Abstract
Lipid modification of the N-terminal Cys residue (N-acyl-S-diacylglyceryl-Cys) has been found to be an essential, ubiquitous, and unique bacterial posttranslational modification. Such a modification allows anchoring of even highly hydrophilic proteins to the membrane which carry out a variety of functions important for bacteria, including pathogenesis. Hence, being able to identify such proteins is of great value. To this end, we have created a comprehensive database of bacterial lipoproteins, called DOLOP, which contains information and links to molecular details for about 278 distinct lipoproteins and predicted lipoproteins from 234 completely sequenced bacterial genomes. The website also features a tool that applies a predictive algorithm to identify the presence or absence of the lipoprotein signal sequence in a user-given sequence. The experimentally verified lipoproteins have been classified into different functional classes and more importantly functional domain assignments using hidden Markov models from the SUPERFAMILY database that have been provided for the predicted lipoproteins. Other features include the following: primary sequence analysis, signal sequence analysis, and search facility and information exchange facility to allow researchers to exchange results on newly characterized lipoproteins. The website, along with additional information on the biosynthetic pathway, statistics on predicted lipoproteins, and related figures, is available at http://www.mrc-lmb.cam.ac.uk/genomes/dolop/.
Essential cellular activities such as adhesion, digestion, transport, sensing, signal transduction, growth, and morphological changes such as spore formation in bacteria, etc., require a class of proteins, called membrane proteins, that work efficiently in aqueous environments while anchored to the hydrophobic membrane that envelops a cell. Organisms have evolved different strategies in the design of their membrane proteins, including the following: (i) transmembrane proteins, in which one or more peptide segments in their helical or beta sheeted structure traverse the width of the membrane to provide anchorage; the loops and parts of the transmembrane segments carry out the relevant function; (ii) proteins with a significant patch of hydrophobic surface which, along with other noncovalent and even ionic interactions, associate either loosely or tightly with the membrane; and (iii) covalent lipid modification of proteins, exo or endo, by fatty acids and other lipid moieties, which provide the hydrophobic anchor either at one end or on the surface of such proteins. The last strategy, particularly suited to hydrophilic proteins, is useful in engineering proteins for anchorage to hydrophobic surfaces.
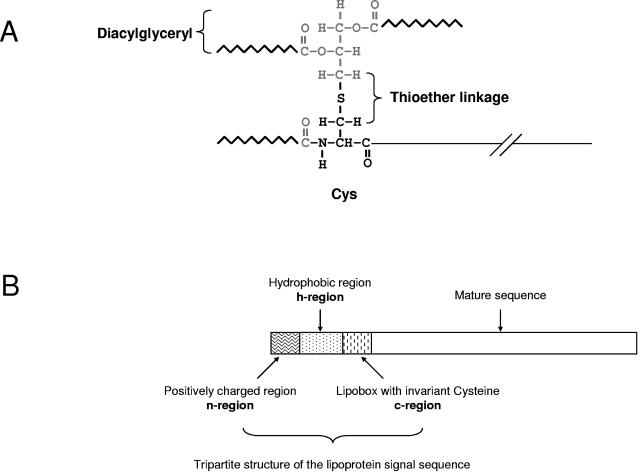
Bacteria, the major class among prokaryotes, possess an interesting N-terminal lipid modification, N-acyl-S-diacylglyceryl-Cys (Fig. 1A), which is unique and ubiquitous among its known members. More than 2,000 such proteins have been identified currently. Three fatty acyl groups at the N terminus which are derived from bacterial phospholipids provide tight anchorage to the membrane surface, allowing the rest of the protein to perform relevant biochemical functions in the aqueous or aqueous-membrane interface. Since its discovery in 1969 (5) in a major outer membrane protein of Escherichia coli called Braun's lipoprotein (named after the discoverer), the same modification in different proteins was seen in a variety of bacteria. The primary structural features required for this modification and the biosynthetic pathway containing three enzymes (the first enzyme in the pathway attaches the diacylglyceryl group from phosphatidyglycerol to the thiol of Cys, the first amino acid after the signal peptide; the second enzyme cleaves off the signal peptide after the initial lipid modification; and the third enzyme acylates the N-terminal amino group with a fatty acid from any available phospholipid) have been elucidated since then (16, 26, 31, 46, 47, 59, 60).
FIG. 1.
(A) The structure of the lipid modification in lipoproteins. The sulfhydryl group of N-terminal cysteine is modified with a diacylglyceryl group attached through a thioether linkage, and the amino group is acylated with a fatty acid. (B) Tripartite structure of the lipoprotein signal sequence. The n-region is made up of five to seven residues and has at least two positively charged residues; the h-region, or the hydrophobic region, is made up of 7 to 22 predominantly hydrophobic and uncharged residues; and the c-region, which has the consensus [LVI][ASTVI][GAS] sequence, along with C, the invariant lipid-modified N-terminal residue in all bacterial lipoproteins, is referred to as the lipobox.
Though by and large the three enzymes are conserved among bacteria and the phospholipid fatty acyl composition is reflected in these lipoproteins, recent findings reveal interesting variations in the theme. Some of the gram-positive eubacteria do not seem to possess the gene (lnt) for the third enzyme responsible for N-acylation of lipoproteins (43, 53, 57). In Borrelia burgdorferi, the second ester-linked fatty acid is just an acetyl group instead of a fatty acyl group (2). Whereas the pathway is essential to gram-negative bacteria, it appears to be nonessential for the gram-positive bacteria, as revealed by resistance to globomycin, an uncompetitive inhibitor of signal peptidase II (the second enzyme), and null mutation studies (11). However, lack of these enzymes does adversely affect survival of these bacteria under certain conditions and their pathogenesis.
There is in fact a renewed interest in lipoproteins from the point of view of their roles in bacterial pathogenesis, as these lipid-modified proteins play a variety of roles in host-pathogen interactions, which necessarily take place in the solid-aqueous interface, from surface adhesion to translocation of virulence factors into the host cytoplasm. Those aiding pathogenesis include PsaA in Streptococcus pneumoniae (4); MxiM, a lipoprotein of the type III secretory pathway in Shigella flexneri important for translocation of invasins (48); MAA1 of Mycoplasma arthritidis, required for adherence to joint tissues early in the infectious process (62); and a gamut of surface lipoproteins specifically expressed by mycoplasmas upon infection (45). Recently, an lsp mutant of Listeria monocytogenes was found to be ineffective in phagosomal escape of bacteria during infection (44). Those that help to activate inflammatory response or evade host defense include lipoproteins released from Enterobacteriaceae that induce cytokine production in the macrophage (66); a 19-kDa lipoprotein of Mycobacteria that elicits antibody and T-cell responses in human and mice and induces innate immune response in dendritic cells and neutrophils (40, 56); LipL41, a surface-exposed lipoprotein of pathogenic Leptospira species (52); and LpK, a lipoprotein from Mycobacterium leprae that induces human interleukin 12 (36). Owing to the above roles in bacterial pathogenesis, lipoproteins are also attractive candidates in vaccine development. For example, Lpp20, a lipoprotein, is a vaccine candidate against Helicobacter pylori (30). In the case of Lyme disease, vaccines based on lipoproteins OspA and DbpA of spirochete Borrelia burgdorferi have been demonstrated to be effective in several animal models (7, 14, 15, 22).
One of the initial focuses of bacterial lipoprotein study was to analyze the signal peptides of experimentally verified lipoproteins and derive primary structure determinants for posttranslational lipid modification. Limited sequence analysis of precursors of only 26 distinct lipoproteins by Hayashi and Wu (23) already indicated a characteristic four-amino-acid sequence at the C-terminal end of the signal peptide including the modifiable Cys. Appropriately this was called the “lipobox,” and site-directed mutagenesis in the region further helped to define the roles of individual amino acids. Later, similar analysis of 75 lipoproteins by Braun and Wu (6) revealed the lipobox consensus sequence L[AS][GA]C. With more reports of experimentally verified lipoproteins, the roles and composition of the lipobox and the signal sequence features such as a stretch of positively charged n-region and uncharged h-region became more accurately defined (Fig. 1B). Accordingly, more robust predictive rules evolved to recognize lipoproteins from the amino acid sequences, mainly deduced from genomic sequences. The first such predictive rule was adapted by the Prosite pattern (PS00013), and later a refined one with better predictive capability was used in the maiden version of DOLOP, the first dedicated website for bacterial lipoproteins (34).
In the past few years there has been intensive bioinformatic analysis of bacterial lipoproteins and comparison of different predictive algorithms (3, 13, 18, 19, 28, 34, 53, 57). Predictive rules that work better for gram-positive bacterial lipoproteins were proposed as G+LPP (53), and recently a trained set of predictive rules was used and an algorithm called LipoP (28) was proposed to predict membrane proteins, lipoproteins, and cellular proteins by looking for signal sequence features. In the last year a detailed comparative analysis of DOLOP and other algorithms was carried out on experimentally verified lipoproteins from one model taxon, E. coli K-12, and a highly fine-tuned algorithm with the best predictive ability was proposed (19). As a result of all these efforts, in the last decade, the numbers of bacterial lipoproteins would cross several thousand, thanks to reliable predictive rules, which are today applied for identifying lipoproteins.
One of the intriguing aspects in the biosynthesis of lipoprotein is its targeting to either the inner or outer membrane. Initial sequence analysis of inner and outer membrane lipoproteins suggested a targeting role for Asp or Ser at the +2 position in the mature sequence (50, 64); Asp led to inner membrane localization, whereas Ser led to outer membrane localization. A series of recent elegant studies by Tokuda and coworkers have led to the identification of outer membrane localization (LOL) machinery for lipoproteins and the effect of amino acids in the vicinity of the modifiable Cys in the mature sequence in their recognition (37, 39, 54, 58, 63, 65). Accordingly, it was realized that Asp at position 2 is not the sole inner membrane retention signal, and amino acid residues at +3 and +4 positions were found to affect the membrane localization (55). The rules for membrane localization are not as straightforward as those of lipid modification to obtain by simple sequence comparison. However, a large database with experimentally verified data on localization could help.
