fosr derivatives were eventually expanded as laboratory stocks. The selective advantage, if any, that is provided by the Fosr island to enable this dominance is currently under investigation.
The identification of hot spots for genetic variation in Wβ enabled certain phage functions to become apparent. In particular, the accumulation of mutations at gp14, combined with its similarity to known tail fibers, led to the demonstration of Gp14 as a host cell binding protein with specificity similar to that of the γ phage host range. In agreement with roles for both Gp14 and Wp14 in host specificity, BLASTP searches indicate a mosaic structure for their coding sequences, composed of a well-conserved phage tail gene (COG4722) internally disrupted by insertion of an ∼800-bp DNA fragment from an unknown source (data not shown). Tail fiber genes are known recombination hot spots that can arise via extensive gene shuffling events which modify binding specificity (41, 62). The identification of host cell binding activities is important, not only from the standpoint of phage biology but also with respect to the development of novel diagnostic tools. Using a fluorescence-based live cell binding system developed here to exploit the GFP-Gp14 fusion, we accurately and rapidly distinguished bona fide B. anthracis from very closely related, atypical B. cereus strains, like RSVF1, that have confounded rapid diagnostic tools in the past (55, 63).
It was surprising to find that a GFP-Wp14 fusion was largely insoluble, considering that the equivalent GFP-Gp14 fusion, differing by only 24 residues, was readily purified in a soluble and fluorescent form. This difference in solubility, like the altered pI values of the Wp14 and Gp14 forms, could reflect a form that more efficiently penetrates the B. anthracis capsule to reach the target receptor (the original basis for identifying γ used by Brown and Cherry in 1955). While the exact Gp14 binding target is unknown, a recent study identified GamR as the γ phage receptor on the surface of B. anthracis (19). A study of the interaction between GamR and Gp14 will next be required, as well as a study of receptor interaction with Gp15, a putative tail protein that has accumulated genetic changes in γ isolates γL, γU, and γC (compared to γd'Herelle and Wβ). Additionally, it will be interesting to determine whether an altered surface distribution of GamR explains the differential binding observed with the GFP-Gp14 fusion on B. anthracis (whole cell) and B. cereus RSVF1 (polar/septal).
Our reference to gp14 as a mutational hot spot is based on the apparent accumulation of point mutations at this locus. Our observation of extensive mutations in orf15 of γL, γU, and γC suggests that this locus may be a hot spot as well. A possible explanation for these clusters of mutations certainly may lie in a mechanism akin to that previously described (23) for the specific introduction of tail fiber gene mutations, although such mechanisms may be rare. Alternatively, it is more likely that the entire γ genome is equally susceptible to mutation but that only events at certain loci, like gp14 or orf15, provide a sufficient selective advantage to always outcompete the remaining phage pool through successive rounds of phage amplification, thus giving the appearance of a hot spot upon sequence analysis. This is consistent with the view of phage evolution in which rampant recombination occurs throughout phage genomes but only those events that do not disrupt functional gene modules or reduce phage viability are maintained (32).
A major reason for our study of B. anthracis phage was an interest in the contribution of lysogeny to the phenotype of an organism primarily considered to be distinguished by its virulence plasmids. In B. anthracis, there are at least four complete or partial prophages encompassing roughly 224 loci, some of which distinguish B. anthracis from closely related B. cereus and B. thuringiensis strains. Taken with the variety of phage infecting B. anthracis identified from the environment and the well-described role of phage in the emergence of virulent forms of low G+C content gram-positive bacterial pathogens, there is a strong impetus for the study of phage contributions to B. anthracis. Here, we have found that Wβ is very similar in gene order and sequence to prophages of B. anthracis and may recombine with these sequences to create a hybrid phage, like γ. Evidence for recombination was observed at a 2,823-bp locus in Wβ that was replaced by a 1,360-bp three-gene island in γ. At least three phage proteins with putative lysogen-converting functions are encoded here, with predicted contributions to spore surface structure (Wp40, the collagen repeat protein similar to a family of spore antigens), vegetative surface structure (Wp41, the ManC-like protein involved in the mannosylation of exopolysaccharide structures), and antibiotic resistance (Gp41, the fosfomycin resistance protein). While these proteins may not be classic virulence factors, they do appear more likely suited for ecological adaptations, perhaps in the soil niche.
