Abstract
Division site placement in Escherichia coli involves interactions of the MinD protein with MinC and MinE and with other MinD molecules to form membrane-associated polymeric structures. In this work, as part of a study of these interactions, we established that heterologous membrane-associated proteins such as MinD can be targeted to the yeast nuclear membrane, dependent only on the presence of a membrane-binding domain and a nuclear targeting sequence. Targeting to the nuclear membrane was equally effective using the intrinsic MinD membrane-targeting domain or the completely unrelated membrane-targeting domain of cytochrome b5. The chimeric proteins differing in their membrane-targeting sequences were then used to establish the roles of membrane association and specificity of the membrane anchor in MinD interactions, using the yeast two-hybrid system. The chimeric proteins were also used to show that the membrane association of MinD and MinE in E. coli cells had no specificity for the membrane anchor, whereas formation of MinDE polar zones and MinE rings required the presence of the native MinD membrane-targeting sequence.
Proper placement of the Escherichia coli division site depends on the coordinate action of the MinC, MinD and MinE proteins (3). This involves the cyclic assembly and disassembly of membrane-associated MinCDE polar zones and MinE rings, which undergo repeated cycles of pole-to-pole oscillations during the cell cycle (reviewed in references 14 and 25). During this process, MinD associates with the E. coli cytoplasmic membrane via a short carboxy-terminal sequence, the membrane-targeting sequence (MTS) (10, 31) (Fig. 1A). The membrane-associated MinD then serves to bring MinC and MinE to the membrane (22, 23).
FIG. 1.
Membrane binding of MinD and cytochrome b5. (A) The MTS of MinD, shown as a cylinder, is an amphipathic α helix oriented parallel to the phospholipid bilayer. Nonpolar amino acid side chains extend downward into the interior of the bilayer. (B) The hydrophobic MBD of cytochrome b5 is deeply inserted in the phospholipid bilayer. (C) Schematic representation of AD-Yfp-MinD (top) and AD-Yfp-MinDΔ10-b5MBD (bottom). The positions of C-terminal MTS of MinD, the b5MBD, and the NLS are indicated.
In vitro and in vivo studies have indicated that MinD self-interacts to form membrane-associated polymers (9, 30) that are organized into a cytoskeletal-like helical structure within the cell (29). This membrane-associated structure may act as a scaffold for membrane assembly of the other Min proteins.
MinD interactions have previously been studied by the yeast two-hybrid system (16, 34), but it is not known whether or not the observed interactions reflected the behavior of the protein in the nucleoplasm or within the yeast nuclear membrane. This reflects a paucity of information about incorporation of exogenous proteins into the inner nuclear membrane of yeast cells. In this article, we show that MinD can be targeted exclusively to the yeast nuclear membrane, dependent on the presence of a high efficiency nuclear localization signal (NLS) and a membrane-targeting domain. Targeting was equally effective when the intrinsic MinD MTS was replaced by the unrelated membrane-binding domain of cytochrome b5. The ability to swap membrane-binding domains made it possible to use the yeast two-hybrid system to establish the role of membrane association and the relative roles of the membrane anchor and extramembranous domain in the MinD-MinD interaction and the interactions of MinD with MinC and MinE. The chimeric proteins were also used to probe the specificity of the membrane-targeting sequence in the membrane association of MinD and MinE in E. coli cells and in formation of MinDE polar zones and MinE rings.
MATERIALS AND METHODS
Strains and growth conditions.
All E. coli strains were grown in LB medium (17), to which 100 μg/ml ampicillin, 25 μg/ml kanamycin, or 30 μg/ml chloramphenicol was added when indicated. All plasmid constructions were done in E. coli DH5α (17) and then transferred to either E. coli RC1 (ΔminCDE) (26) or HL1 (ΔminDE) (8) or to yeast strains AH109 or SFY626 (Clontech). Yeast strains were grown in supplemented synthetic dropout medium (yeast two-hybrid system manual, Clontech).
Plasmids.
Plasmids are listed in Table 1. Primer sequences and details will be provided on request. pATD260 was constructed as pADX1 (16), except a PCR-generated fragment coding for the first 260 amino acids of MinD (MinDΔ10) was used as the insert instead of minD. pADYD1 was constructed by PCR-mediated transfer of the EcoRI/XbaI yfp fragment from pLE7 (29) to pADX1. pADYD260 was constructed as pADYD1, except pATD260 was used as the vector. pADYDCb5 was constructed by a triple-fragment ligation between the large XbaI/BglII fragment from pADYD1, a PCR-generated XbaI/KpnI fragment from pATD260 that codes for MinDΔ10, and the PCR-generated KpnI/BglII fragment coding for the membrane-binding domain of rabbit cytochrome b5 (L91 to D133) (b5MBD) (Swiss-Prot accession number P00169). Two overlapping primers were used to generate the b5MBD fragment. pBDDCb5 was constructed as pADYDCb5, except the pGBKT7 EcoRI/BamHI large fragment was used as the vector and an EcoRI/KpnI PCR-generated fragment from pATD260 encoding MinDΔ10 was used as the insert. pADCb5 was made by ligating the EcoRI/BamHI large fragment of pGADT7 to the EcoRI/BglII PCR-generated b5MBD fragment. pBDCb5 was made as pADCb5, except pGBKT7 was used as a vector. pBDYD1 was constructed by ligating the EcoRI/BglII yfp::minD fragment of pADYD1 to EcoRI/BamHI-digested pGBKT7 vector. pBDCDCb5 was constructed by a triple-fragment ligation between EcoRI/BamHI-digested pGBKT7 vector, the XbaI/BglII b5MBD fragment