Abstract
Streptococcus mutans, the primary etiological agent of human dental caries, has developed multiple mechanisms to colonize and form biofilms on the tooth surface. The brpA gene codes for a predicted surface-associated protein with apparent roles in biofilm formation, autolysis, and cell division. In this study, we used two models to further characterize the biofilm-forming characteristics of a BrpA-deficient mutant, strain TW14. Compared to those of the parent strain, UA159, TW14 formed long chains and sparse microcolonies on hydroxylapatite disks but failed to accumulate and form three-dimensional biofilms when grown on glucose as the carbohydrate source. The biofilm formation defect was also readily apparent by confocal laser scanning microscopy when flow cells were used to grow biofilms. When subjected to acid killing at pH 2.8 for 45 min, the survival rate of strain TW14 was more than 1 log lower than that of the wild-type strain. TW14 was at least 3 logs more susceptible to killing by 0.2% hydrogen peroxide than was UA159. The expression of more than 200 genes was found by microarray analysis to be altered in cells lacking BrpA (P < 0.01). These results suggest that the loss of BrpA can dramatically influence the transcriptome and significantly affects the regulation of acid and oxidative stress tolerance and biofilm formation in S. mutans, which are key virulence attributes of the organism.
Streptococcus mutans, the primary etiological agent of human dental caries, exists almost exclusively in biofilms on tooth surfaces. The pathogenic potential of S. mutans is attributable to potent biofilm-forming abilities, a high capacity to produce acids, a high degree of acid tolerance, and the possession of high-affinity systems for the assimilation of many carbohydrate sources (9).
Biofilms are surface-attached, structurally and compositionally complex bacterial communities (17, 31-33). Accumulating data suggest that bacterial cells in biofilms interact with and coordinate the expression of a wide range of genes in response to evolving environmental conditions, including pH, oxygen, carbon source and nutrient availability, cell density, and the presence of a solid surface. A series of highly coordinated physiological and biochemical functions is required for the bacteria to form mature biofilms in response to environmental cues (17, 31-33). The development of highly structured biofilms, however, gives the adherent populations the flexibility to cope with fluctuating environments and the selective advantages that surface association offers (17).
Recent studies have revealed that several genetic regulatory networks in S. mutans are required for bacterial adherence, biofilm accumulation, and growth under the conditions encountered during biofilm formation. The cell-density-dependent Com system, which is known to regulate genetic competence in S. mutans and other naturally competent streptococci, plays a significant role in biofilm formation and acid tolerance (20, 21). The LuxS enzyme, which is responsible for the production of autoinducer signal molecules that are involved in interspecies communication in a wide range of bacteria, affects acid and oxidative stress tolerance, as well as biofilm accumulation and biofilm structure (24, 32, 34). Likewise, global regulators, including CcpA, RelA, and trigger factor (RopA), and certain two-component signal transduction systems play significant roles in biofilm formation and resistance to acid and oxidative stresses (6, 19, 21, 32-34).
In a previous report, we described the identification of a gene, designated brpA, which encodes a predicted surface-associated protein with high levels of similarity to LytR of Bacillus subtilis and CpsX of Streptococcus agalactiae (32). The BrpA-deficient mutant was found to cause severe defects in biofilm formation on polystyrene surfaces when grown in a glucose-containing, semidefined biofilm medium (32, 36) and displayed increases in autolysis and formed long chains (11, 32). Recently, Nakano et al. found that BrpA deficiency weakened phagocytosis by human polymorphonuclear leukocytes but increased platelet aggregation and the duration of bacteremia, suggesting that BrpA may be involved in systemic virulence in blood (25). In this study, we continued our characterization of the BrpA-deficient mutant by using biofilm models that more closely mimic conditions in the oral cavity and explored the stress tolerance properties of strains lacking BrpA. Microarray analysis was also used to investigate the impact of loss of BrpA on the transcriptome and to begin to understand how BrpA influences biofilm formation and stress tolerance in S. mutans.
MATERIALS AND METHODS
Plasmids, strains, and growth conditions.
Streptococcus mutans UA159 and its derivatives were maintained on brain heart infusion (BHI) medium in a 5% CO2 chamber at 37°C. If necessary, erythromycin (Em, 10 μg/ml) and/or kanamycin (Km, 1 mg/ml) were added to the growth medium. For biofilm formation, S. mutans strains were grown in semidefined biofilm medium (BM) with either 20 mM glucose (BMG) or sucrose (BMS) as the carbohydrate source (22). Solid media were prepared similarly, but agar (Difco Laboratories, Detroit, MI) was added at a concentration of 1.5% (wt/vol). For the construction of green fluorescent protein-tagged strains, plasmid pAYBG754S, which has the gtfB promoter fused to a promoterless gfp gene as a reporter (37), was used to transform S. mutans strains UA159 and TW14, an otherwise isogenic, brpA-deficient mutant that has a portion of its coding sequence, nucleotide (nt) 311 to 581 relative to the start codon, replaced with an erythromycin resistance marker (32). The preparation of competent cells and transformation of S. mutans were done as previously described (10). Transformants carrying the PgtfB::gfp reporter system were isolated on BHI plates with kanamycin (1 mg/ml).
Acid killing and hydrogen peroxide challenge assays.
The ability of bacterial cells to withstand acid and hydrogen peroxide challenges was determined by using procedures described elsewhere (33).
Electron and confocal laser scanning microscopy.
For samples that were to be examined by scanning electron microscopy (SEM), biofilms were grown on hydroxylapatite (HA) disks (0.5” in diameter and 0.04” to 0.06” in thickness, catalog no. 050800-02; HiMed, Inc., Old Bethpage, NY) that were deposited in 24-well culture clusters for 24 h and analyzed by SEM at the University of Florida EM core facility laboratory as previously described (33). For confocal laser scanning microscopy (CLSM), BM with 18 mM glucose and 2 mM sucrose (BMGS) plus kanamycin (100 μg/ml) was used as the growth medium and three-channel flow cells were used to support the growth of biofilms (5). The fully assembled flow cell system (4 mm width × 40 mm length × 1 mm depth; Stovall Life Science, Inc. Greensboro, NC) was maintained in a 37°C warm room. Overnight cultures were transferred to fresh BHI broth and allowed to grow until the optical density at 600 nm (OD600) equaled approximately 0.5. The cultures were then diluted 1:100 in BMGS, and 0.5 ml of the diluted cultures was injected into the flow cell after medium flow was arrested. After 1 h, the medium pump was turned on at a flow rate of 13 ml h−1. Biofilms formed on the coverslips were optically dissected at 4, 16, and 28 h after the initial inoculation by using an MRC-1024 ES confocal laser scanning system (Bio-Rad, Hercules, CA) at the University of Florida optical microscopy facility. Images were obtained using a 60× oil objective plus a 1.5 amplification factor. Simulated xyz three-dimensional images were generated by using MCID Elite 6.0 (Imaging Research, Inc., St. Catharine, Canada). Images were further colorized for display by using Adobe Photoshop.