The finding of phage-encoded fosfomycin resistance is notable for several reasons. Firstly, phage-mediated transfer of Fosr can help account for the intrinsic resistance of B. anthracis to this soil antibiotic. The apparent importance of fosfomycin resistance to B. anthracis certainly challenges the long-held assumption that B. anthracis has no vegetative phase whatsoever in the soil. Also of note was the induction of Wβ from its lysogenic state by fosfomycin. While the significance of this finding for the ecology of Wβ is unclear, it may represent a legitimate mechanism owing to the presence of a near consensus antibiotic-inducible σW promoter upstream of the Wβ lytic pathway genes (data not shown). Again, this is consistent with a more dynamic interaction between B. anthracis vegetative forms and the soil environment than has been previously assumed. The greatest point of interest arising from our finding of phage-encoded fosfomycin resistance is that demonstrable antibiotic resistance can, indeed, be encoded by and transferred directly by phage. Previously, all reports of mobile antibiotic resistance elements have been restricted to plasmid, transposon, gene cassette, and integron vehicles, as well as transduction and transformation. Phage-encoded resistance has not been reported (44). The fact that phage can encode and transfer antibiotic resistance should, however, not be surprising considering the vast array of bacterial virulence and/or fitness factors that are phage encoded, including extracellular toxins, degradative enzymes, mitogenic factors, adhesins, and others (12). Since the evolution of antibiotic resistance has been previously studied primarily with medically relevant pathogens in situations involving potentially large and heterogeneous bacterial populations exposed to the strong selective force of continuous antibiotic usage, it is possible that we have overlooked other methods of genetic exchange, perhaps in the soil milieu, that act to horizontally transfer antibiotic resistance. Indeed, metagenomic analysis of the soil environment shows a reservoir of antibiotic resistance genes with far greater genetic diversity than previously accounted for (53).
Several Wβ loci with possible roles in lysogen conversion have been identified in this study on the basis of their expression in both B. anthracis and B. cereus RSVF1 lysogens. These proteins, including Wp18, Wp25, Wp37, Wp41, and Wp45, lack obvious phage functions and are encoded within regions that harbor lysogen conversion genes in other pathogens. While Wp18 was well expressed under all conditions, Wp45 was restricted to the late-stationary-phase and sporulation cultures, Wp25 was restricted to sporulation, and Wp37 and -41 were restricted to early stationary phase and sporulation. These findings are consistent with multiple distinct roles for these phage genes, which will now require individual mutational analysis to discern. The expression of several of these loci during sporulation certainly suggests contributions to this process, particularly for the Wp37 sigma factor, as has been previously suggested (47).
In summary, we have presented a detailed genomic and physical description of two historically important and highly related B. anthracis phages. We have demonstrated that γ arose from Wβ via changes previously described for the evolution of novel phage types. Additionally, we identified several putative lysogen-converting factors the nature of which must now be investigated. Toward the larger question of phage contributions to the B. anthracis phenotype, we show that gene flux between the chromosome and infecting phage can occur, moving spore antigens and antibiotic resistance mechanisms in the process. On the basis of our work thus far, it seems more likely that phage-mediated conversion in B. anthracis may be manifested as environmental adaptations through modification of the vegetative or spore forms. Detailed studies of other B. anthracis phages isolated directly from the environment are forthcoming and are in agreement with a role for phages in ecological adaptation. Direct demonstration of B. anthracis converting to a form more suitable for survival in the environment is now a high priority.
Supplementary Material
Acknowledgments
We thank Alexander Keynan, Joshua Lederberg, Alex R. Hoffmaster, Richard Calendar, Hans Ackermann, Patrick J. Piggot, and Vasant Kumar for gifts of strains and plasmids; Eleana Sphicas at the Bio-Imaging Resource Center at The Rockefeller University for electron microscopy; Ryann Russell, Adam Pelzek, and Shi Wei Zhu for expert technical assistance; and Derrick E. Fouts and members of the V.A.F. laboratory for discussions.
