Abstract
The iron-dependent transcriptional regulator DtxR from Corynebacterium diphtheriae is the prototype for a family of metal-dependent regulators found in diverse bacterial species. The structure of DtxR and its action as a repressor have been extensively characterized, but little is known about expression of dtxR. In the current study, we investigated transcription of dtxR as well as the sigB and galE genes located immediately upstream and downstream from dtxR, respectively. We identified two promoters that determine transcription of dtxR. The first, located upstream of sigB, appears to be controlled by an extracytoplasmic function σ factor. The second, located in the intergenic region between sigB and dtxR, is similar to promoters used by the primary vegetative σ factors in other actinomycete species. Using quantitative real-time assays, we demonstrated that the number of transcripts initiated upstream from sigB is affected by several environmental factors. In contrast, the presence of sodium dodecyl sulfate was the only factor tested that conclusively affects the number of transcripts initiated in the sigB-dtxR intergenic region. Additionally, we provided evidence for the existence of transcripts that contain sigB, dtxR, and galE. Our studies provide the first quantitative transcriptional analysis of a gene encoding a DtxR family regulator and give new insights into transcriptional regulation in C. diphtheriae.
Transcriptional regulators in the diphtheria toxin repressor (DtxR) family control gene expression in response to changes in the concentration of iron and/or manganese in a wide variety of bacterial species (2, 3, 16, 19, 21, 25, 54, 55). DtxR-like proteins are the predominant iron-dependent regulators in many acid-fast and gram-positive species, whereas the ferric uptake regulator (Fur) is the primary iron-dependent regulator in most gram-negative bacteria (18a). DtxR, the prototype of the family, is best characterized as a regulator that controls transcription of tox, the gene encoding diphtheria toxin in lysogenic strains of Corynebacterium diphtheriae (22). In its dimeric iron-bound form, holo-DtxR binds to an operator sequence within the tox promoter and prevents transcription of tox. In the apo form without bound iron, DtxR is unable to bind its cognate operator sequence, repression is relieved, and transcription of tox proceeds. In addition to controlling expression of tox, holo-DtxR represses transcription of several other genes, most of which are involved in iron uptake and utilization (27, 28, 43, 50, 56). In Mycobacterium species, the DtxR homologue IdeR acts to repress transcription of some genes involved in iron homeostasis and to activate transcription of others (9, 15, 46). The gene encoding DtxR can be inactivated in C. diphtheriae, but the resulting strain has a significant growth defect and is more sensitive to oxidative stress than its wild-type parent (40). Inactivating the gene encoding IdeR in Mycobacterium smegmatis causes a similar phenotype of increased sensitivity to oxidative stress (8), but in Mycobacterium tuberculosis ideR is essential for viability (47). Clearly, DtxR-like proteins are versatile transcriptional regulators that are often required for vital cellular functions.
The DNA binding activity, metal specificity, and physical structure of DtxR have been well characterized. DtxR binds as a dimer to palindromic 19-bp DNA operator sequences within the promoters of the genes that it regulates (51, 59, 60). Transcriptional regulation by DtxR in vivo is observed in response to changing iron concentrations, but sequence-specific binding of DtxR to DNA in vitro can be activated by other divalent transition metals, including cadmium, cobalt, manganese, nickel, and zinc (51, 53, 59). Extensive crystallographic studies have shown that DtxR has three domains (12, 45, 49). Amino acids 1 to 73 form domain 1, which contains a classical helix-turn-helix DNA-binding motif. Domain 2 (amino acids 74 to 140) determines dimerization of DtxR and contains the binding sites for two Fe2+ ions per monomer. The final 86 amino acids of DtxR (residues 141 to 226) comprise domain 3, which contributes ligands to one of the metal binding sites and has topology similar to the SH3 domains found in some eukaryotic signal transduction proteins (44). The first two domains of DtxR are essential for DtxR to function as a transcriptional repressor, while domain 3 is thought to modulate the activity of DtxR at individual promoters (30, 41).
While much research has focused on characterizing the structure and function of DtxR as a transcriptional regulator, little is known about the expression of dtxR. Analysis of RNA isolated from C. diphtheriae showed that the abundance of dtxR transcripts is much less than the abundance of tox transcripts under low-iron conditions, and the prevalence of dtxR transcripts is not affected by the amount of iron in the growth medium (52). This correlates well with the observation that the levels of DtxR in C. diphtheriae are not influenced by available iron (40). Quantitative analysis shows that C. diphtheriae contains approximately 750 dimers of DtxR per cell under both high- and low-iron conditions (39). The C. diphtheriae genome sequence reveals that dtxR is located 224 bp downstream from sigB, which is predicted to encode a sigma factor with homology to σ70-type factors, and 22 bp upstream from galE, which is predicted to encode a UDP-galactose 4-epimerase (4). Previously we reported preliminary evidence indicating that sigB and dtxR as well as dtxR and galE are cotranscribed (40).
The genetic organization of sigB and dtxR in C. diphtheriae is very similar to that in several other actinomycete species including M. tuberculosis (6) and Brevibacterium lactofermentum (37). In M. tuberculosis, B. lactofermentum, and Brevibacterium flavum, sigB is transcribed from a promoter located approximately 25 bp upstream from the coding sequence, and several environmental stress conditions have been shown to affect expression of sigB in these species (17, 23, 37, 38). In addition, in C. diphtheriae and B. lactofermentum, a galE homologue is located immediately downstream from dtxR. The transcripts encoding all three genes in this locus were examined in B. lactofermentum, using Northern blot assays. Two sigB-containing transcripts were identified; one includes only sigB and the other contains sigB as well as a portion of the dtxR homologue dmdR. Similarly, the complete dmdR gene is encoded on two different transcripts: one contains only dmdR and the other includes both dmdR and galE (36).
Although the activity of DtxR has been studied by many researchers, the transcription of dtxR and contiguous genes has not been investigated in C. diphtheriae. In the present study, we investigated the transcriptional regulation of the genes in the dtxR locus with the goals of understanding better the mechanisms of transcriptional regulation in C. diphtheriae and the role of DtxR in the pathogenesis of diphtheria. We showed that the promoter located upstream from sigB functions in C. diphtheriae but not in Escherichia coli, clearly indicating a role for species-specific factors in the regulation of transcription in C. diphtheriae. Furthermore, we demonstrated that transcription at the dtxR locus is affected by environmental factors and that dtxR can be cotranscribed with the genes that surround it. Transcription of dtxR is a complex and multifaceted process that is controlled both by DNA regulatory elements that exert their effects in cis and by soluble regulatory factors that exert their effects in trans.
MATERIALS AND METHODS
Bacterial strains and cultivation conditions.
The toxigenic C. diphtheriae strain C7(β) (1) was isolated after β phage infection of strain C7(−) (13) and has been used extensively for experimental work since the 1950s. E. coli DH5α (Bethesda Research Laboratories, Gaithersburg, Md.) was used as a host for cloning and β-galactosidase assays. C. diphtheriae strains were cultivated in PGT, which is a deferrated casein hydrolysate medium (1) or heart infusion broth (BD-Difco, Franklin Lakes, NJ) supplemented with 0.2% Tween 20 (HIBTW). E. coli strains were cultivated in Luria-Bertani broth (33). Kanamycin was added at a concentration of 20 μg/ml and spectinomycin was added at a concentration of 100 μg/ml for C. diphtheriae. Ampicillin was added at a concentration of 100 μg/ml for E. coli. Medium was deferrated by adding 15 g/liter Chelex-100 (Bio-Rad, Hercules, CA) and mixing it for 2 h followed by filter sterilization.
Construction of ΔdtxR.
First, to eliminate PstI sites in the vector portion of the dtxR-containing plasmid pMS297, the 1,533-bp EcoRI-to-BamHI fragment which contains dtxR (54) was ligated to pBluescript II KS (Stratagene, La Jolla, CA) digested with EcoRI and BamHI to construct pMS297.1. Next pMS297.1 was used as a template in PCR with the primers M13-20 and dtxR1CBE to generate a 671-bp product with a unique PstI recognition site introduced by the primer dtxR1CBE as well as the site for BamHI. The PCR fragment was digested with PstI and BamHI and ligated to the 3,635-bp PstI-to-BamHI fragment of pMS297.1 to construct pCB300. The plasmid pCB300 contains a deletion within dtxR, but the reading frame is shifted +1 from the original dtxR reading frame. To return the gene to its original reading frame, pCB300 was digested with PstI followed by treatment with T4 DNA polymerase to remove the 3′ single-stranded nucleotide overhangs, resulting in the deletion of 4 bp. The molecule was religated to construct pCB300IN, and DNA sequencing was used to confirm the expected DNA sequence within the region encoding the deleted form of dtxR. The plasmid pCB300IN contains a deletion within dtxR that maintains the original reading frame. The restriction enzymes EcoRI and XbaI were used to digest pCB300IN, and the 1,284-bp fragment containing the deleted version of dtxR was ligated to the identically digested vector pK19mobsacB (48) to produce pCB301IN. The plasmid pCB301IN contains an RP4 origin of transfer and was transformed into RP4-mobilizing E. coli S17-1 (58).
Mating of E. coli and C. diphtheriae was performed as described in reference 61 with slight modifications. S17-1/pCB303IN and C. diphtheriae C7(β) were grown overnight, and the cultures were concentrated 10-fold in sterile medium. One-hundred-microliter aliquots of each concentrated sample were spread together on heart infusion agar plates and incubated at 30°C for 16 h. Corynebacterial cointegrates were isolated by plating bacteria scraped from the conjugation plate on heart infusion agar plates supplemented with 30 μg/ml nalidixic acid and 10 μg/ml kanamycin. To resolve the cointegrates, single kanamycin-resistant colonies were picked and grown overnight at 37°C in PGT plus 50 μg/ml ethylenediamine-N,N-diacetic acid (EDDA), and then 100 μl of the overnight culture was plated on heart infusion agar plus 15% sucrose and 10 μg/ml EDDA. The agar plates were incubated at 37°C overnight, and sucrose-resistant colonies were screened for sensitivity to kanamycin (to confirm loss of the integrated plasmid). Finally, the primers dtxRcr1 and dtxRA3 were used in PCR to screen for the subset of colonies in which loss of the integrated plasmid was accompanied by substitution of a deleted copy of dtxR for the wild-type dtxR allele in the chromosome. The nucleotide sequence of the deleted copy of dtxR (ΔdtxR) in the chromosome of C7(β) was confirmed by DNA sequencing.
Growth rate determinations.
The growth rates for selected strains of C. diphtheriae were determined in PGT medium, PGT medium plus 10 mM FeCl3, and HIBTW medium. Strains were inoculated to an absorbance at 600 nm (A600) of approximately 0.1. The cultures were incubated at 37°C with shaking, and the A600 was monitored for 8 to 15 h or until the cells entered stationary phase. The growth rate of each strain during log phase was determined and compared to that of wild-type C7(β).
Measurement of diphtheria toxin and siderophore.
Diphtheria toxin in the supernatant of cultures was determined by using the enzyme-linked immunosorbent assay described previously (40). Siderophore was detected in culture supernatants using a slightly modified version of the CAS assay described previously (40). The CAS assay was adapted so that reactions could be performed in 96-well microtiter plates by altering the total volume of each sample from 1 ml to 0.2 ml. The absorbances of the reaction mixtures were then read in a microplate reader at a wavelength of 630 nm.
