Abstract
Plasmid pXO1 encodes the tripartite anthrax toxin, which is the major virulence factor of Bacillus anthracis. In spite of the important role of pXO1 in anthrax pathogenesis, very little is known about its replication and maintenance in B. anthracis. We cloned a 5-kb region of the pXO1 plasmid into an Escherichia coli vector and showed that this plasmid can replicate when introduced into B. anthracis. Mutational analysis showed that open reading frame 45 (repX) of pXO1 was required for the replication of the miniplasmid in B. anthracis. Interestingly, repX showed limited homology to bacterial FtsZ proteins that are involved in cell division. A mutation in the predicted GTP binding domain of RepX abolished its replication activity. Genes almost identical to repX are contained on several megaplasmids in members of the Bacillus cereus group, including a B. cereus strain that causes an anthrax-like disease. Our results identify a novel group of FtsZ-related initiator proteins that are required for the replication of virulence plasmids in B. anthracis and possibly in related organisms. Such replication proteins may provide novel drug targets for the elimination of plasmids encoding the anthrax toxin and other virulence factors.
Bacillus anthracis is a gram-positive, spore-forming bacterium that is the etiological agent of anthrax in humans (reviewed in references 12, 19, and 25). B. anthracis is genetically very closely related to Bacillus cereus and Bacillus thuringiensis (13, 16, 34), and most of the chromosomal virulence-promoting genes of B. anthracis are also present in the latter organisms. A major difference between B. anthracis and related organisms is the presence of two large plasmids, pXO1 and pXO2, which are required for the virulence of this organism (14, 19, 25, 31, 36). Recently, a strain of B. cereus has been described that contains a plasmid almost identical to pXO1 and causes an anthrax-like disease (15). The pXO1 plasmid (181.6 kb) encodes the anthrax toxin proteins termed the protective antigen, lethal factor, and the edema factor (10, 18, 19, 25). This plasmid also contains genes that are involved in germination of the spores and genes such as atxA and pagR that regulate the expression of the anthrax toxin and other virulence factors (3, 10, 19, 28). Plasmid pXO1 also contains a number of genes resembling those involved in the horizontal transfer of plasmids (9, 19, 28), suggesting that this plasmid may mediate its own transfer to other related organisms.
Very little is known about the replication properties of the pXO1 plasmid. Studies of the identification of the pXO1 replicon have been hampered since this plasmid does not encode proteins that share significant similarity with known replication initiator proteins encoded by other plasmids (5, 6, 17) (http://www.essex.ac.uk/bs/staff/osborn/DPR_home.htm). Understanding the replication properties of pXO1 is critical for analyzing the potential of this plasmid to replicate and transfer the anthrax toxin-encoding genes in nature.
In this study, we describe the isolation of the replication region of the pXO1 plasmid. Interestingly, the putative replication initiator protein (RepX) of pXO1 shares limited homology with the FtsZ proteins of eubacteria that are involved in cell division. Genes nearly identical to repX were also present on plasmids in related organisms such as B. cereus and B. thuringiensis. Our results suggest that RepX may define a novel family of plasmid-encoded initiator proteins involved in the replication of virulence plasmids in B. anthracis and other members of the B. cereus group.
MATERIALS AND METHODS
Strains and plasmid construction.
The B. anthracis Sterne 34F2 strain containing the pXO1 plasmid but lacking pXO2, as well as recombinant plasmids containing various regions of pXO1, was obtained from Theresa Koehler. A 10,138-bp PstI fragment of pXO1 (nucleotides [nt] 52397 to 62534 containing open reading frames [ORFs] 43 to 48 and a portion of ORF 42) was cloned into the PstI site of the Escherichia coli vector pBSCm containing the ColE1 replicon of E. coli and ampicillin and chloramphenicol resistance genes (35) to generate plasmid pC43-48P. The recombinant plasmid was recovered by transforming E. coli DH5α (33). The pC43-48P plasmid was then used as the source for subsequent subclonings. Plasmid pC43-48P was digested with XbaI and BamHI, and the 3,482-bp fragment (pXO1 nt 56685 to 60166 containing ORFs 45 and 46 and a portion of ORFs 44 and 47) was cloned into BamHI- and XbaI-digested pBSCm to generate plasmid pC45-46XB. Plasmid pC43-48P was digested with EcoRV and PstI, and the 3,441-bp fragment (nt 59094 to 62534 containing ORFs 47 