Each bacterium has a common as well as a unique set of lipoproteins, whose numbers vary widely, and their proteomics would be interesting as well as challenging. To aid this study, we have introduced a new feature which provides domain assignments to identified lipoproteins in the updated version of DOLOP, and this paper is meant to (i) propose the refined lipoprotein identification algorithm based on a larger data set, (ii) highlight the updated list of genome-wide prediction of lipoproteins, and (iii) introduce readers to the new feature in the domain search, as it would give a better idea about the relatedness of various lipoproteins in terms of function between themselves and with nonlipoproteins. A case study, where integration of other external information such as gene expression data with information on predicted lipoproteins leads to the identification of differentially expressed lipoproteins under quorum-sensing conditions in Pseudomonas aeruginosa, will also be discussed.
MATERIALS AND METHODS
Creation of the database.
Lipoproteins were obtained from the Swiss-Prot database using a combination of multiple keywords such as “lipid modification,” “lipoprotein,” “N-acyl-S-diacylglyceryl,” etc. Additionally, the literature was searched to identify lipoproteins that would have been potentially missed by the keyword search. From this list of 773 lipoproteins, which included some that were experimentally verified and some that were deduced by the authors based on homology, we grouped them into 278 clusters, where each cluster represented orthologs from different bacterial organisms. One sequence was further chosen to be represented in the database. For a detailed procedure about the database creation step, please refer to the study by Madan Babu and Sankaran (34).
Statistical analysis of the lipoprotein signal sequence.
The first 45 amino acids from each of the 278 lipoprotein sequences were aligned using the T-Coffee multiple sequence alignment tool (41) to identify the consensus sequence. Additionally, in-house PERL scripts were written to calculate the various statistics such as the amino acid charge distribution in the n-region (Fig. 1), the length of the hydrophobic region, and the amino acid choices available in the lipobox sequence.
Prediction of lipoproteins from completely sequenced bacterial genomes.
The complete genome sequences of the 234 organisms listed in Table 1 were downloaded from the NCBI website. A PERL script incorporating the algorithm discussed in Results was developed to predict potential lipoproteins. The script also calculates the fraction of the genome encoding potential lipoproteins. It should be noted that the predicted list does not contain entries that have been predicted to be lipoproteins by the authors of the original study describing the genome sequence, for it can give rise to false positives. This is because the procedure used to assign function by the authors relies on sequence similarity of the mature sequence, and a protein which is lipid modified in one organism need not be modified in another organism. Thus, the predicted lipoproteins were identified purely based on the presence of the lipoprotein signal sequence as discussed above.
TABLE 1.
Number of predicted lipoproteins from 234 predicted bacterial lipoproteins
| Organism name | Phylogenetic group | No. of proteins | No. of predicted lipoproteins froma:
|
|
|---|---|---|---|---|
| DOLOP | LipoP | |||
| Acinetobacter sp. strain ADP1 | Proteobacteria | 3,325 | 68 (2.05) | 102 (3.07) |
| Agrobacterium tumefaciens C58 | Proteobacteria | 4,548 | 29 (0.64) | 29 (0.64) |
| Anaplasma marginale St. Maries | Proteobacteria | 949 | 8 (0.84) | 13 (1.37) |
| Aquifex aeolicus VF5 | Aquificae | 1,529 | 9 (0.59) | 18 (1.18) |
| Azoarcus sp. strain EbN1 | Proteobacteria | 4,133 | 41 (0.99) | 59 (1.43) |
| Bacillus anthracis Ames ancestor | Firmicutes | 5,309 | 110 (2.07) | 150 (2.83) |
| Bacillus anthracis Ames | Firmicutes | 5,311 | 110 (2.07) | 150 (2.82) |
| Bacillus anthracis Sterne | Firmicutes | 5,287 | 111 (2.10) | 154 (2.91) |
| Bacillus cereus ATCC 10987 | Firmicutes | 5,603 | 113 (2.02) | 155 (2.77) |
| Bacillus cereus ATCC 14579 | Firmicutes | 5,234 | 105 (2.01) | 158 (3.02) |
| Bacillus cereus E33L | Firmicutes | 5,134 | 115 (2.24) | 164 (3.19) |
| Bacillus clausii KSM-K16 | Firmicutes | 4,096 | 108 (2.64) | 152 (3.71) |
| Bacillus halodurans C-125 | Firmicutes | 4,066 | 107 (2.63) | 133 (3.27) |
| Bacillus licheniformis ATCC 14580 | Firmicutes | 4,152 | 72 (1.73) | 100 (2.41) |
| Bacillus licheniformis ATCC 14580 | Firmicutes | 4,196 | 73 (1.74) | 103 (2.45) |
| Bacillus subtilis subsp. subtilis 168 | Firmicutes | 4,105 | 70 (1.71) | 103 (2.51) |
| Bacillus thuringiensis serovar Konkukian 97-27 | Firmicutes | 5,117 | 121 (2.36) | 165 (3.22) |
| Bacteroides fragilis NCTC 9343 | Bacteroidetes | 4,189 | 177 (4.23) | 455 (10.86) |
| Bacteroides fragilis YCH46 | Bacteroidetes | 4,578 | 184 (4.02) | 469 (10.24) |
| Bacteroides thetaiotaomicron VPI-5482 | Bacteroidetes | 4,778 | 223 (4.67) | 601 (12.58) |
| Bartonella henselae Houston-1 | Proteobacteria | 1,488 | 36 (2.42) | 39 (2.62) |
| Bartonella quintana Toulouse | Proteobacteria | 1,142 | 18 (1.58) | 26 (2.28) |
| Bdellovibrio bacteriovorus HD100 | Proteobacteria | 3,587 | 147 (4.10) | 240 (6.69) |
| Bifidobacterium longum NCC2705 | Actinobacteria | 1,727 | 26 (1.51) | 32 (1.85) |
| Bordetella bronchiseptica RB50 | Proteobacteria | 4,994 | 83 (1.66) | 92 (1.84) |
| Bordetella parapertussis 12822 | Proteobacteria | 4,185 | 65 (1.55) | 141 (3.37) |
| Bordetella pertussis Tohama I | Proteobacteria | 3,436 | 53 (1.54) | 66 (1.92) |
| Borrelia burgdorferi B31 | Spirochaetes | 851 | 8 (0.94) | 28 (3.29) |
| Borrelia garinii PBi | Spirochaetes | 832 | 8 (0.96) | 25 (3.00) |
| Bradyrhizobium japonicum USDA 110 | Proteobacteria | 8,317 | 45 (0.54) | 55 (0.66) |
| Brucella abortus bv. 1 9-941 | Proteobacteria | 3,085 | 33 (1.07) | 43 (1.39) |
| Brucella melitensis 16M | Proteobacteria | 3,198 | 22 (0.69) | 27 (0.84) |
| Brucella suis 1330 | Proteobacteria | 3,271 | 35 (1.07) | 46 (1.41) |
| Buchnera aphidicola APS | Proteobacteria | 564 | 0 (0.00) | 2 (0.35) |
| Buchnera aphidicola Bp | Proteobacteria | 504 | 1 (0.20) | 3 (0.60) |
| Buchnera aphidicola Sg | Proteobacteria | 546 | 0 (0.00) | 5 (0.92) |
| Burkholderia mallei ATCC 23344 | Proteobacteria | 4,764 | 65 (1.36) | 93 (1.95) |
| Burkholderia pseudomallei K96243 | Proteobacteria | 5,728 | 93 (1.62) | 148 (2.58) |
| Campylobacter jejuni RM1221 | Proteobacteria | 1,838 | 22 (1.20) | 41 (2.23) |
| Campylobacter jejuni subsp. jejuni NCTC 11168 | Proteobacteria | 1,629 | 22 (1.35) | 47 (2.89) |
| “Candidatus Blochmannia floridanus” | Proteobacteria | 583 | 2 (0.34) | 3 (0.51) |
| “Candidatus Blochmannia pennsylvanicus” BPEN | Proteobacteria | 610 | 3 (0.49) | 5 (0.82) |
| “Candidatus Pelagibacter ubique” HTCC1062 | Proteobacteria | 1,354 | 14 (1.03) | 18 (1.33) |
| Caulobacter crescentus CB15 | Proteobacteria | 3,737 | 61 (1.63) | 76 (2.03) |
| Chlamydia muridarum Nigg | Chlamydiae | 904 | 18 (1.99) | 20 (2.21) |
| Chlamydia trachomatis D/UW-3/CX | Chlamydiae | 895 | 14 (1.56) | 20 (2.23) |
| Chlamydophila abortus S26/3 | Chlamydiae | 932 | 19 (2.04) | 24 (2.58) |
| Chlamydophila caviae GPIC | Chlamydiae | 998 | 16 (1.60) | 25 (2.51) |
| Chlamydophila pneumoniae AR39 | Chlamydiae | 1,112 | 15 (1.35) | 27 (2.43) |
| Chlamydophila pneumoniae CWL029 | Chlamydiae | 1,052 | 16 (1.52) | 28 (2.66) |