Resistance to killing by hydrogen peroxide.
Resistance to killing by hydrogen peroxide, H2O2, was assayed as described previously (40). Briefly, in the zone of inhibition assay we measured the diameters of the zones of inhibition of bacterial growth produced when 20 μl of 1 M H2O2 was applied to 0.6-cm-diameter paper disks in the centers of plates containing heart infusion agar and lawns of various strains of C. diphtheriae.
In the percent killing assay cultures were grown in PGT until the absorbance of the culture measured at 600 nm was between 0.5 and 0.8, at which time H2O2 was added to the growth medium at a final concentration of 50 mM and the cultures were then incubated for an additional 30 min. Viable counts from each culture were then determined by plating dilutions of the culture onto heart infusion agar.
Construction of reporter plasmids and β-galactosidase assays.
The expression vector pSPZ that was used to determine β-galactosidase activity encoded by a lacZ reporter gene in C. diphtheriae was constructed by ligating the 1.2-kb FspI-to-NsiI fragment of pJKS1 (40) to the 7.7-kb NsiI to NruI fragment of pKMZ (Y. Qian and R. K. Holmes, unpublished data). This fragment of pKMZ contained transcriptional terminators and the promoterless lacZ gene from pCM502 (50) as well as the stable replication origin of pNG2 (57). The plasmid pSPZ encodes resistance to spectinomycin. To construct the promoter fusion plasmids used in Fig. 4, the following DNA fragments were cloned into either pSPZ (for assays in C. diphtheriae) or pQF50 (for assays in E. coli): dtxRUP, generated by PCR amplifying a region containing the 5′ end of dtxR and a portion of sigB with the primers dtxRPE1 and dtxRH3 (the sequences of all primers are shown in Table 1); dtxRU1, generated by PCR amplifying the region upstream of dtxR with the primers dtxR1 and dtxRPE1; dtxRU2, the portion of dtxRU1 downstream of the NcoI recognition site; dtxRU3, the portion of dtxRU1 upstream of the NcoI recognition site; dtxRU4, generated by PCR amplifying the region upstream of dtxR with the primers dtxR2 and dtxRPE1; sigBUP, generated by amplifying the region upstream of sigB with the primers sigBPEBam and sigBXhoI; and galEUP, generated by amplifying the region upstream of galE with the primers MCS3 and MS5. To ensure that no nucleotide changes had been introduced during cloning and PCR amplification, DNA sequencing was used to confirm the correct sequence of each fragment.
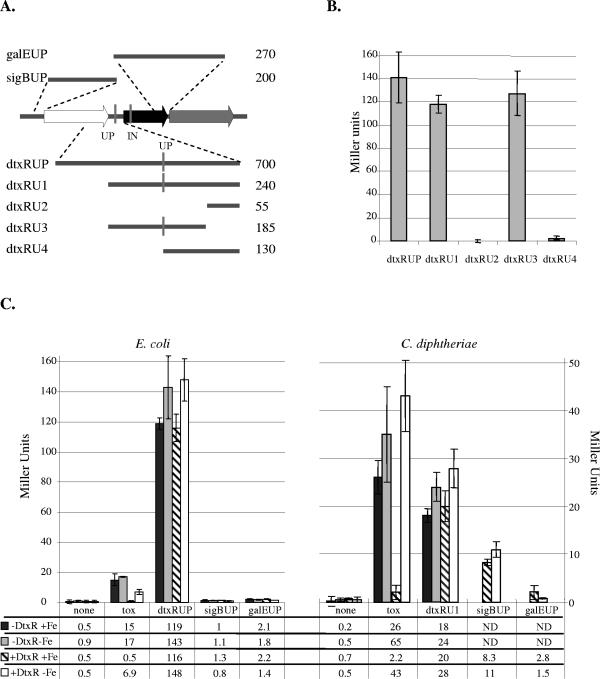
FIG. 4.
A. Maps of the regions from the dtxR locus assayed for promoter activity are shown. The genes and transposon insertions are indicated as in Fig. 1. The name of the DNA fragment is on the left, and the size in base pairs is indicated on the right. B. The promoter activity in E. coli (strain DH5α) of each dtxR promoter fragment listed at the bottom of the chart is indicated. C. The activity of each promoter fragment listed at the bottom of the chart is compared in E. coli (strain DH5α) and in C. diphtheriae [strains C7(β) and C7(β)ΔdtxR]. The DtxR expression phenotype of the host strain as well as the presence or absence of iron (Fe) in the assay medium is shown on the bottom left of each chart. All values are shown in Miller units, and ND indicates no data. Note the different scales on the vertical axes.
TABLE 1.
Primers and probes
| Name | Sequence, 5′ to 3′ | Binding site and/or purpose | Reference or sourcea |
|---|---|---|---|
| Primers for cloning, screening, and primer extension | |||
| dtxR1 | TAAAGCGCATGCTTAGATATGCC | Upstream of dtxR, cloning | * |
| dtxR2 | CTAGCATGCAACAAGAAAACG | Upstream of dtxR, cloning | * |
| dtxRA3 | CAGACACTTCCTACGTATCCGGC | Downstream of dtxR, RT-PCR | * |
| dtxR1CBE | GCCTGCAGCGCAAAGTACGC | 3′ end of dtxR, cloning | * |
| dtxRcr1 | GTCGATACCACCGAGATGTA | 5′ end of dtxR, RT-PCR | 39 |
| dtxRH3 | GAGCAGGTAAACAAGCTTTCTCG | Middle of sigB, cloning (A, Fig. 1) | * |
| dtxRPE1 | GCAAGTAAAGCTTTGTGGTATCG | 5′ end of dtxR, cloning | * |
| dtxRPE2 | CCTTATAGTGCCATGGAGACTG | Upstream of dtxR, primer extension | * |
| kntsprev | GGTGATCAATTCCTATCAATC | Upstream of dtxR, primer extension | * |
| lacZ2 | GGAAGCAGCCCAGTAGTAGGTTG | Near polylinker in pSPZ and pQF50, cloning | * |
| M13-20 | GTAAAACGACGGCCAGT | pBluscript vectors, cloning | New England Biolabs (Beverly, MA) |
| MCS3 | AACTCGGCGTAGGCAATT | 3′ end of dtxR, cloning | * |
| MS5 | AGAACAGTCGACCAGACACTTCCTA | 5′ end of galE, cloning | * |
| Qflac | CGGCCAGTGAATCCGAATTCCCTTTC | Near polylinker in pSPZ and pQF50, cloning | * |
| sigBPE3 | CCGCGGTCGACGGTCTCCGTGG | 5′ end of sigB, primer extension | * |
| sigBPEBam | GTGGATGGATCCACGTCGGAAGGGC | 5′ end of sigB, cloning | * |
| sigBXhoI | CGGACTGCGATCTCGAGGAAT | Upstream of sigB, cloning | * |
| Primers and probes for qPCR | |||
| dtxR-Afor | CGTCAATTTGGACTGTCTCG | 3′ end of sigB (B, Fig. 1) | * |
| dtxR-Arev | AAGCAAACGCTTTATTGTGC | 3′ end of sigB (d, Fig. 1) | * |
| dtxR-Aprobe | AGAGTTCGCCAAATTGAACG | 3′ end of sigB (SB, Fig. 1) | * |
| dtxR-Bfor | CCAGCACACAACAGTCTCCA | 5′ end of dtxR (C, Fig. 1) | * |
| dtxR-Brev | CAGATTGTTCCAGACGCTCA | 5′ end of dtxR (c, Fig. 1) | * |
| dtxR-Bprobe | CTCTTCGCGCTAGGATCG | 5′ end of dtxR (DR, Fig. 1) | * |
| galE1for | CACACTATTCGTATCGAAGAACTC | 3′ end of dtxR (F, Fig. 1) | * |
| galE1rev | GGAACAGACACTTCCTACGTATCC | 5′ end of galE (b, Fig. 1) | * |
| galE1probe | AGGCGGCGAAATTAGATGAAAC | Intergenic region between dtxR and galE (DG, Fig. 1) | * |
| galE2for | CATACGGCGAACCCGAAACAGTCCCG | 5′ end of galE (G, Fig. 1) | * |
| galE2rev | CGCAGCAAACCCATATGCATGTGC | 5′ end of galE (a, Fig. 1) | * |
| galE2probe | CTGAAGACGCTCCTACCCAC | 5′ end of galE (GE, Fig. 1) | * |
| sigA RTF | GATGAATTTGACGACGACGA | sigA | * |
| sigA RTR | GAAAGCTCCGCATCTTTACG | sigA | * |
| sigA RTP | CGGAGAAGATGACTCTTCCG | sigA | * |
| toxBF-RT | CGCCCTAAATCTCCTGTTTATGTT | tox | 34 |
| toxBR-RT | GTACCCAAGAACGCCTATGGAA | tox | 34 |
| toxB probe | TTCACAGAAGCAGCTCGGAGAAAA TTCATTC | tox | 34 |
*, this work.
In E. coli, promoter activity was measured in low-iron medium consisting of deferrated Luria-Bertani medium plus 100 μM each of MgCl2, MnCl2, ZnCl2, and CaCl2 and in high-iron medium consisting of low-iron medium plus 10 μM FeCl3. In C. diphtheriae, promoter activity was measured in low-iron medium consisting of deferrated PGT and high-iron medium consisting of PGT plus 10 μM FeCl3. Assays were performed on three overnight cultures on a single day, and independent experiments were performed on at least three different days. Units of β-galactosidase activity were determined using the method of Miller (33) in E. coli, and in C. diphtheriae the Miller method was altered slightly by increasing the concentration of sodium dodecyl sulfate (SDS) by 100-fold to permeabilize the cells.
Isolation of RNA.
C. diphtheriae cultures were pelleted by centrifugation and resuspended in acidified phenol (pH 4.3). The samples were then transferred to screw-cap tubes containing 0.1-mm glass beads and diethyl pyrocarbonate-treated water. Each tube was processed in a Minibeater (BioSpec, Bartlesville, OK) for 1 min at 4°C at full speed. The bead-beating process was repeated three times with the samples being placed on ice for 1 min between bead-beater cycles. Samples were next spun in a microcentrifuge at full speed for 8 min, and the top phase was removed to a new tube. The samples were extracted three times with equal volumes of acidified phenol and chloroform. The RNA was precipitated with 3 volumes of 95% ethanol and 0.1 volume of 3 M sodium acetate, pH 4.8. After being washed with 75% ethanol, the RNA was resuspended in diethyl pyrocarbonate-treated water and treated with RQ1 DNase (Promega, Madison, WI) per the manufacturer's instructions for 4 h at 37°C. Following DNase treatment the RNA was once again ethanol precipitated as described above.
RNase protection and primer extension assays.