of pADYDCb5, and the EcoRI/XbaI PCR-generated fragment coding for cfp (cyan fluorescent protein) using pLE18 as the template (29). pBDYDCb5 was constructed by transfer of the EcoRI/BglII fragment of pADYDCb5 to EcoRI/BamHI-digested pGBKT7. pADCDCb5 was constructed by transfer of the EcoRI fragment of pBDCDCb5 to the EcoRI-digested pADYDCb5; the correct orientation of the insert was selected based on KpnI fragment size. pAT1 was constructed by four-fragment ligation between the XbaI/BamHI-digested pYLS68 vector (28), the XbaI/KpnI fragment of pADYDCb5 coding for MinDΔ10, the PCR-generated KpnI/XhoI fragment encoding the bacterial codon-optimized sequence of b5MBD (6), and the XhoI/BamHI PCR-generated MinE-encoding fragment from pYLS68. The native minE ribosome-binding site sequence was appended to the 5′ end of the forward primer to correct for the disruption introduced by deleting the MTS sequence of MinD. The bacterial codon-optimized b5MBD fragment was made by PCR using four overlapping primers. pAT5 was constructed by ligation between the XbaI/BglII MinDΔ10-encoding fragment from pATD260 and the XbaI/BamHI large fragment of pYLS68. pAT6 was constructed by transfer of the EcoRI/XhoI fragment from pAT1 to the EcoRI/SalI-digested pMLB1113 vector (3). pAT7 was constructed by transfer of the EcoRI/HindIII fragment from pYLS95 (29) coding for MinE-Cfp to pSJ4 (26). pADMTSEc was constructed by inserting the EcoRI/BglII PCR-generated E. coli MinD MTS fragment (corresponding to nucleotide [nt] 781 to 811), using pADX1 as template, into EcoRI/BglII-digested pGAD424. pBDMTSEc was constructed as pADMTSEc, except pGBKT7 digested with EcoRI/BamHI was used as a vector. pGADC was constructed by ligation between the EcoRI/BamHI PCR-generated minC fragment and pGADT7. pGBKC was constructed as pGADC, except pGBKT7 was used as a vector. pADCMEc was constructed by three-fragment ligation between the PCR-generated EcoRI/HindIII minC fragment by using pGADC as a template, the HindIII/BglII PCR-generated E. coli MinD MTS by using pADX1 as a template (corresponding to nt 769 to 811), and the EcoRI/BamHI-digested pGADT7 vector. pBDCMEc was constructed as pADCMEc, except pGBKT7 was used as a vector. pADYCMEc was constructed by a four-fragment ligation between the EcoRI/XbaI PCR-generated yfp fragment using pLE7 as a template, the PCR-generated XbaI/HindIII minC fragment by using pGADC as a template, the HindIII/BglII PCR-generated E. coli MinD MTS (corresponding to nt 769 to 811) by using pADX1 as a template, and the EcoRI/BamHI-digested pGADT7 vector. pADCMBs, pADYCMBs, and pBDCMBs were constructed as pADCMEc, pADYCMEc, and pBDCMEc, respectively, except B. subtilis MinD MTS was used (corresponding to nt 742 to 807), three glycines were inserted between MinC and MTSBs, and a BamHI site was used instead of BglII. The Bacillus MinD MTS was PCR generated by using two overlapping primers.
TABLE 1.
Plasmid list
| Plasmida | Characteristics |
|---|---|
| pAT1 | Plac-yfp::minDΔ10::b5MBD-minE::cfp |
| pAT5 | Plac-yfp::minDΔ10 |
| pAT6 | Plac-yfp::minDΔ10::b5MBD |
| pAT7 | Para-minE::cfp |
| pADX1b | PADH1-Gal4AD::minD |
| pATD260 | PADH1-Gal4AD::minDΔ10 |
| pADYD1 | PADH1-Gal4AD::yfp::minD |
| pADYD260 | PADH1-Gal4AD::yfp::minDΔ10 |
| pADYDCb5 | PADH1-Gal4AD::yfp::minDΔ10::b5MBD |
| pADCDCb5 | PADH1-Gal4AD::cfp::minDΔ10::b5MBD |
| pADCb5 | PADH1-Gal4AD::b5MBD |
| pMDB1c | PADH1-Gal4BD::minD |
| pBDYD1 | PADH1-Gal4BD::yfp::minD |
| pBDDCb5 | PADH1-Gal4BD::minDΔ10::b5MBD |
| pBDYDCb5 | PADH1-Gal4BD::yfp::minDΔ10::b5MBD |
| pBDCDCb5 | PADH1-Gal4BD::cfp::minDΔ10::b5MBD |
| pBDCb5 | PADH1-Gal4BD::b5MBD |
| pADMTSEc | PADH1-Gal4AD::MTSEc |
| pBDMTSEc | PADH1-Gal4BD::MTSEc |
| pGADC | PADH1-Gal4AD::minC |
| pGBKC | PADH1-Gal4BD::minC |
| pADCMEc | PADH1-Gal4AD::minC::MTSEc |
| pADYCMEc | PADH1-Gal4AD::yfp::minC::MTSEc |
| pBDCMEc | PADH1-Gal4BD::minC::MTSEc |
| pADCMBs | PADH1-Gal4AD::minC::MTSBs |
| pADYCMBs | PADH1-Gal4AD::yfp::minC::MTSBs |
| pBDCMBs | PADH1-Gal4BD::minC::MTSBs |
Yeast two-hybrid system experiments.
Plasmids were transferred from E. coli DH5α to yeast strains AH109 or SFY626 by using the lithium acetate method (7). Protein interactions in SFY626 cotransformants able to grow in the absence of leucine and tryptophan were measured by determining the β-galactosidase activity (19; yeast two-hybrid system manual, Clontech). The ability to grow in the absence of histidine and adenine of AH109 cotransformants was also used to indicate protein interactions.
Fluorescence microscopy.
E. coli cells containing plasmids coding for Yfp- and Cfp-labeled proteins were grown in the presence of 10 μM of IPTG (isopropyl β-D-thiogalactoside), and labeled proteins were detected by fluorescence microscopy as previously described (29). Images were not subjected to deconvolution processing. To check for MinDE polar zones and MinE-ring formation in the presence of MinDΔ10-b5MBD, RC1/pAT1 was grown in the presence of IPTG (10, 20, 50, and 100 μM) and RC1/pAT6/pAT7 was grown in the presence of 10 μM IPTG and L-arabinose (0.00005%, 0.0001%, and 0.0025%). To test for MinC-induced septation inhibition, HL1/pAT1 and HL1/pYLS68 were grown in the presence of IPTG (10, 20, 50, and 100 μM). For study of protein localization in yeast, AH109 cells containing plasmids encoding Yfp and/or Cfp-labeled proteins were grown overnight in the absence of leucine and tryptophan at 30°C, diluted 20 times in the same medium, and examined by fluorescence microscopy after 4 to 5 h growth at 30°C. For DAPI (4,′6′-diaminido-2-phenylindole) staining, cells were fixed in presence of 0.2% glutaraldehyde and 2% formaldehyde for 20 min and then stained with 2 μg/ml DAPI for 10 min on ice and washed with saline solution before microscopy.
