Abstract
Enteropathogenic Escherichia coli (EPEC) secretes many Esps (E. coli-secreted proteins) and effectors via the type III secretion (TTS) system. We previously identified a novel needle complex (NC) composed of a basal body and a needle structure containing an expandable EspA sheath-like structure as a central part of the EPEC TTS apparatus. To further investigate the structure and protein components of the EPEC NC, we purified it in successive centrifugal steps. Finally, NCs with long EspA sheath-like structures could be separated from those with short needle structures on the basis of their densities. Although the highly purified NC appeared to lack an inner ring in the basal body, its core structure, composed of an outer ring and a central rod, was observed by transmission electron microscopy. Matrix-assisted laser desorption ionization-time-of-flight mass spectrometry, Western blot, and immunoelectron microscopic analyses revealed that EscC was a major protein component of the outer ring in the core basal body. To investigate the mechanisms of assembly of the basal body, interactions between the presumed components of the EPEC TTS apparatus were analyzed by a glutathione S-transferase pulldown assay. The EscC outer ring protein was associated with both the EscF needle protein and EscD, a presumed inner membrane protein. EscF was also associated with EscJ, a presumed inner ring protein. Furthermore, escC, escD, and escJ mutant strains were unable to produce the TTS apparatus, and thereby the secretion of the Esp proteins and Tir effector was abolished. These results indicate that EscC, EscD, and EscJ are required for the formation of the TTS apparatus.
Enteropathogenic Escherichia coli (EPEC) is a major cause of diarrhea in young children (12). This pathogen induces a characteristic histopathological lesion referred to as an attaching/effacing lesion, which is defined by the intimate attachment of bacteria to the epithelial surface and the effacement of host cell microvilli (25). Factors responsible for the formation of attaching/effacing lesions are encoded by a 35-kbp locus designated LEE (24), which encodes the following components: (i) the type III secretion (TTS) apparatus (15), (ii) E. coli-secreted proteins (EspA, EspB, and EspD) and several effectors, including Tir, (iii) type III-specific chaperones, and (iv) regulators (9). LEE is highly conserved among enterohemorrhagic E. coli, rabbit EPEC, and Citrobacter rodentium.
TTS systems are found in many other gram-negative bacterial species, and they constitute a strategy for the delivery of effectors into host cells, a process which in turn disrupts host physiology and thereby contributes to the disease process (14). Unlike the sec-dependent secreted proteins, the effector does not have a typical signal sequence at its N terminus, and it is directly delivered via the TTS system without N-terminal processing in the bacterial periplasm. Analysis by transmission electron microscopy (TEM) revealed that the supermolecular structures of the TTS apparatus in EPEC (8, 31), Salmonella enterica serovar Typhimurium (18, 20-22), and Shigella flexneri (2, 32) are highly conserved with respect to each other and that their shapes are similar to that of the flagellar basal body complex. The core TTS apparatus, which is referred to as a needle complex (NC), is composed of two distinct portions: (i) a needle structure that extrudes from the bacterial outer membrane and functions as an injector of effectors into host cells and (ii) a cylindrical basal body that is similar to the flagellar basal body and functions as a channel that spans the outer and inner membranes of the bacterium as well as the periplasmic region. The basal body is further divided into three major portions: (i) an outer ring, (ii) an inner ring, and (iii) a presumed central rod (22) that can connect the outer and inner rings to build a channel.
Although the supermolecular structure of the EPEC NC is similar in shape to that of the NCs of both Salmonella and Shigella, a unique extracellular appendage in the needle structure of the EPEC NC was discovered previously (31). The EPEC needle structure is composed of a thin needle (neck portion) and an expandable sheath-like structure (31). EPEC EscF is predicted to be an 8-kDa protein and may polymerize to form the thin needle in the needle structure. EscF shows homology to PrgI (24% identity) in the Salmonella pathogenicity island 1, Shigella MxiH (25% identity), and Yersinia enterocolitica YscF (20% identity), which are major components of the thin and stiff needle structures of the Salmonella, Shigella, and Yersinia NCs, respectively (21, 26, 32). EscF is required for NC formation and the secretion of the Esp proteins (31); this observation agrees with findings of secretion-defective phenotypes of Salmonella prgI and Shigella mxiH mutant strains (21, 32). EspA is predicted to be a 20-kDa protein and is secreted via the EPEC TTS system (17). We previously demonstrated that EspA is directly associated with the tip of the putative EscF needle and polymerizes into an expandable filamentous structure, referred to as the sheath-like structure (31). The EPEC needle structure, including the EspA sheath-like structure, extended to a length of more than 600 nm and was 10 times longer than the Shigella needle (45 nm) (31). Three-dimensional structure analysis of the EspA filament revealed that the structure consists of a helical tube with a diameter of 12 nm enclosing a central channel with a diameter of 2.5 nm (7). Recent TEM analysis of purified EspA revealed that EspA alone is able to polymerize into irregular short filaments (33). On the other hand, the widths of the outer and inner membrane rings in the basal body of the EPEC NC are estimated to be 17 and 18 nm, respectively, and the height of the basal body is 31 nm (31). However, the molecular composition of the EPEC NC basal body remains unclear.
From the results of a membrane fractionation study (13), yeast two-hybrid analysis (5), whole mutation analyses of LEE (10), and computer modeling predictions, several proteins are thought to be components for the EPEC TTS apparatus. The outer ring protein of the basal body has been suggested to be EscC, according to a membrane fractionation study (13), and this hypothesis received further support from a demonstration of its similarity to the YscC protein (31.1% identity) in the Yersinia TTS system, which forms a ring-shaped oligomeric complex with a diameter of 20 nm in the outer membrane (4, 19). EscC belongs to a member of the secretin superfamily, which participates in the delivery of large molecules through the outer membrane by the formation of a channel. EscC is predicted to be synthesized as a 56-kDa preprotein possessing a signal sequence that is cleavable with type I signal peptidase after amino acid residue 19. Then, the EscC preprotein most likely undergoes signal peptide cleavage after its export across the inner membrane by a sec-dependent secretion pathway, thus generating a mature 54-kDa protein. Indeed, a mature form of Salmonella InvG, an EscC orthologue, starts at amino acid residue 25, suggesting that its preprotein is cleaved by type I signal peptidase in order to reach maturation (20). Recently, the association of EscC with EscD was suggested by use of a yeast two-hybrid system (5). EscD is predicted to be a 45-kDa protein and shows amino acid sequence similarity to Yersinia YscD, a bacterial inner membrane protein (27).
In Salmonella, PrgK and PrgH together form the inner membrane ring of the TTS apparatus (18). Although a PrgH orthologue has not been found in EPEC, a PrgK orthologue, EscJ, is thought to form a portion of the inner ring. Molecular modeling using the 1.8-Å crystal structure of EscJ suggests that EscJ oligomerizes to form a large 24-subunit ring structure (34). Furthermore, a membrane fraction study indicates that EscJ localizes mainly to the inner membrane (34). These findings suggest that the inner ring of the EPEC TTS apparatus contains the EscJ multimeric complex. EscJ is predicted to be produced as a 21-kDa preprotein with a lipoprotein signal sequence that can be cleaved after amino acid residue 19 by type II signal peptidase. After sec-dependent translocation, the EscJ preprotein may undergo lipid modification and signal peptide cleavage to form a mature 19-kDa lipoprotein, which may be anchored to the inner membrane via its N-terminal lipid moiety (34). Although several Esc proteins, namely, EscR, EscS, EscT, and EscU, are predicted to be components of the basal body, their precise functions and localizations in the EPEC TTS apparatus remain unclear.
In this study, we analyzed the structures of NCs isolated from EPEC by using centrifugation techniques, including CsCl density gradient centrifugation. Furthermore, protein components of NCs were examined by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS), Western blotting, and immunoelectron microscopy. In addition, the protein-protein interactions required for assembly of the EPEC TTS apparatus were analyzed using a glutathione S-transferase (GST) pulldown assay, and confirmation of the putative components of the TTS apparatus needed for NC assembly, as well as those needed for the secretion of Esp proteins and the Tir effector, was carried out by the construction of deletion mutants.
MATERIALS AND METHODS
Bacterial strains and growth media.
EPEC was grown in Dulbecco's modified Eagle's medium (DMEM) at 37°C, as previously described (31). Shigella was grown in Luria-Bertani (LB) broth at 37°C. For details regarding the strains and their genotypes, see Table 1.