RNase protection assays were performed using the RPAIII RNase protection kit (Ambion, Austin, TX) per the manufacturer's instructions. The gene-specific probes for dtxR consisted of single-stranded RNA generated in vitro using T7 RNA polymerase (Fermentas, Hanover, MD) per the manufacturer's protocol. The construction of the plasmids used as templates in the transcription reaction is described below. To construct pBSdtxR1, the 1,400-bp PCR product containing dtxR, generated in PCR with the primers dtxRH3 and dtxRA3, was digested with HindIII and AgeI and ligated to pBluescript II KS (Stratagene, La Jolla, CA) digested with HindIII and XmaI. All plasmids used as transcription templates were derived from pBSdtxR1. To construct pBSdtxR1-1, pBSdtxR1 was digested with BamHI and Tth111I; to construct pBSdtxR1-2, pBSdtxR1 was digested with BamHI and NcoI; and finally to construct pBSdtxR1-3, pBSdtxR1 was digested with BamHI and PstI. The ends of the resulting linear DNA fragments were blunted using T4 DNA polymerase, and each fragment was circularized with T4 ligase to construct the corresponding plasmid. The plasmid pBSdtxR1-3 was digested with NcoI and used as a template in the in vitro transcription reaction with [32P]CTP to generate probe 1 (shown in Fig. 5). The plasmid pBSdtxR1-1 was digested with XmnI and used to generate probe 2, and the plasmid pBSdtxR1-2 was digested with PvuII and used to generate probe 3. Each probe contains a portion of the dtxR sequence as shown in Fig. 5 as well as a 5′ end identical to the polylinker region of the pBluescript II KS vector. Each 32P-labeled probe was purified on a 6% denaturing acrylamide gel before it was used in the RNase protection assay.
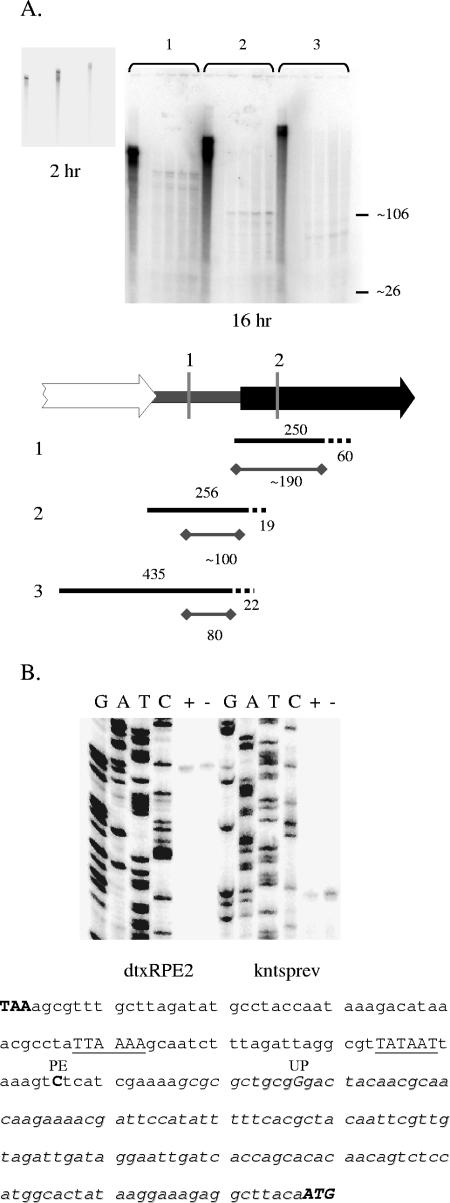
FIG. 5.
A. RNase protection assays. The exposure time is indicated below each image. The positions of the xylene cyanol (∼106 bases) and bromophenol blue (∼26 bases) dyes on the gel are indicated on the right. For each probe (1 to 3) the first lane contains a reaction mixture in which nuclease was not included. The reaction mixtures were loaded in the following order: 2 μl probe plus yeast tRNA, 2 μl probe plus yeast tRNA, 2 μl probe plus 1 μg C7(β) RNA, 2 μl probe plus 2.5 μg C7(β) RNA, 10 μl probe plus 1 μg C7(β) RNA, 10 μl probe plus 2.5 μg C7(β) RNA. Under the gel images is a diagram indicating the input probe sizes and region protected from digestion. The input probe is shown as a black line (the dashed portion of the line is complementary to plasmid vector sequences rather than C. diphtheriae sequences). The region of each probe protected in the RNase assay is shown as a gray line with a diamond at each end. All sizes are shown in bases. B. Primer extension assays. The primer used to generate the sequencing ladder and the primer extension products is indicated. The lanes containing primer extension products are labeled + for template RNA isolated from high-iron cultures and − for template RNA isolated from low-iron cultures. The DNA sequence of the region between sigB and dtxR is shown below the images of the gels. The TAA stop codon of sigB and the ATG start codon of dtxR are capitalized and shown in boldface. The approximate region protected in the RNase assay is italicized. The site of the transposon insertion in C7(β)dtxR-TnUP is labeled UP. The base as the start site of dtxR transcription is labeled PE. The predicted −10 and −35 sequences of P-dtxR are capitalized and underlined.
Primers used in the primer extension assays were labeled with [32P]ATP using T4 polynucleotide kinase (Fermentas) per the manufacturer's protocol. Each primer was purified away from unincorporated nucleotide using a G25 Sepharose spin column. The labeled primers were used in both the sequencing and the primer extension reactions. Sequencing was performed using the fmol DNA cycle sequencing system (Promega, Madison, WI) per the manufacturer's instructions. The primer extension reactions were performed with Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA) per the manufacturer's protocol. Sequencing reactions and primer extension reactions were run on a 6% acrylamide urea denaturing gel as shown in Fig. 5 and 6.
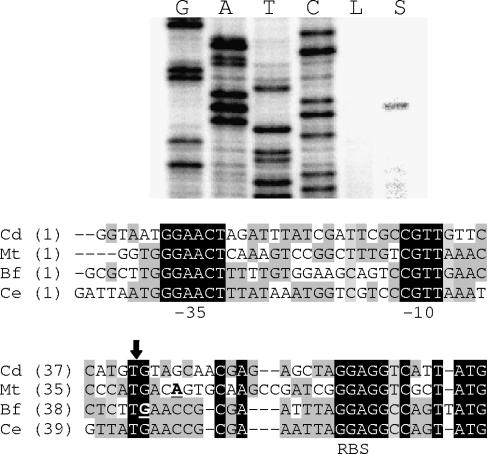
FIG. 6.
The gel image includes the results of primer extension assays. The primer sigBPE3 was used to generate both the sequencing ladder (G, A, T, C) and the primer extension products. The lanes containing primer extension reaction mixtures are labeled L for template RNA isolated from low-iron cultures and S for template RNA isolated from low-iron cultures containing SDS. The DNA sequences of the regions upstream of sigB in C. diphtheriae (Cd), M. tuberculosis (Mt), B. flavum (Bf), and Corynebacterium efficiens (Ce) are shown below the image of the gel. The T residue corresponding to the start of the sigB transcript in C. diphtheriae is indicated by the vertical arrow. The underlined boldface bases in the Mt and Bf sequences indicate the previously reported transcript starts in those bacteria. The sequences corresponding to −10 and −35 sequences used by ECF sigma factors σE and σH are labeled. The proposed ribosome binding site is labeled RBS.
Generation and detection of cDNA.
To generate cDNA, 1 μg of total RNA was used as a template in reaction mixtures containing a primer specific for the gene of interest and Superscript III reverse transcriptase (Invitrogen). The reaction conditions were as recommended by the manufacturer in a total volume of 20 μl. The cDNA was detected using PCR. For standard PCR, 5 μl of the reverse transcriptase reaction product was used as a template with gene-specific primers and Taq DNA polymerase. Reactions were performed using standard PCR conditions in a thermocycler.
For real-time quantitative reverse transcriptase PCR (qRT-PCR) 1 μl of the reverse transcriptase reaction product was used as a template in reactions using Cepheid Omnimix beads, primers specific for the gene of interest, and a probe synthesized by Integrated DNA Technologies, Coralville, IA, that included 5′ 6-carboxyfluorescein and 3′ black hole quencher 1 modifications. The final reaction volume was 25 μl. Reactions were performed in a Cepheid Smartcycler II using the following conditions: 75°C for 75 s and then 50 cycles of 95°C for 5 s and 59°C for 40 s. Fluorescence was monitored and recorded. DNA containing the gene of interest was used to generate a standard curve for each probe using these conditions. The primers and probes used to detect transcripts containing the gene of interest are listed in Table 1.
Stress conditions.
C. diphtheriae cultures were grown in low-iron PGT medium until the absorbance at 600 nm was 4 to 5, at which point the cultures were exposed to the particular stress for 35 min. For acid stress, the pH of the medium was adjusted to 4.5 with HCl; for cold stress, the cultures were shifted to 15°C; for ethanol stress, a final concentration of 5% ethanol was added; for H2O2 stress, a final concentration of 10 mM hydrogen peroxide was added; for heat stress, the cultures were shifted to 45°C; for salt stress, a final concentration of 2.5% NaCl was added; and for SDS stress, a final concentration of 0.05% sodium dodecyl sulfate was added. Following exposure to each stress condition, the cells were pelleted and RNA was harvested as described above.
RESULTS
Construction of C7(β)ΔdtxR.
Previously, we identified two transposon mutants of C. diphtheriae C7(β), designated C7(β)18.5 and C7(β)3B11, both of which show altered expression of dtxR (40). C7(β)3B11 (hereafter referred to as the dtxR-TnUP strain) has a transposon inserted 121 bp upstream of the dtxR ATG start codon and produces threefold-less DtxR than C7(β) does. C7(β)18.5 (hereafter referred to as the dtxR-TnIN strain) has a transposon inserted within dtxR (Fig. 1) and fails to produce any detectable DtxR. To eliminate possible phenotypic consequences of polar effects caused by these transposon insertions, we constructed C7(β)ΔdtxR, which has an in-frame deletion from bp 220 to bp 481 within dtxR (bp 1 is the A of the ATG start codon). This deletion removes the coding sequence for 87 amino acids from Ala74 through Pro160, corresponding to all of domain 2 and part of domain 3. Western blots of extracts from C7(β)ΔdtxR and C7(β)dtxR-TnIN using polyclonal anti-DtxR144+ (41) showed no immunoreactive DtxR, but DtxR was easily detected in Western blots with extracts from wild-type C7(β) and C7(β)dtxR-TnUP (data not shown).
FIG. 1.
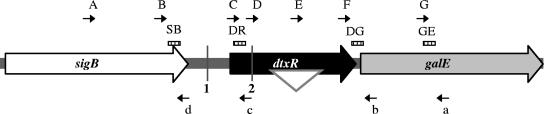
The region of the C. diphtheriae C7(β) chromosome that includes sigB, dtxR, and galE is depicted. The region of dtxR deleted in the ΔdtxR strain is shown as a white inverted triangle within dtxR. The locations of the transposon insertions in C7(β)dtxR-TnUP labeled as 1 and in C7(β)dtxR-TnIN labeled as 2 are shown as vertical gray lines. The positions of primer binding sites are shown as small black arrows labeled A, B, C, D, E, F, G, a, b, c, and d. The fluorescent probes used in qRT-PCR assays are shown as striped rectangles labeled SB, DR, DG, and GE. The sequences of the primers and probes are given in Table 1.
Phenotypes of dtxR mutant strains.