RESULTS
Targeting MinD to the yeast nuclear membrane.
To determine whether heterologous membrane-associated proteins such as MinD can be targeted to the yeast nuclear membrane, we inserted the coding sequence for yellow fluorescent protein (Yfp) between MinD and the GAL4 transcriptional activation domain (AD) (Fig. 1C) or the GAL4 DNA-binding domain (BD) in the standard yeast two-hybrid vectors pGAD424 and pGBKT7 (Clontech). This led to formation of Yfp-labeled chimeric proteins that also contained an NLS at the amino-terminus of the fusion proteins.
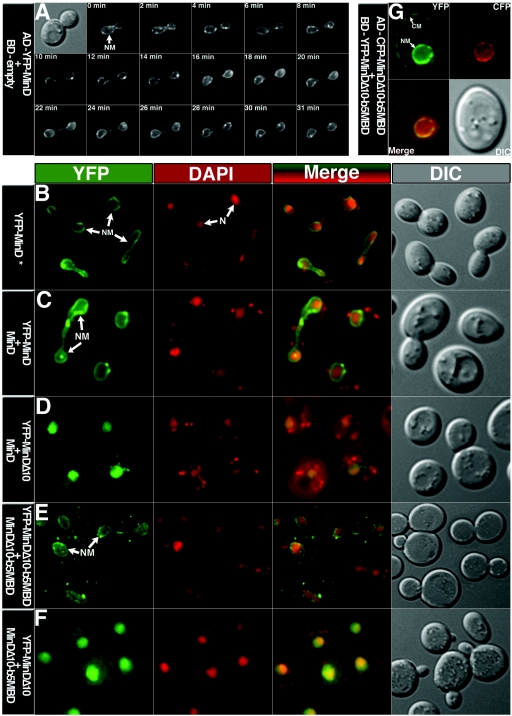
When expressed in yeast cells, Yfp-MinD was exclusively located in the nuclear membrane when present in the activating domain construct containing the simian virus 40 (SV40) NLS. The fluorescent Yfp-MinD was visible as a circumferential envelope surrounding the DAPI-stained nuclear DNA of each cell (Fig. 2B). The localization of Yfp-MinD in the nuclear membrane was observed at all stages of the division cycle and Yfp-MinD remained within the membrane of dividing nuclei of septating cells (Fig. 2A). Yfp-MinD was also targeted to the nuclear membrane when present in the BD-Yfp-MinD fusion that contained the Gal4 NLS instead of the SV40 NLS. In this case, Yfp-MinD was not exclusively targeted to the nuclear membrane, and some of the labeled protein was also present in the cytoplasmic membrane (data not shown). This presumably reflects the difference in efficiency of targeting proteins to the nucleus by the SV40 and Gal4 NLS (20). These results showed that an exogenous membrane-associated protein can be targeted to the yeast nuclear membrane and that the efficiencies of targeting the proteins to the nucleus appear to differ depending on the NLS.
FIG. 2.
Localization of fluorescently labeled MinD proteins in yeast AH109 cells. The proteins were expressed from the relevant plasmids (Table 1) as indicated at the left of each series of micrographs (AD fusion protein in the first line and BD fusion protein, when present, in the second line). (A) Time-lapse micrographs showing a dividing nucleus at different stages of separation; time is shown in minutes at the top of each YFP image. (B through F) Columns show yellow fluorescent protein (YFP) images (green), DAPI images (red), merged images (green-red), and differential interference contrast (DIC) images (gray), as indicated. Nuclear membrane (NM) and nucleus (N) are shown by arrows. In panel B, Yfp-MinD is expressed in presence of empty-BD plasmid pGBKT7. (G) The cyan fluorescent protein (CFP) image is shown in red and the YFP image in green.
Role of MTS in targeting MinD to the membrane.
It has previously been shown that MinD associates with phospholipid bilayers through a carboxy-terminal amphipathic helix of 10 residues (Fig. 1A). Membrane binding involves interactions between nonpolar residues on one face of the helix and acyl groups of phospholipids, and may also involve interaction between phospholipid head groups and charged residues on the other face of the helix (10, 31).
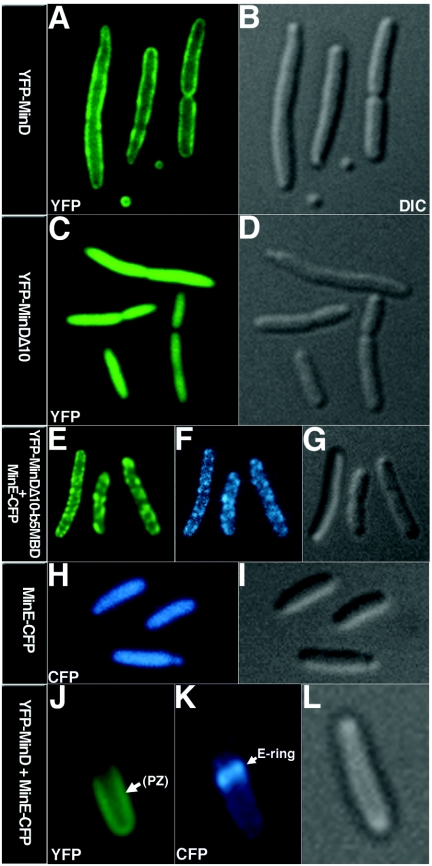
In E. coli cells, Yfp-MinD was localized at the cell periphery (Fig. 3A), whereas truncated MinD (Yfp-MinDΔ10) that lacked the carboxy-terminal MTS was diffusely distributed throughout the cytoplasm (Fig. 3C). This confirmed previous reports that the membrane localization of MinD in E. coli is dependent on the MTS (10, 31).
FIG. 3.
Localization of fluorescently labeled MinD and MinE in E. coli RC1. The proteins were expressed from relevant plasmids (Table 1) as indicated at the left of each image. (A, C, E, and J) Yellow fluorescent protein (YFP) images. MinD polar zone (PZ) is shown by the arrow in panel J. (F, H, and K) Cyan fluorescent protein images of MinE-CFP. MinE ring is shown by the arrow in (K). (B, D, G, I, and L) Differential interference contrast (DIC) images.