RNA extraction.
S. mutans strains were grown in 50 ml of BHI broth and harvested at an OD600 of ≅0.5 by centrifugation at 3,800 × g at 4°C for 5 min. The pellets were quickly resuspended and treated with RNAprotect (QIAGEN, Inc., CA) by using the procedures recommended by the supplier. The treated cells were collected by centrifugation at 3,800 × g at 4°C for 10 min and stored at −80°C. RNA extractions were performed by using hot phenol as previously described (33). To remove all DNA, the purified RNAs were treated with DNaseI (Ambion, Inc., Austin, TX) and retrieved with the RNeasy purification kit (QIAGEN, Inc., CA). To prepare a reference RNA, S. mutans UA159 was grown in 3 liters of BHI until mid-exponential phase (OD600 ≅ 0.5) and total RNA was extracted as described above. The purified RNA was then aliquoted and stored at −80°C until use.
cDNA synthesis, Cy dye coupling and array hybridization.
Array analysis was performed by using the whole-genome S. mutans microarrays that were obtained from PFGRC at TIGR (http://pfgrc.tigr.org). The S. mutans genome array consisted of 70-mer oligonucleotides representing 1,960 open reading frames from strain UA159. The full 70-mer complement was printed four times on the surface of the slides. Experiments, including cDNA synthesis, Cy dye coupling, hybridization, and washing, were performed by using protocols from the PFGRC (http://pfgrc.tigr.org/protocols.shtml) with minor modifications. Briefly, cDNA synthesis was carried out in a total volume of 45 μl by mixing 10 μg total RNA with 3 μg random hexamers (Invitrogen Life Technologies, CA), 9 μl 5× first-strand buffer, 4.5 μl 0.1 M dithiothreitol, 2 μl 12.5 mM dNTP/aa-UTP with a 1.5- to-1 ratio of aa-dUTP to dTTP, and 3 μl Superscript III reverse transcriptase. This mixture was incubated at 42°C for 16 h. Following the completion of cDNA synthesis, RNA was hydrolyzed with NaOH and the aminoallyl-labeled cDNA samples were purified by using the QIAGEN QIAquick PCR purification kit, followed by drying in a speed vacuum. The samples were then resuspended in 5 μl 0.1 M NaCO3 and incubated with 5 μl of Cy dye (resuspended as recommended by the manufacturer) for 2 h in the dark. For all experiments, the reference RNA samples were coupled to Cy5 and both wild-type and the BrpA-deficient mutant RNA samples were coupled to Cy3. Uncoupled Cy dyes were removed by the QIAquick PCR purification kit, and the coupled samples were eluted in 100 μl elution buffer. Each Cy3-labeled experimental sample was mixed with a Cy5-labeled reference sample, and the mixtures were allowed to dry in a speed vacuum. To hybridize, the samples were resuspended in 60 μl hybridization buffer and heated at 90°C for 10 min. Following a quick centrifugation, the Cy dye-coupled probes were applied to microarray slides and incubated at 42°C in a water bath for 17 h. Slides were then washed and scanned by using an Axon GenePix 4000B scanner (Axon Instruments, Foster City, CA).
Array data normalization and statistical analysis.
Data were collected from at least four separate array slides, which contained four copies of the genome, with RNA isolated from four independent experiments. The raw data were loaded into the TIGR Spotfinder program (http://www.tigr.org/software) and further normalized with the TIGR microarray data analysis system (http://www.tigr.org/software) using LOWESS iterative log mean centering parameters with the default settings, followed by in-slide replicate analysis. Spots that were missing or labeled as “bad” during the upstream processes in 50% of the slides were cut off in the output files. The ratios of channel A over channel B were then converted to log2 values and analyzed with BRB array tools (version 3.01, developed by Richard Simon and Amy Peng Lam, National Cancer Institute, Bethesda, MD; http://linus.nci.nih.gov/BRB-ArrayTools.html). A pairwise Student's t test was used to analyze the mean log ratios of the BrpA-deficient strain and the wild type, and genes that were differentially expressed with the significance level of a P value of <0.001 were selected.
Real-time PCR.
RNA was isolated as described above, and cDNA was synthesized with 1 μg of total RNA by using the iScript cDNA synthesis kit (Bio-Rad Laboratories, CA) with random nonamers as primers by following the procedures recommended by the manufacturer. Real-time PCR was carried out with a Bio-Rad iCycler by using gene-specific primers (Table 1) and procedures that were detailed elsewhere (2).
TABLE 1.
Primers used in this study
| Primer | DNA sequence (5′→3′)a | Application | Amplicon (bp) |
|---|---|---|---|
| For real-time PCR | |||
| AtpD-F | CGGATGCGTGTTGCTCTTACTG | atpD fragment | 164 |
| AtpD-R | GGCTGATAACCAACGGCTGATG | ||
| RecA-F | GCCTATGCTGCTGCTCTTGG | recA fragment | 139 |
| RecA-R | CTGCCACTGAGTCAACAACAAC | ||
| Sod-F | GCAGTGCTAAGACTCCCGAATC | sod fragment | 149 |
| Sod-R | TTGCGGAAGTGTGAGATTGGC | ||
| Nox-F | GGGTTGTGGAATGGCACTTTGG | nox fragment | 128 |
| Nox-R | CAATGGCTGTCACTGGCGATTC | ||
| Dpr-F | TATGCGTGGTTCAGGCTTCC | dpr fragment | 110 |
| Dpr-R | CGCCACCGATAGTAATCAAGC | ||
| FruC-F | AGCAGTGAAGCATCATCAAGTG | fruC fragment | 106 |
| FruC-R | TACCGCCACCGATAACGAATG | ||
| For PCR amplification | |||
| BrpA55 | AGCTCAGATAAGGCTGAGCTCCTA | 5′ Fragment for brpA deletion | 860 |
| BrpA53 | ATAAGCTTCCTCCTAAGGTAAACATCAAGA | ||
| BrpA35 | TAGAATTCCCAACAACCTGCTGCAGGAA | 3′ Fragment for brpA deletion | 744 |
| BrpA33 | AAACCGTCTTTCATGCCCATGTGCAT | ||
| BrpAc5′ | TAGAATTCGAGCATGACTGAGCAGGAAGT | For brpA complementation | 1,626 |
| BrpAc3′ | TAGGATCCAAGGTGAAAGGGTATAACTG |
The incorporated restriction sites are underlined.