| Chlamydophila pneumoniae J138 | Chlamydiae | 1,069 | 16 (1.50) | 28 (2.62) |
| Chlamydophila pneumoniae TW-183 | Chlamydiae | 1,113 | 14 (1.26) | 24 (2.16) |
| Chlorobium tepidium TLS | Chlorobia | 2,252 | 15 (0.67) | 33 (1.47) |
| Chromobacterium violaceum ATCC 12472 | Proteobacteria | 4,407 | 84 (1.91) | 91 (2.06) |
| Clostridium acetobutylicum ATCC 824 | Firmicutes | 3,672 | 45 (1.23) | 94 (2.56) |
| Clostridium perfringens 13 | Firmicutes | 2,660 | 46 (1.73) | 75 (2.82) |
| Clostridium tetani E88 | Firmicutes | 2,373 | 21 (0.88) | 68 (2.87) |
| Colwellia psychrerythraea 34H | Proteobacteria | 4,910 | 139 (2.83) | 195 (3.97) |
| Corynebacterium diphtheriae NCTC 13129 | Actinobacteria | 2,272 | 42 (1.85) | 49 (2.16) |
| Corynebacterium efficiens YS-314 | Actinobacteria | 2,950 | 43 (1.46) | 40 (1.36) |
| Corynebacterium glutamicum ATCC 13032 | Actinobacteria | 3,057 | 84 (2.75) | 89 (2.91) |
| Corynebacterium glutamicum ATCC 13032 | Actinobacteria | 2,993 | 78 (2.61) | 83 (2.77) |
| Corynebacterium jeikeium K411 | Actinobacteria | 2,137 | 24 (1.12) | 39 (1.82) |
| Coxiella burnetii RSA 493 | Proteobacteria | 2,010 | 25 (1.24) | 33 (1.64) |
| Dechloromonas aromatica RCB | Proteobacteria | 4,171 | 81 (1.94) | 110 (2.64) |
| Dehalococcoides ethenogenes 195 | Chloroflexi | 1,580 | 18 (1.14) | 29 (1.84) |
| Dehalococcoides sp. strain CBDB1 | Chloroflexi | 1,458 | 13 (0.89) | 21 (1.44) |
| Deinococcus radiodurans R1 | Deinococcus-Thermus | 2,997 | 45 (1.50) | 50 (1.67) |
| Desulfotalea psychrophila LSv54 | Proteobacteria | 3,116 | 44 (1.41) | 23 (0.74) |
| Desulfovibrio vulgaris subsp. vulgaris Hildenborough | Proteobacteria | 3,379 | 38 (1.12) | 28 (0.83) |
| Ehrlichia canis Jake | Proteobacteria | 925 | 14 (1.51) | 19 (2.05) |
| Ehrlichia ruminantium Gardel | Proteobacteria | 950 | 6 (0.63) | 10 (1.05) |
| Ehrlichia ruminantium Welgevonden | Proteobacteria | 958 | 6 (0.63) | 10 (1.04) |
| Ehrlichia ruminantium Welgevonden | Proteobacteria | 888 | 9 (1.01) | 12 (1.35) |
| Enterococcus faecalis V583 | Firmicutes | 3,113 | 64 (2.06) | 81 (2.60) |
| Erwinia carotovora subsp. atroseptica SCRI1043 | Proteobacteria | 4,472 | 112 (2.50) | 126 (2.82) |
| Escherichia coli CFT073 | Proteobacteria | 5,379 | 86 (1.60) | 85 (1.58) |
| Escherichia coli K-12 | Proteobacteria | 4,237 | 86 (2.03) | 103 (2.43) |
| Escherichia coli O157:H7 | Proteobacteria | 5,253 | 116 (2.21) | 139 (2.65) |
| Escherichia coli O157:H7 EDL933 | Proteobacteria | 5,324 | 98 (1.84) | 127 (2.39) |
| Francisella tularensis subsp. tularensis SCHU S4 | Proteobacteria | 1,603 | 38 (2.37) | 56 (3.49) |
| Fusobacterium nucleatum subsp. nucleatum ATCC 25586 | Fusobacteria | 2,067 | 27 (1.31) | 39 (1.89) |
| Geobacillus kaustophilus HTA426 | Firmicutes | 3,498 | 61 (1.74) | 75 (2.14) |
| Geobacter sulfurreducens PCA | Proteobacteria | 3,446 | 56 (1.63) | 22 (0.64) |
| Gloeobacter violaceus PCC 7421 | Cyanobacteria | 4,430 | 37 (0.84) | 45 (1.02) |
| Gluconobacter oxydans 621H | Proteobacteria | 2,432 | 41 (1.69) | 58 (2.38) |
| Haemophilus ducreyi 35000HP | Proteobacteria | 1,717 | 41 (2.39) | 45 (2.62) |
| Haemophilus influenzae 86-028NP | Proteobacteria | 1,791 | 44 (2.46) | 59 (3.29) |
| Haemophilus influenzae Rd KW20 | Proteobacteria | 1,657 | 39 (2.35) | 48 (2.90) |
| Helicobacter hepaticus ATCC 51449 | Proteobacteria | 1,875 | 19 (1.01) | 59 (3.15) |
| Helicobacter pylori 26695 | Proteobacteria | 1,576 | 14 (0.89) | 37 (2.35) |
| Helicobacter pylori J99 | Proteobacteria | 1,491 | 16 (1.07) | 39 (2.62) |
| Idiomarina loihiensis L2TR | Proteobacteria | 2,628 | 73 (2.78) | 95 (3.61) |
| Lactobacillus acidophilus NCFM | Firmicutes | 1,864 | 36 (1.93) | 43 (2.31) |
| Lactobacillus johnsonii NCC 533 | Firmicutes | 1,821 | 25 (1.37) | 40 (2.20) |
| Lactobacillus plantarum WCFS1 | Firmicutes | 3,009 | 51 (1.69) | 52 (1.73) |
| Lactococcus lactis subsp. lactis II1403 | Firmicutes | 2,321 | 32 (1.38) | 34 (1.46) |
| Legionella pneumophila Lens | Proteobacteria | 2,878 | 45 (1.56) | 62 (2.15) |
| Legionella pneumophila Paris | Proteobacteria | 3,027 | 47 (1.55) | 66 (2.18) |
| Legionella pneumophila subsp. pneumophila Philadelphia | Proteobacteria | 2,942 | 40 (1.36) | 58 (1.97) |
| Leifsonia xyli subsp. xyli CTCB07 | Actinobacteria | 2,030 | 24 (1.18) | 30 (1.48) |
| Leptospira interrogans serovar Copenhageni Fiocruz L1-130 | Spirochaetes | 3,658 | 25 (0.68) | 163 (4.46) |
| Leptospira interrogans serovar Lai 56601 | Spirochaetes | 4,727 | 23 (0.49) | 149 (3.15) |
| Listeria innocua Clip11262 | Firmicutes | 2,968 | 62 (2.09) | 70 (2.36) |
| Listeria monocytogenes 4b F2365 | Firmicutes | 2,821 | 58 (2.06) | 65 (2.30) |
| Listeria monocytogenes EGD-e | Firmicutes | 2,846 | 63 (2.21) | 69 (2.42) |
| Mannheimia succiniciproducens MBEL55E | Proteobacteria | 2,380 | 51 (2.14) | 70 (2.94) |
| Mesoplasma florum L1 | Firmicutes | 682 | 21 (3.08) | 12 (1.76) |
| Mesorhizobium loti MAFF303099 | Proteobacteria | 6,743 | 50 (0.74) | 58 (0.86) |
| Methylococcus capsulatus Bath | Proteobacteria | 2,959 | 49 (1.66) | 63 (2.13) |
| Mycobacterium avium subsp. paratuberculosis K-10 | Actinobacteria | 4,350 | 49 (1.13) | 75 (1.72) |
| Mycobacterium bovis AF2122/97 | Actinobacteria | 3,920 | 50 (1.28) | 67 (1.71) |
| Mycobacterium leprae TN | Actinobacteria | 1,605 | 16 (1.00) | 24 (1.50) |
| Mycobacterium tuberculosis CDC1551 | Actinobacteria | 4,189 | 43 (1.03) | 65 (1.55) |
| Mycobacterium tuberculosis H37Rv | Actinobacteria | 3,991 | 51 (1.28) | 69 (1.73) |
| Mycoplasma gallisepticum R | Firmicutes | 726 | 14 (1.93) | 43 (5.92) |
| Mycoplasma genitalium G-37 | Firmicutes | 484 | 11 (2.27) | 18 (3.72) |
| Mycoplasma hyopneumoniae 232 | Firmicutes | 691 | 9 (1.30) | 34 (4.92) |
| Mycoplasma hyopneumoniae 7448 | Firmicutes | 663 | 7 (1.06) | 28 (4.22) |
| Mycoplasma hyopneumoniae J | Firmicutes | 665 | 8 (1.20) | 26 (3.91) |
| Mycoplasma mobile 163K | Firmicutes | 633 | 19 (3.00) | 10 (1.58) |
| Mycoplasma mycoides subsp. mycoides SC PG1 | Firmicutes | 1,016 | 40 (3.94) | 39 (3.84) |
| Mycoplasma penetrans HF-2 | Firmicutes | 1,037 | 60 (5.79) | 51 (4.92) |
| Mycoplasma pneumoniae M129 | Firmicutes | 689 | 38 (5.52) | 44 (6.39) |
| Mycoplasma pulmonis UAB CTIP | Firmicutes | 782 | 23 (2.94) | 44 (5.63) |
| Mycoplasma synoviae 53 | Firmicutes | 672 | 5 (0.74) | 18 (2.68) |
| Neisseria gonorrhoeae FA 1090 | Proteobacteria | 2,002 | 58 (2.90) | 71 (3.55) |
| Neisseria meningitidis MC58 | Proteobacteria | 2,079 | 69 (3.32) | 79 (3.80) |
| Neisseria meningitidis Z2491 | Proteobacteria | 2,065 | 62 (3.00) | 72 (3.49) |
| Nitrobacter winogradskyi Nb-255 | Proteobacteria | 3,122 | 20 (0.64) | 34 (1.09) |
| Nitrosomonas europaea ATCC 19718 | Proteobacteria | 2,461 | 44 (1.79) | 68 (2.76) |
| Nocardia farcinica IFM 10152 | Actinobacteria | 5,683 | 63 (1.11) | 97 (1.71) |
| Nostoc sp. strain PCC 7120 | Cyanobacteria | 5,366 | 40 (0.75) | 85 (1.58) |
| Oceanobacillus iheyensis HTE831 | Firmicutes | 3,500 | 107 (3.06) | 131 (3.74) |
| Onion yellows phytoplasma OY-M | Firmicutes | 754 | 3 (0.40) | 2 (0.27) |
| Parachlamydia sp. strain UWE25 | Chlamydiae | 2,031 | 16 (0.79) | 23 (1.13) |
| Pasteurella multocida subsp. multocida Pm70 | Proteobacteria | 2,015 | 53 (2.63) | 66 (3.28) |
| Photobacterium profundum SS9 | Proteobacteria | 5,424 | 104 (1.92) | 151 (2.78) |
| Photorhabdus luminescens subsp. laumondii TTO1 | Proteobacteria | 4,683 | 73 (1.56) | 88 (1.88) |
| Porphyromonas gingivalis W83 | Bacteroidetes | 1,909 | 26 (1.36) | 65 (3.40) |
