Abstract
Bacterial growth on a surface often involves the production of a polysaccharide-rich extracellular matrix that provides structural support for the formation of biofilm communities. In Salmonella, cellulose is one of the major constituents of the biofilm matrix. Its production is regulated by CsgD and the diguanylate cyclase AdrA that activates cellulose synthesis at a posttranscriptional level. Here, we studied a collection of Escherichia coli isolates, and we found that the ability to produce cellulose is a common trait shared by more than 50% of the tested strains. We investigated the genetic determinants of cellulose production and its role in biofilm formation in the commensal strain E. coli 1094. By contrast with the Salmonella cellulose regulatory cascade, neither CsgD nor AdrA is required in E. coli 1094 to regulate cellulose production. In this strain, an alternative cellulose regulatory pathway is used, which involves the GGDEF domain protein, YedQ. Although AdrA1094 is functional, it is weakly expressed in E. coli 1094 compared to YedQ, which constitutively activates cellulose production under all tested environmental conditions. The study of cellulose regulation in several other E. coli isolates showed that, besides the CsgD/AdrA regulatory pathway, both CsgD-independent/YedQ-dependent and CsgD-independent/YedQ-independent pathways are found, indicating that alternative cellulose pathways are common in E. coli and possibly in other cellulose-producing Enterobacteriaceae.
Bacterial colonization and survival in different ecological niches often involve growth on a surface and formation of multicellular biofilm communities. Whereas bacterial envelope proteins and fimbriae have been shown to play critical roles in this process, the production of an extracellular matrix is also recognized as a key element in determining the mature biofilm architecture (61).
The biofilm matrix is a complex hydrated milieu that contains proteins, DNA, RNA, ions, and polysaccharidic polymers (9, 61, 62). These polymers are very diverse and include components such as β-1,6-N-acetyl-d-glucosamine polymer (PIA in Staphylococcus spp. or PGA in Escherichia coli), colanic acid (E. coli), and alginate-, glucose-, and mannose-rich components (Pseudomonas aeruginosa and Bacillus subtilis), as well as cellulose (Salmonella, E. coli, and Pseudomonas fluorescens) (7, 8, 19, 20, 27, 44, 58, 59, 64, 68). Polysaccharides are often involved in the establishment of productive cell-to-cell contacts that contribute to the formation of biofilms at liquid/solid interfaces, of pellicles at air/liquid interfaces, of cell aggregates and clumps in liquid cultures, and of wrinkled colony morphology on agar plates. Evidence for a structural role of some of these matrix polysaccharides is accumulating, and the regulation of the production of these exopolysaccharides is being actively investigated for different bacteria (9, 33, 55).
Cellulose production is a widespread phenomenon in Enterobacteriaceae, including Salmonella enterica serovar Typhimurium, Salmonella enterica subsp. enterica serovar Enteritidis, E. coli, Citrobacter spp., and Enterobacter spp. (49, 58, 67, 68). In association with the production of curli fibers, cellulose synthesis in Salmonella has been shown to be a primary cause of biofilm formation and to lead to a distinctive phenotype on agar plates, the red, dry, and rough (rdar) morphotype in LB medium at 28°C (58, 68). The genetic dissection of the rdar morphotype in Salmonella showed that both curli fibers and cellulose production are positively regulated by CsgD (AgfD in Salmonella), a transcriptional regulator belonging to the LuxR family (47, 51). The roles of CsgD in these two pathways can be monitored by the absorption of the Congo red dye (CR; indicative of curli and cellulose production) or the fixation of calcofluor (CF; indicative of cellulose production). In Salmonella serovar Typhimurium and Salmonella serovar Enteritidis, the ability to bind CF depends on the expression of two divergent cellulose synthesis operons, bcsABZC and bcsEFG, that are constitutively expressed (58, 68). The positive regulation of cellulose production by CsgD is mediated through the transcriptional regulation of adrA. AdrA, in turn, activates cellulose synthesis at a posttranscriptional level by controlling the synthesis of cyclic diguanylate (c-di-GMP). In Gluconacetobacter xylinus, it has been suggested that c-di-GMP binds to BcsB, promoting an allosteric change of the protein conformation that leads to its activation (36). Recently, a PilZ domain proposed to be part of the c-di-GMP binding protein has been identified in several bacterial cellulose synthases, including BscA, although direct evidence for c-di-GMP binding is still missing (2). In addition to being subject to several global regulators, such as OmpR, CpxR, H-NS, and IHF, depending on environmental conditions (25, 31), CsgD expression is also controlled by RpoS through the positive action of the transcriptional regulator MlrA (see Fig. 6A) (11, 46).
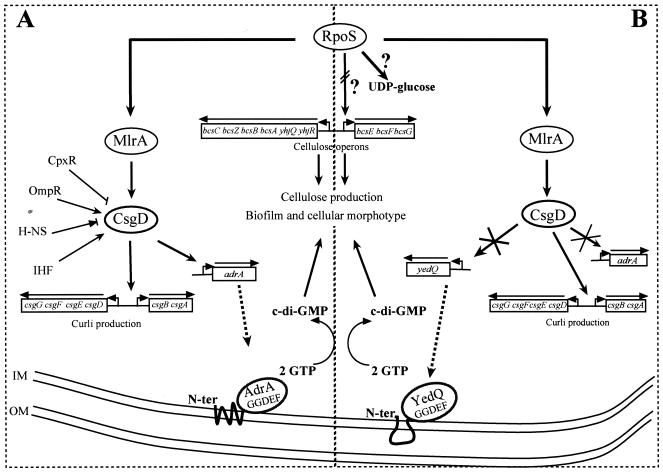
FIG. 6.
Models for cellulose production regulation in Salmonella and E. coli. Schematic regulation of cellulose production via a CsgD/AdrA pathway in Salmonella and some E. coli strains (A, in LB at 28°C) and via a CsgD-independent and YedQ-dependent pathway in E. coli 1094 (B, minimal or LB medium, 30 or 37°C). In both cases, the RpoS sigma factor regulates csgD expression via MlrA. Although CsgD is the central regulator for curli and cellulose production in Salmonella (A), it regulates only curli synthesis in E. coli 1094 (B). In both pathways, the cellulose operons bcsA-C and bcsE-G, as well as the production of c-di-GMP, are required for cellulose synthesis, biofilm formation, and expression of the cellular morphotype. In the case of E. coli 1094, RpoS regulates cellulose production via a yet-uncharacterized pathway. Production of c-di-GMP depends on the diguanylate cyclase activity of the GGDEF domain of the AdrA (A) or the YedQ (B) protein. Both proteins are membrane proteins, but they differ in their N-terminal domains. Both proteins are schematically represented with their N-terminal domains anchored at the membrane (IM, inner membrane; OM, outer membrane).