We compared wild-type C7(β) with the dtxR transposon insertion and internal-deletion mutants with respect to growth rate, iron regulation of diphtheria toxin and siderophore production, and susceptibility to H2O2. The growth rates were determined in three different media: HIBTW, PGT, and PGT supplemented with 10 μM FeCl3 (for components of each medium see Materials and Methods). The doubling times of the mutants in each medium were normalized to the doubling time of C7(β) in the same medium. C7(β)dtxR-TnUP had a doubling time of 1.1 to 1.3 times that of C7(β), whereas C7(β)dtxR-TnIN and C7(β)ΔdtxR had normalized doubling times of 1.2 to 1.5 times that of C7(β). In each medium, C7(β) had the shortest doubling time, and C7(β)dtxR-TnUP grew faster than C7(β)dtxR-TnIN and C7(β)ΔdtxR. In PGT plus 10 μM FeCl3, the differences among strains were greatest and C7(β)dtxR-TnIN and C7(β)ΔdtxR exhibited their longest doubling times of 80 to 90 min [1.5 times that of C7(β)]. Following transformation with pJKSdtxR, which determines production of approximately five times the amount of DtxR found in C7(β) (40), the growth rate for all mutant strains was comparable to that of C7(β).
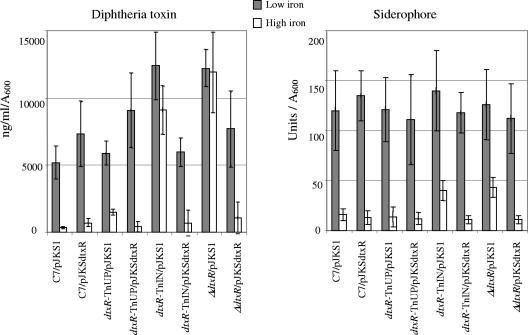
Next, we examined the regulation of diphtheria toxin and siderophore production by iron (Fig. 2). Under high-iron conditions, C7(β)ΔdtxR and C7(β)dtxR-TnIN failed to repress production of diphtheria toxin, whereas C7(β)dtxR-TnUP showed a slight defect in repression compared with C7(β). The amounts of toxin produced under low-iron conditions were consistently greater for C7(β)ΔdtxR and C7(β)dtxR-TnIN than for C7(β)dtxR-TnUP and C7(β), suggesting that growth under low-iron conditions does not result in complete inactivation of wild-type DtxR in the intracellular milieu. The repression ratio (the amount of toxin produced under low-iron conditions divided by the amount produced under high-iron conditions) ranged from 10 to 20 for C7(β), from 4 to 12 for C7(β)dtxR-TnUP, and from 0.5 to 2 for C7(β)ΔdtxR and C7(β)dtxR-TnIN. When pJKSdtxR was present, toxin production by all strains was usually repressed under high-iron conditions to levels comparable with those of C7(β), although some C7(β)ΔdtxR and C7(β)dtxR-TnIN transformants carrying pJKSdtxR continued to produce two to five times more toxin than C7(β) under high-iron conditions. Both C7(β) and dtxR mutant strains produced large and comparable amounts of siderophore under low-iron conditions (Fig. 2). Under high-iron conditions production of siderophore was strongly repressed in C7(β) and C7(β)dtxR-UP. Although C7(β)ΔdtxR and C7(β)dtxR-TnIN produced significantly less siderophore under high-iron conditions than under low-iron conditions, the amounts of siderophore that they produced under high-iron conditions were two to three times greater than those for C7(β) and C7(β)dtxR-UP (Fig. 2). The presence of pJKSdtxR in C7(β)ΔdtxR and C7(β)dtxR-TnIN restored their ability to repress siderophore production under high-iron conditions,. These observations confirm previous findings with the transposon mutants (40), and they provide strong evidence that down-regulation of siderophore production in C. diphtheriae under high-iron growth conditions involves both DtxR-dependent and DtxR-independent mechanisms.
FIG. 2.
The ability of each strain to repress diphtheria toxin production in the presence of iron is shown on the left. The nanograms of diphtheria toxin detected in an ELISA per milliliter of culture per absorbance of the culture at a wavelength of 600 nm is indicated. The ability of each strain to repress production of siderophore in the presence of iron is shown on the right. Siderophore units detected in a CAS assay per absorbance of the culture at a wavelength of 600 nm are indicated. All strains are lysogens of corynebacteriophage β. In both charts the error bars indicate the standard deviations of the values from three independent cultures subjected to the appropriate assay.
The final DtxR-dependent phenotypes that we examined were susceptibility to H2O2 in disk diffusion tests for growth inhibition performed on agar medium and in tests for killing performed in liquid medium. As previously reported (40), the phenotypes of C7(β)dtxR-TnUP and C7(β) were indistinguishable in these tests (data not shown). In contrast, the diameters of the zones of inhibition for C7(β)ΔdtxR and C7(β)dtxR-TnIN carrying the pJKS1 vector plasmid vector were approximately 20% greater than that for C7(β)/pJKS1. In addition the phenotype of C7(β)ΔdtxR was identical to that observed previously for C7(β)dtxR-TnIN in that both mutant strains were more susceptible than wild type to killing by H2O2 in liquid medium (data not shown and reference 40). When pJKSdtxR was present instead of pJKS1, the resistance of the mutant strains to killing by H2O2 was comparable to that of C7(β). Taken together, these findings demonstrated that the phenotype of C7(β)ΔdtxR is very similar to that of C7(β)dtxR-TnIN, and they indicate that possible polar effects of the transposon insertion in C7(β)dtxR-TnIN do not contribute significantly to the DtxR-dependent phenotypes tested in these experiments.
Transcripts containing dtxR.
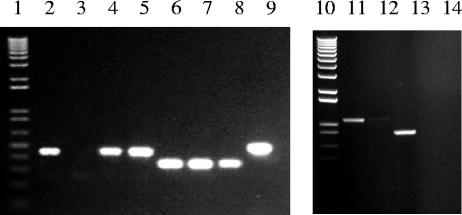
Previously, we reported preliminary evidence for a transcript that includes the 3′ end of sigB and the 5′ end of dtxR (40). Here, we sought to confirm and extend this observation and also to investigate whether the galE gene, which starts only 22 bp downstream from the dtxR translational stop codon, can be cotranscribed with dtxR. Whole-cell RNAs from wild-type and mutant strains of C. diphtheriae were used as templates in reverse transcription reactions to generate cDNAs that initiated within dtxR and galE, respectively. The resulting cDNAs were used as templates to generate double-stranded PCR products that were analyzed by electrophoresis on agarose gels. With primers A and c, which are specific for the 3′ end of sigB and the 5′ end of dtxR, respectively (Fig. 1), a product of the expected size was detected with RNA from strains C7(β), C7(β)dtxR-TnIN, and C7(β)ΔdtxR, confirming the existence of a transcript extending from sigB into dtxR. No transcript was detected with primers A and c when RNA from C7(β)dtxR-TnUP was used as template because the transposon insertion in C7(β)dtxR-TnUP interrupts the chromosomal segment between primers A and c (Fig. 3). When primers specific for a dtxR-galE transcript were used [primers D and b for C7(β)ΔdtxR and primers E and b for all other strains (Fig. 1)], products of the expected sizes were detected in all strains, demonstrating the existence of a transcript that extends from dtxR into galE (Fig. 3). Finally we used primer b (which is identical to the 5′ end of galE) and primer B (which is identical to the 3′ end of sigB) to detect a transcript that includes all three genes shown in Fig. 1 (Fig. 3). Detection of these transcripts was not dependent on whether the cells from which RNA was isolated were starved for iron or not. In addition, control reactions without the reverse transcription step showed no products, indicating that the RNA samples were not contaminated with chromosomal DNA (data not shown and Fig. 3).
FIG. 3.
Products of RT-PCR are shown. Lanes 1 and 10 contain a 1-kb Plus DNA ladder (Invitrogen), and lanes 2 to 9 and 11 to 14 contain the products of the RT-PCR performed with template RNA isolated from the following strains and the primers shown in Fig. 1: lane 2, C7(β), primers A and c; lane 3, C7(β)dtxR-TnUP, primers A and c; lane 4, C7(β)dtxR-TnIN primers A and c; lane 5, C7(β)ΔdtxR, primers A and c; lane 6, C7(β), primers E and b; lane 7, C7(β)dtxR-TnIN, primers E and b; lane 8, C7(β)dtxR-TnUP, primers E and b; lane 9, C7(β)ΔdtxR, primers D and b; lane 11, C7(β), primers B and b; lane 12, C7(β), primers B and b with no reverse transcriptase added; lane 13, C7(β)ΔdtxR, primers B and b; lane 14, C7(β)ΔdtxR, primers B and b with no reverse transcriptase added.
We attempted to detect dtxR-containing transcripts by using a dtxR-specific probe in Northern blots. As a control, we used a tox-specific probe and demonstrated a 1.8-kb tox-containing RNA transcript in whole-cell RNA isolated from C7(β) that was not present in RNA from C7(−), a nontoxinogenic strain that does not contain the tox gene encoded by phage β (13). As expected, the tox transcript was detected only when strains were grown under low-iron conditions (data not shown). When we used the dtxR-specific probe in the Northern blots, we sometimes detected a very faint band of approximately 1.7 kb, consistent with a transcript that included dtxR and galE, but we were unable to reproduce this result consistently. The occasional detection of this transcript did not depend on whether the RNA was isolated from iron-starved or iron-replete cells. These findings suggested that dtxR-specific transcripts in cells grown under low-iron or high-iron conditions were much less abundant than tox-specific transcripts in cells grown under low-iron conditions.
Transcriptional fusions.
We began our study of the promoter(s) responsible for transcription of dtxR, sigB, and galE in C. diphtheriae by cloning several DNA fragments that included portions of the sigB-dtxR intergenic region from C7(β) and testing them for promoter activity in both E. coli and C. diphtheriae (Fig. 4). To assay the promoter activity of each DNA fragment, we used the transcriptional fusion vectors pQF50 (11) and pSPZ for assays in E. coli and C. diphtheriae, respectively. The pSPZ vector contains an origin of replication for C. diphtheriae and a promoterless lacZ reporter gene with upstream transcriptional terminators (see Materials and Methods). In both vectors, the DNA fragment of interest was cloned immediately upstream from lacZ, and promoter activity was determined by measuring β-galactosidase activity.
The largest clone, dtxRUP, included the final 465 bp of sigB, the 224-bp intergenic region between dtxR and sigB, and the first 25 bp of dtxR (Fig. 4A). The dtxRUP clone produced approximately 140 units of β-galactosidase activity in E. coli, and the activity of this promoter was not affected by the presence of either DtxR or iron (Fig. 4B and C). Under identical conditions, regulation of the tox promoter by DtxR and iron was confirmed with the positive control plasmid pQFtox (53). We constructed several subclones of the dtxRUP fragment to further localize the functional promoter (Fig. 4A). Subclone dtxRU1 has the same 3′ end as dtxRUP, but its 5′ end is the stop codon for sigB and it includes the entire sigB-dtxR intergenic region. Subclones dtxRU2 and dtxRU3 contain nonoverlapping upstream and downstream fragments of dtxRU1. Subclone dtxRU4 extends from the site of the transposon insertion in C7(β)dtxR-TnUP to the start codon of dtxR. Each of these subclones in pQF50 was tested for promoter activity in E. coli (Fig. 4B). The results localized the active dtxR promoter, P-dtxR, to a region within dtxRU3, demonstrated that dtxRU2 and dtxRU4 have no detectable promoter activity, and indicated that P-dtxR is more than 100 bp upstream from the translational start site of dtxR. We also tested pQF50 clones containing the DNA segments upstream of sigB (sigBUP) and galE (galEUP) (Fig. 4A) and demonstrated that they both lacked promoter activity in E. coli (Fig. 4C).