Construction of complemented strains.
For the complementation of the BrpA-deficient mutant, a strain designated TW14D, which had the entire brpA coding sequence deleted, was generated by using a PCR-ligation-mutagenesis strategy (34). Briefly, an 860-bp 5′ fragment (nt 874 to 14 upstream of the start codon) and a 643-bp 3′ fragment (encompassing 39 nt upstream and 601 nt downstream of the TAA stop codon) were amplified by PCR by using the primers listed in Table 1. The fragments were digested with the appropriate restriction enzymes and ligated to an erythromycin resistance (Ermr) gene (34). The ligation mixture was then used to transform S. mutans UA159, and the deletional-replacement mutation of brpA as a result of double crossover homologous recombination was confirmed by PCR. To complement the BrpA-deficient mutants, a fragment of 1,626 bp that contains the entire brpA coding sequence plus 344 bp upstream of the start codon was directly cloned into the shuttle vector pDL278 (2). After confirming by DNA sequencing that the amplification of the insert was correct, the resulting plasmid, pDL278:brpAc, was used to transform S. mutans strains TW14 and TW14D, resulting in complementing strains TW14C and TW14DC, respectively. The possession of pDL278:brpAc in these complementing strains was further verified by PCR.
RESULTS AND DISCUSSION
BrpA deficiency causes defects in biofilm accumulation.
It was previously reported that the brpA gene (for biofilm regulatory protein) of S. mutans encodes a predicted surface-associated protein that plays a major role in biofilm formation (32), cell division, and autolysis (11, 32). BrpA deficiency does not have any major impact on growth rates when cells are grown in BHI or BM medium, but in broth cultures the strain lacking BrpA, TW14 (31) forms chains that are more than twice the cell number of that of the parent strain, UA159 (32). On polystyrene surfaces, TW14 failed to accumulate and form mature biofilms (32, 36), and such defects appeared to be related primarily to cell accumulation and biofilm development rather than initial adherence. When pDL278:brpAc, which contains the intact brpA with its apparent cognate promoter, was introduced into the BrpA-deficient mutants TW14C and TW14DC, the transformants were shown to have similar chain lengths and to form biofilms in an amount and of an appearance similar to that of S. mutans UA159 (data not shown). These results confirm that the phenotypes that were observed with strain TW14 resulted from BrpA deficiency and not from polar effects on linked genes.
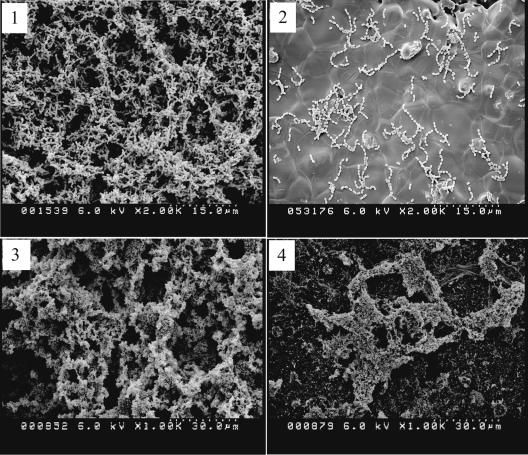
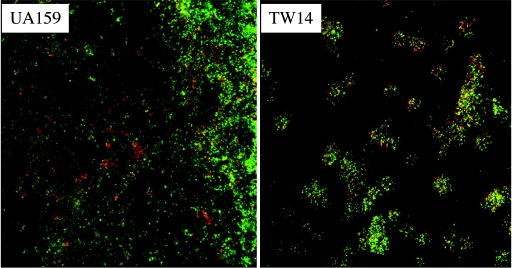
In this study, we used hydroxylapatite disks, an in vitro model of tooth enamel, and scanning electron microscopy to further characterize the BrpA-deficient mutant. TW14 bound to the HA disks, albeit less effectively than did UA159, forming scattered long chains and microcolonies on the HA disk surfaces (Fig. 1). However, similar to what we observed in the microtiter model, the mutant was not able to accumulate and develop into multicellular, three-dimensional biofilms (Fig. 1). When sucrose was provided as the carbohydrate source, both UA159 and TW14 formed better biofilms than they did in medium with glucose as the supplemental carbohydrate source but the depth of biofilms formed by the mutant was still less than that of strain UA159. Also, the biofilms formed by TW14 were thin and scattered relative to the evenly distributed biofilms formed by the wild-type strain. There were also more apparently broken cells and cell debris in TW14 biofilms than in UA159 biofilms under SEM at higher magnifications (data not shown), which perhaps was related to the increased autolysis of TW14 (11, 32). In an attempt to evaluate the viability of the biofilm populations, we used the LIVE/DEAD BacLight fluorescent dye (Molecular Probes, OR) and CLSM to analyze biofilms formed on 16-well Lab-Tek chamber slides (Nalge Nunc International, IL) by using the method of Hazlett et al. (15). It was found that in comparison to the wild-type strain, TW14 on glass slides had more dead cells scattered throughout the biofilms (Fig. 2). Considering the fact that the BrpA deficiency increases autolysis (11, 32), the observation that TW14 had decreased viability is not totally surprising. Decreases in viability could also contribute to the observed defects in biofilm formation by TW14.
FIG. 1.