TABLE 1.
Strains used in this study
| Organism and strain | Genotype/description | Reference or source |
|---|---|---|
| EPEC | ||
| E2348/69 | Prototype EPEC O126:H6 strain | 16 |
| ILS002 | E2368/69 ΔespA | 23 |
| ILS003 | E2348/69 ΔescC | This study |
| ILS004 | E2348/69 ΔescD | This study |
| ILS005 | E2348/69 ΔescJ | This study |
| E. coli | ||
| BL21(DE3) | F−ompT hsdSBgal dcm (DE3) | Invitrogen |
| Sm10λpir | Permissive strain for replication of pCVD442 | 11 |
| DH5α | hsdR17 deoR thi-1 phoA supE44 λ− | Invitrogen |
Plasmids and oligonucleotides.
The plasmids and oligonucleotides used in this study are listed in Tables 2 and 3, respectively.
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Reference or source |
|---|---|---|
| pABB-CRS2 | Positive-selection suicide vector | 31 |
| pDONR201 | Gateway cloning vector | Invitrogen |
| pDONR-escC | escC cloned into pDONR201 | This study |
| pDONR-escD | escD cloned into pDONR201 | This study |
| pDONR-escJ | escJ cloned into pDONR201 | This study |
| pDONR-GST-escD | escD cloned into pDONR201 | This study |
| pDONR-GST-escF | escF cloned into pDONR201 | This study |
| pDONR-escD-V5 | escD cloned into pDONR201 | This study |
| pDONR-escF-V5 | escF cloned into pDONR201 | This study |
| pABB-ΔescC | escC deletion in pABB-CRS2 | This study |
| pABB-ΔescD | escD deletion in pABB-CRS2 | This study |
| pABB-ΔescJ | escJ deletion in pABB-CRS2 | This study |
| pTrc99A | Prokaryotic expression vector | Pharmacia |
| pABB-Trc99cm | pTrc99A-derived vector | 31 |
| p99-escC | escC cloned into pABB-Trc99cm | This study |
| p99-escD | escD cloned into pABB-Trc99cm | This study |
| p99-escJ | escJ cloned into pABB-Trc99cm | This study |
| p99-espA | espA cloned into pABB-Trc99cm | 31 |
| pGEX-6P-1 | GST fusion protein expression vector | Amersham |
| pGEX-escC | escC cloned into pGEX-6P-1 | This study |
| pGEX-escJ | escJ cloned into pGEX-6P-1 | This study |
| pDEST15 | GST fusion protein expression vector | Invitrogen |
| p15-escD | escD cloned into pDEST15 | This study |
| p15-escF | escF cloned into pDEST15 | This study |
| pBAD-DEST49 | V5-tagged protein expression vector | Invitrogen |
| p99ccdB-V5 | V5-tagged protein expression vector derived from pTrc99A | This study |
| p99-escD-V5 | escD cloned into p99ccdB-V5 | This study |
| p99-escF-V5 | escF cloned into p99ccdB-V5 | This study |
TABLE 3.
Oligonucleotides used in this study
| Oligonucleotide | Sequencea | Cleavage site |
|---|---|---|
| B1-escC | 5′-AAAAAGCAGGCTGCTGCTATTTGTTTGACAGTGGCA-3′ | |
| B2-escC | 5′-AGAAAGCTGGGTAACGTTACGTTTTTATCAAGAATCG-3′ | |
| R1-escC | 5′-GAAGATCTTTACATTACACAATTCGTCCTA-3′ | BglII |
| R2-escC | 5′-GAAGATCTTGAATGATGATTTCTCTGGCGA-3′ | BglII |
| B1-escD | 5′-AAAAAGCAGGCTTCATGTTATCCTCATATAAAATTA-3′ | |
| B2-escD | 5′-AGAAAGCTGGGTTTAATACGACAGTGGAATATGTA-3′ | |
| R1-escD | 5′-CCGCTCGAGTGGAGTTTAATCCGTGGTTG-3′ | XhoI |
| R2-escD | 5′-CCGCTCGAGCCGCAATTAATGATGTCTTAACG-3′ | XhoI |
| B1-escJ | 5′-AAAAAGCAGGCTCATATCCCGTTAGCATCA-3′ | |
| B2-escJ | 5′-AGAAAGCTGGGTTGGCAAAGATACCACTTC-3′ | |
| R1-escJ | 5′-GAAGATCTTATTGCGCGAACTTAATCCTC-3′ | BglII |
| R2-escJ | 5′-GAAGATCTAATACCCGGTGTTATCGATTG-3′ | BglII |
| B1-espAcom | 5′-AAAAAGCAGGCTATTATAAGGAGGATGTTATGTG-3′ | |
| B2-espAcom | 5′-AGAAAGCTGGGTCACAGACTGGATATCGTT-3′ | |
| B1-escCcom | 5′-AAAAAGCAGGCTGATGCCGCCAACACACT-3′ | |
| B2-escCcom | 5′-AGAAAGCTGGGTGGTCATTGAGATCTTGTTGTGTC-3′ | |
| B1-escDcom | 5′-AAAAAGCAGGCTCATTAGCCATTGGAAACTCAC-3′ | |
| B2-escDcom | 5′-AGAAAGCTGGGTGTATCCTGGTAATCAAGTCTCT-3′ | |
| B1-escJcom | 5′-AAAAAGCAGGCTCATATCCCGTTAGCATCA-3′ | |
| B2-escJcom | 5′-AGAAAGCTGGGTTGGCAAAGARACCACTTC-3′ | |
| 5GST-escC | 5′-CGGGATCCGCCCCGTCCTCTCTGGAA-3′ | BamHI |
| 3GST-escC | 5′-CGGAATTCTTATTCGCTAGATGCAGATTTTAT-3′ | EcoRI |
| 5GST-escJ | 5′-CGGGATCCTGATAAGAGCAATTGTATACAG-3′ | BamHI |
| 3GST-escJ | 5′-CGGAATTCTTACCCGTCCTGTCCTGAGGAT-3′ | EcoRI |
| 5GST-escD | 5′-AAAAAGCAGGCTTCATGTTATCCTCATATAAAATA-3′ | |
| 3GST-escD | 5′-AGAAAGCTGGGTTTAATACGACAGTGGAATATGTA-3′ | |
| 5GST-escF | 5′-AAAAAGCAGGCTTCCTGGAAGTTCTGTTCCAGGGGCCCATGAATTTATC TGAAATTAC-3′ | PreScission protease |
| 3GST-escF | 5′-AGAAAGCTGGGTTTAAAAACTACGGTTAGAAATG-3′ | |
| R1V5HIS | 5′-GCGAGCTCACAAGTTTGTACAAAAAAGCTGAA-3′ | SacI |
| R2V53 | 5′-GCTCTAGATTAACCGGTACGCGTAGAATCGAG-3′ | XbaI |
| B1-escD-V5 | 5′-AAAAAGCAGGCTCCATATTTTTTCTCATTGTTCAACC-3′ | |
| B2-escD-V5 | 5′-AGAAAGCTGGGTTATACGACAGTGGAATATGTATAGA-3′ | |
| B1-escF-V5 | 5′-AAAAAGCAGGCTGTTCGTAATGAACGTAAATAGTTAA-3′ | |
| B2-escF-V5 | 5′-AGAAAGCTGGGTTAAAACTACGGTTAGAAATGGTTGA-3′ |
Underlining indicates restriction enzyme sites or, for 5GST-escF, a region encoding a recognition site for PreScission protease.
Cloning and construction of nonpolar mutants.