In yeast cells, the fluorescently labeled full-length MinD, in AD-Yfp-MinD, was targeted to the nuclear membrane when expressed in the presence (Fig. 2C) or the absence of BD-MinD (Fig. 2B). In contrast, when the MTS was absent, in AD-Yfp-MinDΔ10, the labeled protein was absent from the nuclear membrane and was diffusely distributed in the nucleoplasm (Fig. 2D). Thus, the localization of MinD to the yeast nuclear membrane is dependent on the MTS.
The failure of MinDΔ10 to associate with the nuclear membrane was accompanied by loss of MinD-MinD interaction in the two-hybrid system, as shown by the absence of interaction between MinDΔ10 and full-length MinD (Table 2), suggesting that membrane association may play a role in the MinD-MinD interaction. Significantly, although MinDΔ10 was unable to interact with MinD, it retained its ability to interact with MinC and MinE1-31, the MinD-binding domain of MinE (15) (Table 2). This shows that the loss of MinD-MinD interaction associated with the deletion of the MTS was not due to global unfolding of MinDΔ10 but was due to the specific absence of the membrane-targeting sequence. This also confirmed previous evidence that interaction between MinDΔ10 and MinC is independent of the MinD MTS (10). Significant MinD-MinE and MinD-MinC interaction occurred in the absence of the MinD MTS. This indicates that the interaction of MinD with MinC and MinE does not require that MinD be membrane-associated, although membrane association may increase the efficiency of the interactions since the presence of MTS resulted in twofold increase in β-galactosidase activity (Table 2).
TABLE 2.
MinD interactions in the yeast two-hybrid systema
| GAL4 AD fused to: | GAL4 BD fused to:
|
|||
|---|---|---|---|---|
| MinD | MinDΔ10 | MinC | MinE1-31 | |
| MinD | 100 ± 3.5 | 0.74 ± 0.16 | 100 ± 1.4 | 100 ± 3.1 |
| MinDΔ10 | 0.38 ± 0.06 | 4.1 ± 0.24 | 49 ± 4 | 41 ± 0.2 |
| MinC | 12.8 ± 1 | 113.2 ± 23 | ND | ND |
| Nothing | 0.4 ± 0.08 | 0.55 ± 0.11 | 0.09 ± 0.02 | 0.6 ± 0 |
Interaction is indicated as β-galactosidase activity; relative activity is shown as the percentage of MinD-MinD interaction (190 Miller units) (for MinD and MinDΔ10 columns), MinD-MinC interaction (1,453 Miller units) (for the MinC column), and MinD-MinE1-31 interaction (250 Miller units) (for the MinE1-31 column). The interactions were further confirmed by the ability of AH109 cotransformed with each of the paired constructs to grow in the absence of histidine and adenine. All cotransformants grew in the absence of histidine and adenine except for the six pairs with β-galactosidase relative activity less than 0.75. ND, not determined.
Substitution of the MinD MTS by the cytochrome b5 membrane-binding domain.
To further define the role of the MinD membrane-binding sequence, we replaced the MTS of MinD with an unrelated membrane-binding domain, the membrane-anchoring sequence of rabbit cytochrome b5, to construct a chimeric MinD protein (MinDΔ10-b5MBD). In contrast to the amphipathic α-helical MinD MTS which appears to orient on the surface of the lipid bilayer (Fig. 1A), the cytochrome b5 membrane-binding domain dips deeply into the phospholipid bilayer, acting as a hydrophobic integral membrane anchor (33) (Fig. 1B). We used Yfp to fluorescently label the MinDΔ10-b5MBD (Fig. 1C).
Localization studies showed that the cytochrome b5 membrane-binding domain was fully effective in targeting the MinDΔ10 protein to the nuclear membrane (Fig. 2E). This was true both in nondividing cells and in dividing cells in which the nuclei were at different stages of separation. Thus, the cytochrome b5 membrane-binding domain was able to replace the MinD membrane-targeting sequence in restoring nuclear membrane binding to the truncated MinDΔ10 protein.
MinD molecules that contained the membrane-binding domain of MinD or cytochrome b5 were unable to recruit MinDΔ10 to the nuclear membrane. Thus, MinDΔ10 (AD-Yfp-MinDΔ10) was not localized in the nuclear membrane when coexpressed with either BD-MinD (Fig. 2D) or BD-MinDΔ10-b5MBD (Fig. 2F). This indicates that membrane-associated MinD is incapable of stable interaction with nucleoplasmic MinD.
As expected, in double-label experiments with AD-Cfp-MinDΔ10-b5MBD and BD-Yfp-MinDΔ10-b5MBD the labeled proteins were both targeted to the nuclear membrane (Fig. 2G). In this case a fraction of the Yfp-labeled protein was also present in the cytoplasmic membrane, presumably reflecting the difference in efficiency of nuclear targeting by the Gal4 NLS and the SV40 NLS in the BD and AD constructs, respectively (20).
MinD-MinD interactions.
To determine whether the nuclear membrane localization was required for MinD to interact with other MinD molecules in a reaction that normally leads to formation of MinD polymers (9), MinD-MinD interactions were examined in different MinD constructs in the two-hybrid system. The MinD-MinD interaction in the two-hybrid system was lost when either of the interacting proteins was devoid of a membrane-binding domain (Table 2). This was consistent with the inability of MinD containing a membrane-binding domain to recruit MinDΔ10 to the nuclear membrane, as discussed above. When both partners lacked a membrane-binding domain, 96% of the MinD-MinD interaction was lost (Tables 2 and 3). These results indicate either that the MinD MTS sequence includes specific determinants for MinD-MinD interaction or that both partners must be bound to the membrane in order to significantly interact.
TABLE 3.