This research was supported by a grant from the Northeast Biodefense Center (NBC) to R.S. and grants from the Defense Advanced Research Projects Agency (DARPA) and USPHS grant AI057472 to V.A.F.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abshire, T. G., J. E. Brown, and J. W. Ezzell. 2005. Production and validation of the use of gamma phage for identification of Bacillus anthracis. J. Clin. Microbiol. 43:4780-4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ackermann, H. W., R. R. Azizbekyan, H. P. Emadi Konjin, M. M. Lecadet, L. Seldin, and M. X. Yu. 1994. New Bacillus bacteriophage species. Arch. Virol. 135:333-344. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Armstrong, R. N. 2000. Mechanistic diversity in a metalloenzyme superfamily. Biochemistry 39:13625-13632. [DOI] [PubMed] [Google Scholar]
- 6.Baek, K., S. Svenningsen, H. Eisen, K. Sneppen, and S. Brown. 2003. Single-cell analysis of lambda immunity regulation. J. Mol. Biol. 334:363-372. [DOI] [PubMed] [Google Scholar]
- 7.Bardell, D. 1982. An 1898 report by Gamaleya for a lytic agent specific for Bacillus anthracis. J. Hist. Med. Allied Sci. 37:222-225. [DOI] [PubMed] [Google Scholar]
- 8.Bergman, N. H., and B. J. Akerley. 2003. Position-based scanning for comparative genomics and identification of genetic islands in Haemophilus influenzae type b. Infect. Immun. 71:1098-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bone, E. J., and D. J. Ellar. 1989. Transformation of Bacillus thuringiensis by electroporation. FEMS Microbiol. Lett. 49:171-177. [DOI] [PubMed] [Google Scholar]
- 10.Boydston, J. A., P. Chen, C. T. Steichen, and C. L. Turnbough, Jr. 2005. Orientation within the exosporium and structural stability of the collagen-like glycoprotein BclA of Bacillus anthracis. J. Bacteriol. 187:5310-5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown, E. R., and W. B. Cherry. 1955. Specific identification of Bacillus anthracis by means of a variant bacteriophage. J. Infect. Dis. 96:34-39. [DOI] [PubMed] [Google Scholar]
- 12.Brussow, H., C. Canchaya, and W. D. Hardt. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68:560-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brussow, H., and F. Desiere. 2001. Comparative phage genomics and the evolution of Siphoviridae: insights from dairy phages. Mol. Microbiol. 39:213-222. [DOI] [PubMed] [Google Scholar]
- 14.Bushman, F. 2002. Lateral DNA transfer. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Cao, M., B. A. Bernat, Z. Wang, R. N. Armstrong, and J. D. Helmann. 2001. FosB, a cysteine-dependent fosfomycin resistance protein under the control of σW, an extracytoplasmic-function σ factor in Bacillus subtilis. J. Bacteriol. 183:2380-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chibani-Chennoufi, S., A. Bruttin, M. L. Dillmann, and H. Brussow. 2004. Phage-host interaction: an ecological perspective. J. Bacteriol. 186:3677-3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 18.Cowles, P. B. 1931. A bacteriophage for B. anthracis. J. Bacteriol. 21:161-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davison, S., E. Couture-Tosi, T. Candela, M. Mock, and A. Fouet. 2005. Identification of the Bacillus anthracis γ phage receptor. J. Bacteriol. 187:6742-6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Palmenaer, D., C. Vermeiren, and J. Mahillon. 2004. IS231-MIC231 elements from Bacillus cereus sensu lato are modular. Mol. Microbiol. 53:457-467. [DOI] [PubMed] [Google Scholar]