Finally, we tested clone dtxRU1 in pSPZ for promoter activity in wild-type C. diphtheriae C7(β) and in the C7(β)ΔdtxR mutant under high- and low-iron growth conditions, and we performed similar experiments in C7(β) with the sigBUP and galEUP clones (Fig. 4C). Clone dtxRU1 produced 20 to 30 Miller units of β-galactosidase activity, and its P-dtxR promoter was not regulated by DtxR or iron. In control experiments, however, the P-tox promoter did show the expected pattern of DtxR-dependent repression by iron. As in E. coli, the galEUP clone had no promoter activity. Although the sigBUP clone did not show any detectable promoter activity in E. coli (Fig. 4C), it did exhibit significant promoter activity in C. diphtheriae under both high-iron and low-iron growth conditions (Fig. 4C). We named the promoter contained on the sigBUP clone P-sigB.
Identification of the 5′ ends of transcripts encoding dtxR and sigB.
First, we used RNase protection assays to identify the 5′ end of the transcript that initiates between sigB and dtxR. Three different 32P-labeled single-stranded RNA probes were prepared that were complementary to different regions of the transcript within and upstream of dtxR. These probes were then hybridized with RNA from C7(β) grown under high- or low-iron conditions, and the reaction products were digested with a mixture of RNase A and RNase T1 and run on a 6% denaturing acrylamide gel. A scan of the gel obtained with a phosphoimager (Bio-Rad, Hercules, CA) is shown in Fig. 5A, along with a diagram that aids in the interpretation of the gel. The entire C. diphtheriae-specific region contained in probe 1 was protected from digestion, indicating that transcription initiated upstream of this probe's binding site. On the other hand, the ends of the C. diphtheriae-specific regions contained within probes 2 and 3 were not protected from digestion, indicating that probes 2 and 3 both overlapped the 5′ end of a dtxR-containing transcript. The sizes of the protected RNA fragments indicated that the 5′ end of that dtxR-containing transcript was located near the site of the transposon insertion in strain C7(β)dtxR-TnUP.
To localize the end of the transcript more precisely, we used RNA isolated from C7(β) as template in primer extension assays with primers that were labeled with 32P at their 5′ ends. The resulting DNA products were run on 6% denaturing polyacrylamide gels next to sequencing ladders that were generated with the same primers (Fig. 5B). The results demonstrated that the 5′ end of the dtxR-containing transcript that initiates between sigB and dtxR is the C residue (note that the sequence in the figure is of the noncoding DNA strand) located 142 bp upstream of the dtxR ATG. The same 5′ end was identified using two different primers, and the detection of the primer extension product did not depend on whether the cells from which the RNA was isolated were starved for iron. In addition, we isolated RNA from E. coli cells containing the plasmid pQFdtxRUP (described above) and confirmed that the 5′ end of the dtxR transcript was the same in E. coli as in C. diphtheriae (data not shown). The DNA sequence of the sigB-dtxR intergenic region is also shown in Fig. 5B, and an E. coli σ70-like −10 promoter-like sequence is properly positioned to direct initiation of transcription at the base identified by the primer extension experiments.
We also used primer extension assays to determine the 5′ end of the transcript that initiates upstream from sigB, within the sigBUP DNA fragment shown in Fig. 4A. Using RNA isolated from wild-type C7(β) grown under our standard high-iron and low-iron conditions (PGT medium with or without 10 μM of added FeCl3), we were unable to detect any primer extension products. However, since transcription of sigB can be induced in Mycobacterium by stresses such as exposure to SDS (32), we also exposed C7(β) to SDS, isolated the RNA, and used that RNA in primer extension reactions with a sigB-specific primer. Figure 6 shows that the 5′ end of the sigB-containing transcript is the T residue located 26 bp upstream from the ATG start codon for sigB (as in Fig. 5, the sequencing ladder shown is for the noncoding strand). Figure 6 also compares the sigB promoter region of C. diphtheriae with homologous sigB promoters that are known to be regulated by extracytoplasmic function (ECF) sigma factors in several other actinomycete species. As will be discussed later, the hypothesis that sigB transcription is dependent on a corynebacterial ECF sigma factor is consistent with the observation that the sigBUP fragment has promoter activity only in C. diphtheriae and not in E. coli.
Quantitative analysis of transcripts containing dtxR, sigB, galE, and tox.
We used real-time qRT-PCR to determine the relative amounts of dtxR-, sigB-, galE-, and tox-specific RNA transcripts and to investigate further the transcriptional regulation of these genes. We determined that transcripts specific for sigA (the primary σ70-type sigma factor in C. diphtheriae) represented a constant fraction of total RNA during exponential growth, as reported previously for the closely related species M. tuberculosis (7) and B. lactofermentum (38), and we measured the other specific transcripts relative to the amount of sigA-specific transcripts. We examined tox-containing transcripts to confirm that the growth conditions used were sufficient to achieve transcriptional repression under high-iron conditions and derepression under low-iron conditions for this known DtxR-regulated gene. The positions of the probes and primers used for dtxR transcripts (probe DR, primers C and c), sigB transcripts (probe SB, primers B and d), galE transcripts (probe GE, primers G and a), and transcripts that span the dtxR-galE intergenic region (probe DG, primers F and b) are shown in Fig. 1, and the probes used for sigA and tox transcripts are described in Materials and Methods.
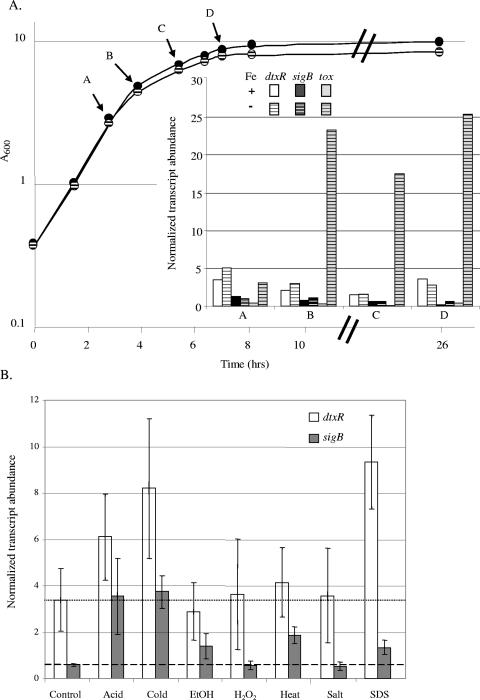
First, we determined the levels of tox, dtxR, and sigB transcripts at several points on the growth curves of C. diphtheriae, under low- and high-iron conditions. The quality of the RNA from cultures incubated longer than 15 h, as judged by smearing on ethidium bromide-stained agarose gels, was poor, so we did not include such samples in our analysis. The levels of each of these specific transcripts were usually significantly lower in samples from overnight cultures than in samples from log-phase and early-stationary-phase cultures (data not shown). The levels of transcripts for sigB and dtxR did not vary by more than 20% during the transition from log-phase growth to stationary phase and were similar under high-iron and low-iron conditions (Fig. 7A). In contrast, tox-containing transcripts did increase dramatically as cells growing under low-iron conditions exited from log phase, but there was no corresponding increase in tox-containing transcripts in cells growing under high-iron conditions. To investigate whether tox transcription was also regulated by growth phase, independently of its DtxR-dependent regulation by iron, we measured tox transcription under high- and low-iron conditions in strain C7(β)ΔdtxR at points on the growth curves corresponding with A to D in Fig. 7A (data not shown). During log-phase growth (point A), the levels of tox transcripts under both high- and low-iron growth conditions were approximately 25 units, comparable to the maximum levels observed during late log phase and early stationary phase under low-iron growth conditions in wild-type C7(β) (points B, C, and D in Fig. 7A). As the C7(β)ΔdtxR cells transitioned into stationary phase, the levels of tox transcripts increased progressively to approximately 65 units under both low- and high-iron conditions (data not shown). In the absence of DtxR-dependent regulation, therefore, the levels of tox transcripts did increase by two- to threefold, by mechanisms not yet determined, as the bacteria exited from log phase and entered stationary phase.
FIG. 7.
(A) The growth curves of C7(β) in PGT (low-iron conditions, striped circles) and PGT plus 10 μM FeCl3 (high-iron conditions, solid circles) media are shown. The abundance of transcripts measured by qRT-PCR with the probes indicated, normalized versus the abundance of sigA transcripts, is shown in the inset graph. The RNA used as a template in the qRT-PCR was isolated from samples of cultures harvested at the time points indicated in the growth curve (A, B, C, and D). Values obtained with RNA from high-iron cultures are shown by the solid bars, and values from low-iron RNA samples are shown by the striped bars. (B) Induction of specific transcripts (defined as normalized transcript abundance under stress conditions/normalized transcript abundance under control conditions) after exposure of C7(β) to indicated stress conditions is shown in the graph. The mean levels of sigB transcripts and dtxR transcripts under the low-iron control conditions are indicated by the dashed and dotted horizontal lines, respectively. The standard deviations between the values for three independent cultures are shown by the error bars. EtOH, ethanol.
Next, we used qRT-PCR to compare the levels of tox-, sigB-, and dtxR-specific transcripts in C. diphtheriae strains C7(β), C7(β)dtxR-TnUP, C7(β)dtxR-TnIN, and C7(β)ΔdtxR under low- and high-iron conditions equivalent to point C on the growth curves in Fig. 7A (Table 2). In wild-type C7(β), expression of tox was strongly repressed by iron (repression ratios of 19 to 37). In contrast, transcript levels for sigB and dtxR were not significantly affected by iron (repression ratios of 0.8 to 1.5), and dtxR-specific transcripts were approximately threefold more abundant than sigB-specific transcripts. In addition, tox-specific transcripts were at least sixfold more abundant than dtxR transcripts under low-iron conditions, and they were threefold less abundant under high-iron conditions. Repression of tox transcription under high-iron conditions in C7(β)dtxR-TnUP was less stringent and more variable (repression ratios of 4 to 30) than it was in wild-type C7(β), consistent with the phenotype described previously for C7(β)dtxR-TnUP and its lower intracellular level of DtxR. In C7(β) dtxR-TnIN and C7(β)ΔdtxR, transcription of tox was not iron regulated (repression ratios of 0.6 to 1.7), and the abundance of tox transcripts under both high- and low-iron conditions was two to three times greater than in C7(β) under low-iron conditions, consistent with incomplete derepression of tox transcription in C7(β) under low-iron conditions. The abundance of sigB transcripts in all three mutants was comparable to that in wild-type C7(β). The average abundance of dtxR transcripts was comparable in C7(β) and C7(β)dtxR-TnIN, trended toward slightly lower values in C7(β)dtxR-TnUP, and was approximately twofold greater in C7(β)ΔdtxR. Although the lower average levels of dtxR transcription in C7(β)dtxR-TnUP were consistent with its observed phenotype of lower DtxR production (40), the differences from wild type were not statistically significant. The dtxR transcripts in C7(β)ΔdtxR differed in structure from those in the other strains because of the internal deletion in dtxR located downstream from the location of the DR probe (Fig. 1), and this difference may have affected stability or production of dtxR transcripts in C7(β)ΔdtxR.
TABLE 2.