Interactions of MinDΔ10-b5MBD in the yeast two-hybrid system
| GAL4 AD fused to: | GAL4 BD fused to: | Activitya |
|---|---|---|
| Yfp-MinD | MinD | 100 ± 6 |
| Yfp-MinDΔ10 | MinD | 0.8 ± 0.1 |
| Yfp-MinDΔ10-b5MBD | MinDΔ10-b5MBD | 58 ± 2.2 |
| Yfp-MinDΔ10 | MinDΔ10-b5MBD | 1 ± 0.8 |
| Yfp-MinDΔ10-b5MBD | MinDΔ10 | 2 ± 0.4 |
| Yfp-MinD | MinDΔ10-b5MBD | 8 ± 0.7 |
| YFP-MinDΔ10-b5MBD | MinD | 5 ± 0.5 |
| b5MBD | MinDΔ10-b5MBD | 1 ± 0.2 |
| Yfp-MinDΔ10-b5MBD | b5MBD | 1 ± 0.4 |
| Yfp-MinD | MTS | 0.6 ± 0.2 |
| MTS | MinD | 0.8 ± 0.2 |
| Yfp-MinDΔ10 | MinDΔ10 | 3.9 ± 0.6 |
| Yfp-MinD | MinC | 100 ± 15 |
| Yfp-MinDΔ10-b5MBD | MinC | 111.6 ± 3.8 |
| Yfp-MinD | MinE1-31 | 100 ± 16 |
| Yfp-MinDΔ10-b5MBD | MinE1-31 | 165.4 ± 8.3 |
| Nothing | MinD | 1 ± 0.2 |
β-Galactosidase activity is normalized to interactions between Yfp-MinD and MinD (67 Miller units) (for rows 1 through 12 and 17), Yfp-MinD and MinC (1,117 Miller units) (for rows 13 and 14), and Yfp-MinD and MinE1-31 (34 Miller units) (for rows 15 and 16). The interactions were further confirmed by the ability of AH109 cotransformants to grow in the absence of histidine and adenine; growth was observed only in the strains indicated on rows 1, 3, 6, 7, and 12 through 16.
Evidence that the MinD MTS does not directly participate in the MinD-MinD interaction came from two-hybrid experiments. Thus, when both partners contained the cytochrome b5 MBD in place of the native MinD MTS, the extent of self-interaction was only slightly lower than the MinD-MinD interaction observed when both partners contained the native MinD MTS (Table 3). Neither the cytochrome b5 membrane-binding domain alone nor the MinD MTS alone interacted with MinDΔ10-b5MBD or with MinD, respectively (Table 3), showing that the self-interactions of MinDΔ10-b5MBD and of MinD were not merely due to interaction between the cytochrome b5 or MinD membrane-binding sequences. The absence of interaction between MinDΔ10-b5MBD or MinD and their respective membrane anchor was not due to breakdown of the proteins as shown by Western blots of total yeast extract (data not shown). This experiment also confirmed the previous observation (Table 2) that MinD-MinD interaction was lost when either partner lacked a membrane anchoring sequence (Table 3).
The fact that the interacting MinD partners can contain the unrelated cytochrome b5 MBD and the failure of the MinD MTS to interact with full-length MinD argue against the idea that the MinD MTS participates directly in the MinD-MinD interaction. Taken together with the observation that significant MinD-MinD interaction requires that both interacting partners contain a membrane-anchoring domain, this indicates that membrane binding is a prerequisite for MinD-MinD interaction and strongly implies that the only role of the MTS in the MinD-MinD interaction is to attach MinD to the membrane.
Interestingly, the interaction between wild-type MinD and MinDΔ10-b5MBD was significantly weaker than the interaction between partners that contained the same membrane-binding domain (Table 3). This suggests that heterologous membrane anchors within membrane-associated MinD molecules may affect the relative orientations of the remainder of the proteins, thereby affecting the efficiency of interaction between the two MinD moieties.
MinC and MinE1-31 interacted with MinDΔ10-b5MBD as well as with full-length MinD (Table 3) indicating that the b5MBD anchor did not interfere with the interactions between MinDΔ10-b5MBD and the other Min proteins.
MinD self-interaction and membrane association.
Specific interaction between membrane-bound MinD molecules reflects collisions due to random lateral diffusion of the proteins within the two-dimensional membrane structure. There was a 25-fold increase in MinD-MinD interaction when both MinD molecules were membrane associated (Tables 2 and 3). This could reflect conformational changes in MinD, induced by the membrane association, that increased the self-association properties of MinD. Alternatively, the effect of membrane association might be explained by the increased probability of collision between MinD molecules because of the higher effective concentration of molecules located within a two-dimensional membrane compartment than within a three-dimensional aqueous compartment (27). To evaluate the effect of the increase in collisional interaction that occurs when a self-interacting soluble protein is associated with a membrane in a manner similar to MinD, we tethered MinC to the yeast nuclear membrane by appending to its carboxy terminus the MinD MTS sequence from E. coli (MTSEc) or B. subtilis (MTSBs). MinC is normally a cytoplasmic protein when expressed in the absence of MinD (22). As shown in Fig. 4A through D, the AD-Yfp-MinC-MTSEc and AD-Yfp-MinC-MTSBs proteins were exclusively targeted to the yeast nuclear membrane by the membrane-binding extensions. Tethering MinC to the nuclear membrane increased the MinC-MinC interaction approximately twofold over the self-interaction of MinC without the MTS extension, as measured by increase in β-galactosidase activity (Table 4). We presume that the approximately twofold increase in activity predominantly reflected the increased probability of collision between membrane-associated molecules compared with non-membrane-associated molecules. This contrasts with the approximately 25-fold increase in MinD-MinD interaction that occurred when MinD was membrane associated (Tables 2 and 3).
FIG. 4.
Targeting of MinC to nuclear membrane and cell division phenotype of ΔminDE strains. (A through D) Yeast AH109 cells expressed the AD fusion protein shown together with BD-MinC-MTSEc (A) or BD-MinC-MTSBs (C). The yeast nuclear membrane (NM) is shown by the arrows. (A and C) YFP image; (B and D) DIC image. (E through H) HL1 E. coli cells (minC+ ΔminDE) expressed the plasmid-encoded proteins shown at the left. (E and G) Cells were grown in presence of 0.4% glucose to suppress expression of the plasmid-encoded proteins. (F and H) Cells were grown in presence of 10 μM IPTG to induce expression of the plasmid-encoded proteins. minC was expressed constitutively from the chromosome under control of the normal min promoter.
TABLE 4.