Owing to the high genetic conservation of the cellulose synthesis and regulatory genes, it has been suggested that the Salmonella cellulose regulatory cascade could be common to all cellulose-producing Enterobacteriaceae, including E. coli (46, 5). However, some deviations from this regulatory network were found in several Salmonella strains. Romling et al. observed that in Salmonella serovar Enteritidis, csgD expression was only partially dependent on RpoS, and in one of the Salmonella serovar Enteritidis strains, cellulose synthesis was uncoupled from csgD expression in LB medium (49). Similarly, in Salmonella serovar Enteritidis, but in minimal ATM medium at 37°C, cellulose production and biofilm formation were not affected by mutations in the regulatory protein RpoS, OmpR, or CsgD (58). Under these conditions, cellulose synthesis has proven to be independent of adrA but dependent on a putative diguanylate cyclase, stm1987 (23). These observations suggested the existence of alternative cellulose regulatory pathways in Salmonella and other eubacteria.
Here, we have investigated cellulose and biofilm formation in a collection of E. coli isolates. We show that cellulose synthesis is the primary cause of biofilm formation and of the expression of multicellular behavior (rdar morphotype) in the commensal E. coli strain 1094. In this strain, cellulose synthesis does not require CsgD or AdrA, which is indicative of an alternative CsgD-independent cellulose regulatory pathway. We identified the genetic determinant involved in this pathway and provided evidence of the existence of alternative cellulose regulatory networks in E. coli. These findings indicate that the regulation of cellulose production in E. coli is more complex than previously recognized.
MATERIALS AND METHODS
Bacterial strains, plasmids, and liquid growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1 (see also Table S1 in the supplemental material). (Supplemental material may also be found at http://www.pasteur.fr/recherche/unites/Ggb/supmet.html.) All experiments were performed in 0.4% glucose M63B1 minimal medium or in LB medium at 30°C or 37°C as specified. All liquid cultures were shaken, agitated cultures. Antibiotics were added when required at the following concentrations: ampicillin, 100 μg ml−1; apramycin, 30 μg ml−1; chloramphenicol, 25 μg ml−1; spectinomycin, 50 μg ml−1; zeocin, 30 μg ml−1; and kanamycin, 50 μg ml−1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| 1094 | E. coli commensal strain isolated from a healthy male adult; phylogenetic group A as detected by PCR | Gift; C. Le Bouguenec |
| 1094bcsC | ΔbcsC::Km Kmr | This study |
| 1094bcsA-C | ΔbcsA-C::Km Kmr | This study |
| 1094bcsEFG | ΔbcsE-G::Km Kmr | This study |
| 1094csgD | ΔcsgD::aadA Specr | This study |
| 1094adrA | ΔadrA::aadA Specr | This study |
| 1094mlrA | ΔmlrA::aac Aprar | This study |
| 1094yedQ | ΔyedQ::zeo Zeocinr | This study |
| 1094rpoS | ΔrpoS::Km Kmr | This study |
| 1094yedQ-uidA | uidA transcriptional fusion placed under the control of the yedQ promoter activity; Zeocinr | This study |
| 1094csgD-pgi | ΔcsgD::aadA; Specr with a transposon inserted into pgi; Kmr | This study |
| 1125 | E. coli commensal strain | Gift; C. Le Bouguenec |
| 1125bcsC | ΔbcsC::aac Aprar | This study |
| 1125csgD | ΔcsgD::aadA Specr | This study |
| 1125yedQ | ΔyedQ::zeo Zeocinr | This study |
| 1125adrA | ΔadrA::aadA Specr | This study |
| 1125yedQ adrA | ΔadrA::aadA Specr ΔyedQ::zeo Zeocinr | This study |
| DSM6601 | E. coli commensal strain | Laboratory collection |
| DSM6601bcsC | ΔbcsC::Km Kmr | This study |
| DSM6601csgD | ΔcsgD::aadA Specr | This study |
| DSM6601yedQ | ΔyedQ::zeo Zeocinr | This study |
| 55989 | E. coli entero-aggregative pathogenic strain | Gift; C. Le Bouguenec |
| 55989bcsC | ΔbcsC::Km Kmr | This study |
| 55989csgD | ΔcsgD::aadA Specr | This study |
| 55989yedQ | ΔyedQ::zeo Zeocinr | This study |
| 55989adrA | ΔadrA::aadA Specr | This study |
| MG1655 | λ− rph-1 | Laboratory collection |
| MG1655csgD | ΔcsgD::aadA Specr | This study |
| PHL818 | MG1655ompR234 malT::Tn10 Tetr | 43 |
| PHL818csgD | ΔcsgD::aadA Specr | This study |
| 3934 | Salmonella serovar Enteritidis clinical isolate | 58 |
| 3934adrA | ΔadrA::Km | 23 |
| S17-1λpir | RP4-2Tc::mu km::Tn7 λpir; Pir-dependent replication | Laboratory collection |
| EcoR collection | E. coli strains listed in Table S1 in the supplemental material | 41 |
| Plasmids | ||
| pSC189 | Ampr Kmr | 13 |
| pKOBEGA | Arabinose-inducible λ Red recombinase expression plasmid [oriR101 repA101(Ts) ParaB-gam-bet-exo Ampr] | 12 |
| pKOBEG | Arabinose-inducible λ Red recombinase expression plasmid [oriR101 repA101(Ts) ParaB-gam-bet-exo Cmr] | 12 |
| pRS415 | Ampr pMB1 ori; promoterless-lacZ plasmid | 57 |
| pRSpyedQ::lacZ | lacZ fusion placed under the transcriptional control of the yedQ promoter from strain 1094 in pRS415; pMB1 ori | This study |
| pRSpadrA::lacZ | lacZ fusion placed under the transcriptional control of the adrA promoter from strain 1094 in pRS415; pMB1 ori | This study |
| pZE12-gfp | Green fluorescent protein placed under the control of the synthetic lacp promoter on the pZE vector used as a cloning vector; Ampr ColE1 ori | Gift; C. C. Guet |
| pZE12-yedQ | yedQ of strain 1094 placed under the control of the synthetic lacp promoter on the pZE vector; Ampr ColE1 ori | This study |
| pZE12-yedQgaaef | yedQgaaef (mutation in the GGDEF domain) placed under the control of the synthetic lacp promoter on the pZE vector; Ampr ColE1 ori | This study |
| pZE12-adrA | adrA of strain 1094 placed under the control of the synthetic lacp promoter on the pZE vector; Ampr ColE1 ori | This study |
| pZE1Pr-adrA | adrA placed under the control of its own promoter on the pZE vector; Ampr ColE1 ori | This study |
| pSTC14 | rpoS placed under the control of the lacp promoter on the pACYC177 vector; Ampr p15A ori | 40 |
Generation of deletion and uidA fusion mutants in E. coli.
The deletion mutants in the different strains of E. coli and the pyedQ-uidA fusion mutant in E. coli 1094 were generated by the λ-red linear DNA gene inactivation method using the three-step PCR procedure as described previously (12, 17; for details, see http://www.pasteur.fr/recherche/unites/Ggb/3SPCRprotocol.html). In the pyedQ-uidA fusion mutant in E. coli 1094, yedQ was replaced by uidA. The primers used to delete the genes presented in this study are listed in Table S2 in the supplemental material. All constructs were checked by PCR with specific primers (see Table S2 in the supplemental material).