| Prochlorococcus marinus MIT 9313 | Cyanobacteria | 2,265 | 16 (0.71) | 22 (0.97) |
| Prochlorococcus marinus NATL2A | Cyanobacteria | 1,890 | 12 (0.63) | 17 (0.90) |
| Prochlorococcus marinus subsp. marinus CCMP1375 | Cyanobacteria | 1,882 | 12 (0.64) | 16 (0.85) |
| Prochlorococcus marinus subsp. pastoris CCMP1986 | Cyanobacteria | 1,712 | 9 (0.53) | 10 (0.58) |
| Propionibacterium acnes KPA171202 | Actinobacteria | 2,297 | 50 (2.18) | 55 (2.39) |
| Pseudomonas aeruginosa PAO1 | Proteobacteria | 5,567 | 113 (2.03) | 186 (3.34) |
| Pseudomonas fluorescens Pf-5 | Proteobacteria | 6,137 | 141 (2.30) | 182 (2.97) |
| Pseudomonas putida KT2440 | Proteobacteria | 5,350 | 74 (1.38) | 118 (2.21) |
| Pseudomonas syringae pv. phaseolicola 1448A | Proteobacteria | 4,982 | 94 (1.89) | 128 (2.57) |
| Pseudomonas syringae pv. syringae B728a | Proteobacteria | 5,090 | 113 (2.22) | 151 (2.97) |
| Pseudomonas syringae pv. tomato DC3000 | Proteobacteria | 5,470 | 101 (1.85) | 151 (2.76) |
| Psychrobacter arcticum 273-4 | Proteobacteria | 2,120 | 44 (2.08) | 70 (3.30) |
| Ralstonia eutropha JMP134 | Proteobacteria | 5,846 | 97 (1.66) | 128 (2.19) |
| Ralstonia solanacearum GMI1000 | Proteobacteria | 3,440 | 47 (1.37) | 80 (2.33) |
| Rhodopirellula baltica SH 1 | Planctomycetes | 7,325 | 46 (0.63) | 86 (1.17) |
| Rhodopseudomonas palustris CGA009 | Proteobacteria | 4,813 | 36 (0.75) | 51 (1.06) |
| Rickettsia conorii Malish 7 | Proteobacteria | 1,374 | 16 (1.16) | 23 (1.67) |
| Rickettsia felis URRWXCal2 | Proteobacteria | 1,400 | 19 (1.36) | 22 (1.57) |
| Rickettsia prowazekii Madrid E | Proteobacteria | 835 | 9 (1.08) | 18 (2.16) |
| Rickettsia typhi Wilmington | Proteobacteria | 838 | 10 (1.19) | 8 (0.95) |
| Salmonella enterica subsp. enterica serovar Choleraesuis | Proteobacteria | 4,445 | 94 (2.11) | 110 (2.47) |
| Salmonella enterica subsp. enterica serovar Paratyphi A ATCC | Proteobacteria | 4,093 | 103 (2.52) | 109 (2.66) |
| Salmonella enterica subsp. enterica serovar Typhi CT18 | Proteobacteria | 4,395 | 101 (2.30) | 116 (2.64) |
| Salmonella enterica subsp. enterica serovar Typhi Ty2 | Proteobacteria | 4,318 | 101 (2.34) | 114 (2.64) |
| Salmonella enterica serovar Typhimurium LT2 | Proteobacteria | 4,425 | 104 (2.35) | 111 (2.51) |
| Shewanella oneidensis MR-1 | Proteobacteria | 4,324 | 95 (2.20) | 146 (3.38) |
| Shigella flexneri 2a 2457T | Proteobacteria | 4,068 | 73 (1.79) | 85 (2.09) |
| Shigella flexneri 2a 301 | Proteobacteria | 4,182 | 76 (1.82) | 87 (2.08) |
| Shigella sonnei Ss046 | Proteobacteria | 4,223 | 81 (1.92) | 97 (2.30) |
| Silicibacter pomeroyi DSS-3 | Proteobacteria | 3,810 | 42 (1.10) | 56 (1.47) |
| Sinorhizobium meliloti 1021 | Proteobacteria | 3,341 | 33 (0.99) | 45 (1.35) |
| Staphylococcus aureus subsp. aureus COL | Firmicutes | 2,615 | 47 (1.80) | 65 (2.49) |
| Staphylococcus aureus subsp. aureus MRSA252 | Firmicutes | 2,656 | 50 (1.88) | 61 (2.30) |
| Staphylococcus aureus subsp. aureus MSSA476 | Firmicutes | 2,579 | 49 (1.90) | 36 (1.40) |
| Staphylococcus aureus subsp. aureus Mu50 | Firmicutes | 2,697 | 55 (2.04) | 72 (2.67) |
| Staphylococcus aureus subsp. aureus MW2 | Firmicutes | 2,632 | 49 (1.86) | 66 (2.51) |
| Staphylococcus aureus subsp. aureus N315 | Firmicutes | 2,588 | 53 (2.05) | 70 (2.70) |
| Staphylococcus epidermidis ATCC 12228 | Firmicutes | 2,419 | 42 (1.74) | 52 (2.15) |
| Staphylococcus epidermidis RP62A | Firmicutes | 2,494 | 45 (1.80) | 58 (2.33) |
| Staphylococcus haemolyticus JCSC1435 | Firmicutes | 2,676 | 40 (1.49) | 56 (2.09) |
| Staphylococcus saprophyticus subsp. saprophyticus ATCC 15305 | Firmicutes | 2,446 | 36 (1.47) | 46 (1.88) |
| Streptococcus agalactiae 2603V/R | Firmicutes | 2,124 | 31 (1.46) | 45 (2.12) |
| Streptococcus agalactiae NEM316 | Firmicutes | 2,094 | 32 (1.53) | 42 (2.01) |
| Streptococcus mutans UA159 | Firmicutes | 1,960 | 25 (1.28) | 20 (1.02) |
| Streptococcus pneumoniae R6 | Firmicutes | 2,043 | 31 (1.52) | 39 (1.91) |
| Streptococcus pneumoniae TIGR4 | Firmicutes | 2,094 | 37 (1.77) | 40 (1.91) |
| Streptococcus pyogenes M1 GAS | Firmicutes | 1,697 | 27 (1.59) | 30 (1.77) |
| Streptococcus pyogenes MGAS10394 | Firmicutes | 1,886 | 24 (1.27) | 28 (1.48) |
| Streptococcus pyogenes MGAS315 | Firmicutes | 1,865 | 28 (1.50) | 30 (1.61) |
| Streptococcus pyogenes MGAS5005 | Firmicutes | 1,865 | 29 (1.55) | 31 (1.66) |
| Streptococcus pyogenes MGAS6180 | Firmicutes | 1,894 | 28 (1.48) | 31 (1.64) |
| Streptococcus pyogenes MGAS8232 | Firmicutes | 1,845 | 28 (1.52) | 31 (1.68) |
| Streptococcus pyogenes SSI-1 | Firmicutes | 1,861 | 24 (1.29) | 25 (1.34) |
| Streptococcus thermophilus CNRZ1066 | Firmicutes | 1,915 | 21 (1.10) | 25 (1.31) |
| Streptococcus thermophilus LMG 18311 | Firmicutes | 1,889 | 22 (1.16) | 28 (1.48) |
| Streptomyces avermitilis MA-4680 | Actinobacteria | 7,577 | 80 (1.06) | 140 (1.85) |
| Streptomyces coelicolor A3(2) | Actinobacteria | 7,769 | 96 (1.24) | 172 (2.21) |
| Symbiobacterium thermophilum IAM 14863 | Actinobacteria | 3,337 | 55 (1.65) | 58 (1.74) |
| Synechococcus elongatus PCC 6301 | Cyanobacteria | 2,525 | 18 (0.71) | 33 (1.31) |
| Synechococcus sp. strain WH 8102 | Cyanobacteria | 2,517 | 16 (0.64) | 29 (1.15) |
| Synechocystis sp. strain PCC 6803 | Cyanobacteria | 3,167 | 24 (0.76) | 39 (1.23) |
| Thermoanaerobacter tengcongensis MB4 | Firmicutes | 2,588 | 32 (1.24) | 50 (1.93) |
| Thermobifida fusca YX | Actinobacteria | 3,110 | 25 (0.80) | 55 (1.77) |
| Thermosynechococcus elongatus BP-1 | Cyanobacteria | 2,475 | 11 (0.44) | 20 (0.81) |
| Thermotoga maritima MSB8 | Thermotogae | 1,858 | 16 (0.86) | 18 (0.97) |
| Thermus thermophilus HB27 | Deinococcus-Thermus | 1,982 | 20 (1.01) | 26 (1.31) |
| Thermus thermophilus HB8 | Deinococcus-Thermus | 1,973 | 23 (1.17) | 23 (1.17) |
| Thiobacillus denitrificans ATCC 25259 | Proteobacteria | 2,827 | 44 (1.56) | 69 (2.44) |
| Treponema denticola ATCC 35405 | Spirochaetes | 2,767 | 52 (1.88) | 14 (0.51) |
| Treponema pallidum subsp. pallidum Nichols | Spirochaetes | 1,036 | 16 (1.54) | 31 (2.99) |
| Tropheryma whipplei TW08/27 | Actinobacteria | 783 | 9 (1.15) | 11 (1.40) |
| Tropheryma whipplei Twist | Actinobacteria | 808 | 9 (1.11) | 10 (1.24) |
| Ureaplasma parvum serovar 3 ATCC 700970 | Firmicutes | 614 | 16 (2.61) | 25 (4.07) |
| Vibrio cholerae O1 bv. eltor N16961 | Proteobacteria | 3,835 | 54 (1.41) | 82 (2.14) |
| Vibrio fischeri ES114 | Proteobacteria | 3,747 | 104 (2.78) | 145 (3.87) |
| Vibrio parahaemolyticus RIMD 2210633 | Proteobacteria | 4,832 | 118 (2.44) | 163 (3.37) |
| Vibrio vulnificus CMCP6 | Proteobacteria | 4,488 | 86 (1.92) | 113 (2.52) |
| Vibrio vulnificus YJ016 | Proteobacteria | 4,955 | 101 (2.04) | 151 (3.05) |
| Wigglesworthia glossinidia | Proteobacteria | 611 | 3 (0.49) | 8 (1.31) |
| Wolbachia endosymbiont TRS of Brugia malavi | Proteobacteria | 805 | 4 (0.50) | 8 (0.99) |
| Wolbachia sp. | Proteobacteria | 1,195 | 5 (0.42) | 13 (1.09) |
| Wolinella succinogenes DSM 1740 | Proteobacteria | 2,043 | 18 (0.88) | 46 (2.25) |
| Xanthomonas axonopodis pv. citri 306 | Proteobacteria | 4,312 | 92 (2.13) | 136 (3.15) |
| Xanthomonas campestris pv. campestris 8004 | Proteobacteria | 4,273 | 101 (2.36) | 140 (3.28) |
| Xanthomonas campestris pv. campestris ATCC 33913 | Proteobacteria | 4,181 | 95 (2.27) | 136 (3.25) |
| Xanthomonas oryzae pv. oryzae KACC10331 | Proteobacteria | 4,637 | 58 (1.25) | 85 (1.83) |
| Xylella fastidiosa 9a5c | Proteobacteria | 2,766 | 47 (1.70) | 57 (2.06) |
| Xylella fastidiosa Temecula1 | Proteobacteria | 2,034 | 43 (2.11) | 57 (2.80) |