SEM analysis of biofilms by S. mutans strains. Biofilms of S. mutans strains UA159 (panels 1 and 3) and TW14 (panels 2 and 4) were allowed to form for 24 h on hydroxylapatite disks that were deposited in 24-well cell culture clusters in semidefined BM medium with 20 mM glucose (panels 1 and 2) or sucrose (panels 3 and 4) as the supplemental carbohydrate sources. Data presented here are representative of three independent experiments.
FIG. 2.
Live/dead staining of biofilms. S. mutans strains UA159 and TW14 were grown in BM with 18 mM glucose and 2 mM sucrose (BMSG) as the supplemental carbohydrate sources in 16-well Lab-Tek chamber slides, stained with LIVE/DEAD BacLight fluorescent dye, and analyzed with CLSM. Data presented here are representative of three independent experiments.
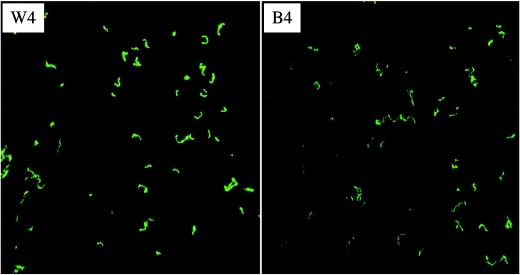
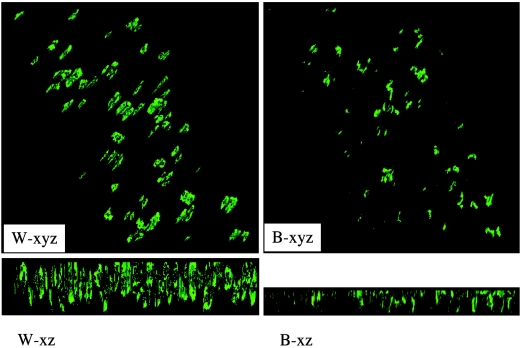
Flow cells take advantage of constant medium flow to impart a shear force to the microbes residing on the glass slips imbedded in the flow cell chambers and offer simple and effective means to study cell accumulation and biofilm structures (12). Flow cells are especially useful for more closely mimicking the natural conditions on tooth surfaces, where constant saliva flow and mechanical shear force could have a significant impact on bacterial adherence, cell-cell interactions, and biofilm accumulation. Three-channel flow cells were chosen to further characterize the role of BrpA in biofilm formation. To allow visualization of the bacterial biofilms by CLSM, we used a green fluorescent protein gene (gfp) as a reporter (37). The PgtfB::gfp system is stable in S. mutans grown in the presence of low levels of kanamycin selective pressure. As demonstrated by comparative analysis of images that were obtained by CLSM and phase-contrast microscopy, gfp was expressed well in the adherent biofilm cells on the glass surfaces, which is consistent with what was observed on polystyrene surfaces by Yoshida and Kuramitsu (37). When glucose was provided as the carbohydrate source, S. mutans UA159 was found to bind to the coverslips in the flow cell chambers, but binding was very limited (data not shown). No adherent TW14 cells were observed in flow cells when glucose alone was the carbohydrate source. Therefore, BMGS, which contains 18 mM glucose and 2 mM sucrose, was used to grow S. mutans strains in flow cells. When grown on BMGS, both UA159 and TW14 were able to bind to the glass surfaces and form microcolonies within 4 h of inoculation. In contrast to the more evenly distributed microcolonies of the wild type, the BrpA-deficient mutant formed sparse colonies of various sizes (Fig. 3). The average depth of biofilms, as estimated by optical sectioning with CLSM at the 4-h time point, was 10.7 μm for the wild type and 10.0 μm for the mutant. The wild-type strain continued to grow and accumulate as the experiments went on, forming thick biofilms after 28 h, with an average depth of 18.3 μm (Fig. 3). The BrpA-deficient strain, on the other hand, did not accrue nearly as much biomass, with an average depth of biofilms measured at 12.6 μm. These results further support the notion that BrpA deficiency causes defects in biofilm maturation (32). Although we previously showed that growth in suspension was not affected in TW14, we cannot exclude that BrpA deficiency impacts the growth of adherent population.
FIG.3.
CLSM analysis of biofilms grown in flow cells. S. mutans strains UA159 and TW14 were grown in three-channel flow cells on BMGS. Upper panels show compressed z sections of 4-h biofilms of UA159 (W4) and its BrpA-deficient mutant, TW14 (B4), respectively. Lower panels show the xyz and xz image reconstructions of 28-h biofilms of the wild type (W-xyz and -xz) and the BrpA mutant (B-xyz and -xz), respectively. Dimensions of the regions displayed are 512 by 512 μm. Data presented here are representative of three independent experiments.
Recently, Chatfield et al. reported on the characterization of the brpA gene, which they designated lytR, and its role in adherence and biofilm formation on hydroxylapatite surfaces (11). Contrary to data presented here and to the findings of Yoshida and Kuramitsu (36), which support a clear role for BrpA in biofilm maturation, Chatfield and coworkers reported no significant differences between the wild-type and the BrpA-deficient strains in terms of biofilms formed (11). In S. mutans, there are two distinct mechanisms to colonize tooth surfaces, sucrose-independent adherence primarily mediated by the high-affinity adhesin SpaP and the sucrose-dependent pathway involving the glucosyltransferases B, C, and D, their cohesive extracellular polysaccharide products, and glucan binding proteins (4, 8, 15, 27). Considering that Chatfield and coworkers used saliva-coated HA disks and a rich medium containing 1% sucrose, the high level of intercellular adherence mediated through glucans would likely have masked the biofilm formation defect of the brpA mutant. In fact, most biofilm-defective mutants of S. mutans do not display the defect when high concentrations of sucrose are present (8, 13, 34).
BrpA is required for stress tolerance.