MinC-MinC interactions with or without the MinD MTS
| GAL4 AD fused to: | GAL4 BD fused to: | Activitya |
|---|---|---|
| MinC | MinC | 100 ± 9.7 |
| MinC-MTSEc | MinC-MTSEc | 233 ± 6.0 |
| MinC-MTSBs | MinC-MTSBs | 184.5 ± 8.7 |
| MinC-MTSBs | MinC-MTSEc | 146.2 ± 3 |
| MinC | Nothing | 0.33 ± 0.07 |
| MinC-MTSEc | Nothing | 0.32 ± 0.08 |
| MinC-MTSEc | MTSEc | 0.34 ± 0.14 |
β-Galactosidase activity is normalized to MinC self-interaction (78 Miller units).
Localization of MinDΔ10-b5MBD in E. coli.
The membrane-binding domain of cytochrome b5 was as effective as the MinD MTS in directing MinDΔ10 to the nuclear membrane and in restoring MinD self-interaction to MinDΔ10. In E. coli, MinD when expressed by itself localizes to the cell periphery by binding to the membrane. When coexpressed with MinE, MinD redistributes into a peripheral horseshoe-like structure at the cell pole, the polar zone (reviewed in reference 14 and 25). To determine whether the cytochrome b5 membrane-binding domain can replace the native MinD membrane-targeting sequence in supporting a normal pattern of MinD localization in E. coli cells, a bacterial codon-optimized cytochrome b5 membrane-binding domain was attached in-frame to the carboxy terminus of truncated MinDΔ10. Yfp-MinDΔ10-b5MBD and MinE-Cfp were then coexpressed in double-label experiments in E. coli Δmin cells (Fig. 3E through G). Under these conditions, Yfp-MinDΔ10-b5MBD was not only peripherally localized in the E. coli cells but also formed long-range coiled structures winding around the cell that resembled the cytoskeleton-like MinD spiral structures that have previously been described with normal MinD (Fig. 3E) (29). In contrast, Yfp-MinDΔ10 showed a diffuse localization pattern (Fig. 3C), reflecting the loss of its membrane anchor. This confirms that the cytochrome b5 membrane-binding domain can bring MinD to the E. coli membrane. The ability of the markedly dissimilar cytochrome b5 MBD to substitute for native MinD MTS in supporting the association of MinD with the E. coli membrane shows that the membrane association of MinD in E. coli has no specificity for the MinD MTS. The total lack of specificity for the membrane anchor extends the previous report that the membrane-targeting sequence of FtsA, which is structurally similar to MinD MTS, can direct MinDΔ10 to the E. coli membrane (21).
The membrane-associated MinDΔ10-b5MBD was also capable of recruiting MinE to the membrane, as shown by the peripheral localization pattern of MinE-Cfp (Fig. 3F), whereas, as expected, MinE-Cfp was diffusely localized when expressed in the absence of MinDΔ10-b5MBD or MinD (Fig. 3H). However, it was striking that MinE rings and MinD-MinE polar zones were not formed when Yfp-MinDΔ10-b5MBD and MinE-Cfp were coexpressed (Fig. 3E and F). In this regard, MinDΔ10-b5MBD differed significantly from MinD that contains its normal membrane-targeting sequence, which induces formation of polar zones and MinE rings when coexpressed with MinE (Fig. 3J and K) (5, 8, 24). The failure of MinDΔ10-b5MBD to induce the formation of MinDE polar zones and E-rings held true over a wide range of MinE expression (data not shown).
Formation of MinCDE polar zones is believed to be necessary to keep the MinC division inhibitor away from midcell, thereby preventing inhibition of septation at the normal midcell division site. Consistent with this idea, coexpression of Yfp-MinD and MinE-Cfp in a minC+ ΔminDE strain gave a relatively normal cell division pattern (Fig. 4H), whereas coexpression of Yfp-MinDΔ10-b5MBD and MinE-Cfp under the same conditions led to the formation of long nonseptate filaments (Fig. 4F). The filamentation phenotype presumably reflects the fact that MinDΔ10-b5MBD, in the presence of MinE, was associated with the membrane around the entire cell instead of forming polar zones. Since MinC generally accompanies MinD in its membrane distribution, this would permit MinC to block division at all sites instead of restricting the inhibition of septation to the polar regions. As expected, repression of yfp::minDΔ10::b5MBD and minE::cfp by growth in the presence of glucose reversed the division block (Fig. 4E), reflecting the fact that MinC expressed at normal levels in the absence of MinD does not block division (2).
DISCUSSION
Nuclear membrane targeting.
This study showed directly that heterologous membrane-associated proteins can be targeted to the yeast nuclear membrane, where their interactions can be studied by the powerful Gal4 two-hybrid system. It was not previously known whether yeast two-hybrid interactions observed with membrane-associated proteins represented their behavior within the nuclear membrane or within the nucleoplasm. This is an important distinction, since the conformations and behaviors of many membrane proteins are significantly affected when they are membrane associated. Nuclear membrane localization had only two requirements, a suitable nuclear localization signal and a nonspecific membrane-binding domain.
The membrane-targeting sequence of the bacterial MinD protein and the membrane-binding domain of the eukaryotic cytochrome b5 protein functioned equally well in targeting Yfp-labeled MinD chimeras to the nuclear membrane, and the membrane-associated proteins functioned similarly in the two-hybrid protein interaction system. The MinD and cytochrome b5 membrane-targeting sequences differ significantly in their normal membrane targets and in their mechanisms of membrane association. The MinD protein containing its native MTS is normally targeted to the inner surface of the bacterial cytoplasmic membrane (4). The MinD MTS consists of a 10-amino-acid amphipathic helix that is located on the surface of the membrane, oriented parallel to the bilayer surface (Fig. 1A), and the membrane association is mediated by interactions of hydrocarbon residues of the bilayer with nonpolar amino acid side chains that extend downward from one face of the amphipathic helix into the bilayer structure (10, 31). In contrast, cytochrome b5 is normally targeted by its MBD to the membrane of the endoplasmic reticulum of eukaryotic cells (1). The b5MBD consists of a 43-amino-acid segment composed of 56% nonpolar residues (12) and extends completely into the membrane (Fig. 1B), where essentially all of the hydrophobic residues can interact with hydrocarbon chains of the bilayer as an integral membrane anchor. The major difference of sequence and mechanism of membrane binding for the two membrane attachment domains probably exclude the possibility that nuclear membrane localization requires that the membrane-targeting domains recognize specific targets within the nuclear membrane.