Molecular biology procedures.
Standard techniques were used for cloning, DNA analysis, PCR, electroporation, and conjugation, as described previously (54). As the genome sequence for 1094 is currently unavailable, E. coli 1094 genomic sequences were determined from PCR products amplified from the bacterial genome with proofreading polymerase. All DNA sequencing was performed by Genome Express SA, France.
Congo red and calcofluor phenotype assays.
Two microliters of an overnight (o/n) culture grown at 37°C in LB medium (with added antibiotics when needed) was spotted onto LB plates (without NaCl) containing 0.004% CR and 0.002% brilliant blue (referred to as CR plates) or onto LB or M63B1-0.4% glucose (M63B1-glu) plates containing 0.02% CF (Sigma reference F-3543) and 1 mM HEPES (referred to as CF-LB or CF-M63 plates, respectively). The spotted drops were allowed to dry, and the plates were incubated for 24 to 48 h at 30 or 37°C. Red or pink colonies on CR plates indicated the binding of CR. Fluorescence of a colony under UV light indicated the binding of CF on CF-LB or CF-M63B1 plates.
Plasmid construction.
pZE12-yedQ and pZE12-adrA, which encode YedQ and AdrA, respectively, placed under the control of a synthetic lacp promoter (34), were constructed as follows. The yedQ and adrA open reading frames were amplified from the E. coli 1094 chromosome and cloned into pZE12-gfp by replacing the gfp gene with yedQ or adrA. A similar procedure was used to clone the promoter region and the entire adrA gene from the E. coli 1094 chromosome into pZE12-gfp. The resulting plasmid, pZE1Pr-adrA, encodes adrA placed under the control of its own promoter. The second glycine and first glutamic acid from the GGEEF motif of YedQ were both mutated to alanine, leading to a GAAEF motif. First, we amplified from pZE12-yedQ the yedQ sequence between two FspI sites using the primer FspI-GAEEF-5, which contains two mismatches compared to the wild-type sequence of yedQ from E. coli 1094 and FspI-GAEEF-3. The amplified PCR products were then used to replace the wild-type GGEEF motif of YedQ with a GAAEF motif in pZE12-yedQ, leading to plasmid pZE12-yedQgaaef. DNA fragments corresponding to 247 bp preceding the ATG start codon of yedQ or adrA of E. coli strain 1094 were amplified, and the PCR products were cloned into pRS145 to create pRSpyedQ::lacZ and pRSpadrA::lacZ plasmids, which carried a transcriptional fusion between the promoterless reporter gene lacZ and the yedQ or adrA promoter, respectively. The integrity of all the cloned fragments was verified by sequencing. The sequences of the primers used for plasmid construction can be found in Table S2 in the supplemental material.
Biofilm formation assay.
All experiments were performed at least in triplicate in M63B1 minimal medium with 0.4% glucose at 37°C. Sixty-milliliter microfermentors containing a removable glass slide were configured as continuous-flow culture bioreactors with a 40-ml h−1 flow rate, which minimizes planktonic growth of the bacteria as described previously (26; http://www.pasteur.fr/recherche/unites/Ggb/matmet.html). The equivalent of 1 optical density at 600 nm (OD600) unit from o/n bacterial cultures grown in 0.4% glucose M63B1 minimal medium supplemented with the appropriate antibiotics was used to inoculate the microfermentors. The bacteria were then cultivated for 24 h. Pictures of the microfermentors were taken before resuspension of the bacterial biofilm. The biomass was estimated by measuring the OD600 of the biofilm after resuspension in the microfermentor by vigorous shaking (the biofilm is weakly attached and easily detached from the surface) and further vortexing once the resuspended biofilm was transferred in a 15-ml tube.
Mutagenesis.
CF-negative mutants were screened after mutagenesis with the ori6K plasmid pSC189 carrying the kanamycin-resistant Mariner transposon described previously (13). pSC189 was conjugated from strain S17-1 λpir (pSC189) into strain 1094csgD. The loss of CF-binding ability was screened under UV light at 30°C on CF-LB plates containing kanamycin (selection of the transposon insertion) and spectinomycin (selection for 1094csgD).
Multiplex PCR.
Two PCRs were performed for each mutant: one used the primers pairs yhjL.ext5/yhjL.500-3 and cell.2-3/yhjO.ext5, leading to the amplification in the wild-type strain 1094 of 4,266-bp and 3,161-bp fragments, respectively, while the second PCR used the primers pairs cell. 1-3/cell.1-5, yhjO.int3/cell.3-5, and cell.4-3/cell.4-5, leading to the amplification of 3,180-bp, 1,692-bp, and 3,456-bp PCR products, respectively. The primer sequences are listed in Table S2 in the supplemental material.
Arbitrary PCR.
To determine the transposon insertion sites in the studied mutants, an arbitrary PCR was used as described previously (14, 18). This method involves a first round of PCR using a primer specific for the right end of the transposon (IR2) and an arbitrary primer (ARB1 or ARB6). A second PCR is then performed on the product from the first PCR, using a primer specific for the rightmost end of the transposon (IR2-60-5) and a primer that is identical to the 5′ end of the arbitrary primer (ARB2). Arbitrary PCR primer sequences are listed in Table S2 in the supplemental material.
β-Galactosidase assays.
To determine the β-galactosidase enzyme activities of strains carrying transcriptional lacZ fusions, pRSpyedQ-lacZ and pRSpadrA-lacZ, cultures were grown o/n (stationary-phase samples) or until the OD600 reached 0.3 to 0.5 (exponential-phase samples) in LB or M63B1-glu medium at 37°C or 30°C. To avoid formation of clumps in the cultures grown in M63B1 medium, 3 μg ml−1 of purified endo-β-1,4-glucanase from Clostridium thermocellum (30) was added to the medium. Biofilms were grown for 24 h in M63B1- glu plus 50 μg ml−1 ampicillin medium. The enzyme activity was assayed at least in triplicate for each strain under each condition as described previously (37).
Sequence analysis.
Amino acid sequence comparisons were performed using the Clustal program package (28). Homology searches were performed using BLAST 2.0 (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi?). DNA sequence manipulations were done with DNA Strider 1.3 (35).
RESULTS
Calcofluor binding is a widespread phenotype in natural E. coli isolates.