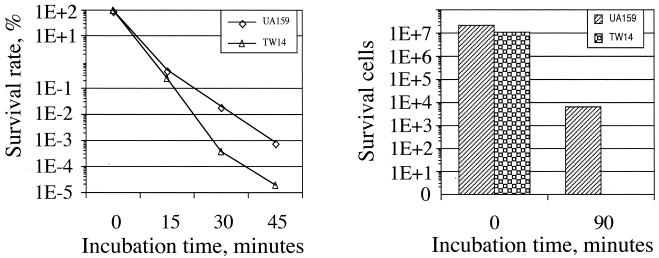
Accumulating data suggest that biofilm formation is a highly regulated process and that formation of mature biofilms requires the cooperation of a wide range of gene products, including those that are involved in acid and oxidative stress tolerance (17, 31, 32). When grown in BHI adjusted to pH 5.0, the wild-type strain UA159 had a final OD600 of 0.49 after 48 h, but the BrpA-deficient mutant TW14 achieved an optical density of only 0.24, indicative of a role for BrpA in acid tolerance. The stress tolerance properties of strains UA159 and TW14 were further analyzed by acid killing and hydrogen peroxide challenge (33). As shown in Fig. 4, BrpA deficiency significantly increases the susceptibility of S. mutans both to acid killing and to oxidative stress induced by hydrogen peroxide. The survival rate of TW14 was 1 log lower than that for UA159 when incubated in buffer at pH 2.8 for 45 min. As expected, the possession of brpA in trans was able to restore the capacity to tolerate acid stress in TW14 (data not shown). Similar observations of diminished acid tolerance in strains lacking BrpA were also made by Nakano et al. (25). When challenged by incubation in 0.2% hydrogen peroxide for 90 min, the survival rate of TW14 was at least 3 logs lower than that of the wild-type strain (Fig. 4). Increases of autolysis of TW14 as a result of BrpA deficiency (11, 32) could contribute, at least in part, to the observed increases in susceptibility of TW14 to acid and oxidative stresses.
FIG. 4.
Acid killing (left) and hydrogen peroxide challenge (right) assays. For acid tolerance assays, S. mutans strains were grown in BHI until mid-exponential phase (OD600 ≅ 0.3), harvested by centrifugation, washed once with 0.1 M glycine buffer (pH 7.0), and then subjected to acid killing at pH 2.8 for 45 min. Survival cells were plated on BHI agar plates in triplicate, and the results were expressed as survival rate over time. For hydrogen peroxide challenge, the properly washed cells were resuspended in 0.1 M glycine buffer (pH 7.0) containing 58.8 mM (0.2%, wt/vol) hydrogen peroxide for 2 h. The survival cells were then plated on BHI agar plates in triplicate. Data presented here are representative of at least three independent experiments.
Transcription profiling.
Both SEM analysis of biofilms grown on HA disks and CLSM analysis of biofilms grown on glass coverslips in flow cells showed that BrpA deficiency in S. mutans caused major defects in biofilm formation, especially when glucose is the carbohydrate source. Such defects appear to be related to biofilm accumulation, although the mutant did have a moderately diminished capacity to bind to the surfaces compared to that of the parental strain (Fig. 1 through 3). The formation of longer chains by TW14 may enhance detachment or reduce adherence, especially when cells were grown under conditions where shearing force was a factor. The decreased viability (Fig. 2) and the weakened tolerance to acid in particular (Fig. 4) as a result of BrpA deficiency could also have an impact on the capacity of the mutant to form mature biofilms since development of pH gradients in maturing biofilms may place an acid stress on the organisms. The effects of increased autolysis notwithstanding, the basis for diminished biofilm formation and stress tolerance by TW14 may in fact be more complex and related to global effects on gene expression in the mutant.
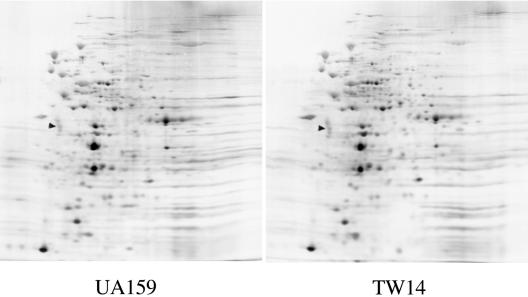
Two-dimensional gel electrophoresis of whole-cell extracts of UA159 and TW14 provided evidence that BrpA deficiency may have global effects on gene expression in S. mutans. A comparison of the protein profiles of silver-stained two-dimensional gels of UA159 and TW14 revealed that the quantities of more than 100 proteins were altered in response to BrpA deficiency (Fig. 5). These results suggest that changes in protein expression in response to loss of BrpA are substantial and could account for the defects in biofilm formation and stress tolerance.
FIG. 5.
Silver-stained two-dimensional gels of UA159 and TW14. S. mutans strains were grown in BHI to mid-exponential phase (OD600 ≅ 0.3). Clarified whole-cell lysates (50 μg) were subjected to two-dimensional electrophoresis using 2% ampholines at pH 4 to 8 (33). The triangles indicate the internal standard, tropomyosin, with a pI of 5.2 and a molecular weight of 32,700.
To further investigate the scope of effects of loss of BrpA, RNA was isolated from early-exponential-phase (OD600 ≅ 0.3) cultures of strains UA159 and TW14 grown in BHI and expression profiles were analyzed by DNA microarrays. More than 41 genes were identified as up- or down-regulated by a factor of at least 1.5-fold and a P value of <0.001 in response to BrpA deficiency (Tables 2 and 3). It is apparent that the differences in expression levels of the affected genes were small compared to those of highly regulated genes in eubacteria, so the loss of BrpA does not radically alter the transcriptome. However, on closer examination, the changes in gene expression induced by BrpA deficiency have the potential to profoundly affect cellular physiology and to account for the observed phenotypes.
TABLE 2.