To construct the EPEC escC mutant, the 2.0-kbp DNA fragment encoding EscC and its flanking regions was amplified by PCR with the primers B1-escC and B2-escC, using EPEC E2348/69 genomic DNA as the template. Likewise, to construct EscD and EscJ mutants, the 1.2- and 1.2-kbp DNA fragments encoding EscD and EscJ and their respective flanking regions were amplified by PCR with the following sets of primer pairs: B1-escD and B2-escD and B1-escJ and B2-escJ. The resulting PCR products were cloned into pDONR201 to obtain pDONR-escC, pDONR-escD, and pDONR-escJ by means of adapter PCR and site-specific recombination techniques using the Gateway cloning system (Invitrogen). By using circular pDONR-escC, -escD, and -escJ as the DNA templates, inverse PCR was carried out with the following sets of primer pairs, respectively: R1-escC and R2-escC, R1-escD and R2-escD, and R1-escJ and R2-escJ. The resulting PCR products were digested with the respective restriction enzymes, which were generated in R1 and R2 primers and self-ligated to obtain pDONR-ΔescC, pDONR-ΔescD, and pDONR-ΔescJ, respectively. pDONR-ΔespC contained the stop codon and the BglII restriction site in addition to a 180-bp deletion starting 3 bp downstream from the escC start codon. pDONR-ΔescD contained the stop codon and the XhoI restriction site in addition to a 70-bp deletion starting 565 bp downstream from the escD start codon. pDONR-ΔescJ contained the stop codon and the BglII restriction site in addition to a 148-bp deletion starting 196 bp downstream from the escJ start codon. pDONR-ΔescC, pDONR-ΔescD, and pDONR-ΔescJ were then mixed with a positive suicide vector, pABB-CRS2 (31), to obtain pABB-ΔescC, pABB-ΔescD, and pABB-ΔescJ, respectively, by the Gateway cloning system. These plasmids were introduced separately into E. coli SM10λpir and transconjugated into the EPEC wild-type (WT) strain (nalidixic acid resistant), as described previously (11). The resulting mutant strains were designated strains ΔescC, ΔescD, and ΔescJ, respectively.
For complementation of the espA defect in EPEC, p99-espA was constructed by cloning the DNA fragment containing espA into pABB-Trc99cm (31), which can replicate in EPEC. A DNA fragment of espA was amplified by PCR with the primer pair B1-espAcom and B2-espAcom, using EPEC genomic DNA as the template. Likewise, DNA fragments for escC, escD, and escJ were amplified by PCR with the following sets of primer pairs, respectively: B1-escCcom and B2-escCcom, B1-escDcom and B2-escDcom, and B1-escJcom and B2-escJcom. The resulting PCR products of espA, escC, escD, and escJ were mixed with pDONR201, and then each recombinant plasmid was mixed with pABB-Trc99cm (31) to obtain p99-espA, p99-escC, p99-escD, and p99-escD, respectively, by the Gateway cloning system. These p99 series were used for complementation of strains ΔespA, ΔescC, ΔescD, and ΔescJ.
For the pulldown assay, each DNA fragment encoding EscC, EscD, EscF, and EscJ was amplified by PCR using EPEC E2348/69 genomic DNA as the template with the following sets of primer pairs, respectively: 5GST-escC and 3GST-escC, 5GST-escD and 3GST-escD, 5GST-escF and 3GST-escF, and 5GST-escJ and 3GST-escJ. PCR products of escC or escD were digested with BamHI and EcoRI and were then inserted into the BamHI and EcoRI sites of pGEX-6P-1 to obtain pGEX-escC or pGEX-escJ, respectively. PCR products of escD or escF were mixed with pDONR201 to obtain pDONR-GST-escD or pDONR-GST-escF, respectively, and then each recombinant was mixed with pDEST15 to obtain p15-espD or p15-escF, respectively, by using site-specific recombination techniques and the Gateway cloning system. For the addition of the V5 epitope tag at the C terminus, a plasmid, p99ccdB-V5, was constructed from pTrc99A and pBAD-DEST49. The 1.8-kbp DNA fragment encoding the chloramphenicol resistance gene, the ccdB gene for negative selection, the gene encoding the V5 epitope tag, and specific recombination sites (attR1 and attR2) was amplified by PCR with R1V5HIS and R2V53 primers, using pBAD-DEST49 (Invitrogen) as the DNA template. The resulting PCR product was digested with SacI and XbaI and then inserted into the SacI and XbaI sites of pTrc99A to obtain p99ccdB-V5. To construct EscD-V5 and EscF-V5, DNA fragments encoding either EspD or EscF were amplified from EPEC E2348/69 genomic DNA by adapter PCR with the following sets of primer pairs, respectively: B1-escD-V5 and B2-escD-V5 and B1-escF-V5 and B2-escF-V5. The resulting PCR products were cloned into pDONR201 to obtain pDONR-escD-V5 and pDONR-escF-V5, respectively, using the Gateway cloning system. pDONR-escD-V5 and pDONR-escF-V5 were then mixed with p99ccdB-V5 to obtain p99-escD-V5 or p99-escF-V5, respectively, using the Gateway cloning system.
Purification of the NCs from EPEC.
Whole-cell lysates were prepared from EPEC WT and mutant strains according to the protocol used for Shigella (32). The whole-cell lysates were clarified by centrifugation at 16,000 × gav (where gav is average gravity) for 20 min at 4°C and were then subjected to ultracentrifugation in a Hitachi RP42 rotor at 80,000 × gav for 1 h at 4°C. The resulting pellets containing the NCs were resuspended in TET buffer (10 mM Tris-HCl [pH 8.0 at 20°C], 1 mM EDTA, and 0.1% Triton X-100). Following centrifugation at 17,000 × gav for 10 min, the resulting supernatant was recovered and is designated NC fraction I (NC fr. I). To further purify the EPEC NCs, NC fr. I (3 ml, 0.3 mg of protein/ml) from a 3-liter culture of the espA mutant harboring cloned espA in trans (ΔespA/A strain) (31) was layered on 1.5 ml of a sucrose cushion (30% [wt/vol] sucrose, 10 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA) and then centrifuged in a Beckman SW55 rotor at 170,000 × gav for 4 h at 4°C. The resulting pellet was resuspended in TET buffer, and then the suspension was centrifuged at 17,000 × gav for 10 min. The resulting supernatant (1.2 ml, 0.05 mg of protein/ml) was recovered and is referred to as NC fraction II (NC fr. II). To isolate the NCs, NC fr. II was subjected to CsCl density gradient centrifugation. NC fr. II (1 ml) was mixed with 4 ml of TET buffer containing 1.9 g of CsCl to give an initial density of 1.28 g/cm3; the sample was then centrifuged to equilibrium in the SW55 rotor at 90,000 × gav for 24 h at 20°C. Twenty fractions (250 μl each) were collected from the top of the gradient, diluted to a volume of 3mlwith TET buffer, and then centrifuged in a Beckman TLA-100.4 rotor at 80,000 × gav for 1 h at 4°C. The respective pellets were resuspended in 0.25 ml of TET buffer. The protein concentrations were determined according to the method of Bradford (3) by using bovine serum albumin as a standard.
Electron microscopy.
Samples were negatively stained with 2% phosphotungstic acid (pH 7.3) in a solution containing 0.2% (wt/vol) sucrose on Butval-98 grids. The samples were then observed under a JEM 1010 transmission electron microscope (JEOL, Tokyo, Japan). For the immunolabeling of the NCs, samples were applied to Butval-98 grids, fixed with 1% formaldehyde in physiological salt solution, and immunolabeled first with affinity-purified anti-EspA or anti-EscC polyclonal antibodies and subsequently with 6-nm colloidal gold-conjugated antibodies against guinea pig immunoglobulin G (Aurion, Wageningen, The Netherlands) at room temperature for 20 min.
MALDI-TOF MS analysis.
After sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out, the protein bands were stained with Coomassie brilliant blue (CBB) and excised from the polyacrylamide gel. Proteins in the excised gel fragments were digested with modified trypsin (sequencing grade; Roche Molecular Biochemicals), and the resulting peptides were analyzed using a Voyager-DE PRO MALDI mass spectrometer (Applied Biosystems). Peptide mass fingerprint searches were performed with the Mascot search program, which is available at the Matrix Science website (www.matrixscience.com).
Preparations of secreted proteins.
EPEC grown in DMEM was removed by centrifugation at 17,000 × gav for 10 min, and the proteins in the supernatants were precipitated by trichloroacetic acid at a concentration of 10% and then analyzed by SDS-PAGE.
GST pulldown assay.