| Yersinia pestis bv. medievalis 91001 | Proteobacteria | 3,895 | 54 (1.39) | 74 (1.90) |
| Yersinia pestis CO92 | Proteobacteria | 3,885 | 72 (1.85) | 85 (2.19) |
| Yersinia pestis KIM | Proteobacteria | 4,086 | 54 (1.32) | 71 (1.74) |
| Yersinia pseudotuberculosis IP 32953 | Proteobacteria | 3,901 | 69 (1.77) | 89 (2.28) |
| Zymomonas mobilis subsp. mobilis ZM4 | Proteobacteria | 1,998 | 21 (1.05) | 29 (1.45) |
Absolute number (% of all proteins encoding lipoproteins).
HMM-based functional assignment.
Proteins are made up of functional and evolutionarily conserved units called domains. The structural classification of proteins database, SCOP, is a collection of such domains that have been observed in naturally occurring protein structures. The procedure to build hidden Markov models (HMMs), which are representations of such domains that capture essential features, and identification of domains in the known and predicted lipoproteins are described by Gough and Chothia (20). The library of such HMMs is made available through the SUPERFAMILY database (21, 35).
RESULTS
Signal sequence analysis of bacterial lipoproteins.
From the time the first version of DOLOP was introduced in 2002, there has been a steady increase in the reports of experimentally verified lipoproteins and a tremendous increase in the reports on deduced lipoproteins using predictive tools. Furthermore, the number of bacterial genomes sequenced has increased from a mere 43 used in the first version to 234 now. These inputs have necessitated updating of the database and the training of the predictive algorithm previously used in DOLOP for better prediction. Since taxon-specific trained predictive methods have also been reported, the database could be utilized more purposefully.
With the advent of genomic study and discovery of new lipoproteins, a large-scale bioinformatics analysis to define the lipoprotein signal sequence was performed to obtain the 278 distinct clusters, where each cluster represents proteins with the same function (34). Our results corroborated the general observations made by previous investigators and also helped to define a more accurate lipoprotein signal sequence. Our studies show that the n-region contains five to seven residues with two positively charged Lys or Arg residues (Fig. 2A). The length of the h-region varies between 7 and 22 residues, with a modal value of 12 residues. The c-region has a consensus [LVI][ASTVI][GAS]C sequence. It is important to mention here that the PS00013 signature provided by Prosite (25) was one of the first available prediction algorithms to identify bacterial lipoproteins. However, the amino acid choices available at each position in the signature sequence are quite broad, thus resulting in a large number of false positives. The results of the statistical analysis of the lipobox are shown in Fig. 2B. The lipid-modifiable Cys (+1 position) is invariant. In about 70% of the cases, the −3 position is Leu (71%), followed by Val (9%) and I (6%). We also see A, F, G, C, and M in the −3 position, but at low frequencies (<5%); therefore, we do not include it in the algorithm. The −2 position is more flexible and can accommodate uncharged, polar, and nonpolar residues Ala (30%), Ser (28%), Thr (12%), Val (10%), and Ile (8%). Again, we do find G, L, and M at low frequencies in this position, but we have not included these amino acids in the predictive algorithm. The −1 position is shared equally by Gly (45%) and Ala (39%); significantly, Ser has been observed in 16% of the cases.
FIG. 2.
(A) Positive charge distribution in the n-region. This graph shows that most lipoproteins have at least two positively charged amino acids in their n-region. (B) Amino acid distribution in the lipobox. Leucine has the highest propensity to occur at the −3 position; alanine and serine at the −2 position; alanine, glycine, or serine at the −1 position; and the invariant cysteine that gets lipid modified. Please refer to the text for details.
Predictive rules for identifying lipoproteins.
The availability of a larger database of experimentally verified lipoproteins has enabled the devising of predictive rules that have been found to be fairly accurate. Reports of identification of putative lipoproteins using this method followed by experimental verification justified the approach (1, 24, 33, 51). Using the currently obtained largest set of 278 distinct lipoproteins, the following predictive rules have been derived. (i) The sequence should start with Met followed by one or more positively charged residues (Lys or Arg) in the first five to seven residues. (ii) The h-region should contain 7 to 22 residues. (iii) The consensus sequence [LVI][ASTVI][GAS][C] should occur within the first 40 residues from the N-terminal end.
A predictive algorithm based on these rules has been incorporated in the website http://www.mrc-lmb.cam.ac.uk/genomes/dolop/analysis.shtml to analyze a user-given query sequence and to pull out probable lipoproteins from completely or partially sequenced bacterial genomes.
Predicted lipoproteins in the completely sequenced bacterial genomes.
In the past few years, the genomic data available have increased enormously, and therefore one of the major updates in DOLOP is the inclusion of a list of predicted lipoproteins from 234 genomes. Since other lipoprotein-predicting tools have also been made available in the literature, we have included a comparative analysis and provided the data in a tabular form (Table 1). There is generally a fair agreement in the number of predicted lipoproteins in a genome between the two methods, with LipoP predicting 20% more in general (it should be noted that our algorithm is more conservative in predicting the lipoprotein signal sequence in comparison to the Prosite pattern or LipoP). For genomes with more than 1,000 open reading frames (ORFs), it was interesting to note that the number of predicted lipoproteins varied enormously between the various bacteria: from as many as 223 lipoproteins for Bacteroides thetaiotaomicron VP3-5482 to as little as 8 to 9 in the case of Aquifex aeolicus VP5, Prochlorococcus marinus subsp. pastoris CCMP 1378. In the case of smaller genomes, two species of Buchnera had no predicted lipoprotein and the third had only one. In others, the number varied from 2 to 180. The plot of the proteome size against the number of predicted lipoproteins revealed a weak, linear correlation (Fig. 3). We had worked out another index of comparison, the percentage of genome coding for lipoproteins, and found that there was no correlation between the proteome size and the fraction of the proteome coding for lipoproteins. In fact, we observed that within the same proteome, the fraction of proteins encoding lipoproteins was fairly conserved. For example, Mycoplasma penetrans showed the highest ratio of 5.79%, followed by Mycoplasma pneumoniae with 5.52%. The ratio of 4.67% is high in the case of Bacteroides, especially from the point of view of its large genome size (4,500 ORFs). For many, the ratio varied typically from 1 to 3%. In E. coli CFT073 and K-12, even though the former has about 1,000 additional genes compared to the latter, there were no additional lipoproteins. Both have 86 predicted lipoproteins. In the case of E. coli O157:H7 and O157:H7 EDL933, for the same genome size there were nine additional lipoproteins. Rhodopirellula baltica is one of the bigger genomes (7,325 ORFs) but contains only 46 lipoproteins.