Up-regulated genes identified by microarray analysis
| Locus | Description and putative functiona | Ratio (mt/wt)b | P value |
|---|---|---|---|
| SMU.600c | Conserved hypothetical protein | 2.5 | 7.8E−4 |
| SMU.915c | Conserved hypothetical protein | 2.2 | 4.8E−4 |
| SMU.1158c | Conserved hypothetical protein | 2.2 | 6.3E−4 |
| SMU.1053 | Conserved hypothetical protein | 2.2 | 6.8E−4 |
| SMU.1479 | Conserved hypothetical protein | 2.0 | 5.7E−4 |
| SMU.1616c | Conserved hypothetical protein | 1.9 | 6.0E−4 |
| SMU.1488c | Conserved hypothetical protein | 1.9 | 4.8E−4 |
| SMU.1597c | Conserved hypothetical protein | 1.9 | 5.4E−4 |
| SMU.290 | Conserved hypothetical protein | 1.8 | 6.2E−4 |
| SMU.2113c | Conserved hypothetical protein | 1.8 | 5.8E−4 |
| SMU.379 | Hypothetical protein | 2.2 | 2.7E−4 |
| SMU.457 | Hypothetical protein | 2.1 | 4.6E−4 |
| SMU.1131c | Hypothetical protein | 2.0 | 4.9E−4 |
| SMU.202c | Hypothetical protein | 1.9 | 8.9E−4 |
| SMU.847c | Hypothetical protein | 1.9 | 9.0E−4 |
| SMU.1772c | Hypothetical protein | 1.9 | 8.3E−4 |
| SMU.529 | Hypothetical protein | 1.9 | 6.7E−4 |
| SMU.1209c | Hypothetical protein | 1.9 | 9.7E−4 |
| SMU.524 | Putative ABC transporter, ATP-binding protein | 1.9 | 3.6E−4 |
| SMU.1078c | Putative ABC transporter, ATP-binding protein | 2.3 | 2.2E−4 |
| SMU.657 | Putative MutG | 2.1 | 5.9E−5 |
| SMU.364 | Glutamine synthetase type 1 glutamate-ammonia ligase | 1.9 | 2.3E−4 |
| SMU.913 | Putative NADP-specific glutamate dehydrogenase | 1.7 | 8.9E−4 |
| SMU.765 | NADH oxidase/alkyl hydroperoxidase peroxide forming | 1.9 | 3.3E−4 |
| SMU.540 | Peroxide resistance protein Dpr | 2.0 | 3.7E−4 |
| SMU.629 | Manganese-type superoxide dismutase SOD | 2.2 | 1.8E−4 |
| SMU.2055 | Putative acetyltransferase | 2.1 | 8.8E−5 |
| SMU.1343c | Polyketide synthase | 2.1 | 3.8E−4 |
| SMU.676 | NADP-dependent glyceraldehydes-3-P dehydrogenase | 1.7 | 9.5E−4 |
| SMU.1747c | Putative phosphatase | 2.2 | 4.9E−4 |
| SMU.1330c | Putative transposase | 2.3 | 4.2E−5 |
The description and putative function of the identified genes are based upon the published S. mutans database (3). Both hypothetical proteins and conserved hypothetical proteins are those of unknown functions, although the conserved hypothetical proteins have homologues identified in other bacteria.
The mt/wt ratio is defined as the expression level of the BrpA-deficient mutant/the expression level of the wild type.
TABLE 3.
Down-regulated genes identified by microarray analysis
| Locus | Description and putative functiona | Ratiob (mt/wt) | P value |
|---|---|---|---|
| SMU.1510 | Putative phenylalanyl-tRNA synthetase (beta subunit) | 0.39 | 8.2E−4 |
| SMU.938 | Putative phosphomevalonate kinase | 0.39 | 1.6E−4 |
| SMU.410 | Putative transcriptional regulator | 0.41 | 2.8E−4 |
| SMU.1017 | Oxaloacetate decarboxylase, sodium ion pump subunit | 0.41 | 1.9E−4 |
| SMU.901 | Putative poly(A) polymerase | 0.41 | 9.5E−5 |
| SMU.886 | Galactokinase, GalK | 0.46 | 7.8E−5 |
| SMU.114 | PTS, fructose-specific IIBC component, FruC | 0.47 | 3.8E−4 |
| SMU.86 | Conserved hypothetical protein | 0.48 | 2.0E−4 |
| SMU.151 | Hypothetical protein | 0.39 | 6.1E−4 |
| SMU.1896c | Hypothetical protein | 0.41 | 2.3E−4 |
The description and putative function of the identified genes are based upon the published S. mutans database (3). Both hypothetical proteins and conserved hypothetical proteins are those of unknown functions, although the conserved hypothetical proteins have homologues identified in other bacteria.
The mt/wt ratio is defined as the expression level of the BrpA-deficient mutant/the expression level of the wild type.
Among the down-regulated genes of established or predicted function were those coding for the IIBC domain of the fructose-specific phosphotransferase system (PTS) and for galactokinase, GalK. IIBCFru is one of the primary enzymes responsible for fructose transport, although the BrpA-deficient mutant, TW14, does not seem to have major defects in growth on glucose or fructose (11, 32). However, the fructose PTS has been shown to have regulatory roles and EIIFru mutants of Streptococcus gordonii have been shown to display biofilm formation defects (21). GalK is part of the Leloir pathway for galactose metabolism. A galK knockout mutation of S. mutans UA159 had a decreased capacity to grow on galactose but had no effect on growth of the organisms on glucose or lactose (1). In Lactococcus lactis subsp. cremoris, the Leloir pathway is also linked to galactose substitution in lipoteichoic acid and to cell wall synthesis (14). A mutation of the gal operon caused deficiencies in cell wall synthesis along with the appearance of long-chain phenotype in cells grown in glucose. As in L. lactis, the gal operon in S. mutans is under the control of carbon catabolite repression and repressed by glucose (1). S. mutans lacking BrpA forms long chains, especially when glucose was the sole carbohydrate source (11, 32), but it is not clear whether down-regulation of GalK contributes to this phenotype.
Among the up-regulated genes were those encoding apparent homologues of the manganese-type superoxide dismutase (SOD), a hydrogen peroxide-forming NADH oxidase (Nox) and peroxide resistance protein (Dpr). In streptococci, Nox, SOD, and Dpr are thought to play major roles in oxidative stress tolerance (16, 23, 26, 35) and are highly inducible by oxidative stresses. Elevated expression of the respective genes in TW14 therefore suggests that BrpA deficiency may signal oxidative stress. The result of hydrogen peroxide challenge assays (Fig. 4) further supports a connection between BrpA-defects and oxidative stress. The actual mechanism of the induction of enzymes involved in oxygen metabolism and the basis for enhanced H2O2 sensitivity remain to be studied, although envelope defects are likely to contribute to the observed phenotypes.