and 48 and a portion of ORF 46) was ligated into pBSCm digested with EcoRV and PstI to generate pC47-48EP. Plasmid pC43-48P was digested with SacI to remove a 3,999-bp fragment containing ORFs 45 to 48, and the DNA with the remaining 6,187-bp region of pXO1 was religated to generate pC43-44S (pXO1 nt 52397 to 58583 containing ORFs 43 and 44 and a portion of ORFs 42 and 45). Similarly, BamHI was used to remove a 2,376-bp fragment containing ORFs 47 and 48 from pC43-48P, and the plasmid with the remaining 7,770-bp region of pXO1 was religated to generate pC43-46B (pXO1 nt 52397 to 60167 containing ORFs 43 to 46 and portions of ORFs 42 and 47). The plasmid pC43-45SMB was generated by deleting a large segment of ORF 42 by digesting pC43-46B with SwaI and MscI and religating the plasmid containing the 5,304-bp region of pXO1 (nt 54863 to 60167). The plasmid pCΔ44 was constructed by introducing a frameshift into ORF 44 (see below) and by digesting the resulting plasmid with XbaI, thus releasing a 297-bp internal fragment from ORF 44 (nt 56388 to 56684); the resulting 5,803 and 3,494-bp fragments were religated, maintaining the correct orientation of the gene in the plasmid (pXO1 nt 54863 to 56387 and 56685 to 60166). For complementation experiments, a new shuttle vector (pBSKm) was produced by inserting a HindIII fragment from pUC4Ωkan (29) containing the aphA3 (Kmr) gene into the HindIII site of pBSIISK. The plasmid pK43-46SM was generated by digesting pC43-46B with SpeI and SwaI to delete the majority of ORF 42, yielding a 5,304-bp region of pXO1 (nt 54863 to 60166), and ligating this fragment to SpeI/SmaI-digested pBSKm. This plasmid was used as the source of RepX for the complementation of plasmids lacking a functional repX gene. The plasmid pXori1 was obtained by digesting pCΔ44mut43 with EcoRV and cloning the 738-bp fragment (nt 55617 to 56355) into the EcoRV site of pBSCm. The plasmid pXori was generated by amplifying a 158-bp region of pXO1 (nt 55726 to 55883) using the following primers containing BamHI linkers: 5′-CCGGATCCGATGCAAATTGTAAATTCATTATC-3′ and 5′-CCGGATCCGGTGTTAGAATAGCGATTGAAC-3′. The reaction mixtures (50 μl) contained 20 mM Tris-HCl (pH 8.8), 10 mM KCl, 10 mM (NH4)2SO4, 2 mM MgSO4, 0.1% Triton X-100, 100 μg/ml nuclease-free bovine serum albumin, 200 μM of each deoxynucleoside triphosphate, 10 ng of plasmid DNA, 1 μM of each primer, and 2.5 units of Pfu polymerase (Stratagene, La Jolla, CA). The conditions of PCR amplification were as follows: 95°C for 3 min; 95°C for 1 min, 65°C for 1 min, and 72°C for 1 min for 25 cycles; and 72°C for 10 min. The PCR product and pBSCm were digested with BamHI, and the DNA was ligated to generate pXori. All the above plasmids were passaged through E. coli GM2163 (dam dcm mutant), which facilitated the subsequent introduction of plasmid DNA into B. anthracis (24). Plasmid DNAs (10 to 15 μg) were then electroporated into the plasmidless B. anthracis strain UM23C1-1 (7, 8, 20) to test for the presence of a functional replicon. Plasmids which did not generate any B. anthracis UM23C1-1 transformants in at least three independent experiments were subsequently electroporated into B. anthracis containing the RepX-expressing pK43-46SM plasmid to test for complementation.
Mutagenesis of various pXO1 ORFs.
Translational frameshifts were introduced into ORFs 43, 44, 45, and 46 using the Stratagene QuikChange kit according to the manufacturer's instructions. ORF 43 was mutagenized using two complementary mutagenic primers containing pXO1 nt 55715 to 55749 but lacking the A residue at nt 55728, which should result in the loss of an SfaNI site. ORF 44 was mutagenized using two complementary mutagenic primers containing pXO1 nt 57160 to 57194 but lacking the A residue at position 5716,7 which is expected to result in the loss of a DraI site. Similarly, ORF 45 was mutagenized using primers containing pXO1 nt 58907 to 58942 but lacking a T at position 58934, resulting in the loss of a StyI site. ORF 46 was mutagenized using primers containing pXO1 nt 59327 to 59359 and introducing GC between nt 59347 and 59348, thus generating a new HindIII site. Use of the above primers is expected to result in frameshifts at amino acid positions 7, 19, 11, and 3 of ORFs 43, 44, 45, and 46, respectively. A GTP binding mutant of ORF 45 was made using primers containing pXO1 nt 58579 to 58611 with bp 58591 to 58593 changed from CAG to GCA, which is expected to result in a Thr→Ala mutation at amino acid 125 and the loss of an AlwNI site. The sequences of the primers for mutagenesis of the various ORFs were as follows: ORF 43, 5′-GATAATGAATTTACAATTTGCTCAACAACGAATG-3′ and 5′-CATTCGTTGTTGAGCAAATTGTAAATTCATTATC-3′; ORF 