E. coli DH5α carrying p99-escD-V5 or p99-escF-V5 was incubated overnight at 30°C with shaking; the overnight cultures were then diluted 1:40 in LB broth containing 50 μg/ml ampicillin and incubated for 4 h at 30°C with shaking. The bacterial culture was further incubated for 4 h at 30°C in the presence of IPTG (isopropyl-β-d-thiogalactopyranoside) at a final concentration of 1 mM. Bacteria were collected by centrifugation at 17,000 × gav for 10 min and suspended in cold bead binding buffer (50 mM potassium phosphate [pH 7.2], 150 mM KCl, 1 mM MgCl2). Bacterial suspensions were sonicated, and the cell debris was removed by centrifugation at 17,000 × gav for 10 min. Triton X-100 and glycerol were added to the supernatants to final concentrations of 1% and 10%, respectively, and their protein concentrations were adjusted to 3.5 mg/ml with cold bead binding buffer. The resulting E. coli whole-cell extracts were used for the GST pulldown assay.
GST fusion proteins of EscC, EscD, EscF, and EscJ were expressed in E. coli BL21(DE3) containing the respective GST expression vectors pGEX-escC, pGEX-escJ, p15-escD, and p15-escF, as listed in Table 2. The whole-cell extracts of the respective bacteria were prepared as described above, and the respective GST fusion proteins were purified from the extracts with glutathione Sepharose 4 Fast Flow (Amersham Biosciences) according to the manufacturer's instructions. The respective GST-EscC, -EscD, -EscF, and -EscJ fusion proteins (20 μg), bound to glutathione beads (20 μl of 50% slurry), were incubated with the whole-cell extract (400 μl) of E. coli DH5α expressing EscC-V5 or EscF-V5 for 4 h at 4°C with gentle shaking. After the beads had been washed three times in cold bead binding buffer, 40 μl of 2× Laemmli sample buffer (200 mM Tris-HCl [pH 6.8], 4 mM EDTA, 4% SDS, 40% glycerol, 10 mM dithiothreitol, 0.02% bromphenol blue) was added to the beads. After the samples were boiled for 5 min, the proteins eluted from the beads were analyzed by 14% SDS-PAGE, followed by immunoblot analysis, as described below.
Antibodies and immunoblotting.
A mouse monoclonal antibody against Tir was kindly provided by B. Brett Finlay (Michael Smith Laboratories, University of British Columbia). The guinea pig polyclonal antibodies against EspA and EscC used here were characterized previously (31). EscC or EscF tagged with V5 epitope was detected by anti-V5 monoclonal antibodies (Invitrogen). For the immunoblot analysis, proteins were resolved by SDS-PAGE and transferred to polyvinylidene difluoride membranes (Immobilon filter; Millipore). The proteins were detected by immunoblotting using an ECL detection kit (Amersham Biosciences).
RESULTS
Purification of the NCs from EPEC.
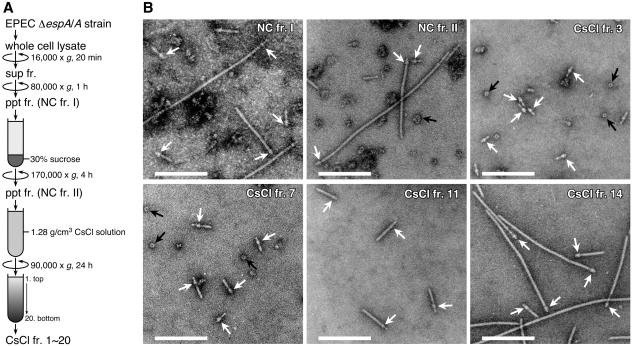
In our previous study, we partially purified the NCs from EPEC, and the supermolecular structure of these NCs was analyzed by TEM (31). In order to further investigate the molecular composition and fine structure of these NCs, we attempted to isolate them from an espA mutant harboring cloned espA in trans (ΔespA/A strain). This strain produces larger amounts of the NC with longer sheath-like structures than does the WT strain (31). However, the supermolecular structure of the basal body is identical in both strains (31). As illustrated in Fig. 1A, the whole-cell lysate of the ΔespA/A strain was clarified by centrifugation at 16,000 × g and then the supernatant fraction was centrifuged at 80,000 × g for 1 h to sediment the NCs. The resulting pellet fraction, NC fr. I, was layered on a 30% sucrose cushion and centrifuged at 170,000 × g for 4 h. The high-molecular-weight complexes, including the NCs, were precipitated through the sucrose cushion and were then resuspended in TET buffer in order to recover the pellet fraction, NC fr. II. Finally, NC fr. II was subjected to CsCl density gradient centrifugation at an initial density of 1.28 g/cm3. After centrifugation, 20 fractions (CsCl frs. 1 to 20) were obtained from the top to the bottom of the gradient. At the respective purification steps, representative fractions were negatively stained, and the resulting samples were observed by TEM (Fig. 1B). As expected (31), a large number of other bacterial components was still observed in NC fr. I, but the number of other components was substantially lower in the case of NC fr. II. After CsCl density gradient centrifugation of NC fr. II, the NCs with various needle lengths were separated into various density fractions from 2 to 15 (e.g., CsCl frs. 3, 7, 11, and 14 [Fig. 1B]). Interestingly, the NCs with shorter needle structures accumulated in the low-density fractions 2 to 4 (e.g., CsCl fr. 3). In contrast, NCs with longer sheath-like structures at the tip of the thin needles accumulated in the high-density fractions 13 to 15 (e.g., CsCl fr. 14). In these high-density fractions, the NCs were purified to apparent homogeneity. Furthermore, the length of the sheath-like structure tended to increase gradually with increases in density of the CsCl fractions (e.g., CsCl frs. 7, 11, and 14). Thus, we established a method of distinguishing between the EPEC NCs showing heterogeneity in terms of the length of the needle structure; here, separation was achieved on the basis of the respective densities of these NCs in CsCl solution.
FIG. 1.
Purification of the EPEC NCs. (A) Purification scheme for the EPEC NCs. Whole-cell lysate was prepared from an EPEC ΔespA/A strain grown in DMEM, and the lysate was clarified by centrifugation. The resulting supernatant fraction (sup fr.) was ultracentrifuged, and then a pellet fraction (ppt fr.) (NC fr. I) was recovered. NC fr. I was layered on a sucrose cushion and then ultracentrifuged. The resulting pellet fraction (NC fr. II) was subjected to CsCl density gradient centrifugation, and then 20 fractions (CsCl frs. 1 to 20) were collected from the top of the gradient. For more details, see Materials and Methods. (B) Electron micrographs of negatively stained EPEC NCs. NC frs. I and II and CsCl fractions 3, 7, 11, and 14 were negatively stained and observed by TEM. White arrows indicate the basal bodies of the EPEC NCs, and black arrows indicate ring structures that were presumed to be the BfpB-BfpG complexes (29). Bar, 250 nm.
Identification of the protein composition of the EPEC NCs.