It is noteworthy that at a significance level of a P value of <0.01, 162 additional genes were identified as differentially expressed in the BrpA-deficient mutant by a factor of at least 1.5-fold (data not shown). Among the down-regulated genes were those that encode glucan-binding protein (GbpD) and recombination protein (RecA), and those for the F1Fo proton-translocating ATPase (F1Fo-ATPase), which are transcribed as an operon (3, 18). GbpD is unique to the mutans streptococci and contains repeated domains commonly found in glucan-binding proteins (30). In fact, GbpD was found to be capable of binding to glucans and the deficiency of GbpD caused decreases in the hydrophobicity of bacterial cells and their binding to hard surfaces. Glucans, especially the water-insoluble α1,3-linked glucose polymers, are the major architectural features of S. mutans biofilms. The ability of S. mutans to produce and to bind to extracellular glucans plays major roles in bacterial adherence and biofilm accumulation when grown on sucrose (4, 15, 27). Thus, the altered expression of GbpD could affect intercellular adhesive interactions that negatively affect biofilm formation. Notably, GbpD has also been shown to have lipase activity and may be involved in membrane biogenesis or turnover (30), which could affect the composition or integrity of the cell envelope and possibly contribute to the stress-sensitive phenotype.
The F1Fo-ATPase is the major enzyme that is responsible for the maintenance of ΔpH in mutans streptococci (7, 29). In S. mutans, the genes for the F1Fo-ATPase are arranged as an operon (3, 18). To verify the down-regulation of F1Fo-ATPase, real-time PCR was conducted with primers that were specific for the beta subunit (atpD), which is the second to the last of the operon. Results showed that as a result of BrpA deficiency, the expression of atpD was decreased by 46% (Table 4), consistent with the result of array analysis. Thus, the down-regulation of F1Fo-ATPase as a result of BrpA deficiency appears to contribute to the enhanced acid sensitivity of the mutant. Recombinant protein RecA, a key enzyme in DNA repair, has also been shown to be involved in acid tolerance in S. mutans (28). The down-regulation of RecA, as shown both by array analysis and real-time PCR, would likely have an impact on the repair of DNA that was damaged by acid and oxidative stresses and therefore may be part of the underlying factors that are responsible for the weakened stress tolerance by TW14.
TABLE 4.
Real-time PCR analysisa
| Gene | UA159 | TW14 | Ratio (mt/wt) |
|---|---|---|---|
| sod | 1.80E + 05 | 4.52E + 05 | 2.5 |
| nox | 1.40E + 05 | 3.40E + 05 | 2.43 |
| dpr | 5.26E + 06 | 8.21E + 06 | 1.56 |
| fruC | 2.66E + 07 | 1.18E + 07 | 0.44 |
| atpD | 1.52E + 07 | 8.19E + 06 | 0.54 |
| recA | 3.71E + 06 | 2.48E + 06 | 0.66 |
Results are expressed as copy numbers in 1 μg of total RNA.
Summary.
S. mutans exists almost exclusively as biofilms on the tooth surface. BrpA, a predicted surface-associated protein, appears to be involved in the regulation of cell division, autolysis, stress tolerance, and biofilm formation. Comparisons of the transcriptomes of the brpA mutant and the parent reveal alterations in the expression of genes that are involved in cell wall biogenesis, stress tolerance, and intercellular adherence. These findings further reinforce the notion that biofilm formation is a highly regulated process and that the ability of bacteria to adjust properly to environmental cues during biofilm maturation has a major impact on their capacity to form mature biofilms. The underlying molecular mechanisms for the observed defects in TW14 are under investigation.
Acknowledgments
This work is supported by National Institute for Dental and Craniofacial Research RO1 DE13239.
We would also like to thank NIDCR and TIGR for the access to S. mutans microarray slides, Tim Vaught and the Optical Microscopy Facility at the University of Florida Brian Institute for their technical assistance with CLSM analysis, Fred Bennett and the EM Core lab of the university for their help with SEM analysis, and Cecilia Lopez at the University of Florida Department of Molecular Genetics and Microbiology for her expertise and support with DNA array analysis.
REFERENCES
- 1.Abranches, J., Y. Y. Chen, and R. A. Burne. 2004. Galactose metabolism by Streptococcus mutans. Appl. Environ. Microbiol. 70:6047-6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn, S. J., J. A. Lemos, and R. A. Burne. 2005. Role of HtrA in growth and competence of Streptococcus mutans UA159. J. Bacteriol. 187:3028-3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ajdic, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 99:14434-14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banas, J. A., and M. M. Vickerman. 2003. Glucan-binding proteins of the oral streptococci. Crit. Rev. Oral Biol. Med. 14:89-99. [DOI] [PubMed] [Google Scholar]
- 5.Beloin, C., J. Valle, P. Latour-Lambert, P. Faure, M. Kzreminski, D. Balestrino, J. A. Haagensen, S. Molin, G. Prensier, B. Arbeille, and J. M. Ghigo. 2004. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol. Microbiol. 51:659-674. [DOI] [PubMed] [Google Scholar]
- 6.Bhagwat, S. P., J. Nary, and R. A. Burne. 2001. Effects of mutating putative two-component systems on biofilm formation by Streptococcus mutans UA159. FEMS Microbiol. Lett. 205:225-230. [DOI] [PubMed] [Google Scholar]
- 7.Bowden, G. H., and I. R. Hamilton. 1998. Survival of oral bacteria. Crit. Rev. Oral Biol. Med. 9:54-85. [DOI] [PubMed] [Google Scholar]
- 8.Bowen, W. H., K. Schilling, E. Giertsen, S. Pearson, S. F. Lee, A. Bleiweis, and D. Beeman. 1991. Role of a cell surface-associated protein in adherence and dental caries. Infect. Immun. 59:4606-4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burne, R. A. 1998. Oral streptococci. Products of their environment. J. Dent. Res. 77:445-452. [DOI] [PubMed] [Google Scholar]
- 10.Burne, R. A., Z. T. Wen, Y. Y. Chen, and J. E. Penders. 1999. Regulation of expression of the fructan hydrolase gene of Streptococcus mutans GS-5 by induction and carbon catabolite repression. J. Bacteriol. 181:2863-2871.10217779 [Google Scholar]