Effects of iron on abundance of tox, sigB, and dtxR transcripts in wild-type and dtxR mutant strains of C. diphtheriaea
| Strain | Value for transcript:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
tox
|
sigB
|
dtxR
|
|||||||
| Low Fe | High Fe | Repression ratio | Low Fe | High Fe | Repression ratio | Low Fe | High Fe | Repression ratio | |
| wt | 13 ± 6 | 0.73 ± 0.7 | 19-37 | 0.67 ± 0.4 | 0.58 ± 0.3 | 0.8-1.5 | 2 ± 0.8 | 1.9 ± 0.5 | 0.9-1.5 |
| UP | 41 ± 51 | 7 ± 11 | 4-30 | 0.80 ± 0.5 | 0.83 ± 0.3 | 0.6-1.1 | 1.7 ± 0.3 | 1.6 ± 0.4 | 0.7-1.2 |
| IN | 34 ± 21 | 29 ± 18 | 0.7-1.7 | 1.1 ± 0.5 | 1.0 ± 0.5 | 0.7-1.7 | 1.8 ± 0.8 | 1.7 ± 0.7 | 0.7-1.5 |
| ΔdtxR | 40 ± 10 | 47 ± 21 | 0.6-1.2 | 1.1 ± 0.5 | 0.90 ± 0.5 | 0.5-1.7 | 3.2 ± 0.8 | 4.1 ± 1.1 | 0.5-1.2 |
The strains from which RNA was isolated are listed in the stub: wt, C7(β); UP, C7(β)dtxR-TnUP; IN, dtxR-TnIN; ΔdtxR, C7(β)ΔdtxR. The level of each transcript was normalized to the level of the sigA transcript under high- or low-iron growth conditions (as described in Materials and Methods), and the data shown are the means and standard deviations for samples isolated from three independent cultures on the same day. The standard deviation of replicate reactions performed with the same input RNA was less than 2%, so the error introduced by the qRT-PCR assay itself was negligible. The range is shown for the repression ratios (low-iron/high-iron) among the three independent cultures.
We also used qRT-PCR to investigate further whether galE is cotranscribed with dtxR or is transcribed from an independent promoter that was not detected in our previous experiments (Fig. 4). We used probe DG with primers F and b to detect transcripts that extend from the 3′ end of dtxR into the 5′ end of galE and probe GE with primers G and a to detect transcripts of an internal region of the coding sequence for galE (Fig. 1). If galE can be transcribed both from a promoter in the dtxR-galE intergenic region and also by extension of dtxR-specific transcripts that initiate further upstream, then the transcripts detected by the GE probe should be more abundant than the transcripts detected by the DG probe. The results of our qRT-PCR assays on RNA from three independent cultures grown under both high- and low-iron conditions revealed that transcripts including the 3′ end of dtxR and the 5′ end of galE were slightly more abundant (1.8 ± 0.6 times), rather than less abundant, than those corresponding to the internal sequence in galE. These results are most consistent with the hypothesis that galE is cotranscribed with dtxR but not also transcribed from a galE-only promoter.
Finally, we tested several different stress conditions, some of which are known to affect transcription of sigB in other actinomycetes, for their effect on transcription of dtxR and sigB in C. diphtheriae. We exposed exponentially growing C. diphtheriae to acid, cold, ethanol, H2O2, heat, salt, or SDS for 35 min and then isolated whole-cell RNA. We assayed each sample for sigB- and dtxR-specific transcripts, once again normalizing them to the amounts of sigA-specific transcripts (Fig. 7B). The mean abundance for transcripts of each gene during growth under low-iron control conditions is shown by the first set of data bars and also by the dashed horizontal line for sigB transcripts and the dotted horizontal line for dtxR transcripts. Under these control conditions, dtxR-specific transcripts were four- to fivefold more abundant than sigB-specific transcripts. Significant increases in mean levels of sigB transcripts were observed after exposure to acid (∼5-fold), cold (∼6-fold), ethanol (∼2-fold), heat (∼3-fold), or SDS (∼2-fold), but no significant increases were seen after exposure to H2O2 or salt. In contrast, significant increases in mean levels of dtxR transcripts were observed only after exposure to acid (∼2-fold), cold (2- to 3-fold), or SDS (∼3-fold). For acid and cold stress conditions, the increase in dtxR transcripts could potentially be due to extension of sigB-containing transcripts. In contrast, under SDS stress conditions, the increase in sigB-containing transcripts was not large enough for extension of sigB-containing transcripts to serve as a possible explanation for the observed increase in dtxR-containing transcripts.
DISCUSSION
Until recently, the molecular and genetic tools required to dissect the nuances of transcriptional regulation in the Actinomycetales order of bacteria were not readily available. Among the actinomycetes, several families in the Corynebacteriaceae suborder (i.e., Mycobacteriaceae, Nocardiaceae, Rhodococcus, and Corynebacteriaceae) contain species classified as human pathogens, so understanding transcriptional regulation in this order of bacteria is of interest not only to microbial geneticists but also to investigators committed to understanding and developing new antimicrobials directed against bacterial pathogens.
While characterizing transcription in and around the chromosomal locus for dtxR, we uncovered several new and fascinating features of transcriptional regulation in Corynebacterium diphtheriae. There are 224 bp between dtxR and the gene encoded immediately upstream, sigB. Although we found no evidence of promoter activity within the 150-bp segment immediately upstream of dtxR, we demonstrated an active promoter further upstream in this intergenic region that directs transcription to initiate at a C located 142 bases upstream of the start codon for dtxR. Transcription initiation controlled by this promoter, P-dtxR, both in E. coli and in C. diphtheriae is not affected by the presence of DtxR or the concentration of iron in the growth medium. In contrast, the promoters from C. diphtheriae that have been characterized previously were chosen for study because they exhibit DtxR-dependent negative regulation by iron both in E. coli and in C. diphtheriae (3, 28, 43, 53, 54), and in the case of the hmuO promoter repression of transcription requires iron-activated DtxR while full activation requires the presence of heme (50). Furthermore, the position of P-dtxR, over 150 bases upstream of dtxR, results in a much larger untranslated region (142 bases) than any of the other transcripts from C. diphtheriae for which start sites have been mapped. For example, the transcriptional start sites for P-tox, IRP1, and the hmuO promoter are 40, 35, and 47 bases upstream from their respective translational start codons (29, 50, 56). The significance of the longer untranslated region in the mRNA transcript that begins at P-dtxR is unknown. We also scanned the region between sigB and dtxR for sites with homology to the consensus DtxR binding site, and one site with greater than 60% identity was identified. This site matches the consensus DtxR binding sequence at 12 of the 19 nucleotide positions and is centered 63 nucleotides upstream of the transcriptional start and 206 nucleotides upstream of the translational start of dtxR. The low homology of this site with the consensus DtxR binding site as well as its location far upstream from P-dtxR make it unlikely to be involved in regulation of transcription of P-dtxR, and we found no evidence of DtxR-dependent transcription in this region.
When the DNA sequence in the P-dtxR region was scanned for promoter-like −10 and −35 elements, candidate sequences were easily identified. A −10 sequence identical to the consensus −10 (TATAAT) for the primary σ70-type promoters in E. coli (σ70) and B. subtilis (σA) is shown in Fig. 5. The TTaAaA −35 sequence matches the TTGACA −35 consensus sequence for primary σ70-type promoters at four of six positions. The spacing between the −10 and −35 sequences of P-dtxR is 20 bases, whereas the spacing for the vast majority of σ70-type promoters is 16 to 18 bases. The sequences in the P-dtxR region were also compared to promoter sequences from other actinomycete species. In Corynebacterium glutamicum, an extended −10 consensus sequence, tgngnTA(c/t)aaTgg (the bases shown as lowercase occur in at least 40% of the promoters and bases shown as uppercase occur in more than 70% of promoters), was identified by analyzing 53 different promoters (42). The P-dtxR −10 promoter sequence gGcGtTATAATta (Fig. 5) matches this consensus sequence at 8 of the 11 positions (ignoring the positions at which any base n is acceptable), including all three bases conserved in greater than 70% of the promoters. The −35 region of C. glutamicum promoters is much less conserved than the −35 regions of E. coli and B. subtilis promoters, and the extended −10 region appears to be more important than the −35 sequence for promoter recognition in that species. In M. tuberculosis, the consensus sequence for promoters recognized by the primary sigma factor σA is TTGAC(A/T)-N17-TATA(A/C)T (62). This consensus is nearly identical to those for E. coli σ70 and B. subtilis σA. Although the sequences in and around P-dtxR appear to be most similar to those observed in C. glutamicum promoters, our experiments demonstrated that P-dtxR can function in E. coli to direct transcription initiating at the same base as in C. diphtheriae, notwithstanding the poor match and spacing of its −35 region with the consensus −35 region for E. coli. Whether the atypical spacing between the −10 and −35 regions of P-dtxR is a factor in determining the abundance of the dtxR-specific transcripts that initiate from P-dtxR remains to be determined.
Transcription of dtxR was unaffected by most growth conditions. The exceptions were exposure to acid, cold, or SDS, each of which caused transcription of dtxR to increase by two- to threefold (Fig. 7B). The increase in dtxR transcription following exposure to acid or cold could be due to the five- to sixfold increase in transcription of sigB under those conditions if most or all of the sigB-containing transcripts extended through dtxR. On the other hand, the increase in sigB transcription after exposure to SDS was too small to serve as the primary mechanism for the concomitant increase in dtxR-containing transcripts. To investigate whether transcription of dtxR in the presence of SDS was the result of the activity of P-dtxR or of a separate SDS-responsive promoter in the sigB-dtxR intergenic region, we repeated the primer extension analysis with RNA isolated under SDS stress conditions. The transcription start site identified with the RNA from SDS-stressed cultures was identical to that found under all other conditions tested (data not shown). These results suggest that the dtxR transcripts observed in the presence of SDS arise primarily as the result of transcription initiating at P-dtxR.
Although multiple mechanisms may mediate the increase in transcription of dtxR in response to the various stress conditions that we studied, it is possible that transcription of dtxR and the galE gene located immediately downstream is affected by changes in the cell surface. The role of the galE gene product in C. diphtheriae physiology has not been determined, but it demonstrates homology with UDP-galactose 4-epimerases of other bacteria. GalE enzymes catalyze the interconversion of UDP-galactose and UDP-glucose. In other bacterial species, UDP-galactose and UDP-glucose are substrates for the production of exopolysaccharides on the surface of the cell (26, 63). Since it is possible that the galE gene product is required to synthesize a cell surface polysaccharide, it is reasonable to speculate that transcription of galE as part of a polycistronic message controlled by P-dtxR or P-sigB or both might be responsive to changes in the cell surface.
In addition to the dtxR-containing transcript that initiates from P-dtxR, our analysis of mRNAs in this region demonstrated that a second transcript spans the 3′ end of sigB and the 5′ end of dtxR and that this transcript must initiate at least 800 bases upstream of dtxR. Since it seemed likely that this second dtxR-containing transcript included sigB, we tested the region upstream of sigB for promoter activity. We determined that the promoter located upstream of sigB was active only in C. diphtheriae and not in E. coli. In addition the number of transcripts containing sigB was induced by several environmental conditions, including exposure to acid, cold, heat, ethanol, or SDS. Exposure to salt or hydrogen peroxide had no effect on transcription of sigB (Fig. 7B). These findings suggested that the promoter upstream of sigB may be recognized by an ECF sigma factor, similar to ones that have been shown to coordinate gene transcription in response to various stress conditions in many other bacterial species (20).