44, 5′-CTCTTTTAACGAAAAGATTTCTGTGTGCCTTTAC-3′ and 5′-GTAAAGGCACACAGAAATCTTTTCGTTAAAAGAG-3′; ORF 45, 5′-GAATCCAAATTTCAAACTAATATTTCCTGGCTTTC-3′ and 5′-GAAAGCCAGGAAATATTAGTTTGAAATTTGGATTC-3′; ORF 46, 5′-GGTGGTGTGAAAGCTTAGTGTCTAATATATCAATG-3′ and 5′-CATTGATATATTAGACACTAAGCTTTCACACCACC-3′; GTP binding mutant of ORF 45, 5′-GTGGTCTTGGTGGAGGAAGCAGAACTGGAGCTC-3′ and 5′-GAGCTCCAGTTCTGCTTCCTCCACCAAGACCAC-3′. The reaction mixtures (50 μl) contained 20 mM Tris-HCl (pH 8.8), 10 mM KCl, 10 mM (NH4)2SO4, 2 mM MgSO4, 0.1% Triton X-100, 100 μg/ml nuclease-free bovine serum albumin, 200 μM of each deoxynucleoside triphosphate, 75 ng of plasmid DNA, 125 ng of each primer, and 2.5 units of Pfu Turbo polymerase (Stratagene, La Jolla, CA). The conditions of PCR amplification were as follows: 95°C for 30 s; 95°C for 30 s, 55°C for 1 min, and 68°C for 10 min for 12 cycles for ORFs 44 and 45, 14 cycles for ORFs 43 and 46, and 16 cycles for the GTP binding mutant; and 68°C for 10 min. The reaction products were treated with 20 units of DpnI for 1 h at 37°C to remove the parental, methylated template DNA, followed by phenol-chloroform-isoamyl alcohol extraction and ethanol precipitation. The mutagenized plasmids were recovered by transforming E. coli DH5α, and miniplasmid preparations were screened by digestion with SfaNI for ORF 43, DraI for ORF 44, StyI for ORF 45, HindIII for ORF 46, and AlwNI for the GTP binding mutant of ORF 45. The sequences of all the above mutants were confirmed by automated DNA sequencing.
RESULTS
Analysis of the pXO1 sequence for the identification of the putative replicon.
Most plasmids with sequenced genomes contain genes encoding a replication initiator protein. An alignment of such genes has shown that various plasmids can be grouped into plasmid families which share significant homologies in their replication initiator genes and the origin of replication (5, 6, 17). However, in our homology searches using a variety of software we failed to identify homologs of known replication initiator proteins in the pXO1 sequence. The genes encoding replication initiator proteins of both chromosome and plasmids are generally located in the vicinity of their replication origins (5, 6, 17). In order to identify the putative replicon of the pXO1 plasmid, we relied upon GC skew, strand-specific biases such as gene orientation, plasmid-specific oligomer skew analysis, and origin comparisons (11, 32, 38) provided by the Genome Atlas Database at http://www.cbs.dtu.dk/services/GenomeAtlas/ to predict the location of an origin of replication (ori). This suggested two possible locations of the putative ori, the region of pXO1 from nt 40000 to nt 60000 and near nt 150000. The pXO1 region near position 150000 falls within the pathogenicity island encoding the anthrax toxin, which is potentially a mobile element and unlikely to contain the ori (27). Therefore, we postulated that the region near position 60000 is more likely to contain the pXO1 ori. A close examination of the pXO1 sequence also showed that the region between nt 50000 and 60000 contained many ORFs that were conserved in other large plasmids of the B. cereus family and therefore may be involved in essential plasmid functions such as replication and maintenance. Based on the above analyses, we focused on this region in our attempts to identify the pXO1 replicon.
Cloning of a 10,138-bp region containing the pXO1 replicon.
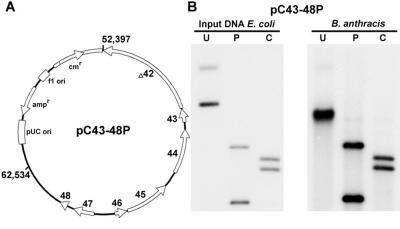
We generated several plasmid constructs that contained sections of the nt 52000 to 62000 region of the pXO1 plasmid. A 10,138-bp PstI fragment of pXO1 (NC_001496; positions 52397 to 62534 containing ORFs 43 to 48 and a portion of ORF 42) was cloned into the E. coli vector pBSCm to generate the plasmid pC43-48P (Fig. 1A). The resulting plasmid was isolated from the dam dcm mutant strain of E. coli GM2163 and electroporated into the plasmid-free B. anthracis strain UM23C1-1. Plasmid DNA from Cmr B. anthracis transformants was isolated and digested with PstI or ClaI. Agarose gel electrophoresis followed by Southern blot analysis showed that both the supercoiled form of the pC43-48P DNA and its restriction digests were identical to the plasmid DNA isolated from E. coli (Fig. 1B). The slight difference in the migration of supercoiled plasmid DNA isolated from E. coli and B. anthracis was seen only when the DNA was prepared from B. anthracis using a maxiprep procedure and not when a miniprep procedure was used (data not shown). No B. anthracis transformants were obtained when the pBSCm plasmid was introduced into B. anthracis. These results suggested that the above 10,138-bp region contains the functional replicon of the pXO1 plasmid.