FIG. 3.
Plot of the proteome size against the number of predicted lipoproteins for the 234 completely sequenced bacterial genomes used in our analysis. Note that there is a positive correlation between the genome size and the number of lipoproteins encoded. Organisms whose predicted number of lipoproteins falls way above or below the linear trend fitted for the observed data are marked on the graph. The large number of lipoproteins seen in Bacteroides corresponds, in large part, to a lineage-specific expansion of predicted lipoproteins with an N-terminal beta-propeller domain, which may form a specialized adhesion module. In Bdellovibrio, several lipoproteins appear to belong to an expansion of peptidases.
Functional assignment to known and predicted lipoproteins.
Rather than just make predictions about which proteins might be lipid modified, we went a step further to provide information about possible functions by identifying protein domains (e.g., P-loop NTP hydrolase domain) in the predicted lipoproteins. To get this information, the bacterial lipoproteins in the database were subjected to a previously described (21) structural domain analysis, which is that used by the SUPERFAMILY database (20, 35). The experimentally verified proteins were analyzed separately from the predicted proteins. The results of the analysis are organized into domain superfamilies and are available at http://supfam.mrc-lmb.cam.ac.uk/SUPERFAMILY/cgi-bin/gen_list.cgi?genome=lp for the predicted lipoproteins and http://supfam.mrc-lmb.cam.ac.uk/SUPERFAMILY/cgi-bin/gen_list.cgi?genome=lq for the experimentally verified ones. The domains in the sequences are detected and classified according to the SCOP (38) classification of domain superfamilies using HMMs (12, 32). This provides, for each sequence, a list of known structural domains and the order in which they occur; this is called the domain architecture of a protein.
In the SCOP classification scheme, proteins are split into domains as minimum functional and evolutionary units, i.e., all domains are observed either on their own or in combination with more than one different partner. The superfamily level of classification groups domains for which there is structural, sequence, and/or functional evidence for a common evolutionary ancestor. The expertly built HMMs in the SUPERFAMILY library are able to detect remote homologies, and they assign known structural domains to half of the total lipoprotein sequence.
The information provided by this analysis reveals the composition of domains, which evolution has selected for use in lipoproteins, and the architectures show how these domain units have been shuffled and recombined to form the larger, more complicated multidomain proteins.
In the example shown in Fig. 4A, we show a predicted lipoprotein represented by its domain architecture as determined above. The individual domains, which go to make up the whole protein, are each independent units, which have been combined in this particular order during evolution, and selected for, to carry out the function of the complete protein. For this particular example shown, there are ten such proteins in the database, all with the same architecture, all in the set of “predicted” lipoproteins. This particular architecture is detected in every staphylococcal genome only once, which suggests that it could be an essential protein with a specific functional role.
FIG. 4.
(A) Domain architecture for the protein gi 21284057 gb NP_647145.1 from Staphylococcus aureus MW2. This architecture contains two domains: a periplasmic metal-binding protein domain and a lipocalin fold metal-binding domain (in that order). In this case the assignments span the entire protein and provide a complete picture of the protein. (B) Screen shot of the output from PSATool. The program calculates molecular weight, amino acid frequency, composition, and weight composition and displays charge distribution and the nature of the sequence. This tool is available for predicted and verified lipoproteins.
Relevance of the database to the study of bacterial pathogenesis.
In the introduction we had highlighted the importance of lipoproteins in pathogenesis, evasion of host defense, elicitation of inflammatory response, and vaccine development. Thus, being able to identify such lipoproteins from the completely sequenced bacterial genomes of many harmful pathogens is an important problem in the postgenomic era (9). Being able to predict them will undoubtedly help us to define candidate proteins to be studied, which will eventually contribute to a better understanding of the molecular events involved in such key processes.
To highlight how one can gain a better understanding about which lipoproteins are differentially expressed in bacteria during the different conditions, we performed the following calculation. (i) Using our method, we first identified the predicted list of Pseudomonas aeruginosa proteins that could potentially be lipid modified. (ii) Next, we identified up-regulated and down-regulated genes in P. aeruginosa under quorum-sensing conditions using the data set that was previously published (49). In their study, Schuster et al. obtained the set of differentially expressed genes under quorum-sensing conditions using microarrays. (iii) By integrating the above two lists of proteins, we predict that at least 10 lipoproteins are up-regulated preferentially under quorum-sensing conditions (Pseudomonas aeruginosa gene identifiers: PA1324, PA1664, PA1666, PA1745, PA1888, PA2414, PA3677, PA3692, PA4208, and PA4876). Since quorum sensing has been shown to be important for the formation of biofilms (10), and hence important during the course of infection in the case of Pseudomonas (8), studying these up-regulated lipoproteins can help us understand the process of biofilm formation much better, and it may eventually lead to a better understanding of the whole process of infection.
DISCUSSION
Lipid modification of proteins is a ubiquitous posttranslational modification successfully evolved by biological systems to carry out a variety of biochemical functions in the aqueous and membrane interface, a challenge common to even man-made applications. In this regard, the comprehensive lipid modification by bacteria at the N-terminal end of a protein is attractive even from a commercial angle, as any protein can be potentially converted to lipoprotein by adequately understanding bacterial lipid modification determinants in a bacterium like E. coli, a popular recombinant host. Recently, we demonstrated such engineering using a nonlipoprotein (29). Further, essential lipoproteins and the pathway enzymes are targets for interfering with bacterial growth and viability. Therefore, the need for an exclusive database for bacterial lipoproteins was felt, and it was introduced in 2002. Subsequently, with the rapid expansion of the bacterial genomic database and reports on the roles of lipoproteins in bacterial homeostasis and pathogenesis, we have undertaken a major update, and this is a report highlighting the various features, especially the functional assignments to predicted lipoproteins, an aspect not well understood or addressed.
Features of the database—genome-wide predicted lipoproteins are useful in proteomics.
The number of current, characteristic lipoproteins has gone up from 199 in the previous version (34) to 278 in this version. Compared to the increase in the number of lipoproteins reported as well as predicted from the genome data, this increase in unique lipoproteins is not high. To make the database functionally relevant, these have been classified as in the previous version according to the information gained from the literature into antigens, adhesins, binding proteins, enzymes, transporters, toxins, surface proteins, interesting factors, and hypothetical. We performed several analyses, one of which was to refine the rule to predict which proteins can be lipid modified. Using this rule, we predicted potential lipoproteins for the 234 completely sequenced bacterial organisms, many of which are important pathogens. When we applied the current DOLOP prediction algorithm to the 81 experimentally verified lipoproteins from E. coli K-12, published by Gonnet et al. (19), 71 are predicted correctly (the number cited by the authors, however, is 51 even though 60 can be readily counted from the data provided in their table and another 11 are predicted correctly when we performed the analysis). Many of the 10 that are not predicted were due to our stringent cutoff applied at the −2 and −3 positions to reduce the false positives as defined previously. Thus, inclusion of minor amino acids like M and A in these positions obviously improved prediction to near 100%, except one in which the lipobox was more internal (51 amino acids inside). The fact that it is an experimentally verified lipoprotein and such internalized lipoboxes were found to be modified in the early investigations does suggest the relevance of increasing the length of the N-terminal sequence for query. But, for the sake of keeping the false positives low, we maintain it at 40 residues. The same analysis with a gram-positive database of experimentally verified lipoproteins reported by Juncker et al. predicted 26 out of 32, and by introducing M and A in the −3 and −2 positions, all were predicted correctly. With such refinements, the new predictive rule used in the current version of DOLOP would be able to predict at an extent seen with the other available algorithms. Though taxon-specific algorithms are obviously the best way to go after prediction, they would require structural data from many lipoproteins belonging to individual taxons, which is a farfetched proposition and beats the necessity for prediction. Therefore a reasonably accurate predictive algorithm as presented here to handle sequence data from a variety of different bacteria is a good first-level bioinformatic tool.
Our analysis shows that there are a large number of uncharacterized lipoproteins even in thoroughly studied bacterial systems. Our results on the comparison of genome size against the predicted number of lipoproteins show that there is a weak positive correlation, indicating that organisms have evolved their own set of lipoproteins to meet their needs. In the case of pathogenic variants, the number could be more or less, but their pathogenic association gives another dimension and a reason to look at them more carefully, as whatever cases have been characterized showed that they were essential for pathogenesis. As illustrated by an example in Results, using comparative proteomics in silico by integrating information about the predicted lipoproteins contained in DOLOP for an organism with other external data, such as gene expression by microarray analysis, one can come up with meaningful predictions. In this regard, the superfamily domain prediction would further aid in short-listing those activities related to the pathogenic aspect being studied.
Features of the database—domain predictions help in functional assignments.
Though lipid modification of proteins is an essential function, not much is known about individual lipoproteins in bacteria in terms of biochemical functions, and their proteome is not adequately investigated. To enhance the utility of the database in terms of functional correlation, a link to the SUPERFAMILY structural domain assignment prediction tool has been provided for each predicted lipoprotein. Information about a protein domain directly provides clues about the actual molecular function and also helps in identifying functionally important residues involved in performing the function. Thus, this feature should help at the first level in obtaining useful information for a suspected biochemical function that may account for an observed phenotype or function or for planning mutation experiments to define the roles. For researchers interested in obtaining basic properties of the predicted lipoprotein, a link to PSAtool has also been provided, which provides information like molecular weight, amino acid composition, and charge distribution for a given sequence (Fig. 4B). This feature, we believe, will help experimental biologists in designing experiments to purify proteins of interest.
Extended structure-function relationship of lipoprotein signal sequences.