In the C. diphtheriae genome there are nine annotated sigma factor genes, all of which are in the σ70 family (4). Using the numbering scheme from the published genome, the primary sigma factor σA is encoded by DIP1406 and σB is encoded by DIP1413. This gene arrangement with sigA and sigB encoded near one another is also seen in other actinomycetes such as M. tuberculosis. In M. tuberculosis, sigB transcription is controlled by two different ECF sigma factors, σE and σH (31, 32). These two ECF sigma factors of M. tuberculosis use the same promoter sequences upstream of sigB, but they differ with respect to the conditions under which they are active. Specifically, the consensus sequence for binding of RNA polymerase holoenzyme including σE is a −35 sequence of GG(A/G)(A/C)C and a −10 sequence of (G/C)GTTG (32), and the sequence recognized by RNA polymerase holoenzyme including σH is a −35 sequence equal to (G/C)GGAAC and a −10 sequence of (G/C)GTT(G/C) (31). σE is required for the increase in sigB expression following exposure to SDS (32), while σH is required for the increase in sigB expression following exposure to heat (31). C. diphtheriae encodes homologues of σE (DIP0994) and σH (DIP0709), and as shown in Fig. 6, the P-sigB promoter contains sequences that are homologous to the consensus sequences determined for these two ECF sigma factors.
Transcription dependent on σH has been demonstrated in C. glutamicum, a species used to produce amino acids on an industrial scale. In C. glutamicum, transcription of four genes whose products have a role in the function of the ATP-dependent protease Clp was induced by heat shock. The promoters for these genes all contain the consensus −10 and −35 sequences (shown in Fig. 6) for σE and σH. Finally, inactivation of sigH prevented induction of transcription of all four genes by heat shock (10). These findings provide clear evidence for ECF sigma factor function in Corynebacterium and support the theory that sigB in C. diphtheriae is likely transcribed by an RNA polymerase holoenzyme that contains either σE or σH.
Transcription of sigB has also been investigated in the actinomycete genus Brevibacterium, which is closely related to Corynebacterium. In both B. flavum and B. lactofermentum, the start site for sigB transcription has been mapped to a G located 24 bp upstream of the ATG, and these start sites are nearly identical to the site we identified in C. diphtheriae (Fig. 6) (17, 38). The DNA sequences upstream of sigB in the closely related species B. flavum, B. lactofermentum, and C. glutamicum are identical, so only the B. flavum sequence was included in Fig. 6. In B. flavum, transcription of sigB was shown to be increased after acid stress, cold shock, and exposure to ethanol, all of which also increased transcription of sigB in C. diphtheriae. In addition, inactivation of sigB in B. flavum caused increased sensitivity to acid, salt, ethanol, heat, and cold stress, indicating that σB likely has a role in the environmental stress response (18). This observation correlates with our findings that the transcription of sigB is increased in response to these environmental stress conditions (Fig. 7B).
All of our data indicate that dtxR and galE are encoded on the same transcript in C. diphtheriae, and we could find no evidence of a galE-only promoter (i.e., a promoter in the intergenic region between dtxR and galE). These findings are in agreement with observations in B. lactofermentum, where dmdR (the dtxR homologue) and galE are cotranscribed (36). The gene arrangement at the dtxR locus is conserved among the members of the genus Corynebacterium for which genome sequencing has been completed (24, 35). Also consistent with our observation that there is a transcript that includes the 3′ end of sigB and the 5′ end of dtxR in C. diphtheriae, a transcript that spans the intergenic region between sigB and dmdR was observed in B. lactofermentum (38). Additionally we provide evidence that in C. diphtheriae there is cotranscription of sigB, dtxR, and galE. There is a difference between the gene arrangement at the ideR locus (equivalent to the dtxR locus) found in the genus Mycobacterium and that present in Corynebacterium. In both M. tuberculosis and Mycobacterium bovis, the gene order of sigB and ideR is identical to that seen in Corynebacterium but the gene immediately downstream of ideR is not galE (5, 14). Instead, a gene of unknown function is encoded downstream of ideR on the opposite DNA strand. Therefore, ideR in Mycobacterium does not appear to be cotranscribed with any other downstream gene.
In summary, we have demonstrated that transcriptional regulation of the genes in the C. diphtheriae dtxR locus is a multifaceted process that is affected by several environmental factors. We have mapped and characterized two new promoters from C. diphtheriae, P-dtxR and P-sigB. Of these, P-sigB is the first promoter from C. diphtheriae likely to be recognized by an RNA polymerase holoenzyme that contains an ECF sigma factor. ECF sigma factors direct transcription in response to extracellular signals, and sigB transcript levels were shown to increase in response to acid stress, ethanol exposure, heat shock, cold shock, or exposure to SDS. Transcription initiation at P-dtxR results in a dtxR-containing transcript that includes 142 bases of untranslated mRNA, the role of which remains to be determined. By analyzing the transcripts that include dtxR, we showed that sigB, dtxR, and galE are cotranscribed. Finally, although transcription of dtxR was not dramatically affected by several extracellular signals, we did observe an increase in dtxR-containing transcripts in response to exposure to acid, cold, or SDS, which may indicate that transcription of these genes is responsive to changes in the cell envelope or surface stress.
Acknowledgments
This work was supported in part by grant number AI14107 from the National Institute of Allergy and Infectious Diseases (to R.K.H.) and by a postdoctoral fellowship awarded to D.M.O. under training grant AI07587 from the National Institute of Allergy and Infectious Diseases.
We thank Farrah M. Clemens for designing the fluorescent probe and primers used to detect sigA-containing transcripts and Mark Oram for critical reading of and advice on the manuscript.
REFERENCES
- 1.Barksdale, L. W., and A. M. J. Pappenheimer. 1954. Phage-host relationships in nontoxigenic and toxigenic diphtheria bacilli. J. Bacteriol. 67:220-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boland, C. A., and W. G. Meijer. 2000. The iron dependent regulatory protein IdeR (DtxR) of Rhodococcus equi. FEMS Microbiol. Lett. 191:1-5. [DOI] [PubMed] [Google Scholar]
- 3.Boyd, J., M. N. Oza, and J. R. Murphy. 1990. Molecular cloning and DNA sequence analysis of a diphtheria tox iron-dependent regulatory element (dtxR) from Corynebacterium diphtheriae. Proc. Natl. Acad. Sci. USA 87:5968-5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cerdeno-Tarraga, A. M., A. Efstratiou, L. G. Dover, M. T. Holden, M. Pallen, S. D. Bentley, G. S. Besra, C. Churcher, K. D. James, A. De Zoysa, T. Chillingworth, A. Cronin, L. Dowd, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Moule, M. A. Quail, E. Rabbinowitsch, K. M. Rutherford, N. R. Thomson, L. Unwin, S. Whitehead, B. G. Barrell, and J. Parkhill. 2003. The complete genome sequence and analysis of Corynebacterium diphtheriae NCTC13129. Nucleic Acids Res. 31:6516-6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 6.Doukhan, L., M. Predich, G. Nair, O. Dussurget, I. Mandic-Mulec, S. T. Cole, D. R. Smith, and I. Smith. 1995. Genomic organization of the mycobacterial sigma gene cluster. Gene 165:67-70. [DOI] [PubMed] [Google Scholar]
- 7.Dubnau, E., P. Fontan, R. Manganelli, S. Soares-Appel, and I. Smith. 2002. Mycobacterium tuberculosis genes induced during infection of human macrophages. Infect. Immun. 70:2787-2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dussurget, O., M. Rodriguez, and I. Smith. 1996. An ideR mutant of Mycobacterium smegmatis has derepressed siderophore production and an altered oxidative-stress response. Mol. Microbiol. 22:535-544. [DOI] [PubMed] [Google Scholar]
- 9.Dussurget, O., J. Timm, M. Gomez, B. Gold, S. Yu, S. Z. Sabol, R. K. Holmes, W. R. Jacobs, and I. Smith. 1999. Transcriptional control of the iron-responsive fxbA gene by the mycobacterial regulator IdeR. J. Bacteriol. 181:3402-3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engels, S., J. E. Schweitzer, C. Ludwig, M. Bott, and S. Schaffer. 2004. clpC and clpP1P2 gene expression in Corynebacterium glutamicum is controlled by a regulatory network involving the transcriptional regulators ClgR and HspR as well as the ECF sigma factor σH. Mol. Microbiol. 52:285-302. [DOI] [PubMed] [Google Scholar]
- 11.Farinha, M. A., and A. M. Kropinski. 1990. Construction of broad-host-range plasmid vectors for easy visible selection and analysis of promoters. J. Bacteriol. 172:3496-3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feese, M. D., E. Pohl, R. K. Holmes, and W. G. J. Hol. 2001. Iron-dependent regulators, p. 850-863. In A. Messerschmidt, R. Huber, R. Poulos, and K. Wieghardt (ed.), Handbook of metalloproteins. John Wiley & Sons, Ltd., Chichester, United Kingdom.