FIG. 1.
Replication of a mini-pXO1 plasmid in B. anthracis. A. The pC43-48P plasmid containing a 10-kb region of pXO1 (positions 52397 to 62534). Plasmid pXO1 ORFs and their direction of transcription are shown. B. Southern blot analysis of plasmid DNA isolated from a B. anthracis strain transformed with the pC43-48P plasmid. Input DNA from E. coli is shown in the left panel, and the DNA isolated from B. anthracis is in the right panel. U, uncut plasmid DNA; P, PstI-cleaved DNA; C, ClaI-cleaved DNA.
Deletion analysis of the pXO1 replicon.
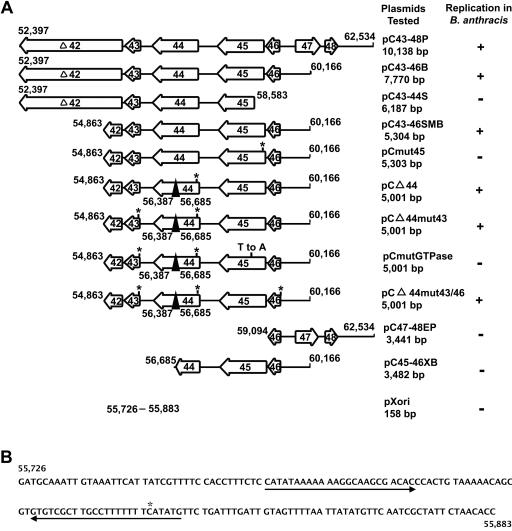
We subcloned various subregions of the 10,138-bp pXO1 replicon into the pBSCm plasmid of E. coli to further identify the elements involved in plasmid replication (Fig. 2A). All constructs were isolated from E. coli GM2163 and electroporated into B. anthracis UM23C1-1. Both pC43-46B (containing a 7,770-bp region of pXO1 from nt 52397 to nt 60166 that includes ORFs 43 to 46 and a majority of ORF 42, 983 amino acids out of 1,109) and pC43-46SMB (containing a 5,304-bp region of pXO1 from position 54863 to 60166 including ORFs 43 to 46 and a small portion of ORF 42, 160 amino acids out of 1,109) yielded Cmr transformants, and analysis of plasmid DNA by agarose gel electrophoresis followed by Southern blot hybridization showed that these plasmids replicated in B. anthracis (Fig. 3 and data not shown). The restriction digests of plasmid DNAs isolated from B. anthracis were also identical to the input plasmid DNA isolated from E. coli (Fig. 3). On the other hand, plasmids pC45-46XB, pC47-48EP, and pC43-44S (Fig. 2A) failed to generate any B. anthracis transformants in at least three independent experiments. The above results showed that the functional pXO1 replicon is present within a 5,304-bp region (positions 54863 to 60166) contained on pC43-46SMB. Generally, for the results shown in Fig. 2A, 10 to 30 B. anthracis transformants were obtained with plasmid constructs that replicated in this organism, while no transformants were obtained with nonreplicating plasmids (in at least three independent experiments).
FIG. 2.
The pXO1 replicon and its putative origin of replication. (A) Regions of the pXO1 replicon cloned into an E. coli plasmid. Boxes with arrows indicate the various ORFs and their directions of transcription. The numbers correspond to the nucleotide coordinates of pXO1. Asterisks correspond to the deletion of 1 bp in various ORFs except for ORF46 in pCΔ44mut43/46, in which 2 bp have been inserted. Plasmid pK43-46SM (not shown) is similar to pC43-46SMB except that it contains a Kmr marker in place of the Cmr gene. The sizes of the pXO1 replication regions and their ability to replicate in a plasmid-negative derivative of B. anthracis are indicated on the right. (B) Nucleotide sequence of the putative origin of replication of pXO1. The arrows show an IR sequence. The asterisk corresponds to an extra C residue in one arm of the IR. The numbers correspond to the nucleotide coordinates of pXO1.
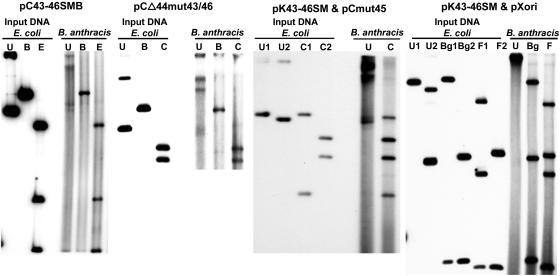
FIG. 3.