It was striking that the proteins containing the membrane anchors appeared to be exclusively targeted to the nuclear membrane when the chimera contained the highly efficient SV40 NLS. The exclusive nuclear membrane targeting of the chimeric protein containing the cytochrome b5 anchor suggests that a rapid transfer of the protein to the nucleus from its site of synthesis in the cytoplasm preempts its normal localization to the endoplasmic reticulum or other cellular membranes. The situation may differ for eukaryotic proteins that are normally cotranslationally inserted into the membrane. Chimeric proteins containing the less efficient NLS from the Gal4 protein were similarly localized to the nuclear membrane but also showed some association with the plasma membrane. This suggests that the yeast two-hybrid system for membrane-associated proteins can be optimized by replacing the Gal4 NLS that is present in the current binding domain plasmids with a more potent NLS.
Membrane interactions of MinD.
The interaction of MinD with other MinD molecules is required for formation of membrane-associated MinD polymeric protofilaments. The fact that strong MinD-MinD interaction in the two-hybrid assay occurred only between membrane-bound molecules suggests that interaction with the membrane may induce or stabilize a MinD conformation that exposes MinD-MinD interaction domains. In the simplest model, collisional interactions subsequent to lateral diffusion within the membrane could then lead to growth of the membrane-associated MinD polymer (Fig. 5).
FIG. 5.
Model for MinD polymer assembly. (a) MinD in the presence of ATP binds the membrane. (b) Membrane-bound MinD becomes competent for MinD-MinD interaction. (c) Lateral diffusion of MinD in the membrane leads to collisional interactions and subsequent growth of the MinD polymer in the membrane.
Some of the 25-fold increase in interaction when MinD became membrane associated could also reflect the higher probability of MinD-MinD collisions within the two-dimensional membrane than within the nucleoplasm. However, parallel studies with MinC tethered to the membrane by the addition of the MinD MTS made it unlikely that this accounted for the magnitude of the observed effect of membrane binding on MinD-MinD interactions. We make the likely assumption that the lateral diffusion rates, and hence the collision rates, are similar for proteins of similar size (24.7 kDa and 29.4 kDa for MinC and MinD, respectively) that are tethered to the membrane in a similar manner. The two-hybrid MinC-MinC interaction was only increased twofold when MinC was tethered to the yeast nuclear membrane by the MinD MTS. The >10-fold difference in the effect of membrane binding on MinC-MinC and MinD-MinD interactions suggested that most of the increase in MinD-MinD interaction was not due to the increased probability of collision between the membrane-associated MinD molecules but instead probably resulted from membrane-induced conformational change in MinD.
In contrast to the MinD-MinD interaction, which required a membrane anchor, interactions between MinD and the other two Min proteins, MinC and MinE, did not require that MinD be membrane associated. Therefore, in E. coli cells, formation of MinD-MinC and MinD-MinE complexes could occur in the cytoplasm prior to membrane entry.
In intact cells, a repetitive membrane assembly and disassembly cycle is responsible for formation of MinDE polar zones and their oscillatory behavior. In the absence of MinE, MinD associates with the membrane as a MinD-ATP complex (9, 13) around the entire periphery of the cell (26). Release of MinD and its redistribution into the polar zones is dependent on MinD-MinE interactions that activate the MinD ATPase and thereby trigger the polarized release of MinD from the membrane.
The cytochrome b5 membrane-binding domain directed MinDΔ10 to the membrane in E. coli cells where the protein formed long-range MinD coiled structures similar to the structures formed by normal MinD. MinD containing the b5MBD also promoted the membrane association of MinE, as does wild-type MinD. However, this did not lead to formation of MinDE polar zones and MinE rings. This may indicate that, in contrast to wild-type MinD, which requires bound ATP to stably associate with the membrane, the large hydrophobic cytochrome b5 membrane-binding domain may be capable of stable membrane binding in the absence of ATP. MinD containing the b5MBD would therefore not be released even in the presence of MinE activation of the MinD ATPase and polar zones would not be formed. Alternatively, the failure to form polar zones when native MTS is replaced by b5MBD may indicate that MinE stimulation of MinD ATPase requires the presence of the native membrane targeting sequence. Further work will be needed to distinguish between these and other possibilities.
Two models have been suggested for the in vivo formation of membrane-associated MinD polymers. In one model, interaction between MinD molecules leading to dimerization occurs in the cytoplasm prior to membrane association and polymerization (10). In the other model, the initial MinD-MinD interactions occur subsequent to membrane association of MinD (32). The idea that MinD dimerization occurs in the cytoplasm prior to entry into the membrane is based on in vitro studies that showed dimer formation in the absence of phospholipid vesicles at protein concentrations of MinD or MinDΔ10 approximately 10- to 15-fold higher than the estimated cellular concentration of MinD (10, 11). The MinD dimerization reaction in the in vitro experiments may correspond to the weak self-interaction of nucleoplasmic MinDΔ10 in the two-hybrid experiments (4% of the activity of membrane-associated full-length MinD). Therefore, if dimer formation in the cytoplasm or nucleoplasm were an obligatory step that preceded membrane association and polymerization, the slow rate of dimer formation would presumably be strongly rate limiting for polymerization. The data seem most easily explained by the idea that the MinD-MinD interactions leading to dimerization and polymerization primarily occur on the membrane surface (Fig. 5). Consistent with the idea that MinD-MinD interactions require a membrane milieu, fluorescence energy transfer between two distinctly labeled MinD molecules in vitro has been shown to occur only in the presence of phospholipid vesicles (18).
Acknowledgments
We thank Mary J. Osborn for helpful discussions.
This work was supported by National Institute of Health Grant GM060632-05.