In order to investigate the occurrence of cellulose, or cellulose-like, extracellular material production in E. coli, we analyzed the phenotypes of 87 natural isolates, including the pathogenic and commensal strains from human and animal sources of the EcoR reference collection (41). These strains were grown on agar plates containing CF, a fluorochrome that binds to (1-3)-β- and (1-4)-β-d-glucopyranosides, such as cellulose, chitin, and succinoglycans (65). Although E. coli K-12 strains (such as MG1655 and TG1) do not bind CF, CF binding is a common phenotype in E. coli strains, since 47 (52.8%) of the tested strains displayed some fluorescence on CF agar plates under at least one of the tested conditions (rich or minimal medium; 30 or 37°C) (see Table S1 in the supplemental material). As extracellular cellulose synthesis plays an important role in biofilm formation in Salmonella, we investigated whether the biofilm phenotype could be correlated with the ability to bind CF in E. coli. Biofilm formation was monitored in continuous-flow culture microfermentors for 17 of the most CF-positive strains and 18 of the CF-negative strains. Eleven out of the 17 CF-positive strains showed a significant biofilm phenotype, and 8 out of 18 of the CF-negative strains showed no biofilm biomass after 48 h (see Table S1 in the supplemental material).
These results indicate that the ability to bind CF is a widespread phenotype among natural E. coli isolates. However, this phenotype does not strictly correlate with the capacity to form a biofilm under our experimental conditions.
Cellulose synthesis is necessary for E. coli 1094 biofilm formation.
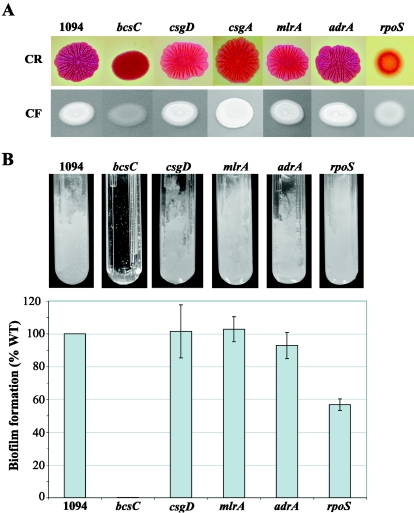
Although E. coli K-12 does not bind CF, it has the homologous genes of the two divergent operons, bcsABZC (formerly yhjONML) and bcsEFG (formerly yhjSTU), which are involved in the synthesis of cellulose in Salmonella (see Fig. 6) (58, 68). These genes could also be amplified by PCR in E. coli strain 1094, an E. coli commensal strain that exhibited strong fluorescence on CF agar plates, a typical rdar morphotype on CR plates, and a strong biofilm phenotype in a microfermentor under all tested conditions (LB; M63B1-glu; 30 or 37°C) (Fig. 1A and B). To test the roles of these genes in the different phenotypes displayed by strain 1094, we deleted bcsC, bcsA-C, or bcsE-G genes in E. coli 1094. These deletions abolished CF binding and led to smooth and red (sar) colonies on CR plates under all tested conditions (Fig. 1A and data not shown). Furthermore, the 1094bcsC mutant lost its ability to form biofilm in microfermentors (Fig. 1B).
FIG. 1.
Cellulose production and biofilm formation in E. coli strain 1094. (A) CR- and CF-binding phenotypes of strains 1094, 1094bcsC, 1094csgD, 1094csgA, 1094mlrA, 1094adrA, and 1094rpoS. Two microliters of an o/n culture was spotted onto CR-LB plates (upper row) and CF-LB plates (lower row) and incubated for 48 h at 30°C (CR) or 37°C (CF). (B) Biofilm formation in microfermentors. The biofilms were grown for 24 h at 37°C in M63B1-glu. The biofilm formation ability of the mutant strains is expressed as a percentage of the 1094 wild-type biofilm, set to 100%. The error bars represent standard errors of the means.
E. coli 1094 also makes small clumps in liquid minimal medium, leading to rapid bacterial aggregation in standing tube cultures. Neither clump formation nor aggregation could be observed with the 1094bcsC mutant (see Fig. S1A in the supplemental material) or when either commercially available cellulase or purified endo-β-1,4-glucanase from C. thermocellum was added to a liquid culture of E. coli 1094 (see Fig. S1B in the supplemental material). Consistent with this, a mature biofilm of strain 1094 was completely disrupted after 16 h of incubation in a buffer containing commercial cellulase (see Fig. S1C in the supplemental material). However, the same treatment had no effect on the biofilm of E. coli K-12 TG1, a strain that forms a strong biofilm due to the expression of the F conjugative pilus and does not produce cellulose (see Fig. S1C in the supplemental material) (26).
Taken together, these results indicate that the extracellular material produced by E. coli 1094 is cellulose and that it depends on the expression of the bcs operons. Furthermore, cellulose production is absolutely required for the expression of mature biofilm and multicellular behavior in E. coli 1094.
CsgD does not regulate cellulose synthesis in E. coli 1094.
In Salmonella, cellulose synthesis generally occurs at 28°C in LB medium and is regulated by CsgD via the MlrA-CsgD-AdrA pathway (hereafter referred to as the CsgD-dependent pathway) (see Fig. 6A). In E. coli 1094, however, cellulose production (determined by CF binding on agar plates) occurs under all tested conditions (30 and 37°C; LB; M63B1-glu), suggesting that the regulation of cellulose synthesis may be different in Salmonella and E. coli 1094. In order to test the contribution of CsgD to cellulose regulation in E. coli 1094, a 1094csgD mutant was grown on CF and CR agar plates. Surprisingly, 1094csgD displayed a wild-type fluorescent phenotype on both LB and minimal-medium agar plates containing CF at both 30°C and 37°C (Fig. 1A), suggesting that CsgD is not involved in the regulation of cellulose synthesis in E. coli 1094. Consistent with this, in E. coli 1094, the deletion of regulatory genes acting either upstream or downstream of csgD (mlrA/yehV and adrA/yaiC, respectively) led to the same phenotype as a csgD mutant on CF plates (Fig. 1A). Moreover, all three mutants exhibited a rough and dry morphotype on CR plates and an almost wild-type biofilm phenotype in microfermentors (Fig. 1A and B). As expected from the loss of curli fiber (which bind CR) production in both 1094csgD and 1094mlrA strains, colonies formed by these strains appeared pink instead of red on CR plates compared to the wild-type 1094 or the 1094adrA strain (Fig. 1A). This phenotype, comparable to the one displayed by a 1094csgA strain (Fig. 1A), is indicative of a reduced ability to bind CR. It also indicates that both the mlrA and csgD genes are expressed and functional in E. coli 1094.
These results demonstrate that mlrA, csgD, and adrA do not regulate cellulose production in E. coli 1094, suggesting the existence of an alternative and CsgD-independent cellulose regulation pathway in this strain.
RpoS participates in both CsgD-dependent and CsgD-independent cellulose pathways.
In Salmonella, RpoS, the main sigma factor of the stationary growth phase, is involved in the CsgD-dependent cellulose regulation pathway via the transcriptional control of MlrA (11). Although neither mlrA, csgD, nor adrA mutants have any impact on cellulose production in E. coli 1094, the deletion of the rpoS gene in E. coli 1094 led to a CF-negative phenotype on LB medium at both 30 and 37°C, as well as a smooth and pink (sap) morphotype on CR plates (Fig. 1A). In minimal medium, E. coli 1094rpoS still displayed a low and inconsistent CF-positive phenotype associated with a reduced but significant biofilm formation phenotype (Fig. 1B). The introduction of a wild-type rpoS allele on the plasmid pSTC14 complemented both the CF-negative phenotype and the sap morphotype (data not shown). Since the pink phenotype is indicative of a lack of curli expression, these results indicate that, in LB medium, RpoS both regulates curli expression and contributes to the CsgD-independent cellulose synthesis regulation pathway. Therefore, although cellulose synthesis in E. coli 1094 is independent of CsgD, it still depends on RpoS.