Previous studies involving detailed site-directed mutagenesis studies of residues in the lipoprotein signal sequence have already led to the elucidation of roles of individual regions as well as the amino acids in the modification. The positive charge at the N-terminal region was found to be important in phospholipid-signal sequence interaction, leading to a complex that is important for the recognition and transport across the inner membrane of gram-negative bacteria (61). Replacement of Gly at the −14 position (inside the h-region) in murein lipoprotein signal sequence with Asp, Glu, or Arg underlined the importance of the uncharged nature of the h-region (27). The −1 position tolerated Ala as well as Gly. Substitution by Ser slowed down lipid modification, and Thr sets the limit (42). In this context, the presence of 16% of lipoproteins in our data set with Ser at the −1 position may be relevant to the homeostasis of bacterial lipid modification in bacteria. The −2 position is the most variable among the lipobox sequences. However, inclusion of charged residues in this region has resulted in deficient lipid modification. In certain mutation studies, it has been found that the unmodified prolipoprotein has been transported and even processed by signal peptidase I, specific for nonlipoprotein signal sequences (17). In certain instances, wherein DOLOP has given false-positive results, a signal peptidase I cleavage sequence was found to lie in the vicinity of the lipobox. As pointed out earlier, the structural determinants required for inner and outer membrane targeting have not yet been fully understood and it is firmly believed that such signals come from the mature sequence in the vicinity of the cleavage site. It is also quite possible that distant primary and secondary structure elements might have a role, as the transport across the two membranes in gram-negative bacteria requires protein machinery and additional protein-protein interactions between the machinery and the lipoprotein. The large set of lipoprotein signal sequences and the genome-wide mature sequence information available in DOLOP should provide a good data set for future analysis.
We see several ways in which our results can be helpful to experimental biologists for carrying out novel research and for prioritizing their experiments. A few instances where our results can be useful include (i) identification of lipoproteins unique to a particular strain; (ii) identification of lipoproteins present in a particular group of pathogens, or organisms which colonize the same ecological niche; (iii) designing microarray experiments focusing on lipoprotein gene expression during different stages of infection; (iv) rapid identification of lipoproteins from two-dimensional gel experiments and mass spectrometric studies; and (v) identification of novel virulence factors.
In conclusion, there is still a huge untapped potential and tremendous scope for analysis and characterization of lipoproteins, and we believe that the results presented here and the database with the various features will serve as useful resources for experimental biologists to address some important questions. In addition, we also offer the possibility for researchers to submit information about newly characterized lipoproteins to our database. This feature also allows researchers to exchange information with the scientific community.
Acknowledgments
M.M.B. and L.A. gratefully acknowledge the intramural research program of the National Institutes of Health for funding their research. K.S. thanks the National Bioinformatics Service, BTIS, Centre for Biotechnology, for providing infrastructure support. K.S. and A.T.S. thank the LG foundation, Chennai, India, for a research career fellowship to A.T.S.
We thank the anonymous referees for helpful comments.
REFERENCES
- 1.Barker, A. P., A. I. Vasil, A. Filloux, G. Ball, P. J. Wilderman, and M. L. Vasil. 2004. A novel extracellular phospholipase C of Pseudomonas aeruginosa is required for phospholipid chemotaxis. Mol. Microbiol. 53:1089-1098. [DOI] [PubMed] [Google Scholar]
- 2.Beermann, C., G. Lochnit, R. Geyer, P. Groscurth, and L. Filgueira. 2000. The lipid component of lipoproteins from Borrelia burgdorferi: structural analysis, antigenicity, and presentation via human dendritic cells. Biochem. Biophys. Res. Commun. 267:897-905. [DOI] [PubMed] [Google Scholar]
- 3.Bendtsen, J. D., T. T. Binnewies, P. F. Hallin, T. Sicheritz-Ponten, and D. W. Ussery. 2005. Genome update: prediction of secreted proteins in 225 bacterial proteomes. Microbiology 151:1725-1727. [DOI] [PubMed] [Google Scholar]
- 4.Berry, A. M., and J. C. Paton. 1996. Sequence heterogeneity of PsaA, a 37-kilodalton putative adhesin essential for virulence of Streptococcus pneumoniae. Infect. Immun. 64:5255-5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun, V., and K. Rehn. 1969. Chemical characterization, spatial distribution and function of a lipoprotein (murein-lipoprotein) of the E. coli cell wall. The specific effect of trypsin on the membrane structure. Eur. J. Biochem. 10:426-438. [DOI] [PubMed] [Google Scholar]
- 6.Braun, V., and H. C. Wu. 1993. Lipoproteins, structure, function, biosynthesis and models for protein export, p. 319-342. In J.-M. Ghuysen and R. Hakenback (ed.), Bacterial cell wall, vol. 27. Elsevier, Amsterdam, The Netherlands. [Google Scholar]
- 7.Chang, Y. F., M. J. Appel, R. H. Jacobson, S. J. Shin, P. Harpending, R. Straubinger, L. A. Patrican, H. Mohammed, and B. A. Summers. 1995. Recombinant OspA protects dogs against infection and disease caused by Borrelia burgdorferi. Infect. Immun. 63:3543-3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 9.Crossman, L., A. Cerdeno-Tarraga, S. Bentley, and J. Parkhill. 2003. Pathogenomics. Nat. Rev. Microbiol. 1:176-177. [DOI] [PubMed] [Google Scholar]
- 10.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 11.Dev, I. K., R. J. Harvey, and P. H. Ray. 1985. Inhibition of prolipoprotein signal peptidase by globomycin. J. Biol. Chem. 260:5891-5894. [PubMed] [Google Scholar]
- 12.Eddy, S. R. 1996. Hidden Markov models. Curr. Opin. Struct. Biol. 6:361-365. [DOI] [PubMed] [Google Scholar]
- 13.Fariselli, P., G. Finocchiaro, and R. Casadio. 2003. SPEPlip: the detection of signal peptide and lipoprotein cleavage sites. Bioinformatics 19:2498-2499. [DOI] [PubMed] [Google Scholar]
- 14.Fikrig, E., S. W. Barthold, F. S. Kantor, and R. A. Flavell. 1990. Protection of mice against the Lyme disease agent by immunizing with recombinant OspA. Science 250:553-556. [DOI] [PubMed] [Google Scholar]
- 15.Fikrig, E., S. R. Telford III, S. W. Barthold, F. S. Kantor, A. Spielman, and R. A. Flavell. 1992. Elimination of Borrelia burgdorferi from vector ticks feeding on OspA-immunized mice. Proc. Natl. Acad. Sci. USA 89:5418-5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gan, K., K. Sankaran, M. G. Williams, M. Aldea, K. E. Rudd, S. R. Kushner, and H. C. Wu. 1995. The umpA gene of Escherichia coli encodes phosphatidylglycerol:prolipoprotein diacylglyceryl transferase (lgt) and regulates thymidylate synthase levels through translational coupling. J. Bacteriol. 177:1879-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghrayeb, J., C. A. Lunn, S. Inouye, and M. Inouye. 1985. An alternate pathway for the processing of the prolipoprotein signal peptide in Escherichia coli. J. Biol. Chem. 260:10961-10965. [PubMed] [Google Scholar]
- 18.Gonnet, P., and F. Lisacek. 2002. Probabilistic alignment of motifs with sequences. Bioinformatics 18:1091-1101. [DOI] [PubMed] [Google Scholar]
- 19.Gonnet, P., K. E. Rudd, and F. Lisacek. 2004. Fine-tuning the prediction of sequences cleaved by signal peptidase II: a curated set of proven and predicted lipoproteins of Escherichia coli K-12. Proteomics 4:1597-1613. [DOI] [PubMed] [Google Scholar]
- 20.Gough, J., and C. Chothia. 2002. SUPERFAMILY: HMMs representing all proteins of known structure. SCOP sequence searches, alignments and genome assignments. Nucleic Acids Res. 30:268-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gough, J., K. Karplus, R. Hughey, and C. Chothia. 2001. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 313:903-919. [DOI] [PubMed] [Google Scholar]
- 22.Hanson, M. S., D. R. Cassatt, B. P. Guo, N. K. Patel, M. P. McCarthy, D. W. Dorward, and M. Hook. 1998. Active and passive immunity against Borrelia burgdorferi decorin binding protein A (DbpA) protects against infection. Infect. Immun. 66:2143-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayashi, S., and H. C. Wu. 1990. Lipoproteins in bacteria. J. Bioenerg. Biomembr. 22:451-471. [DOI] [PubMed] [Google Scholar]
- 24.Howard, M. B., N. A. Ekborg, L. E. Taylor II, R. M. Weiner, and S. W. Hutcheson. 2004. Chitinase B of “Microbulbifer degradans” 2-40 contains two catalytic domains with different chitinolytic activities. J. Bacteriol. 186:1297-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hulo, N., C. J. Sigrist, V. Le Saux, P. S. Langendijk-Genevaux, L. Bordoli, A. Gattiker, E. De Castro, P. Bucher, and A. Bairoch. 2004. Recent improvements to the PROSITE database. Nucleic Acids Res. 32:D134-D137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Innis, M. A., M. Tokunaga, M. E. Williams, J. M. Loranger, S. Y. Chang, S. Chang, and H. C. Wu. 1984. Nucleotide sequence of the Escherichia coli prolipoprotein signal peptidase (lsp) gene. Proc. Natl. Acad. Sci. USA 81:3708-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inouye, S., G. P. Vlasuk, H. Hsiung, and M. Inouye. 1984. Effects of mutations at glycine residues in the hydrophobic region of the Escherichia coli prolipoprotein signal peptide on the secretion across the membrane. J. Biol. Chem. 259:3729-3733. [PubMed] [Google Scholar]