- 11.Chatfield, C. H., H. Koo, and R. G. Quivey, Jr. 2005. The putative autolysin regulator LytR in Streptococcus mutans plays a role in cell division and is growth-phase regulated. Microbiology 151:625-631. [DOI] [PubMed] [Google Scholar]
- 12.Christensen, B. B., C. Sternberg, J. B. Andersen, R. J. Palmer, Jr., A. T. Nielsen, M. Givskov, and S. Molin. 1999. Molecular tools for study of biofilm physiology. Methods Enzymol. 310:20-42. [DOI] [PubMed] [Google Scholar]
- 13.Crowley, P. J., L. J. Brady, S. M. Michalek, and A. S. Bleiweis. 1999. Virulence of a spaP mutant of Streptococcus mutans in a gnotobiotic rat model. Infect. Immun. 67:1201-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grossiord, B. P., E. J. Luesink, E. E. Vaughan, A. Arnaud, and W. M. de Vos. 2003. Characterization, expression, and mutation of the Lactococcus lactis galPMKTE genes, involved in galactose utilization via the Leloir pathway. J. Bacteriol. 185:870-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hazlett, K. R., J. E. Mazurkiewicz, and J. A. Banas. 1999. Inactivation of the gbpA gene of Streptococcus mutans alters structural and functional aspects of plaque biofilm which are compensated by recombination of the gtfB and gtfC genes. Infect. Immun. 67:3909-3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jakubovics, N. S., A. W. Smith, and H. F. Jenkinson. 2002. Oxidative stress tolerance is manganese (Mn2+) regulated in Streptococcus gordonii. Microbiology 148:3255-3263. [DOI] [PubMed] [Google Scholar]
- 17.Jefferson, K. K. 2004. What drives bacteria to produce a biofilm? FEMS Microbiol. Lett. 236:163-173. [DOI] [PubMed] [Google Scholar]
- 18.Kuhnert, W. L., G. Zheng, R. C. Faustoferri, and R. G. Quivey, Jr. 2004. The F-ATPase operon promoter of Streptococcus mutans is transcriptionally regulated in response to external pH. J. Bacteriol. 186:8524-8528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemos, J. A., T. A. Brown, Jr., and R. A. Burne. 2004. Effects of RelA on key virulence properties of planktonic and biofilm populations of Streptococcus mutans. Infect. Immun. 72:1431-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, Y. H., M. N. Hanna, G. Svensater, R. P. Ellen, and D. G. Cvitkovitch. 2001. Cell density modulates acid adaptation in Streptococcus mutans: implications for survival in biofilms. J. Bacteriol. 183:6875-6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, Y.-H., N. Tang, M. B. Aspiras, P. C. Y. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2002. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J. Bacteriol. 184:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loo, C. Y., D. A. Corliss, and N. Ganeshkumar. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 182:1374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marquis, R. E. 1995. Oxygen metabolism, oxidative stress and acid-base physiology of dental plaque biofilms. J. Ind. Microbiol. 15:198-207. [DOI] [PubMed] [Google Scholar]
- 24.Merritt, J., F. Qi, S. D. Goodman, M. H. Anderson, and W. Shi. 2003. Mutation of luxS affects biofilm formation in Streptococcus mutans. Infect. Immun. 71:1972-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakano, K., K. Fujita, K. Nishimura, R. Nomura, and T. Ooshima. 2005. Contribution of biofilm regulatory protein A of Streptococcus mutans, to systemic virulence. Microbes Infect. 7:1246-1255. [DOI] [PubMed] [Google Scholar]
- 26.Nguyen, P. T., J. Abranches, T. N. Phan, and R. E. Marquis. 2002. Repressed respiration of oral streptococci grown in biofilms. Curr. Microbiol. 44:262-266. [DOI] [PubMed] [Google Scholar]
- 27.Ooshima, T., M. Matsumura, T. Hoshino, S. Kawabata, S. Sobue, and T. Fujiwara. 2001. Contributions of three glycosyltransferases to sucrose-dependent adherence of Streptococcus mutans. J. Dent. Res. 80:1672-1677. [DOI] [PubMed] [Google Scholar]
- 28.Quivey, R. G., Jr., R. C. Faustoferri, K. A. Clancy, and R. E. Marquis. 1995. Acid adaptation in Streptococcus mutans UA159 alleviates sensitization to environmental stress due to RecA deficiency. FEMS Microbiol. Lett. 126:257-261. [DOI] [PubMed] [Google Scholar]
- 29.Quivey, R. G., Jr., W. L. Kuhnert, and K. Hahn. 2000. Adaptation of oral streptococci to low pH. Adv. Microb. Physiol. 42:239-274. [DOI] [PubMed] [Google Scholar]
- 30.Shah, D. S., and R. R. Russell. 2004. A novel glucan-binding protein with lipase activity from the oral pathogen Streptococcus mutans. Microbiol. 150:1947-1956. [DOI] [PubMed] [Google Scholar]
- 31.Stanley, N. R., and B. A. Lazazzera. 2004. Environmental signals and regulatory pathways that influence biofilm formation. Mol. Microbiol. 52:917-924. [DOI] [PubMed] [Google Scholar]
- 32.Wen, Z. T., and R. A. Burne. 2002. Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans. Appl. Environ. Microbiol. 68:1196-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wen, Z. T., and R. A. Burne. 2004. LuxS-mediated signaling in Streptococcus mutans is involved in regulation of acid and oxidative stress tolerance and biofilm formation. J. Bacteriol. 186:2682-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wen, Z. T., P. Suntharaligham, D. G. Cvitkovitch, and R. A. Burne. 2005. Trigger factor in Streptococcus mutans is involved in stress tolerance, competence development, and biofilm formation. Infect. Immun. 73:219-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamamoto, Y., K. Fukui, N. Koujin, H. Ohya, K. Kimura, and Y. Kamio. 2004. Regulation of the intracellular free iron pool by Dpr provides oxygen tolerance to Streptococcus mutans. J. Bacteriol. 186:5997-6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshida, A., and H. K. Kuramitsu. 2002. Multiple Streptococcus mutans genes are involved in biofilm formation. Appl. Environ. Microbiol. 68:6283-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshida, A., and H. K. Kuramitsu. 2002. Streptococcus mutans biofilm formation: utilization of a gtfB promoter-green fluorescent protein (PgtfB::gfp) construct to monitor development. Microbiology 148:3385-3394. [DOI] [PubMed] [Google Scholar]