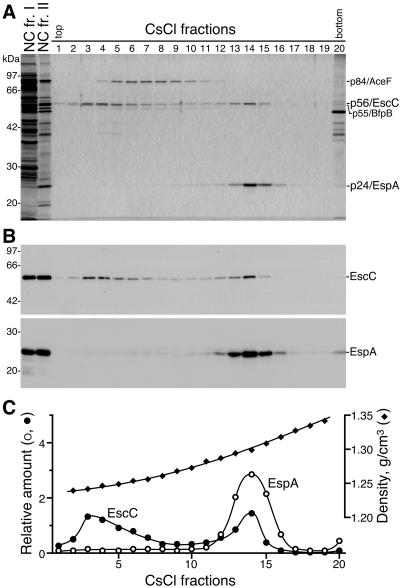
To investigate the protein components of the NCs, CsCl fractions 1 to 20 were analyzed by SDS-PAGE followed by silver staining (Fig. 2A). The NCs appeared to be copurified with a polypeptide with an apparent molecular mass of 56 kDa (p56) (CsCl frs. 2 to 15). Furthermore, a polypeptide with an apparent molecular mass of 24 kDa (p24) was abundant in the high-density fractions 13 to 15, which contained an abundance of NCs with the longer sheath-like structures. To identify p56 and p24, their tryptic fragments were analyzed by MALDI-TOF MS. The peptide mass fingerprints of p56 and p24 were well-matched with the theoretically derived values for EscC (a putative outer ring protein of the EPEC NC basal body) and EspA (a component of the sheath-like structure), respectively (data not shown). Moreover, Western blot analysis revealed that anti-EscC and -EspA antibodies reacted specifically with p56 and p24, respectively, in various fractions (Fig. 2B). From these results, p56 and p24 were identified as EscC and EspA, respectively. In this context, it is of note that EscC (p56) and EspA (p24) migrated more slowly than was expected from their calculated molecular masses (54 and 20 kDa, respectively) (Fig. 2A). To further analyze the distributions of EspA and EscC in the CsCl fractions, the band intensities of the Western blots (Fig. 2B) were measured by using the NIH Image program, and their relative amounts were estimated (Fig. 2C). Interestingly, EscC was distributed throughout various density fractions (frs. 2 to 15), with two apparent peaks at fractions 3 and 14 (Fig. 2B and C). In contrast, EspA was detected primarily in the high-density fractions 13 to 15 (Fig. 2B and C). The distributions of EspA and EscC in the CsCl density gradient were reproducible for three independently purified preparations. These results strongly indicate that the EPEC NCs with various needle lengths have heterogeneous densities; furthermore, these densities might be determined by the number of EspA molecules per basal body possessing a constant number of EscC molecules. Fractions 2 to 8 also contained a polypeptide with an apparent molecular mass of 55 kDa (p55), which migrated close to p56/EscC on the SDS-polyacrylamide gel; this band thereby appeared as a doublet band (Fig. 2A). According to the results of the MALDI-TOF MS analysis, p55 was thought to be an outer membrane lipoprotein, BfpB, which belongs to the secretin superfamily and is a component of a basal body of the type IV bundle-forming pilus (Bfp) (data not shown). BfpB is known to be associated with BfpG to form an outer ring with a diameter of approximately 20 nm (29). Indeed, ring-shaped supermolecular structures, the size and shape of which were quite similar to the previously reported BfpB-BfpG ring, were detected in fractions 2 to 8 (Fig. 1B, CsCl frs. 3 and 7) but not in fractions 9 to 15 (Fig. 1B, CsCl frs. 11 and 14). Thus, EPEC NCs with the longer sheath-like structures among these fractions were completely separated from the components of Bfp by CsCl density gradient centrifugation. In addition, a polypeptide with an apparent molecular mass of 84 kDa (p84), which was present in fractions 4 to 12, was thought to be a dihydrolipoamide acetyltransferase component (AceF) of the pyruvate dehydrogenase complex, based on the MALDI-TOF MS findings (data not shown). However, we were unable to detect any other proteins that were copurified with EscC or EspA, which suggests that the other NC components were not abundant and/or were easily dissociated from the NCs during the bacterial membrane solubilization and purification of the NCs. In addition, it is possible that some integral membrane and/or multimeric proteins comprising the EPEC NC might not be well resolved on a traditional SDS-polyacrylamide gel, thereby reducing the number of proteins available for the MALDI-TOF MS analysis.
FIG. 2.
Identification of EscC and EspA as components of the EPEC NC. (A) NC fr. I (4.5 μg), NC fr. II (0.3 μg), and CsCl fractions 1 to 20 (6 μl each), shown in Fig. 1, were analyzed by 12% SDS-PAGE followed by silver staining. The positions of marker proteins are shown on the left. The positions of p56/EscC, p24/EspA, p84/AceF, and p55/BfpB are shown on the right. (B) Western blot analysis of NC fr. I, NC fr. II, and the CsCl fractions. Samples corresponding to those shown in panel A were immunoblotted with anti-EscC (upper panel) or -EspA (lower panel) antibodies. (C) Distribution of EspA and EscC in the CsCl fractions. Relative amounts of EspA and EscC in the CsCl fractions were estimated from the band densities observed on the Western blots (panel B) by using the NIH Image program. Densities (g/cm3) of the fractions were determined by refractometry.
Supermolecular structures of the EPEC NCs.
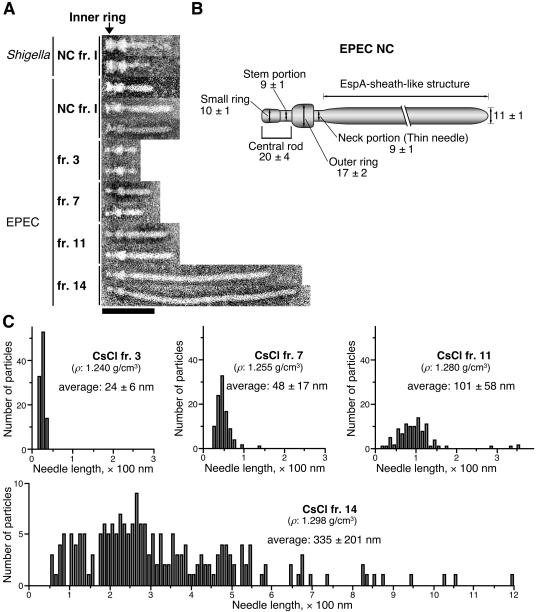
The representative TEM images were obtained from NC fr. I and four CsCl fractions, and a total of 11 images were aligned; the present supermolecular structures were compared with each other and with those of Shigella (Fig. 3A). The TEM images clearly showed that the length of the needle structures containing the EspA sheath-like structures detected in the low-density CsCl fractions (e.g., CsCl frs. 3 and 7) was shorter than that in the high-density fractions (e.g., CsCl frs. 11 and 14) (Fig. 1B and 3A); moreover, these observations agree with the results of the Western blot analysis using anti-EspA antibodies (Fig. 2C). To verify the relationship between the densities of the NCs and their respective needle lengths, we examined the distribution of needle length in randomly chosen NCs from CsCl fractions 3, 7, 11, and 14 (Fig. 3C) (100 particles of NCs were measured from frs. 3, 7, and 11, and 200 particles were measured from fr. 14). The NCs with a buoyant density in CsCl of 1.240 g/cm3 (CsCl fr. 3) had shorter needles, and the needle length appeared to be of an almost fixed size (average, 24 nm). In response to the elongation of the EspA sheath-like structure, the density of the NCs appeared to gradually increase. For example, NCs with densities of 1.255 (CsCl fr. 7) and 1.280 (CsCl fr. 11) g/cm3 had needle structures with average lengths of 48 and 101 nm, respectively. The NCs with a density of 1.298 g/cm3 (CsCl fr. 14) had longer needles of various lengths (average, 335 nm). These findings indicate that the observed increase in the density of the NCs was dependent on the length of the EspA sheath-like structure. Regarding cases in which the needle structure of the NC was longer than 100 nm, the majority appeared to accumulate in the high-density fractions 13 to 15.
FIG. 3.
Supermolecular structures of the EPEC NCs. (A) Alignment of the EPEC and Shigella NCs detected in NC frs. I and CsCl fractions 3, 7, 11, and 14. Bar, 100 nm. (B) Presumed model of the EPEC NC core structure. The size of each structure is given in nanometers. (C) Distribution of needle length in the NCs, as detected in CsCl fractions 3, 7, 11, and 14. The densities (ρ, g/cm3) of the CsCl fractions are shown in each panel. The average needle lengths were determined by measuring those of 100 (frs. 3, 7, and 11) or 200 (fr. 14) particles. The values represent means ± standard deviations.
The purpose of purifying the EPEC NC was to identify all of the protein components of the TTS apparatus. However, the inner ring of the EPEC NC appears to be unstable during bacterial membrane solubilization and/or during precipitation of the NC through the sucrose cushion. Thus, NCs with the inner rings were still observed, but only in small amounts, in NC fr. I. However, we detected only NCs without the inner rings in NC fr. II and the CsCl fractions. In contrast, we detected the typical form of the Shigella NC (Fig. 3A), which was recovered according to the same methods of membrane solubilization and preparation of NC fr. I. Unlike the EPEC NC, most of the Shigella NCs had a typical inner ring. These observations suggest that the components and stability of the EPEC NC may differ from those of the Shigella NC, which has a more rigid inner ring structure. Improvement of the conditions for bacterial membrane solubilization is therefore still necessary for the further analysis of the inner ring portion of the EPEC NC. However, taking into account the instability of the EPEC NC basal body, we were able to dissect a central rod of the basal body of the EPEC NC for the first time; this central rod is typically covered by the inner ring that is predicted to contain a multimeric complex of EscJ (34) (Fig. 3A). We measured each of 10 NCs from three CsCl fractions (frs. 3, 7 and 14; a total of 30 NCs), and each part was estimated as shown in Fig. 3B. The length of the central rod, which contained a small ring-like structure (10 nm in width), was estimated to be 20 nm. The width of the stem portion of the central rod was estimated to be 9 nm, which is identical in size to the neck portion (9 nm in width) of the needle structure.