- 28.Juncker, A. S., H. Willenbrock, G. Von Heijne, S. Brunak, H. Nielsen, and A. Krogh. 2003. Prediction of lipoprotein signal peptides in gram-negative bacteria. Protein Sci. 12:1652-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamalakkannan, S., V. Murugan, M. V. Jagannadham, R. Nagaraj, and K. Sankaran. 2004. Bacterial lipid modification of proteins for novel protein engineering applications. Protein Eng. Des. Sel. 17:721-729. [DOI] [PubMed] [Google Scholar]
- 30.Keenan, J., J. Oliaro, N. Domigan, H. Potter, G. Aitken, R. Allardyce, and J. Roake. 2000. Immune response to an 18-kilodalton outer membrane antigen identifies lipoprotein 20 as a Helicobacter pylori vaccine candidate. Infect. Immun. 68:3337-3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kobayashi, T., M. Nishijima, Y. Tamori, S. Nojima, Y. Seyama, and T. Yamakawa. 1980. Acyl phosphatidylglycerol of Escherichia coli. Biochim. Biophys. Acta 620:356-363. [PubMed] [Google Scholar]
- 32.Krogh, A., M. Brown, I. S. Mian, K. Sjolander, and D. Haussler. 1994. Hidden Markov models in computational biology. Applications to protein modeling. J. Mol. Biol. 235:1501-1531. [DOI] [PubMed] [Google Scholar]
- 33.Leduc, I., P. Richards, C. Davis, B. Schilling, and C. Elkins. 2004. A novel lectin, DltA, is required for expression of a full serum resistance phenotype in Haemophilus ducreyi. Infect. Immun. 72:3418-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madan Babu, M., and K. Sankaran. 2002. DOLOP—database of bacterial lipoproteins. Bioinformatics 18:641-643. [DOI] [PubMed] [Google Scholar]
- 35.Madera, M., C. Vogel, S. K. Kummerfeld, C. Chothia, and J. Gough. 2004. The SUPERFAMILY database in 2004: additions and improvements. Nucleic Acids Res. 32:D235-D239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maeda, Y., M. Makino, D. C. Crick, S. Mahapatra, S. Srisungnam, T. Takii, Y. Kashiwabara, and P. J. Brennan. 2002. Novel 33-kilodalton lipoprotein from Mycobacterium leprae. Infect. Immun. 70:4106-4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Masuda, K., S. Matsuyama, and H. Tokuda. 2002. Elucidation of the function of lipoprotein-sorting signals that determine membrane localization. Proc. Natl. Acad. Sci. USA 99:7390-7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murzin, A. G., S. E. Brenner, T. Hubbard, and C. Chothia. 1995. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247:536-540. [DOI] [PubMed] [Google Scholar]
- 39.Narita, S., K. Kanamaru, S. Matsuyama, and H. Tokuda. 2003. A mutation in the membrane subunit of an ABC transporter LolCDE complex causing outer membrane localization of lipoproteins against their inner membrane-specific signals. Mol. Microbiol. 49:167-177. [DOI] [PubMed] [Google Scholar]
- 40.Neufert, C., R. K. Pai, E. H. Noss, M. Berger, W. H. Boom, and C. V. Harding. 2001. Mycobacterium tuberculosis 19-kDa lipoprotein promotes neutrophil activation. J. Immunol. 167:1542-1549. [DOI] [PubMed] [Google Scholar]
- 41.Notredame, C., D. G. Higgins, and J. Heringa. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302:205-217. [DOI] [PubMed] [Google Scholar]
- 42.Pollitt, S., S. Inouye, and M. Inouye. 1986. Effect of amino acid substitutions at the signal peptide cleavage site of the Escherichia coli major outer membrane lipoprotein. J. Biol. Chem. 261:1835-1837. [PubMed] [Google Scholar]
- 43.Pugsley, A. P. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 57:50-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reglier-Poupet, H., C. Frehel, I. Dubail, J. L. Beretti, P. Berche, A. Charbit, and C. Raynaud. 2003. Maturation of lipoproteins by type II signal peptidase is required for phagosomal escape of Listeria monocytogenes. J. Biol. Chem. 278:49469-49477. [DOI] [PubMed] [Google Scholar]
- 45.Rosengarten, R., and K. S. Wise. 1990. Phenotypic switching in mycoplasmas: phase variation of diverse surface lipoproteins. Science 247:315-318. [DOI] [PubMed] [Google Scholar]
- 46.Sankaran, K., S. D. Gupta, and H. C. Wu. 1995. Modification of bacterial lipoproteins. Methods Enzymol. 250:683-697. [DOI] [PubMed] [Google Scholar]
- 47.Sankaran, K., and H. C. Wu. 1994. Lipid modification of bacterial prolipoprotein. Transfer of diacylglyceryl moiety from phosphatidylglycerol. J. Biol. Chem. 269:19701-19706. [PubMed] [Google Scholar]
- 48.Schuch, R., and A. T. Maurelli. 1999. The Mxi-Spa type III secretory pathway of Shigella flexneri requires an outer membrane lipoprotein, MxiM, for invasin translocation. Infect. Immun. 67:1982-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schuster, M., C. P. Lostroh, T. Ogi, and E. P. Greenberg. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185:2066-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seydel, A., P. Gounon, and A. P. Pugsley. 1999. Testing the ‘+2 rule’ for lipoprotein sorting in the Escherichia coli cell envelope with a new genetic selection. Mol. Microbiol. 34:810-821. [DOI] [PubMed] [Google Scholar]
- 51.Sha, J., A. A. Fadl, G. R. Klimpel, D. W. Niesel, V. L. Popov, and A. K. Chopra. 2004. The two murein lipoproteins of Salmonella enterica serovar Typhimurium contribute to the virulence of the organism. Infect. Immun. 72:3987-4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shang, E. S., T. A. Summers, and D. A. Haake. 1996. Molecular cloning and sequence analysis of the gene encoding LipL41, a surface-exposed lipoprotein of pathogenic Leptospira species. Infect. Immun. 64:2322-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sutcliffe, I. C., and D. J. Harrington. 2002. Pattern searches for the identification of putative lipoprotein genes in gram-positive bacterial genomes. Microbiology 148:2065-2077. [DOI] [PubMed] [Google Scholar]
- 54.Tanaka, K., S. I. Matsuyama, and H. Tokuda. 2001. Deletion of lolB, encoding an outer membrane lipoprotein, is lethal for Escherichia coli and causes accumulation of lipoprotein localization intermediates in the periplasm. J. Bacteriol. 183:6538-6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Terada, M., T. Kuroda, S. I. Matsuyama, and H. Tokuda. 2001. Lipoprotein sorting signals evaluated as the LolA-dependent release of lipoproteins from the cytoplasmic membrane of Escherichia coli. J. Biol. Chem. 276:47690-47694. [DOI] [PubMed] [Google Scholar]
- 56.Thoma-Uszynski, S., S. M. Kiertscher, M. T. Ochoa, D. A. Bouis, M. V. Norgard, K. Miyake, P. J. Godowski, M. D. Roth, and R. L. Modlin. 2000. Activation of toll-like receptor 2 on human dendritic cells triggers induction of IL-12, but not IL-10. J. Immunol. 165:3804-3810. [DOI] [PubMed] [Google Scholar]
- 57.Tjalsma, H., and J. M. van Dijl. 2005. Proteomics-based consensus prediction of protein retention in a bacterial membrane. Proteomics 17:4472-4482. [DOI] [PubMed] [Google Scholar]
- 58.Tokuda, H., and S. Matsuyama. 2004. Sorting of lipoproteins to the outer membrane in E. coli. Biochim. Biophys. Acta 1694:IN1-IN9. [PubMed] [Google Scholar]
- 59.Tokunaga, M., J. M. Loranger, S. Y. Chang, M. Regue, S. Chang, and H. C. Wu. 1985. Identification of prolipoprotein signal peptidase and genomic organization of the lsp gene in Escherichia coli. J. Biol. Chem. 260:5610-5615. [PubMed] [Google Scholar]
- 60.Tokunaga, M., J. M. Loranger, and H. C. Wu. 1984. A distinct signal peptidase for prolipoprotein in Escherichia coli. J. Cell. Biochem. 24:113-120. [DOI] [PubMed] [Google Scholar]
- 61.Vlasuk, G. P., S. Inouye, H. Ito, K. Itakura, and M. Inouye. 1983. Effects of the complete removal of basic amino acid residues from the signal peptide on secretion of lipoprotein in Escherichia coli. J. Biol. Chem. 258:7141-7148. [PubMed] [Google Scholar]
- 62.Washburn, L. R., E. J. Miller, and K. E. Weaver. 2000. Molecular characterization of Mycoplasma arthritidis membrane lipoprotein MAA1. Infect. Immun. 68:437-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yakushi, T., K. Masuda, S. Narita, S. Matsuyama, and H. Tokuda. 2000. A new ABC transporter mediating the detachment of lipid-modified proteins from membranes. Nat. Cell Biol. 2:212-218. [DOI] [PubMed] [Google Scholar]
- 64.Yamaguchi, K., F. Yu, and M. Inouye. 1988. A single amino acid determinant of the membrane localization of lipoproteins in E. coli. Cell 53:423-432. [DOI] [PubMed] [Google Scholar]
- 65.Yokota, N., T. Kuroda, S. Matsuyama, and H. Tokuda. 1999. Characterization of the LolA-LolB system as the general lipoprotein localization mechanism of Escherichia coli. J. Biol. Chem. 274:30995-30999. [DOI] [PubMed] [Google Scholar]
- 66.Zhang, H., D. W. Niesel, J. W. Peterson, and G. R. Klimpel. 1998. Lipoprotein release by bacteria: potential factor in bacterial pathogenesis. Infect. Immun. 66:5196-5201. [DOI] [PMC free article] [PubMed] [Google Scholar]