Southern blot analysis of plasmid DNA isolated from B. anthracis strains transformed with various pXO1 miniplasmids. Input DNAs from E. coli are shown on the left, and the DNA isolated from B. anthracis is on the right in each panel. U, uncut plasmid DNA; B, BamHI; C, ClaI; E, EcoRI; Bg, BglI; F, FspI. The two extreme right panels show a B. anthracis strain containing a functional pXO1 replicon expressing RepX (pK43-46SM) that can complement the replication of the pCmut45 (third panel from left) or pXori (fourth panel from left) plasmids in trans. U1 and C1, uncut or ClaI-digested pK43-46SM DNA, respectively; U2, uncut pCmut45 (third panel) or pXori (fourth panel) DNAs; C2, ClaI-digested pCmut45 DNA (third panel). In the fourth panel, Bg1 and F1 represent pK43-46SM DNA digested with BglI and FspI, respectively, while Bg2 and F2 represent pXori DNA digested with the same enzymes.
The product of ORF 45 is required for pXO1 replication.
Experiments were carried out to identify the gene encoding the replication initiator protein of plasmid pXO1. The pC43-46SMB plasmid which replicates in B. anthracis contains two major pXO1 ORFs, 44 and 45 (Fig. 2A). In addition, it contains two smaller ORFs, 43 and 46 (encoding putative proteins of 128 and 119 amino acids, respectively), and two truncated ORFs, 42 and 47 (Fig. 2A). We, therefore, considered it highly likely that ORF 44 or 45 may encode the essential replication initiator protein of pXO1. To identify this protein, we generated the deletion mutant pCΔ44, which lacks 99 amino acids (positions 180 to 278) and also contains a translational frameshift in ORF44. We also introduced a frameshift in ORF 45 in a separate plasmid, pCmut45. The above mutant plasmids were isolated from E. coli and introduced into B. anthracis. Cmr B. anthracis transformants were obtained with plasmid pCΔ44 but not with pCmut45 in five independent experiments. The presence of the appropriate plasmid DNA in B. anthracis transformed with pCΔ44 was confirmed by agarose gel analysis and Southern blot hybridization (data not shown). To rule out the involvement of smaller ORFs 43 and 46 in pXO1 replication, we also generated frameshift mutations in these ORFs in the context of the above pCΔ44 plasmid and found that the pCΔ44mut43/46 plasmid was able to replicate in B. anthracis (Fig. 3).
The above studies suggested that ORF 45 (encoding RepX) is required for pXO1 replication. Homology searches revealed that RepX has similarity to the GTPases of the FtsZ family (see below). We generated a Thr→Ala mutation at amino acid position 4 of the putative GTP binding domain (GGGTGT/SG) of repX using pCΔ44mut43 as a template. In the case of E. coli FtsZ, such a mutation is lethal and leads to a marked decrease in its GTPase activity although the organism retains its ability to form multimers (26). The resulting plasmid, pCmutGTPase, did not generate any transformants in B. anthracis in five independent experiments. However, both pCmut45 and pCmutGTPase replicated when introduced into a B. anthracis strain containing the Kmr plasmid pK43-46SM, which differs from pC43-46SMB only with respect to the resistance marker used (Fig. 3 and data not shown). Taken together, the above results suggest that ORF 45 encodes the essential RepX protein that may correspond to the replication initiator protein of plasmid pXO1, and GTP binding/hydrolysis may be important for its function.
Identification of the putative origin of replication of plasmid pXO1.
The plasmid pC45-46XB containing the repX gene (Fig. 2A) does not replicate in B. anthracis, suggesting that it may lack the origin of replication of pXO1. To test this possibility, the Cmr plasmid pXori (containing nt 55726 to 55883; Fig. 2B), which does not replicate in B. anthracis UM23C1-1, was introduced into a B. anthracis strain containing pK43-46SM that expresses RepX. Agarose gel electrophoresis and Southern blot hybridization of plasmid DNAs isolated from Kmr and Cmr doubly resistant colonies and digested with two different restriction enzymes showed the presence of both pXori and pK43-46SM plasmids (Fig. 3). Note that only nicked open-circular DNA was observed in the lane containing undigested plasmid DNA from B. anthracis. In similar experiments, pK43-46SM failed to complement the replication of the pC45-46XB plasmid (not shown). Plasmid pXori1 (containing a 738-bp fragment, Fig. 2A; nt 55617 to 56355), which is unable to replicate in B. anthracis, was also found to replicate in the presence of the pK43-46SM plasmid (data not shown). The above results suggested that the functional pXO1 origin is contained within a 158-bp region (nt 55726 to 55883) present in pXori. Furthermore, these results also showed that the RepX protein can act in trans and support the replication of an origin-containing plasmid.