Localization of EscC into the basal body.
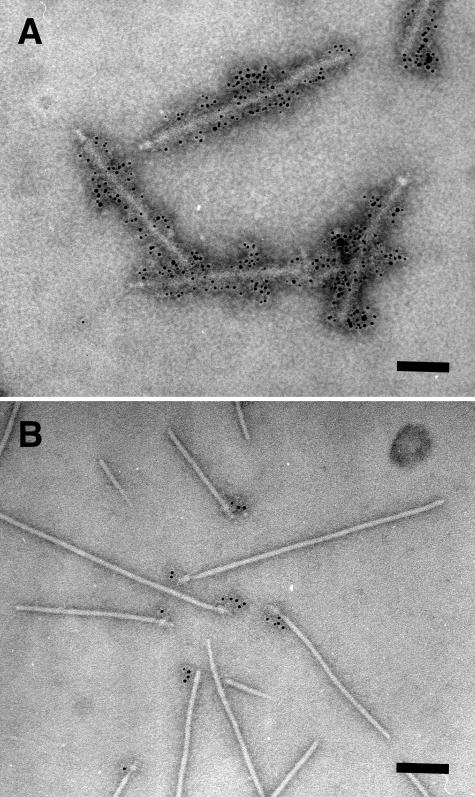
In order to elucidate the localization of EscC in the EPEC NC, highly purified NCs (CsCl fr. 14) were stained with immunogold-labeled anti-EscC antibodies and observed by TEM (Fig. 4). As expected from our previous results obtained using partially purified NCs (31), the sheath-like structures of the NCs were stained specifically with the anti-EspA antibodies (Fig. 4A). In contrast, the outer rings of the NCs were clearly stained with anti-EscC antibodies, indicating that EscC oligomerizes to form the outer ring of the NC (Fig. 4B).
FIG. 4.
Localization of EspA and EscC in the EPEC NC. The highly purified NCs in CsCl fraction 14 (Fig. 1) were analyzed by immunoelectron microscopy. (A) EspA was detected with immunogold-labeled anti-EspA antibodies. Sheath-like structures coated with 6-nm gold particles were observed. (B) EscC was detected with immunogold-labeled anti-EscC antibodies. In contrast to EspA localization in the sheath, EscC was localized in the basal body of the EPEC NC. Bar, 100 nm.
Composition and assembly of the EPEC NC.
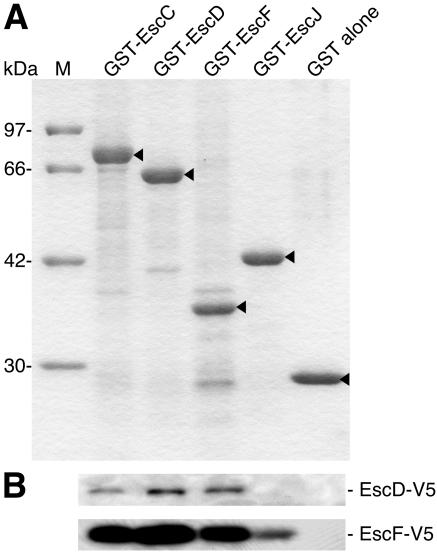
The TEM view showed that the basal body appears to be unstable, specifically in terms of the inner ring of the basal body (Fig. 1B and 3A). For this reason, we were unable to determine all of the components of the EPEC TTS apparatus by MALDI-TOF MS analysis. In our previous study, we demonstrated that EscF is required for NC formation and for the secretion of the Esp proteins (31). However, we could not detect EscF in the CsCl fractions by immunoblotting (data not shown); this result was most likely due to the low abundance of EscF in the NC and/or the low quality of our anti-EscF antibody. To further analyze the protein-protein interactions required to assemble the EPEC TTS apparatus, we performed a GST pulldown assay using purified GST-EscD (a presumed component of the basal body), GST-EscJ (a presumed component of the inner ring), GST-EscC (the outer ring protein), and GST-EscF (the needle protein) as the probes (Fig. 5A). Whole-cell extracts were prepared from E. coli expressing EscD or EscF tagged with the V5 epitope at the C terminus (EscD-V5 or EscF-V5), and then the extracts were incubated with the respective GST probes bound on glutathione beads. Afterwards, the V5 epitope-tagged proteins coprecipitated with the respective GST probes were detected by Western blotting using anti-V5 epitope antibodies (Fig. 5B). EscD-V5 was associated with GST-EscD as well as with GST-EscC and GST-EscF but not with GST alone. In this experiment, we could not detect the interaction between EscD and EscJ. In contrast, EscF-V5 was associated with GST-EscF, GST-EscC, GST-EscD, and GST-EscJ but not with GST alone. Although the GST pulldown assay may not be a suitable procedure for multimeric and/or membrane-associated proteins, these results suggest that the presumed components of the EPEC TTS apparatus, EscD, EscJ, and EscF, are associated with other components of the EPEC NC.
FIG. 5.
Interactions between the putative components of the EPEC TTS apparatus. (A) Purified GST-Esc fusion proteins were analyzed by 12% SDS-PAGE followed by CBB staining. Arrowheads indicate presumed bands corresponding to the respective GST fusion proteins. Lane M indicates the size markers. (B) Interactions of Esc proteins in a GST pulldown assay. Whole-cell extracts from E. coli expressing EscD or EscF tagged with the V5 epitope (EscD-V5 or EscF-V5) were incubated with each agarose-immobilized GST-Esc protein. Proteins bound to the beads were resolved by 14% SDS-PAGE and then analyzed by Western blotting using anti-V5 monoclonal antibodies.
Effects of mutations in the presumed basal component genes on secretion.
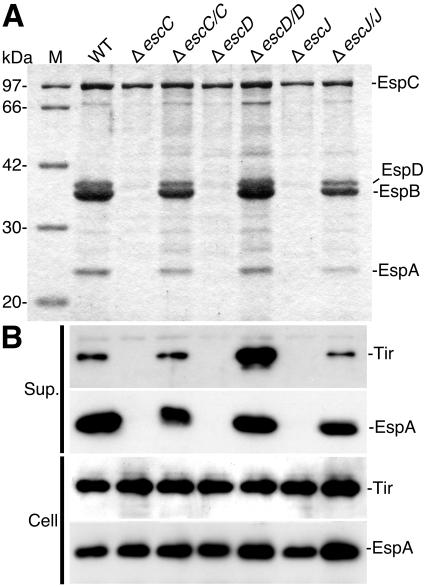
We have demonstrated that a mutant strain unable to synthesize EscF, a putative component of the NC thin needle, failed to produce a functional TTS system, thereby preventing the secretion of type III secreted proteins (31). To investigate the effects of escC, escD, and escJ mutations on type III secretion, the secreted proteins from the respective mutant strains (ΔescC, ΔescD, and ΔescJ) were analyzed by 12% SDS-PAGE. As shown in Fig. 6A, the secretion of EspA, EspB, and EspD, but not that of EspC (a non-TTS system-secreted protein), was blocked by each of the respective mutations in escC, escD, or escJ. Complementation of the escC, escD, and escJ mutants with the respective genes in trans restored the secretion of all of the type III secreted proteins. Therefore, all mutations were nonpolar and could be complemented. To analyze the effects of the above-described mutations on the expression of type III secreted proteins, the secreted proteins and whole-cell extracts from the mutant and complemented strains were analyzed by Western blotting using anti-Tir and anti-EspA antibodies (Fig. 6B). Although neither the production of EspA nor that of Tir in the bacteria was affected by any of the mutations in escC, escD, or escJ, their secretion into the culture supernatants was blocked, indicating that EscC, EscD, and EscJ are required for the secretion of the type III secreted proteins.
FIG. 6.
Effect of esc mutations on secretion of the Esp proteins and Tir effector. (A) Secreted-protein profiles of WT EPEC and various mutant strains. Bacteria were grown in DMEM, and the secreted proteins were resolved by 12% SDS-PAGE and stained with CBB. (B) Immunoblot analysis of Tir and EspA in the culture supernatants (Sup.) and whole-cell extracts (Cell). Anti-EspA and -Tir antibodies were used for immunodetection. Lanes M and WT indicate the size markers and secreted proteins prepared from the EPEC wild type, respectively.