Screening for E. coli 1094 mutants impaired in cellulose production.
In order to identify the genetic determinants of the production of cellulose in E. coli 1094, we performed a Mariner transposon mutagenesis on 1094csgD. We screened for CF-negative mutants on CF-LB agar plates at 30°C and identified 17 independent nonfluorescent mutants out of 7,000 clones.
To directly identify expected CF-negative mutants due to the insertion of the transposon in the cellulose genes, we performed a multiplex PCR on the two divergent bcsABZC and bcsEFG cellulose operons. A transposon mutant exhibiting a non-wild-type multiplex PCR profile indicates that, in this mutant, the transposon was inserted in one of the cellulose genes. We found that 11 out of 17 independent mutations were indeed located in the bcsABZC operon and 1 in the bcsEFG operon. These mutants displayed a CF-negative phenotype and a white and smooth phenotype on Congo red plates (data not shown). The natures of the five remaining CF-negative mutants carrying a transposon insertion located outside of the cellulose operons were determined by sequencing the regions adjacent to the transposon insertion site. The five mutants corresponded to insertions in four different loci belonging to three functional classes and are presented in Table 2.
TABLE 2.
Phenotypes and molecular analysis of E. coli 1094csgD CF-binding-deficient mutants
| COG group or strain | Insertion sitea | Gene | BLAST description | Phenotypee
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| M63B1-glu + CFb
|
LB + CFb
|
LB + CRc
|
Biofilmd | |||||||
| 30°C | 37°C | 30°C | 37°C | 30°C | 37°C | |||||
| Information storage/processing | 2865100 | rpoS | RNA polymerase sigma factor | +/− | +/− | − | − | sap | sap | + |
| Metabolism | 4232307 | pgi | Phosphoglucose isomerase | +++ | +++ | − | − | sar | sar | ND |
| 1291485 | galU | UDP-glucose-1-P uridylyltransferase | − | − | − | − | sar | sar | − | |
| 1290696 | galU | UDP-glucose-1-P uridylyltransferase | − | − | − | − | sar | sar | − | |
| Cellular processes | 2024925 | yedQ | Unknown | − | − | − | − | cdar | cdar | − |
| E. coli 1094csgD | +++ | +++ | +++ | +++ | rdar | rdar | ++ | |||
Insertion site in bp compared to MG1655 sequence.
Fluorescence under UV light was examined after growth on minimal (M63B1-glu) or rich (LB) agar plates containing CF at 30°C or 37°C for 48 h. The intensity of the fluorescence was estimated visually. The highest fluorescence is expressed as +++; no fluorescence as −.
Morphotype on LB-CR plates without NaCl. cdar, circular, dry, and red.
Biofilm in microfermentor at 37°C in M63B1-glu medium. The biofilm was grown for 24 hours. Strong biofilm, ++; medium biofilm, +; poor biofilm, −; ND, not determined.
Phenotypes compared to 1094csgD levels.
Unsurprisingly, one mutant exhibiting a CF-negative phenotype on LB medium was identified in the rpoS gene. Two insertions were in galU (encoding a glucose-1-P-uridylyltransferase that catalyzes the conversion of glucose-1-P into UDP-glucose [UDP-Glc]) and one in pgi (encoding a phosphoglucose isomerase that catalyzes the interconversion of glucose-6-P and fructose-6-P). These mutants are involved in the synthesis of UDP-Glc, the building block of the cellulose polymer. The pgi mutant led to a CF-binding-deficient mutant on CF-LB plates but displayed a highly CF-positive phenotype on CF-M63-glucose plates. The latter phenotype is probably due to enhanced production of UDP-Glc from the conversion of glucose-6-P into glucose-1-P by the phosphoglucomutase Pgm. This correlates with the production of cellulase-sensitive bacterial aggregates when the pgi insertion mutant is grown in liquid M63B1-glucose culture, suggesting that it overproduces cellulose (see Fig. S1B in the supplemental material).
Finally, one insertion was found in the yedQ gene. This uncharacterized gene (1,695 bp) is predicted to encode a putative protein of 564 amino acids (aa) (64.11 kDa) with an unknown function. Domain analysis using the protein family (Pfam) database indicated the presence of a signal peptide (aa 1 to 45), two potential transmembrane domains (aa 20 to 42 and 358 to 380), and, most interestingly, a carboxy-terminal GGDEF domain (aa 394 to 557), shared by proteins that display proven or suspected c-di-GMP synthase activity, such as AdrA in Salmonella (Fig. 2A) or PleD of Caulobacter crescentus (42, 56). Interestingly, yedQ is highly similar (64.7% amino acid identity) to GcpA (STM1987), a regulator of cellulose synthesis in minimal media at 37°C identified recently in Salmonella serovar Typhimurium (23).
FIG. 2.
YedQ contains a GGDEF domain. (A) Sequence alignment of the GGDEF domains of AdrA-Salty and AdrA-1094 from Salmonella and E. coli 1094, respectively, YedQ-1094 from E. coli 1094, and STM1987-Salty (GcpA) from Salmonella. Stars under the sequences identify residues conserved among all the aligned sequences. The GGDEF domain is boxed, and the GGDEF motif is shaded. The two black arrowheads show the positions of the mutated residues in the YedQgaaef mutant. The alignment was produced using the program Multalin version 5.4.1 (15). # indicates conservation of a charged amino acid; ! indicates conservation of hydrophobic amino acids. (B) Schematic domain structures of YedQ and AdrA proteins from domain analysis using the Pfam database. Black box, signal sequence; white boxes, potential transmembrane domains; dark-gray boxes, GGDEF domains; light-gray box, MASE2 domain. The size of the protein is indicated in residue numbers; the sizes of the GGDEF and MASE2 domains are indicated under the domains by residue numbers in the protein.
YedQ is a key GGDEF regulator of cellulose synthesis in E. coli 1094.