DISCUSSION
We have identified the replicon of the pXO1 plasmid to within a 5.3-kb region since plasmid pC43-46SMB (pXO1 nt 54863 to 60166) replicated autonomously in B. anthracis (Fig. 2A and 3). Within this region, the product of ORF 45, RepX, is essential for pXO1 replication and may correspond to its replication initiator protein (see below). Since plasmid pC45-46XB containing an intact ORF 45 did not replicate in B. anthracis (Fig. 2A), it suggested that the pXO1 origin of replication may require sequences upstream of position 56685. Therefore, we tested the ability of plasmid pXori1 containing an upstream 738-bp region of pXO1 (nt 55617 to 56355) to replicate in the presence of the RepX-expressing plasmid pK43-46SM. Plasmid pXori1 replicated in B. anthracis (data not shown), demonstrating that the functional origin of replication of pXO1 is contained within the above 738-bp region. This region contains an inverted repeat (IR) of 24 nt (Fig. 2B), which is a common feature of origins of replication. Plasmid pXori containing a 158-bp region (nt 55726 to 55883) that includes the above IR was also found to replicate in the presence of the RepX-expressing plasmid (Fig. 3). These data showed that the functional pXO1 ori is contained within a 158-bp region (nt 55726 to 55883). The requirement of the above IR sequence in pXO1 origin function is currently unknown and will be the subject of future study. A few plasmids such as R6K and F contain multiple replicons/origins of replication. Whether the replicon identified in this study represents the only replicon/origin of plasmid pXO1 is currently unknown. Extensive homology searches by our laboratory as well as by others have failed to identify significant similarity between pXO1-encoded proteins and initiator proteins that have been shown to be involved in the initiation of plasmid or chromosome DNA replication (5, 6, 17). Interestingly, plasmids in several members of the B. cereus group which are closely related to B. anthracis contain sequences that are nearly identical to the repX gene. Two fully sequenced plasmids of B. cereus, pBCXO1 and pBC10987, which are very similar to pXO1, contain orthologs of repX with 99.8% and 98.4% identity, respectively (15, 30). Furthermore, DNA hybridization and PCR studies have shown that the repX gene is also contained on two megaplasmids of approximately 330 kb present in B. cereus 43881 and B. thuringiensis 33679 (2, 28). We propose that RepX defines a novel family of replication initiator proteins that are involved in the replication of megaplasmids of B. anthracis and other members of the B. cereus group. Orthologs of various plasmid initiator proteins are spread throughout the bacterial kingdom (5, 6, 17; http://www.essex.ac.uk/bs/staff/osborn/DPR_home.htm). Therefore, it is intriguing that RepX is restricted to B. anthracis and closely related organisms. In addition to a plasmid-encoded initiator protein, plasmid replication requires a large number of chromosome-encoded proteins (for a review, see reference 6). It is possible that one or more proteins required for the replication of pXO1-like plasmids are present only in B. anthracis and other members of the B. cereus group. This may explain why such plasmids encoding the anthrax toxin and other virulence factors are not widely distributed in nature. Future studies should reveal the molecular basis for the predicted narrow host range of such plasmids.
An alignment of the RepX protein using the BLAST (1), ClustalW, and EMBOSS programs (global, conserved domains and pairwise/local alignments) revealed that it shares limited similarity with the FtsZ proteins of several bacteria. For example, RepX is approximately 20 to 22% identical and 32 to 38% similar to the chromosome-encoded FtsZ proteins of E. coli, Bacillus subtilis, B. anthracis, and Pyrococcus abyssi. The RepX protein consists of 435 amino acids while the various FtsZ proteins range in size from 350 to 400 amino acids (22). Several regions of identity/similarity between RepX and FtsZ proteins are located between amino acids 1 and 320 of RepX (Fig. 4). The predicted folded structure of RepX using the ESyPred3D program (21; data not shown) is very similar to the structure of Methanococcus jannaschii FtsZ determined by X-ray crystallography (23). Bacterial FtsZ proteins are functional homologs of eukaryotic tubulin and are critical for cell division (for reviews, see references 4 and 37). FtsZ forms a dynamic ring structure (Z-ring) in a GTP-dependent manner that mediates cell division (4). The Z-ring then recruits cellular proteins such as FtsA, ZipA, MinC, and others that promote cytokinesis (4, 37). The FtsZ and tubulin proteins contain a conserved guanine nucleotide binding motif, GGGTGT/SG (4, 37). This motif is totally conserved in the RepX protein (amino acids 122 to 128; Fig. 4), and we have purified the RepX protein as a fusion with the maltose binding protein epitope at its amino-terminal end and found that it has a robust GTPase activity (data not shown). Our mutational and complementation analyses showed that GTP binding and/or GTPase activity of RepX is critical for its function as a replication protein (Fig. 2A and data not shown). Since pXO1 is not essential for B. anthracis growth (18, 19) and the chromosome of this organism encodes the FtsZ protein, RepX is unlikely to share the cell division function of the FtsZ proteins. The carboxyl-terminal region of FtsZ is involved in protein-protein interactions that play important roles in the recruitment of proteins to the septal ring (37). This region of RepX does not share significant homology with FtsZ proteins and may be involved in its replication-specific function. Since pXO1 is a low-copy-number plasmid (one or two per chromosome), it is possible that RepX domains that are similar to FtsZ may position the replicated pXO1 plasmids at the septal ring and promote its segregation to the daughter cells. Future studies should reveal the precise role of RepX in plasmid pXO1 replication and segregation. RepX may provide a novel drug target for the elimination of anthrax toxin-producing plasmids from B. anthracis and other organisms into which pXO1 may either transfer naturally or be introduced intentionally by bioterrorists.