Interestingly, when the escD clone was introduced into the escD mutant strain in trans (ΔescD/D), secretion of the Tir effector was greatly increased compared to Tir secretion from ΔescC/C and ΔescJ/J. In contrast, the level of EspA secretion in ΔescD/D was almost same as that in both ΔescC/C and ΔescJ/J. Our findings suggest that EscD may independently regulate the levels of secretion of the Esp proteins and the Tir effector. However, ΔescD/D has not yet been well characterized. In particular, it will still be necessary to compare the levels of EscD expression and NC formation in ΔescD/D with those of the other strains.
DISCUSSION
We established a method for the purification of EPEC NCs in successive centrifugal steps (Fig. 1). At the final step of purification, EPEC NCs were separated into various density fractions by equilibrium CsCl density gradient centrifugation (Fig. 1B). The NCs with the shorter needle structures were fractionated into the low-density fractions 2 to 4 (ρ = 1.23 to 1.25 g/cm3). Although the densities of the NCs gradually increased to some extent with elongation of the sheath-like needle structures, the majority of the NCs with needle lengths of more than 100 nm were found to have accumulated in the high-density fractions 13 to 15 (ρ = 1.29 to 1.30 g/cm3). EspA was more abundant in the high-density fractions than in the low-density fractions (Fig. 2), which was consistent with the observation that the EspA sheath-like needle structures of the NCs in the high-density fractions were longer than those in the low-density fractions (Fig. 3). These findings indicate that the length of the sheath-like structure is correlated with the amount of EspA attached to the NC. The present results also suggest that NCs with a very small amount of EspA have an inherent buoyant density (ρ = 1.23 to 1.25 g/cm3) in the CsCl gradient and that the densities of NCs gradually increase with increases in the number of EspA molecules in NCs of up to 1.29 to 1.30 g/cm3. The density of NCs with the longer EspA sheath-like structure is likely to be comparable to that of the EspA filaments. In other words, NCs with longer EspA sheath-like structures may behave as the EspA filaments, and, when observed in terms of a CsCl gradient, the density of the EspA filaments may not be affected by their length. The needle length of the EPEC NC was almost fixed (24 nm) in CsCl fraction 3, which contained relatively little EspA (Fig. 2B). Interestingly, the shape of the needle tip of the EPEC NC in the low-density fractions differed somewhat from that of the needle tip of the Shigella NC, i.e., certain appendages appeared to be attached to the needle tip (Fig. 3A). One hypothesis that could account for these findings would be that a small amount of EspA binds to the tip of the EscF thin needle.
In this study, we clearly demonstrated by immunoelectron microscopy using anti-EscC antibodies that EscC is a major protein component of the NC outer ring with a diameter of 17 nm (Fig. 3B and 4B). EscC was detected in the outer membrane fraction (13) and was required for the secretion of the Esp proteins and Tir (Fig. 6). EscC shares 31.1% identity with Yersinia YscC, which belongs to the family of secretins, and forms a ring-shaped oligomeric complex (diameter, 20 nm) (4, 19). The YscC ring-like oligomer is thought to function as a transport channel for macromolecules in the outer membrane. However, the localizations of both EscC and YscC in their respective NCs have not yet been characterized. Here, we carried out an immunoelectron microscopic analysis to demonstrate that EscC is indeed localized in the NC as the outer ring.
The supermolecular structure of the central rod beneath the EscC outer ring could be observed in this study due to the instability of the structure composed of inner membrane proteins (Fig. 3B). The width (9 nm) of the stem in the central rod was identical to that (9 nm) of the neck portion in the needle, suggesting that EscF, a putative major component of the thin needle structure, or an EscF-like protein may form the central rod passing through the EscC outer ring. Although we were unable to identify the component(s) of the central rod, which contained a small ring structure, in the NCs examined here, the composition of the EPEC NC was predicted based on the results of a GST pulldown assay (Fig. 5). As expected, EscF associates with itself, suggesting that EscF may polymerize and thereby construct the thin hollow needle. We also detected the EscF-EscC interaction, which suggested that the EscF needle is sustained by the EscC outer ring. Interestingly, we detected an interaction of EscF with EscJ, which has been predicted to form a periplasmic portion of the inner ring (34). Our present results indicate that EscJ interacts with EscF; this interaction may be required to connect the EscJ inner ring to the external needle structure. Crystal packing analysis and molecular modeling suggest that EscJ oligomerizes to form a large 24-subunit ring structure with a diameter of 18 nm and a height of 5.2 nm. These estimated values are well matched to our previous report for the inner ring with a diameter of 18.1 ± 2.5 nm from electron microscope images of partially purified EPEC NCs (31). The central channel of the modeled EscJ ring is surrounded by the prominent negatively charged trench about 3.2 nm in height and 1.1 nm in width at the periplasmic side (34), and this trench may function as the binding region for the central rod (34). From the results of the GST pulldown assay, we speculate that the EscF thin needle also functions as the central rod. Although the PrgJ rod connecting the InvG outer ring to the PrgH-PrgK inner ring has been considered based on structural analysis of the Salmonella NC (22), no orthologue of PrgJ has been identified in EPEC. Furthermore, a PrgH orthologue, MxiG, was previously identified as a component of the NC basal body in Shigella. Again, no orthologue of PrgH has been found in EPEC or in Yersinia. These findings indicate that structures of both the central rod and the basal body of EPEC are somewhat different from those of Salmonella and Shigella.
A recent study of the Shigella NC assembly using in vivo binding assays demonstrated that MxiD, an EscC orthologue, interacted with MxiJ, an EscJ orthologue (30). From those findings (30), a segment of MxiD is thought to extend into the periplasm, where it can associate with inner membrane proteins such as MxiJ to build or sustain the basal body. Based on the sequence similarities, EscJ is thought to be associated with EscC to form the basal body. However, Creasey et al. (5) detected the EscC-EscD interaction but not the EscC-EscJ interaction by use of a yeast two-hybrid system. In this study, we showed the EscC-EscD and EscD-EscF interactions by using a GST pulldown assay (Fig. 5). EscD shows amino acid sequence similarity to Yersinia YscD, the bacterial inner membrane protein (27), but has no orthologue in the Shigella and Salmonella TTS systems. In addition, EscD contains a putative transmembrane domain (120 to 141 amino acids) at its N terminus (as predicted by TMpred [http://www.ch.embnet.org/software/TMPRED_form.html]). Although the correct localization of EscD in the EPEC NC remains unclear, the transmembrane domain of EscD may associate with the inner membrane and a segment of EscD may be located in the periplasm in order to sustain the central rod, where it can interact with EscC and also with EscF.
We demonstrated that EscC, EscD, and EscJ are required for the secretion of both the Esp proteins and the Tir effector (Fig. 6). These results were supported by mutation analyses of EPEC and/or C. rodentium LEE (6, 10, 13, 34) and could be predicted from the similarity of these proteins to YscC, YscD, and YscJ, which are required for the secretion of Yop proteins in Yersinia (1, 27). To demonstrate the direct contribution of EscC, EscD, and EscJ to the NC assembly, NC fractions were prepared from all of the mutant strains and observed by TEM. Although the putative BfpB-BfpG ring was observed in all of the mutant strains, no NCs were detected in the case of the escC, escD, and escJ mutants (data not shown). In contrast, the NCs were restored in complemented strains (ΔescC/C, ΔescD/D, and ΔescJ/J), indicating that EscC, EscD, and EscJ are all required for EPEC NC assembly (data not shown). In addition, wild-type EPEC was found to accumulate actin beneath the adherent bacteria by the translocation of Tir via the TTS system (28). In contrast, the escC, escD, and escJ mutant strains did not induce actin cytoskeletal rearrangement (data not shown).
In this study, we isolated the core TTS apparatus from EPEC and revealed the localization of EscC to the outer ring and the structure of the central rod in the TTS apparatus by electron microscopy. Furthermore, we showed that EscC, EscD, and EscJ are required for the formation of the functional TTS apparatus. Biochemical and genetic studies to identify all structural proteins of the EPEC TTS system and to evaluate their precise roles in assembly of the TTS apparatus are under way.
Acknowledgments
This research was partially supported by the Ministry of Education, Science, Sports, and Culture of Japan through Grants-in-Aid for Scientific Research (C, 16590370 [2004]), for Young Scientists (B, 14770123 [2002 to 2003]), for Scientific Research on Priority Areas (14021109 [2002]), and for COE research. Support was also received in the form of operating grants from the All Kitasato Project Study and a Kitasato University Research Grant for Young Researchers (2002 to 2004). T.M. is a research fellow of the Japan Society for the Promotion of Science.