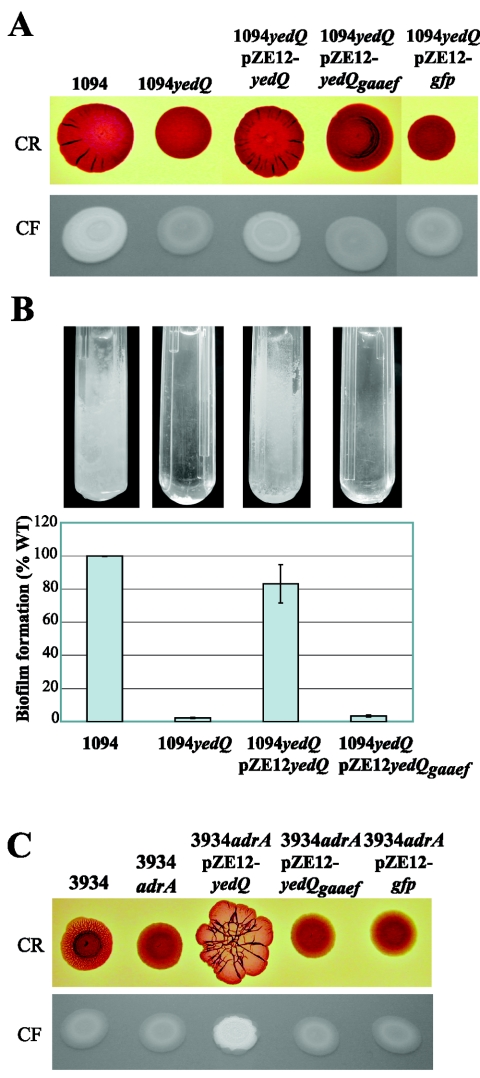
In order to confirm the role of YedQ in cellulose regulation, we performed the deletion of the yedQ gene in E. coli 1094. 1094yedQ displayed a CF-negative phenotype on LB and M63-CF plates at both 30 and 37°C (Fig. 3A) and was also severely impaired in biofilm formation in microfermentors (Fig. 3B). The yedQ knockout in E. coli 1094 was successfully complemented by the introduction of plasmid-borne yedQ under the control of an inducible lac promoter (pZE12-yedQ) but not by the parental vector (Fig. 3A). These results demonstrate that YedQ regulates the production of cellulose under all tested conditions in strain E. coli 1094.
FIG. 3.
YedQ is the regulator for cellulose production in E. coli 1094. (A) CR- and CF-binding phenotypes of strains 1094, 1094yedQ, 1094yedQ(pZE12-yedQ), 1094yedQ(pZE12-yedQgaaef), and 1094yedQ(pZE12-gfp). Two microliters of an o/n culture was spotted onto CR-LB (upper row) and CF-LB (lower row) plates and incubated for 24 h at 30°C. Similar phenotypes were obtained at 37°C and on CF-M63B1 plates. (B) Biofilm formation in microfermentors. The biofilms were grown for 24 h at 37°C in M63B1-glu. The biofilm formation ability of 1094yedQ and complemented mutants is expressed as a percentage of the 1094 wild-type biofilm, set to 100%. The error bars represent standard errors of the means. (C) Complementation of the CR- and CF-binding phenotypes of the Salmonella serovar Enteritidis adrA (3934adrA) mutant by pZE12-yedQ. Two microliters of o/n cultures was spotted onto CR-LB (upper row) and CF-LB (lower row) plates and incubated for 24 h at 30°C.
The presence of a GGEEF motif in the C-terminal part of YedQ (Fig. 2) is strongly indicative of c-di-GMP synthase activity, common to known regulators of cellulose synthesis (50). This suggested that the motif might be critical for YedQ function. To test this hypothesis, the yedQ GGEEF motif was mutagenized to GAAEF in pZE12-yedQ. When introduced into E. coli 1094yedQ, the resulting plasmid-borne yedQgaaef failed to restore any of the CF- or CR-binding and biofilm formation phenotypes (Fig. 3A and B). These results indicate that the YedQ GGEEF motif is essential for the function of the protein. Moreover, pZE12-yedQ, but not pZE12-yedQgaaef, could complement the rdar and CF-binding defects of a Salmonella serovar Enteritidis adrA mutant (Fig. 3C), suggesting that, like Salmonella AdrA, YedQ has c-di-GMP synthase activity.
Altogether, these results indicate that YedQ is a new GGDEF regulator of cellulose synthesis in E. coli.
RpoS does not regulate yedQ expression in E. coli 1094.
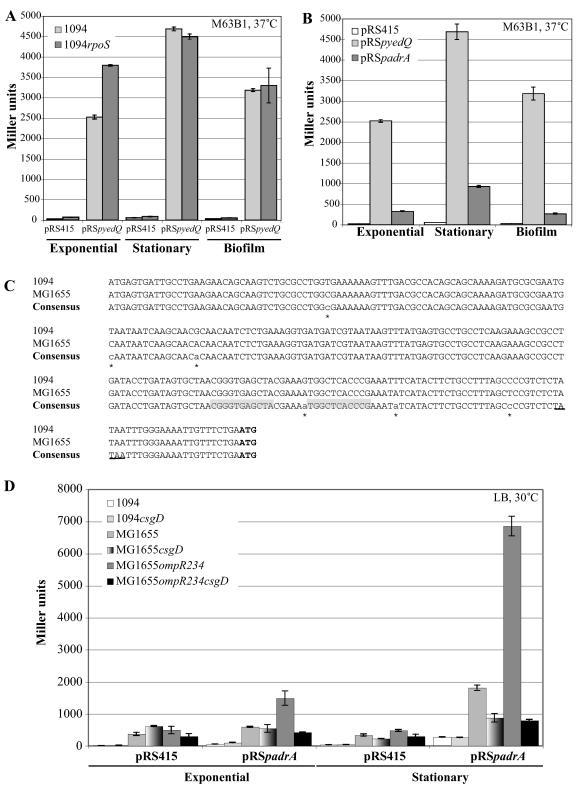
Because of the functional similarity between YedQ and AdrA, we tested whether they could share an RpoS transcriptional control and if the CF- and CR-negative phenotypes displayed by 1094rpoS in LB could be due to the lack of yedQ expression. Neither CF nor CR binding could be restored in 1094rpoS(pZE12yedQ) in LB medium, and the introduction of yedQ in a 1094rpoS strain led to irregular levels of CF binding in minimal medium (data not shown).
To further study the regulation of yedQ transcription, the activity of a plasmid-borne yedQ-lacZ transcriptional fusion (pRSpyedQ::lacZ) introduced in either E. coli 1094 or 1094rpoS was tested under various conditions, including 30°C and 37°C and exponential and stationary phases in liquid rich and minimal media, as well as biofilm. These experiments showed that the transcription of yedQ is only moderately affected by the tested environmental conditions (twofold increase in minimal medium at stationary phase compared to LB) (Fig. 4A; see also Fig. S2A and B in the supplemental material). Moreover, the rpoS mutation did not significantly affect the expression of the yedQ-lacZ fusion (Fig. 4A; see also Fig. S2A and B in the supplemental material). Therefore, in contrast to Salmonella adrA expression, neither RpoS nor CsgD directly regulates the expression of yedQ. In an attempt to identify regulators of yedQ activity, we also mutagenized the strain 1094ΔyedQ::uidA. This strain carries a chromosomal pyedQ-uidA (β-glucuronidase) transcriptional fusion and is pale blue to white on LB plates containing 5-bromo-4-chloro-3-indolyl-beta-d-glucuronic acid (a chromogenic substrate for UidA) using the plasmid pSC189 bearing a Mariner-based transposon. Since the extremities of the transposon have strong constitutive promoter-out activity (A. Roux and J. M. Ghigo, unpublished data), this mutagenesis allowed us to screen both for the inactivation of yedQ repressor and for the promoter-out-mediated activation of yedQ activators by looking for blue colonies due to pyedQ derepression or activation on LB-5-bromo-4-chloro-3-indolyl-beta-d-glucuronic acid at 30°C. This new screen failed to identify any regulators of yedQ expression. Altogether, these results suggest that yedQ is constitutively expressed under all tested conditions in E. coli 1094.