FIG. 4.
Alignment of RepX (ORF 45) with various FtsZ proteins. Amino acid numbering corresponds to that of RepX. The shaded regions indicate amino acid identity, while those in boxes indicate similarity. The various motifs of FtsZ are shown based on the crystal structure of the Methanococcus jannaschii FtsZ protein (23).
Acknowledgments
We thank Theresa Koehler and Agathe Bourgogne for providing B. anthracis strains and plasmids. We also thank Syam Anand for useful comments on the manuscript and members of our laboratory for helpful discussions.
This work was supported in part by grant AI57974 from the National Institute of Allergy and Infectious Diseases to S.A.K. E. Tinsley was supported by NIH training grant T32 AI49820 (Molecular Microbial Persistence and Pathogenesis).
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Berry, C., S. O'Neil, E. Ben-Dov, A. F. Jones, L. Murphy, M. A. Quail, M. T. G. Holden, D. Harris, A. Zaritsky, and J. Parkhill. 2002. Complete sequence and organization of pBtoxis, the toxin-coding plasmid of Bacillus thuringiensis subsp. israelensis. Appl. Environ. Microbiol. 68:5082-5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bourgogne, A., M. Drysdale, S. G. Hilsenbeck, S. N. Peterson, and T. M. Koehler. 2003. Global effects of virulence gene regulators in a Bacillus anthracis strain with both virulence plasmids. Infect. Immun. 71:2736-2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carballido-Lopez, R., and J. Errington. 2003. A dynamic bacterial cytoskeleton. Trends Cell Biol. 13:577-583. [DOI] [PubMed] [Google Scholar]
- 5.Chattoraj, D. K. 2000. Control of plasmid DNA replication by iterons: no longer paradoxical. Mol. Microbiol. 37:467-476. [DOI] [PubMed] [Google Scholar]
- 6.del Solar, G., R. Giraldo, M. J. Ruiz-Echevarria, M. Espinosa, and R. Diaz-Orejas. 1998. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 62:434-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunny, G. M., L. N. Lee, and D. J. LeBlanc. 1991. Improved electroporation and cloning vector system for gram-positive bacteria. Microbiology 57:1194-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Green, B. D., L. Battisti, T. M. Koehler, C. B. Thorne, and B. E. Ivins. 1985. Demonstration of a capsule plasmid in Bacillus anthracis. Infect. Immun. 49:291-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Green, B. D., L. Battisti, and C. B. Thorne. 1989. Involvement of Tn4430 in transfer of Bacillus anthracis plasmids mediated by Bacillus thuringiensis plasmid pXO12. J. Bacteriol. 171:104-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guidi-Rontani, C., Y. Periera, S. Ruffie, J.-C. Sirard, M. Weber-Levy, and M. Mock. 1999. Identification and characterization of a germination operon on the virulence plasmid pXO1 of Bacillus anthracis. Mol. Microbiol. 33:407-414. [DOI] [PubMed] [Google Scholar]
- 11.Hallin, P. F., and D. W. Ussery. 2004. CBS Genome Atlas Database: a dynamic storage for bioinformatic results and sequence data. Bioinformatics 20:3682-3686. [DOI] [PubMed] [Google Scholar]
- 12.Hanna, P. 1998. Anthrax pathogenesis and host response. Curr. Top. Microbiol. Immunol. 225:13-35. [DOI] [PubMed] [Google Scholar]
- 13.Helgason, E., O. A. Okstad, D. A. Caugant, H. A. Johansen, A. Fouet, M. Mock, I. Hegna, and A. B. Kolsto. 2000. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis: one species on the basis of genetic evidence. Appl. Environ. Microbiol. 66:2627-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henderson, I., D. Yu, and P. C. Turnbull. 1995. Differentiation of Bacillus anthracis and other ‘Bacillus cereus group’ bacteria using IS231-derived sequences. FEMS Microbiol. Lett. 128:113-118. [DOI] [PubMed] [Google Scholar]
- 15.Hoffmaster, A. R., J. Ravel, D. A. Rasko, G. D. Chapman, M. D. Chute, C. K. Marston, B. K. De, C. T. Sacchi, C. Fitzgerald, L. W. Mayer, et al. 2004. Identification of anthrax toxin genes in a Bacillus cereus associated with an illness resembling inhalation anthrax. Proc. Natl. Acad. Sci. USA 101:8449-8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensen, G. B., N. Fisker, T. Sparso, and L. Andrup. 2005. The possibility of discriminating within the Bacillus cereus group using gyrB sequencing and PCR-RFLP. Int. J. Food Microbiol. 104:113-120. [DOI] [PubMed] [Google Scholar]
- 17.Khan, S. A. 1997. Rolling-circle replication of bacterial plasmids. Microbiol. Mol. Biol. Rev. 61:442-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koehler, T. M. 2000. Bacillus anthracis, p. 519-528. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. ASM Press, Washington, D.C.