- 21.Desiere, F., W. M. McShan, D. van Sinderen, J. J. Ferretti, and H. Brussow. 2001. Comparative genomics reveals close genetic relationships between phages from dairy bacteria and pathogenic streptococci: evolutionary implications for prophage-host interactions. Virology 288:325-341. [DOI] [PubMed] [Google Scholar]
- 22.d'Herelle, F. H. 1917. Sur un microbe invisible antagoniste des bacilles dysenteriques. C. R. Acad. Sci. 165:373-375. [Google Scholar]
- 23.Doulatov, S., A. Hodes, L. Dai, N. Mandhana, M. Liu, R. Deora, R. W. Simons, S. Zimmerly, and J. F. Miller. 2004. Tropism switching in Bordetella bacteriophage defines a family of diversity-generating retroelements. Nature 431:476-481. [DOI] [PubMed] [Google Scholar]
- 24.Dwyer, K. G., J. M. Lamonica, J. A. Schumacher, L. E. Williams, J. Bishara, A. Lewandowski, R. Redkar, G. Patra, and V. G. DelVecchio. 2004. Identification of Bacillus anthracis specific chromosomal sequences by suppressive subtractive hybridization. BMC Genomics 5:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ekwunife, F. S., J. Singh, K. G. Taylor, and R. J. Doyle. 1991. Isolation and purification of cell wall polysaccharide of Bacillus anthracis (ΔSterne). FEMS Microbiol. Lett. 66:257-262. [DOI] [PubMed] [Google Scholar]
- 26.Fouts, D. E., D. A. Rasko, R. Z. Cer, L. Jiang, N. B. Federova, A. Shvartsbeyn, J. J. Vamathevan, L. Tallon, R. Althoff, T. S. Arbogast, D. W. Fadrosh, T. D. Read, and S. R. Gill. Sequencing Bacillus anthracis typing phages gamma and Cherry reveals a common ancestry. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 27.Gamaleya, N. F. 1898. Bacteriolysins—ferments destroying bacteria. Russ. Arch. Pathol. Clin. Med. Bacteriol. 6:607-613. [Google Scholar]
- 28.Gardy, J. L., C. Spencer, K. Wang, M. Ester, G. E. Tusnady, I. Simon, S. Hua, K. deFays, C. Lambert, K. Nakai, and F. S. Brinkman. 2003. PSORT-B: improving protein subcellular localization prediction for gram-negative bacteria. Nucleic Acids Res. 31:3613-3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Helgason, E., O. A. Okstad, D. A. Caugant, H. A. Johansen, A. Fouet, M. Mock, I. Hegna, and Kolsto. 2000. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis—one species on the basis of genetic evidence. Appl. Environ. Microbiol. 66:2627-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hendlin, D., E. O. Stapley, M. Jackson, H. Wallick, A. K. Miller, F. J. Wolf, T. W. Miller, L. Chaiet, F. M. Kahan, E. L. Foltz, H. B. Woodruff, J. M. Mata, S. Hernandez, and S. Mochales. 1969. Phosphonomycin, a new antibiotic produced by strains of Streptomyces. Science 166:122-123. [DOI] [PubMed] [Google Scholar]
- 32.Hendrix, R. W., J. G. Lawrence, G. F. Hatfull, and S. Casjens. 2000. The origins and ongoing evolution of viruses. Trends Microbiol. 8:504-508. [DOI] [PubMed] [Google Scholar]
- 33.Hirokawa, T., S. Boon-Chieng, and S. Mitaku. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378-379. [DOI] [PubMed] [Google Scholar]
- 34.Inal, J. M., and K. V. Karunakaran. 1996. φ20, a temperate bacteriophage isolated from Bacillus anthracis exists as a plasmidial prophage. Curr. Microbiol. 32:171-175. [DOI] [PubMed] [Google Scholar]
- 35.Ishikawa, J., and K. Hotta. 1999. FramePlot: a new implementation of the frame analysis for predicting protein-coding regions in bacterial DNA with a high G+C content. FEMS Microbiol. Lett. 174:251-253. [DOI] [PubMed] [Google Scholar]
- 36.Ivins, B. E., J. W. Ezzell, Jr., J. Jemski, K. W. Hedlund, J. D. Ristroph, and S. H. Leppla. 1986. Immunization studies with attenuated strains of Bacillus anthracis. Infect. Immun. 52:454-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jensen, G. B., B. M. Hansen, J. Eilenberg, and J. Mahillon. 2003. The hidden lifestyles of Bacillus cereus and relatives. Environ. Microbiol. 5:631-640. [DOI] [PubMed] [Google Scholar]