FIG. 4.
Transcriptional regulation of yedQ and adrA in strain 1094. (A) yedQ expression was studied in wild-type E. coli 1094 and the 1094rpoS mutant by measuring the β-galactosidase activity of a yedQ::lacZ fusion carried on the pRSpyedQ::lacZ plasmid. The cloning vector pRS415 was used as a negative control in both strains. Light-gray bars, β-galactosidase activity in 1094; dark-gray bars, β-galactosidase activity in 1094rpoS. The error bars represent standard errors of the means. (B) yedQ and adrA expression levels were compared in strain E. coli 1094 by measuring the β-galactosidase activities of a yedQ::lacZ and an adrA::lacZ fusion carried, respectively, on the pRSpyedQ::lacZ and pRSpadrA::lacZ plasmids. The cloning vector pRS415 was used as a negative control. White bars, pRS415 β-galactosidase activity; light-gray bars, pRSpyedQ::lacZ β-galactosidase activity; dark-gray bars, pRSpadrA::lacZ β-galactosidase activity. (C) Sequence alignment of the adrA promoters of E. coli strains 1094 and MG1655. The putative CsgD-binding sequences are shaded, and the −10 box is underlined on the consensus sequence. The sequence differences among E. coli strains are indicated by stars. The ATG start codon is in boldface letters. (D) adrA1094 expression was studied in the wild-type E. coli strains 1094, MG1655, and MG1655ompR234 (PHL818) and their respective csgD mutants by measuring the β-galactosidase activity of an adrA::lacZ fusion carried on the pRSpadrA::lacZ plasmid. The bar shades for β-galactosidase activities in the different strains are as indicated in the graph legend. All the measurements were done at least in triplicate at 37°C in M63B1 medium supplemented with 0.4% glucose (A and B) or at 30°C in LB medium (D).
YedQ and AdrA are both potentially functional in E. coli 1094.
We hypothesized that the reason why E. coli 1094 uses YedQ rather than AdrA to regulate cellulose production could be either a lack of expression or functionality of AdrA or because YedQ fulfills specific functions in E. coli 1094. To investigate this, we first tested the expression and functionality of the E. coli 1094 adrA gene by cloning it into pZE1Pr-adrA (expressing adrA under the control of its own promoter) and pZE12-adrA (expressing adrA under the control of an inducible lac promoter). These two constructs were tested in complementation experiments with the CF-negative Salmonella serovar Enteritidis adrA mutant as a heterologous host. As shown in Table 3, adrA1094 expressed from both plasmids could restore the CF phenotype of Salmonella serovar Enteritidis adrA, showing that both the adrA1094 promoter and the protein AdrA1094 are functional. AdrA1094 is likely to display a diguanylate synthase activity in this strain.
TABLE 3.
Complementation by AdrA
| Strain | Phenotype/morphotype
|
|||||
|---|---|---|---|---|---|---|
| M63B1-glu + CFa
|
LB + CFa
|
LB − NaCl + CRb
|
||||
| 30°C | 37°C | 30°C | 37°C | 30°C | 37°C | |
| 1094 | +++ | +++ | +++ | +++ | rdar | rdar |
| 1094yedQ | − | − | − | +/− | cdar | cdar |
| 1094yedQ(pZE1Pr-adrA)c | +++ | +++ | ++ | ++ | rdar | rdar |
| 1094yedQ(pZE12-adrA)c | +++ | +++ | ++ | ++ | rdar | rdar |
| 3934d | +/− | +/− | rdar | |||
| 3934adrA | − | − | sar | |||
| 3934adrA(pZE1Pr-adrA) | +++ | +++ | rdar | |||
| 3934adrA(pZE12-adrA) | +++ | +++ | rdar | |||
Fluorescence under UV light was examined visually after growth on minimal or rich medium agar plates containing CF at 30°C or 37°C for 48 h. The highest fluorescence is expressed as +++; no fluorescence as −.
Morphotype on CR plates. cdar, circular, dry and red.
pZE1Pr-adrA, expression of adrA1094 under the control of its own promoter; pZE12-adrA, expression of adrA1094 under the control of the synthetic lac promoter.
Salmonella serovar Enteritidis strain 3934.
Therefore, we tested whether adrA1094 could restore the ability of E. coli 1094yedQ to bind CF and CR. Both pZE12-adrA and pZE1Pr-adrA complemented the yedQ defect, showing that AdrA is also functional in 1094 when expressed from a multicopy plasmid (Table 3).
We then compared the expression levels of yedQ and adrA in E. coli 1094 using a plasmid-borne transcriptional lacZ fusion to either the yedQ or the adrA promoter (pRSpyedQ::lacZ and pRSpadrA::lacZ, respectively). These comparisons were performed in minimal or rich medium at 30°C or 37°C in exponential- and stationary-phase cultures or biofilm. In strain 1094, adrA is 5- to 16-fold less expressed than yedQ, depending on the environmental conditions (Fig. 4B; see also Fig. S2C and D in the supplemental material).
We did not observe a decrease in the expression of the padrA-lacZ fusion in the 1094csgD mutant, suggesting that under these conditions, adrA expression does not depend on CsgD (Fig. 4D). Since csgD is expressed and functional in E. coli 1094 (see above), the low level of adrA1094 expression could result from the inability of its promoter to be recognized and activated by CsgD. The comparison of the adrA promoter sequences of E. coli strains MG1655 and 1094 showed only a few differences (Fig. 4C). To assess if these differences could affect the capacity of the adrA1094 promoter to be activated by CsgD, we introduced the adrA-lacZ fusion in the strains MG1655 and MG1655ompR234 (which up-regulates csgD) and their respective csgD mutants (10). As shown in Fig. 4D, adrA expression was reduced in an MG1655csgD mutant but was significantly increased by the ompR234 mutation in MG1655 (Fig. 4D). The adrA1094-lacZ fusion activity was also severely reduced in the double ompR234-csgD mutant (Fig. 4D), showing that the adrA promoter of strain 1094 can be recognized and activated by the CsgD protein of MG1655.
Taken together, these results demonstrated that AdrA is potentially functional in strain 1094. Moreover, although it can be activated by CsgD in E. coli MG1655, the adrA1094 promoter is not regulated by CsgD in E. coli 1094, and its expression level is low compared to YedQ expression.
CsgD-independent cellulose production is found in several natural E. coli isolates.