- 19.Koehler, T. M. 2002. Bacillus anthracis genetics and virulence gene regulation. Curr. Top. Microbiol. Immunol. 271:143-164. [DOI] [PubMed] [Google Scholar]
- 20.Koehler, T. M., Z. Dai, and M. Kaufman-Yarbray. 1994. Regulation of the Bacillus anthracis protective antigen gene: CO2 and a trans-acting element activate transcription from one of two promoters. J. Bacteriol. 176:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lambert, C., N. Leonard, X. De Bolle, and E. Depiereux. 2002. ESyPred3D: prediction of protein 3D structures. Bioinformatics 8:1250-1256. [DOI] [PubMed] [Google Scholar]
- 22.Lee, K. N., I. Padmalayam, B. Baumstark, S. L. Baker, and R. F. Massung. 2003. Characterization of the ftsZ gene from Ehrlichia chaffeensis, Anaplasma phagocytophilum, and Rickettsia rickettsii, and use as a differential PCR target. DNA Cell Biol. 22:179-186. [DOI] [PubMed] [Google Scholar]
- 23.Lowe, J., and L. A. Amos. 1998. Crystal structure of the bacterial cell-division protein FtsZ. Nature 391:203-206. [DOI] [PubMed] [Google Scholar]
- 24.Marrero, R., and S. L. Welkos. 1995. The transformation frequency of plasmids into Bacillus anthracis is affected by adenine methylation. Gene 152:75-78. [DOI] [PubMed] [Google Scholar]
- 25.Mock, M., and A. Fouet. 2001. Anthrax. Annu. Rev. Microbiol. 55:647-671. [DOI] [PubMed] [Google Scholar]
- 26.Mukherjee, A., K. Dai, and J. Lutkenhaus. 1993. Escherichia coli cell division protein FtsZ is a guanine nucleotide binding protein. Proc. Natl. Acad. Sci. USA 90:1053-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okinaka, R. T., K. Cloud, O. Hampton, A. R. Hoffmaster, K. K. Hill, P. Keim, T. M. Koehler, G. Lamke, S. Kumano, J. Mahillon, D. Manter, Y. Martinez, D. Ricke, R. Svensson, and P. J. Jackson. 1999. Sequence and organization of pXO1, the large Bacillus anthracis plasmid harboring the anthrax toxin genes. J. Bacteriol. 18:6509-6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pannucci, J., R. T. Okinaka, R. Sabin, and C. R. Kuske. 2002. Bacillus anthracis pXO1 plasmid sequence conservation among closely related bacterial species. J. Bacteriol. 184:134-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perez-Casal, J., M. G. Caparon, and J. R. Scott. 1991. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J. Bacteriol. 173:2617-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rasko, D. A., J. Ravel, O. A. Okstad, E. Helgason, R. Z. Cer, L. Jiang, K. A. Shores, D. E. Fouts, N. J. Tourasse, S. V. Angiuoli, J. Kolonay, W. C. Nelson, A. B. Kolsto, C. M. Fraser, and T. D. Read. 2004. The genome sequence of Bacillus cereus ATCC 10987 reveals metabolic adaptations and a large plasmid related to Bacillus anthracis pXO1. Nucleic Acids Res. 32:977-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Read, T. D., S. L. Salzberg, M. Pop, M. Shumway, L. Umayam, L. Jiang, E. Holtzapple, J. D. Busch, K. L. Smith, J. M. Schupp, D. Solomon, P. Keim, and C. M. Fraser. 2002. Comparative genome sequencing for discovery of novel polymorphisms in Bacillus anthracis. Science 296:2028-2033. [DOI] [PubMed] [Google Scholar]
- 32.Rocha, E. P. C. 2004. The replication-related organization of bacterial genomes. Microbiology 150:1609-1627. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Schuch, R., D. Nelson, and V. A. Fischetti. 2002. A bacteriolytic agent that detects and kills Bacillus anthracis. Nature 418:884-889. [DOI] [PubMed] [Google Scholar]
- 35.Tinsley, E., A. Naqvi, A., Bourgogne, T. M. Koehler, and S. A. Khan. 2004. Isolation of a minireplicon of the virulence plasmid pXO2 of Bacillus anthracis and characterization of the plasmid-encoded RepS replication protein. J. Bacteriol. 186:2717-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vander Auwera, G. A., L. Andrup, and J. Mahillon. 2005. Conjugative plasmid pAW63 brings new insights into the genesis of the Bacillus anthracis virulence plasmid pXO2 and of the Bacillus thuringiensis plasmid pBT9727. BMC Genomics 6:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weiss, D. S. 2004. Bacterial cell division and the septal ring. Mol. Microbiol. 54:588-597. [DOI] [PubMed] [Google Scholar]
- 38.Worning, P., J. L. Jensen, P. F. Hallin, H.-H. Staerfeldt, and D. W. Ussery. 2006. Origin of replication in circular prokaryotic chromosomes. Environ. Microbiol. 8:353-361. [DOI] [PubMed] [Google Scholar]