- 38.Jensen, S. O., and P. R. Reeves. 2001. Molecular evolution of the GDP-mannose pathway genes (manB and manC) in Salmonella enterica. Microbiology 147:599-610. [DOI] [PubMed] [Google Scholar]
- 39.Kaneko, Y., K. Yanagihara, Y. Miyazaki, K. Tsukamoto, Y. Hirakata, K. Tomono, J. Kadota, T. Tashiro, I. Murata, and S. Kohno. 2003. Effects of DQ-113, a new quinolone, against methicillin- and vancomycin-resistant Staphylococcus aureus-caused hematogenous pulmonary infections in mice. Antimicrob. Agents Chemother. 47:3694-3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keim, P., and K. L. Smith. 2002. Bacillus anthracis evolution and epidemiology. Curr. Top. Microbiol. Immunol. 271:21-32. [DOI] [PubMed] [Google Scholar]
- 41.Lucchini, S., F. Desiere, and H. Brussow. 1999. Comparative genomics of Streptococcus thermophilus phage species supports a modular evolution theory. J. Virol. 73:8647-8656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lukashin, A. V., and M. Borodovsky. 1998. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 26:1107-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lupas, A., M. Van Dyke, and J. Stock. 1991. Predicting coiled coils from protein sequences. Science 252:1162-1164. [DOI] [PubMed] [Google Scholar]
- 44.Martinez, J. L., and F. Baquero. 2002. Interactions among strategies associated with bacterial infection: pathogenicity, epidemicity, and antibiotic resistance. Clin. Microbiol. Rev. 15:647-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCloy, E. 1951. Unusual behaviour of a lysogenic Bacillus strain. J. Gen. Microbiol. 5:xiv-xv. [PubMed] [Google Scholar]
- 46.McCloy, E. W. 1951. Studies on a lysogenic Bacillus strain. I. A bacteriophage specific for Bacillus anthracis. J. Hyg. 49:114-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Minakhin, L., E. Semenova, J. Liu, A. Vasilov, E. Severinova, T. Gabisonia, R. Inman, A. Mushegian, and K. Severinov. 2005. Genome sequence and gene expression of Bacillus anthracis bacteriophage Fah. J. Mol. Biol. 354:1-15. [DOI] [PubMed] [Google Scholar]
- 48.Mock, M., and A. Fouet. 2001. Anthrax. Annu. Rev. Microbiol. 55:647-671. [DOI] [PubMed] [Google Scholar]
- 49.Nagy, E., B. Pragai, and G. Ivanovics. 1976. Characteristics of phage AP50, an RNA phage containing phospholipids. J. Gen. Virol. 32:129-132. [DOI] [PubMed] [Google Scholar]
- 50.Read, T. D., S. N. Peterson, N. Tourasse, L. W. Baillie, I. T. Paulsen, K. E. Nelson, H. Tettelin, D. E. Fouts, J. A. Eisen, S. R. Gill, E. K. Holtzapple, O. A. Okstad, E. Helgason, J. Rilstone, M. Wu, J. F. Kolonay, M. J. Beanan, R. J. Dodson, L. M. Brinkac, M. Gwinn, R. T. DeBoy, R. Madpu, S. C. Daugherty, A. S. Durkin, D. H. Haft, W. C. Nelson, J. D. Peterson, M. Pop, H. M. Khouri, D. Radune, J. L. Benton, Y. Mahamoud, L. Jiang, I. R. Hance, J. F. Weidman, K. J. Berry, R. D. Plaut, A. M. Wolf, K. L. Watkins, W. C. Nierman, A. Hazen, R. Cline, C. Redmond, J. E. Thwaite, O. White, S. L. Salzberg, B. Thomason, A. M. Friedlander, T. M. Koehler, P. C. Hanna, A. B. Kolsto, and C. M. Fraser. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423:81-86. [DOI] [PubMed] [Google Scholar]
- 51.Redmond, C., I. Henderson, P. C. B. Turnbull, and J. Bowen. 1996. Phage from different strains of Bacillus anthracis. Salisbury Med. Bull. Spec. Suppl. 87:60-63. [Google Scholar]
- 52.Ren, D., Y. Wang, Z. Wang, J. Cui, H. Lan, and J. Zhou. 2003. Complete DNA sequence and analysis of two cryptic plasmids isolated from Lactobacillus plantarum. Plasmid 50:70-73. [DOI] [PubMed] [Google Scholar]