REFERENCES
- 1.Allaoui, A., R. Schulte, and G. R. Cornelis. 1995. Mutational analysis of the Yersinia enterocolitica virC operon: characterization of yscE, F, G, I, J, K required for Yop secretion and yscH encoding YopR. Mol. Microbiol. 18:343-355. [DOI] [PubMed] [Google Scholar]
- 2.Blocker, A., N. Jouihri, E. Larquet, P. Gounon, F. Ebel, C. Parsot, P. Sansonetti, and A. Allaoui. 2001. Structure and composition of the Shigella flexneri ‘needle complex’, a part of its type III secreton. Mol. Microbiol. 39:652-663. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Burghout, P., R. van Boxtel, P. Van Gelder, P. Ringler, S. A. Muller, J. Tommassen, and M. Koster. 2004. Structure and electrophysiological properties of the YscC secretin from the type III secretion system of Yersinia enterocolitica. J. Bacteriol. 186:4645-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Creasey, E. A., R. M. Delahay, S. J. Daniell, and G. Frankel. 2003. Yeast two-hybrid system survey of interactions between LEE-encoded proteins of enteropathogenic Escherichia coli. Microbiology 149:2093-2106. [DOI] [PubMed] [Google Scholar]
- 6.Crepin, V. F., S. Prasannan, R. K. Shaw, R. K. Wilson, E. Creasey, C. M. Abe, S. Knutton, G. Frankel, and S. Matthews. 2005. Structural and functional studies of the enteropathogenic Escherichia coli type III needle complex protein EscJ. Mol. Microbiol. 55:1658-1670. [DOI] [PubMed] [Google Scholar]
- 7.Daniell, S. J., E. Kocsis, E. Morris, S. Knutton, F. P. Booy, and G. Frankel. 2003. 3D structure of EspA filaments from enteropathogenic Escherichia coli. Mol. Microbiol. 49:301-308. [DOI] [PubMed] [Google Scholar]
- 8.Daniell, S. J., N. Takahashi, R. Wilson, D. Friedberg, I. Rosenshine, F. P. Booy, R. K. Shaw, S. Knutton, G. Frankel, and S. Aizawa. 2001. The filamentous type III secretion translocon of enteropathogenic Escherichia coli. Cell. Microbiol. 3:865-871. [DOI] [PubMed] [Google Scholar]
- 9.Dean, P., M. Maresca, and B. Kenny. 2005. EPEC's weapons of mass subversion. Curr. Opin. Microbiol. 8:28-34. [DOI] [PubMed] [Google Scholar]
- 10.Deng, W., J. L. Puente, S. Gruenheid, Y. Li, B. A. Vallance, A. Vazquez, J. Barba, J. A. Ibarra, P. O'Donnell, P. Metalnikov, K. Ashman, S. Lee, D. Goode, T. Pawson, and B. B. Finlay. 2004. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. USA 101:3597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donnenberg, M. S., and J. B. Kaper. 1992. Enteropathogenic Escherichia coli. Infect. Immun. 60:3953-3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gauthier, A., J. L. Puente, and B. B. Finlay. 2003. Secretin of the enteropathogenic Escherichia coli type III secretion system requires components of the type III apparatus for assembly and localization. Infect. Immun. 71:3310-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jarvis, K. G., J. A. Giron, A. E. Jerse, T. K. McDaniel, M. S. Donnenberg, and J. B. Kaper. 1995. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc. Natl. Acad. Sci. USA 92:7996-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kenny, B., R. Devinney, M. Stein, D. J. Reinscheid, E. A. Frey, and B. B. Finlay. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511-520. [DOI] [PubMed] [Google Scholar]
- 17.Kenny, B., L. C. Lai, B. B. Finlay, and M. S. Donnenberg. 1996. EspA, a protein secreted by enteropathogenic Escherichia coli, is required to induce signals in epithelial cells. Mol. Microbiol. 20:313-323. [DOI] [PubMed] [Google Scholar]
- 18.Kimbrough, T. G., and S. I. Miller. 2000. Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc. Natl. Acad. Sci. USA 97:11008-11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koster, M., W. Bitter, H. de Cock, A. Allaoui, G. R. Cornelis, and J. Tommassen. 1997. The outer membrane component, YscC, of the Yop secretion machinery of Yersinia enterocolitica forms a ring-shaped multimeric complex. Mol. Microbiol. 26:789-797. [DOI] [PubMed] [Google Scholar]
- 20.Kubori, T., Y. Matsushima, D. Nakamura, J. Uralil, M. Lara-Tejero, A. Sukhan, J. E. Galan, and S. I. Aizawa. 1998. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280:602-605. [DOI] [PubMed] [Google Scholar]
- 21.Kubori, T., A. Sukhan, S. I. Aizawa, and J. E. Galan. 2000. Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc. Natl. Acad. Sci. USA 97:10225-10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marlovits, T. C., T. Kubori, A. Sukhan, D. R. Thomas, J. E. Galan, and V. M. Unger. 2004. Structural insights into the assembly of the type III secretion needle complex. Science 306:1040-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuzawa, T., A. Kuwae, S. Yoshida, C. Sasakawa, and A. Abe. 2004. Enteropathogenic Escherichia coli activates the RhoA signaling pathway via the stimulation of GEF-H1. EMBO J. 23:3570-3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moon, H. W., S. C. Whipp, R. A. Argenzio, M. M. Levine, and R. A. Giannella. 1983. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect. Immun. 41:1340-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mueller, C. A., P. Broz, S. A. Muller, P. Ringler, F. Erne-Brand, I. Sorg, M. Kuhn, A. Engel, and G. R. Cornelis. 2005. The V-antigen of Yersinia forms a distinct structure at the tip of injectisome needles. Science 310:674-676. [DOI] [PubMed] [Google Scholar]
- 27.Plano, G. V., and S. C. Straley. 1995. Mutations in yscC, yscD, and yscG prevent high-level expression and secretion of V antigen and Yops in Yersinia pestis. J. Bacteriol. 177:3843-3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosenshine, I., M. S. Donnenberg, J. B. Kaper, and B. B. Finlay. 1992. Signal transduction between enteropathogenic Escherichia coli (EPEC) and epithelial cells: EPEC induces tyrosine phosphorylation of host cell proteins to initiate cytoskeletal rearrangement and bacterial uptake. EMBO J. 11:3551-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidt, S. A., D. Bieber, S. W. Ramer, J. Hwang, C. Y. Wu, and G. Schoolnik. 2001. Structure-function analysis of BfpB, a secretin-like protein encoded by the bundle-forming-pilus operon of enteropathogenic Escherichia coli. J. Bacteriol. 183:4848-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuch, R., and A. T. Maurelli. 2001. MxiM and MxiJ, base elements of the Mxi-Spa type III secretion system of Shigella, interact with and stabilize the MxiD secretin in the cell envelope. J. Bacteriol. 183:6991-6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sekiya, K., M. Ohishi, T. Ogino, K. Tamano, C. Sasakawa, and A. Abe. 2001. Supermolecular structure of the enteropathogenic Escherichia coli type III secretion system and its direct interaction with the EspA-sheath-like structure. Proc. Natl. Acad. Sci. USA 98:11638-11643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamano, K., S. Aizawa, E. Katayama, T. Nonaka, S. Imajoh-Ohmi, A. Kuwae, S. Nagai, and C. Sasakawa. 2000. Supramolecular structure of the Shigella type III secretion machinery: the needle part is changeable in length and essential for delivery of effectors. EMBO J. 19:3876-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yip, C. K., B. B. Finlay, and N. C. Strynadka. 2005. Structural characterization of a type III secretion system filament protein in complex with its chaperone. Nat. Struct. Mol. Biol. 12:75-81. [DOI] [PubMed] [Google Scholar]
- 34.Yip, C. K., T. G. Kimbrough, H. B. Felise, M. Vuckovic, N. A. Thomas, R. A. Pfuetzner, E. A. Frey, B. B. Finlay, S. I. Miller, and N. C. Strynadka. 2005. Structural characterization of the molecular platform for type III secretion system assembly. Nature 435:702-707. [DOI] [PubMed] [Google Scholar]