- 13.Freeman, V. J. 1951. Studies on the virulence of bacteriophage-infected strains of Corynebacterium diphtheriae. J. Bacteriol. 61:675-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garnier, T., K. Eiglmeier, J. C. Camus, N. Medina, H. Mansoor, M. Pryor, S. Duthoy, S. Grondin, C. Lacroix, C. Monsempe, S. Simon, B. Harris, R. Atkin, J. Doggett, R. Mayes, L. Keating, P. R. Wheeler, J. Parkhill, B. G. Barrell, S. T. Cole, S. V. Gordon, and R. G. Hewinson. 2003. The complete genome sequence of Mycobacterium bovis. Proc. Natl. Acad. Sci. USA 100:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gold, B., G. M. Rodriguez, S. A. Marras, M. Pentecost, and I. Smith. 2001. The Mycobacterium tuberculosis IdeR is a dual functional regulator that controls transcription of genes involved in iron acquisition, iron storage and survival in macrophages. Mol. Microbiol. 42:851-865. [DOI] [PubMed] [Google Scholar]
- 16.Gunter-Seeboth, K., and T. Schupp. 1995. Cloning and sequence analysis of the Corynebacterium diphtheriae dtxR homologue from Streptomyces lividans and S. pilosus encoding a putative iron repressor protein. Gene 166:117-119. [DOI] [PubMed] [Google Scholar]
- 17.Halgasova, N., G. Bukovska, J. Timko, and J. Kormanec. 2001. Cloning and transcriptional characterization of two sigma factor genes, sigA and sigB, from Brevibacterium flavum. Curr. Microbiol. 43:249-254. [DOI] [PubMed] [Google Scholar]
- 18.Halgasova, N., G. Bukovska, J. Ugorcakova, J. Timko, and J. Kormanec. 2002. The Brevibacterium flavum sigma factor SigB has a role in the environmental stress response. FEMS Microbiol. Lett. 216:77-84. [DOI] [PubMed] [Google Scholar]
- 18a.Hantke, K. 2001. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4:172-177. [DOI] [PubMed] [Google Scholar]
- 19.Hardham, J. M., L. V. Stamm, S. F. Porcella, J. G. Frye, N. Y. Barnes, J. K. Howell, S. L. Mueller, J. D. Radolf, G. M. Weinstock, and S. J. Norris. 1997. Identification and transcriptional analysis of a Treponema pallidum operon encoding a putative ABC transport system, an iron-activated repressor protein homolog, and a glycolytic pathway enzyme homolog. Gene 197:47-64. [DOI] [PubMed] [Google Scholar]
- 20.Helmann, J. D. 2002. The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 46:47-110. [DOI] [PubMed] [Google Scholar]
- 21.Hill, P. J., A. Cockayne, P. Landers, J. A. Morrissey, C. M. Sims, and P. Williams. 1998. SirR, a novel iron-dependent repressor in Staphylococcus epidermidis. Infect. Immun. 66:4123-4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmes, R. K. 2000. Biology and molecular epidemiology of diphtheria toxin and the tox gene. J. Infect. Dis. 181:S156-S167. [DOI] [PubMed] [Google Scholar]
- 23.Hu, Y., and A. R. M. Coates. 1999. Transcription of two sigma 70 homologue genes, sigA and sigB, in stationary-phase Mycobacterium tuberculosis. J. Bacteriol. 181:469-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalinowski, J., B. Bathe, D. Bartels, N. Bischoff, M. Bott, A. Burkovski, N. Dusch, L. Eggeling, B. J. Eikmanns, L. Gaigalat, A. Goesmann, M. Hartmann, K. Huthmacher, R. Kramer, B. Linke, A. C. McHardy, F. Meyer, B. Mockel, W. Pfefferle, A. Puhler, D. A. Rey, C. Ruckert, O. Rupp, H. Sahm, V. F. Wendisch, I. Wiegrabe, and A. Tauch. 2003. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J. Biotechnol. 104:5-25. [DOI] [PubMed] [Google Scholar]
- 25.Kitten, T., C. L. Munro, S. M. Michalek, and F. L. Macrina. 2000. Genetic characterization of a Streptococcus mutans LraI family operon and role in virulence. Infect. Immun. 68:4441-4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kleerebezem, M., R. van Kranenburg, R. Tuinier, I. C. Boels, P. Zoon, E. Looijesteijn, J. Hugenholtz, and W. M. de Vos. 1999. Exopolysaccharides produced by Lactococcus lactis: from genetic engineering to improved rheological properties? Antonie Leeuwenhoek 76:357-365. [PubMed] [Google Scholar]
- 27.Kunkle, C. A., and M. P. Schmitt. 2003. Analysis of the Corynebacterium diphtheriae DtxR regulon: identification of a putative siderophore synthesis and transport system that is similar to the Yersinia high-pathogenicity island-encoded yersiniabactin synthesis and uptake system. J. Bacteriol. 185:6826-6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, J. H., T. Wang, K. Ault, J. Liu, M. P. Schmitt, and R. K. Holmes. 1997. Identification and characterization of three new promoter/operators from Corynebacterium diphtheriae that are regulated by the diphtheria toxin repressor (DtxR) and iron. Infect. Immun. 65:4273-4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leong, D., and J. R. Murphy. 1985. Characterization of the diphtheria tox transcript in Corynebacterium diphtheriae and Escherichia coli. J. Bacteriol. 163:1114-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Love, J. F., J. C. vanderSpek, V. Marin, L. Guerrero, T. M. Logan, and J. R. Murphy. 2004. Genetic and biophysical studies of diphtheria toxin repressor (DtxR) and the hyperactive mutant DtxR(E175K) support a multistep model of activation. Proc. Natl. Acad. Sci. USA 101:2506-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manganelli, R., M. I. Voskuil, G. K. Schoolnik, E. Dubnau, M. Gomez, and I. Smith. 2002. Role of the extracytoplasmic-function σ factor σH in Mycobacterium tuberculosis global gene expression. Mol. Microbiol. 45:365-374. [DOI] [PubMed] [Google Scholar]
- 32.Manganelli, R., M. I. Voskuil, G. K. Schoolnik, and I. Smith. 2001. The Mycobacterium tuberculosis ECF sigma factor σE: role in global gene expression and survival in macrophages. Mol. Microbiol. 41:423-437. [DOI] [PubMed] [Google Scholar]
- 33.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 34.Mothershed, E. A., P. K. Cassiday, K. Pierson, L. W. Mayer, and T. Popovic. 2002. Development of a real-time fluorescence PCR assay for rapid detection of the diphtheria toxin gene. J. Clin. Microbiol. 40:4713-4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishio, Y., Y. Nakamura, Y. Kawarabayasi, Y. Usuda, E. Kimura, S. Sugimoto, K. Matsui, A. Yamagishi, H. Kikuchi, K. Ikeo, and T. Gojobori. 2003. Comparative complete genome sequence analysis of the amino acid replacements responsible for the thermostability of Corynebacterium efficiens. Genome Res. 13:1572-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oguiza, J. A., A. T. Marcos, M. Malumbres, and J. F. Martin. 1996. The galE gene encoding the UDP-galactose 4-epimerase of Brevibacterium lactofermentum is coupled transcriptionally to the dmdR gene. Gene 177:103-107. [DOI] [PubMed] [Google Scholar]
- 37.Oguiza, J. A., A. T. Marcos, M. Malumbres, and J. F. Martin. 1996. Multiple sigma factor genes in Brevibacterium lactofermentum: characterization of sigA and sigB. J. Bacteriol. 178:550-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oguiza, J. A., A. T. Marcos, and J. F. Martin. 1997. Transcriptional analysis of the sigA and sigB genes of Brevibacterium lactofermentum. FEMS Microbiol. Lett. 153:111-117. [DOI] [PubMed] [Google Scholar]
- 39.Oram, D. M., A. Avdalovic, and R. K. Holmes. 2004. Analysis of genes that encode DtxR-like transcriptional regulators in pathogenic and saprophytic corynebacterial species. Infect. Immun. 72:1885-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oram, D. M., A. Avdalovic, and R. K. Holmes. 2002. Construction and characterization of transposon insertion mutations in Corynebacterium diphtheriae that affect expression of the diphtheria toxin repressor (DtxR). J. Bacteriol. 184:5723-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oram, D. M., L. M. Must, J. K. Spinler, E. M. Twiddy, and R. K. Holmes. 2005. Analysis of truncated variants of the iron dependent transcriptional regulators from Corynebacterium diphtheriae and Mycobacterium tuberculosis. FEMS Microbiol. Lett. 243:1-8. [DOI] [PubMed] [Google Scholar]
- 42.Patek, M., J. Nesvera, A. Guyonvarch, O. Reyes, and G. Leblon. 2003. Promoters of Corynebacterium glutamicum. J. Biotechnol. 104:311-323. [DOI] [PubMed] [Google Scholar]
- 43.Qian, Y., J. H. Lee, and R. K. Holmes. 2002. Identification of a DtxR-regulated operon that is essential for siderophore-dependent iron uptake in Corynebacterium diphtheriae. J. Bacteriol. 184:4846-4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qiu, X., E. Pohl, R. K. Holmes, and W. G. Hol. 1996. High-resolution structure of the diphtheria toxin repressor complexed with cobalt and manganese reveals an SH3-like third domain and suggests a possible role of phosphate as co-corepressor. Biochemistry 35:12292-12302. [DOI] [PubMed] [Google Scholar]
- 45.Qiu, X., C. L. Verlinde, S. Zhang, M. P. Schmitt, R. K. Holmes, and W. G. Hol. 1995. Three-dimensional structure of the diphtheria toxin repressor in complex with divalent cation co-repressors. Structure 3:87-100. [DOI] [PubMed] [Google Scholar]
- 46.Rodriguez, G. M., B. Gold, M. Gomez, O. Dussurget, and I. Smith. 1999. Identification and characterization of two divergently transcribed iron regulated genes in Mycobacterium tuberculosis. Tuber. Lung Dis. 79:287-298. [DOI] [PubMed] [Google Scholar]
- 47.Rodriguez, G. M., M. I. Voskuil, B. Gold, G. K. Schoolnik, and I. Smith. 2002. ideR, an essential gene in Mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect. Immun. 70:3371-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schafer, A., J. Kalinowski, and A. Puhler. 1994. Increased fertility of Corynebacterium glutamicum recipients in intergeneric matings with Escherichia coli after stress exposure. Appl. Environ. Microbiol. 60:756-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schiering, N., X. Tao, H. Zeng, J. R. Murphy, G. A. Petsko, and D. Ringe. 1995. Structures of the apo- and the metal ion-activated forms of the diphtheria tox repressor from Corynebacterium diphtheriae. Proc. Natl. Acad. Sci. USA 92:9843-9850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmitt, M. P. 1997. Transcription of the Corynebacterium diphtheriae hmuO gene is regulated by iron and heme. Infect. Immun. 65:4634-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmitt, M. P., and R. K. Holmes. 1993. Analysis of diphtheria toxin repressor-operator interactions and characterization of a mutant repressor with decreased binding activity for divalent metals. Mol. Microbiol. 9:173-181. [DOI] [PubMed] [Google Scholar]
- 52.Schmitt, M. P., and R. K. Holmes. 1991. Characterization of a defective diphtheria toxin repressor (dtxR) allele and analysis of dtxR transcription in wild-type and mutant strains of Corynebacterium diphtheriae. Infect. Immun. 59:3903-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmitt, M. P., and R. K. Holmes. 1994. Cloning, sequence, and footprint analysis of two promoter/operators from Corynebacterium diphtheriae that are regulated by the diphtheria toxin repressor (DtxR) and iron. J. Bacteriol. 176:1141-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmitt, M. P., and R. K. Holmes. 1991. Iron-dependent regulation of diphtheria toxin and siderophore expression by the cloned Corynebacterium diphtheriae repressor gene dtxR in C. diphtheriae C7 strains. Infect. Immun. 59:1899-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schmitt, M. P., M. Predich, L. Doukhan, I. Smith, and R. K. Holmes. 1995. Characterization of an iron-dependent regulatory protein (IdeR) of Mycobacterium tuberculosis as a functional homolog of the diphtheria toxin repressor (DtxR) from Corynebacterium diphtheriae. Infect. Immun. 63:4284-4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmitt, M. P., B. G. Talley, and R. K. Holmes. 1997. Characterization of lipoprotein IRP1 from Corynebacterium diphtheriae, which is regulated by the diphtheria toxin repressor (DtxR) and iron. Infect. Immun. 65:5364-5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Serwold-Davis, T. M., and N. B. Groman. 1986. Mapping and cloning of Corynebacterium diphtheriae plasmid pNG2 and characterization of its relatedness to plasmids from skin coryneforms. Antimicrob. Agents Chemother. 30:69-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering—transposon mutagenesis in Gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 59.Tao, X., J. Boyd, and J. R. Murphy. 1992. Specific binding of the diphtheria tox regulatory element DtxR to the tox operator requires divalent heavy metal ions and a 9-base-pair interrupted palindromic sequence. Proc. Natl. Acad. Sci. USA 89:5897-5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tao, X., and J. R. Murphy. 1992. Binding of the metalloregulatory protein DtxR to the diphtheria tox operator requires a divalent heavy metal ion and protects the palindromic sequence from DNase I digestion. J. Biol. Chem. 267:21761-21764. [PubMed] [Google Scholar]
- 61.Ton-That, H., and O. Schneewind. 2003. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol. Microbiol. 50:1429-1438. [DOI] [PubMed] [Google Scholar]
- 62.Unniraman, S., M. Chatterji, and V. Nagaraja. 2002. DNA gyrase genes in Mycobacterium tuberculosis: a single operon driven by multiple promoters. J. Bacteriol. 184:5449-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Virji, M. 1997. Post-translational modifications of meningococcal pili. Identification of common substituents: glycans and alpha-glycerophosphate—a review. Gene 192:141-147. [DOI] [PubMed] [Google Scholar]