- 53.Riesenfeld, C. S., R. M. Goodman, and J. Handelsman. 2004. Uncultured soil bacteria are a reservoir of new antibiotic resistance genes. Environ. Microbiol. 6:981-989. [DOI] [PubMed] [Google Scholar]
- 54.Rohwer, F. 2003. Global phage diversity. Cell 113:141. [DOI] [PubMed] [Google Scholar]
- 55.Schuch, R., D. Nelson, and V. A. Fischetti. 2002. A bacteriolytic agent that detects and kills Bacillus anthracis. Nature 418:884-889. [DOI] [PubMed] [Google Scholar]
- 56.Schuch, R., R. C. Sandlin, and A. T. Maurelli. 1999. A system for identifying post-invasion functions of invasion genes: requirements for the Mxi-Spa type III secretion pathway of Shigella flexneri in intercellular dissemination. Mol. Microbiol. 34:675-689. [DOI] [PubMed] [Google Scholar]
- 57.Smith, R. F., B. A. Wiese, M. K. Wojzynski, D. B. Davison, and K. C. Worley. 1996. BCM Search Launcher—an integrated interface to molecular biology data base search and analysis services available on the World Wide Web. Genome Res. 6:454-462. [DOI] [PubMed] [Google Scholar]
- 58.Steichen, C., P. Chen, J. F. Kearney, and C. L. Turnbough, Jr. 2003. Identification of the immunodominant protein and other proteins of the Bacillus anthracis exosporium. J. Bacteriol. 185:1903-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stragier, P., C. Bonamy, and C. Karmazyn-Campelli. 1988. Processing of a sporulation sigma factor in Bacillus subtilis: how morphological structure could control gene expression. Cell 52:697-704. [DOI] [PubMed] [Google Scholar]
- 60.Sullivan, M. B., M. L. Coleman, P. Weigele, F. Rohwer, and S. W. Chisholm. 2005. Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol. 3:e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sylvestre, P., E. Couture-Tosi, and M. Mock. 2002. A collagen-like surface glycoprotein is a structural component of the Bacillus anthracis exosporium. Mol. Microbiol. 45:169-178. [DOI] [PubMed] [Google Scholar]
- 62.Tetart, F., C. Desplats, and H. M. Krisch. 1998. Genome plasticity in the distal tail fiber locus of the T-even bacteriophage: recombination between conserved motifs swaps adhesin specificity. J. Mol. Biol. 282:543-556. [DOI] [PubMed] [Google Scholar]
- 63.Turnbull, P. C. B. 1999. Definitive identification of Bacillus anthracis—a review. J. Appl. Microbiol. 87:237-240. [DOI] [PubMed] [Google Scholar]
- 64.Twort, F. W. 1915. An investigation on the nature of ultra-microscopic viruses. Lancet ii:1241-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ventura, M., A. Bruttin, C. Canchaya, and H. Brussow. 2002. Transcription analysis of Streptococcus thermophilus phages in the lysogenic state. Virology 302:21-32. [DOI] [PubMed] [Google Scholar]
- 66.Walter, M. H., and D. D. Baker. 2003. Three Bacillus anthracis bacteriophages from topsoil. Curr. Microbiol. 47:55-58. [DOI] [PubMed] [Google Scholar]
- 67.Watanabe, T., A. Morimoto, and T. Shiomi. 1975. The fine structure and the protein composition of gamma phage of Bacillus anthracis. Can. J. Microbiol. 21:1889-1892. [DOI] [PubMed] [Google Scholar]
- 68.Weinbauer, M. G., and F. Rassoulzadegan. 2004. Are viruses driving microbial diversification and diversity? Environ. Microbiol. 6:1-11. [DOI] [PubMed] [Google Scholar]
- 69.Weitz, J. S., H. Hartman, and S. A. Levin. 2005. Coevolutionary arms races between bacteria and bacteriophage. Proc. Natl. Acad. Sci. USA 102:9535-9540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu, J., R. W. Hendrix, and R. L. Duda. 2004. Conserved translational frameshift in dsDNA bacteriophage tail assembly genes. Mol. Cell 16:11-21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.