REFERENCES
- 1.Borgese, N., I. A. Gazzoni, M. Barberi, S. Colombo, and E. Pedrazzini. 2001. Targeting of a tail-anchored protein to endoplasmic reticulum and mitochondrial outer membrane by independent but competing pathways. Mol. Biol. Cell 12:2482-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Boer, P., R. E. Crossley, and L. I. Rothfield. 1992. Roles of MinC and MinD in the site-specific septation block mediated by the MinCDE system of Escherichia coli. J. Bacteriol. 174:63-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Boer, P. A. J., R. E. Crossley, and L. I. Rothfield. 1989. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56:641-649. [DOI] [PubMed] [Google Scholar]
- 4.de Boer, P. A. J., R. E. Crossley, A. R. Hand, and L. I. Rothfield. 1991. The MinD protein is a membrane ATPase required for the correct placement of the Escherichia coli division site. EMBO J. 10:4371-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu, X., Y.-L. Shih, Y. Zhang, and L. I. Rothfield. 2001. The MinE ring required for proper placement of the division site is a mobile structure that changes its cellular location during the Escherichia coli division cycle. Proc. Natl. Acad. Sci. USA 98:980-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.George, S. K., L. Najera, R. P. Sandoval, C. Countryman, R. W. Davis, and G. M. Ihler. 1989. The hydrophobic domain of cytochrome b5 is capable of anchoring beta-galactosidase in Escherichia coli membranes. J. Bacteriol. 171:4569-4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gietz, R., and R. Woods. 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350:87-96. [DOI] [PubMed] [Google Scholar]
- 8.Hale, C., H. Meinhardt, and P. de Boer. 2001. Dynamic localization cycle of the cell division regulator MinE in Escherichia coli. EMBO J. 20:1563-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu, Z., E. Gogol, and J. Lutkenhaus. 2002. Dynamic assembly of MinD on phospholipid vesicles regulated by ATP and MinE. Proc. Natl. Acad. Sci. USA 99:6761-6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu, Z., and J. Lutkenhaus. 2003. A conserved sequence at the C-terminus of MinD is required for binding to the membrane and targeting MinC to the septum. Mol. Microbiol. 47:345-355. [DOI] [PubMed] [Google Scholar]
- 11.Hu, Z., C. Saez, and J. Lutkenhaus. 2003. Recruitment of MinC, an inhibitor of Z-ring formation, to the membrane in Escherichia coli: role of MinD and MinE. J. Bacteriol. 185:196-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kondo, K., S. Tajima, R. Sato, and K. Narita. 1979. Primary structure of the membrane-binding segment of rabbit cytochrome b5. J. Biochem. 86:1119-1128. [DOI] [PubMed] [Google Scholar]
- 13.Lackner, L., D. Raskin, and P. de Boer. 2003. ATP-dependent interactions between Escherichia coli Min proteins and the phospholipid membrane in vitro. J. Bacteriol. 185:735-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lutkenhaus, J. 2002. Dynamic proteins in bacteria. Curr. Opin. Microbiol. 5:548-552. [DOI] [PubMed] [Google Scholar]
- 15.Ma, L., G. F. King, and L. Rothfield. 2003. Mapping the MinE site involved in interaction with the MinD division site selection protein. J. Bacteriol. 185:4948-4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma, L., G. F. King, and L. Rothfield. 2004. Positioning of the MinE binding site on the MinD surface suggests a plausible mechanism for activation of the Escherichia coli MinD ATPase during division site selection. Mol. Microbiol. 54:99-108. [DOI] [PubMed] [Google Scholar]
- 17.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 18.Mileykovskaya, E., I. Fishov, X. Fu, B. D. Corbin, W. Margolin, and W. Dowhan. 2003. Effects of phospholipid composition on MinD-membrane interactions in vitro and in vivo. J. Biol. Chem. 278:22193-22198. [DOI] [PubMed] [Google Scholar]
- 19.Miller, J. H. 1992. A short course on bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Peng, G., and J. E. Hopper. 2000. Evidence for Gal3p's cytoplasmic location and Gal80p's dual cytoplasmic-nuclear location implicates new mechanisms for controlling Gal4p activity in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:5140-5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pichoff, S., and J. Lutkenhaus. 2005. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol. Microbiol. 55:1722-1734. [DOI] [PubMed] [Google Scholar]
- 22.Raskin, D., and P. de Boer. 1999. MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J. Bacteriol. 181:6419-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raskin, D., and P. de Boer. 1997. The MinE ring: an FtsZ-independent cell structure required for selection of the correct division site in E. coli. Cell 91:685-694. [DOI] [PubMed] [Google Scholar]
- 24.Raskin, D., and P. de Boer. 1999. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl. Acad. Sci. USA 96:4971-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rothfield, L. I., Y.-L. Shih, and G. F. King. 2001. Polar explorers: membrane proteins that determine division site placement. Cell 106:13-16. [DOI] [PubMed] [Google Scholar]
- 26.Rowland, S. L., X. Fu, M. A. Sayed, Y. Zhang, W. R. Cook, and L. I. Rothfield. 2000. Membrane redistribution of the Escherichia coli MinD protein induced by MinE. J. Bacteriol. 182:613-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saffman, P., and M. Delbruck. 1975. Brownian motion in biological membranes. Proc. Natl. Acad. Sci. USA 72:3111-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shih, Y.-L., X. Fu, G. F. King, T. Le, and L. I. Rothfield. 2002. Division site placement in E. coli: mutations that prevent formation of the MinE ring lead to loss of the normal midcell arrest of growth of polar MinD membrane domains. EMBO J. 21:3347-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shih, Y.-L., T. Le, and L. Rothfield. 2003. Division site selection in Escherichia coli involves dynamic redistribution of Min proteins within coiled structures that extend between the two cell poles. Proc. Natl. Acad. Sci. USA 100:7865-7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suefuji, K., R. Valluzzi, and D. RayChaudhuri. 2002. Dynamic assembly of MinD into filament bundles modulated by ATP, phospholipids, and MinE. Proc. Natl. Acad. Sci. USA 99:16776-16781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szeto, T., S. Rowland, L. Rothfield, and G. F. King. 2002. Membrane localization of MinD is mediated by a C-terminal motif that is conserved across eubacteria, archaea, and chloroplasts. Proc. Natl. Acad. Sci. USA 99:15693-15698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szeto, T. H., S. Rowland, C. Habrukowich, and G. F. King. 2003. The MinD membrane targeting sequence is a transplantable lipid-binding helix. J. Biol. Chem. 279:40050-40056. [DOI] [PubMed] [Google Scholar]
- 33.Vergeres, G., and L. Waskell. 1995. Cytochrome b5, its functions, structure and membrane topology. Biochimie 77:604-620. [DOI] [PubMed] [Google Scholar]
- 34.Zhou, H., and J. Lutkenhaus. 2004. The switch I and II regions of MinD are required for binding and activating MinC. J. Bacteriol. 186:1546-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]