To investigate whether E. coli isolates other than 1094 may also display a CsgD-independent cellulose regulation pathway, we studied three other CF-positive E. coli strains from our collection: strains 1125, DSM6601, and 55989 (Table 1; see also Table S1 in the supplemental material). These three strains appear to produce cellulose, as a deletion of the bcsC gene led to a CF-negative phenotype on CF agar plates and a sar phenotype on CR plates (Fig. 5).
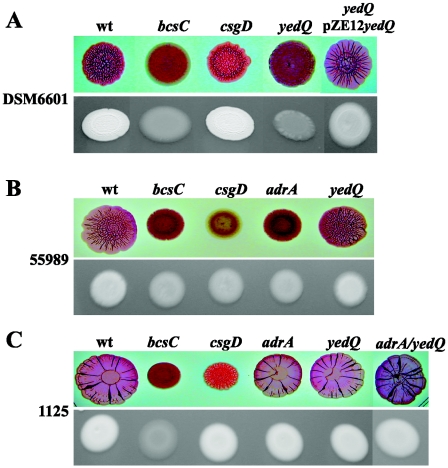
FIG. 5.
CsgD-independent and -dependent cellulose synthesis regulation in E. coli strains. CR-binding (upper rows) and CF-binding (lower rows) phenotypes of the wild type and deletion mutants of strains DSM6601 (A), 55989 (B), and 1125 (C). Two microliters of an o/n culture was spotted onto CR-LB and CF-LB plates and incubated for 48 h at 30°C.
We first introduced a csgD mutation in the 1125, DSM6601, and 55989 strains and tested the abilities of the mutant strains to bind CF and CR. As shown in Fig. 5, only 55989csgD displayed a CF-negative phenotype and completely lost its ability to bind CR. Moreover, the CF-negative phenotype and sar morphotype exhibited by the 55989adrA mutant indicated that 55989 shows a CsgD/AdrA-dependent cellulose synthesis regulation (Fig. 5B). By contrast, DSM6601csgD and 1125csgD remained CF positive and exhibited the rough morphotype on CR-LB plates, suggesting that their cellulose regulation pathway could be YedQ dependent. We therefore introduced a yedQ mutation in the 1125, DSM6601, and 55989 strains and tested the abilities of these mutants to bind CF and CR. As expected from a CsgD-dependent strain, 55989yedQ remained CF positive and exhibited the rdar morphotype on CR-LB plates. The CsgD-independent strain, DSM6601yedQ, however, lost its ability to bind CF and exhibited a smooth morphotype. This defect could be complemented by the introduction of pZE12-yedQ (Fig. 5A).
Interestingly, 1125yedQ remained CF positive and exhibited an rdar morphotype on CR-LB plates (Fig. 5C). To exclude the possibility that AdrA could regulate cellulose synthesis independently of CsgD in this strain, the adrA mutation was introduced in 1125. This mutant displayed wild-type phenotypes on CF and CR plates (Fig. 5C). Finally, in order to check that the CF-positive phenotype exhibited by either the 1125adrA or 1125yedQ mutant did not result from a cross-complementation by yedQ or adrA, respectively, the double mutant 1125adrA-yedQ was created. It exhibited the same phenotypes as the wild type (Fig. 5C).
These results demonstrate that CsgD-independent and YedQ-dependent cellulose synthesis is not restricted to the E. coli 1094 strain and also operates in DSM6601. Moreover, since neither YedQ, CsgD, nor AdrA is required to bind CF or CR in E. coli 1125, this suggests that the regulation of the cellulose production in this strain uses a different genetic pathway, possibly involving one of the 17 other GGDEF proteins encoded by genes identified in the E. coli genome (21).
DISCUSSION
The production of polysaccharide polymers in the extracellular biofilm matrix is recognized as central in the development of bacterial biofilms (9). Among the identified polysaccharide components of the biofilm matrix, cellulose, a β-1-4-d-glucose polymer, has been shown to contribute to the formation of biofilm in Salmonella and Pseudomonas by promoting cell contacts at both liquid and solid interfaces (58, 59, 68).
Although laboratory E. coli K-12 derivative strains do not produce cellulose, the polymer has been detected in a few natural E. coli isolates (68). Our analysis of a collection of 87 E. coli strains that did not undergo significant laboratory subcultivation showed that cellulose production is a common phenotype in E. coli. This trait was not, however, strictly associated with strong biofilm formation capacities, suggesting that cellulose synthesis is a biofilm determinant of some but not all E. coli strains. Nevertheless, as CF is also a nonspecific indicator of chitin and succinoglycan polymers, we cannot exclude the possibility that some of these biofilm-negative but CF-positive strains do not produce bona fide cellulose, a hypothesis that could be tested by the systematic introduction of mutations in the cellulose operons of these strains. We also observed that half of the non-CF-binding strains that were assessed for biofilm formation were indeed able to form a biofilm. This suggests that, in these strains, other proteinaceous components (such as pili or curli) or other exopolysaccharides, such as colanic acid or poly-1,6-GlcNAc (PGA), could be involved in biofilm formation (4, 16, 44, 64).
To investigate the link between cellulose production and biofilm formation in E. coli, the commensal E. coli strain 1094, which exhibits both rdar and strong CF-binding phenotypes and forms a biofilm, was studied in more detail. As observed with Salmonella (23, 48, 58, 68), cellulose is required for biofilm formation in E. coli strain 1094. However, unlike in Salmonella, curli production is not necessary for the expression of a mature biofilm and multicellular behavior. Indeed, whereas the deletion of the cellulose operons abolished these phenotypes, the 1094csgD or 1094csgA mutant behaved like the wild-type strain.
Considering the conservation of the genes involved in cellulose production and regulation in Salmonella and E. coli, it was reasonable to assume that CsgD and AdrA could play roles in cellulose production in E. coli (46). Indeed, during the course of this work, a study of cellulose and curli expression in E. coli showed that a representative fecal isolate of E. coli regulates cellulose synthesis via CsgD (5). However, the analysis of the genetic basis of cellulose synthesis in the commensal E. coli strain 1094 allowed us to show that neither CsgD, AdrA, nor MlrA is involved in this process in E. coli 1094.
Using a genetic screen based on the ability of the cellulose-producing strain 1094csgD to bind the fluorescent dye CF, we identified five mutants defective for CF binding located outside of the cellulose operons. Three of these transposition mutants corresponded to insertions in the glucose-1-P-uridylyltransferase (galU) and phosphoglucose isomerase (pgi) genes. Because these enzymes play a direct or indirect role in the synthesis of UDP-glucose, the sugar subunit of cellulose, both galU and pgi are likely to have an effect on cellulose synthesis per se, but not on its regulation. galU has been shown to be necessary for biofilm formation and biosynthesis of the exopolysaccharide of a phage-resistant rugose variant of Vibrio cholerae and to affect E. coli early adhesion (24, 38). A pgi mutant has been isolated from a mutagenesis screen performed on a CF-positive Salmonella serovar Enteritidis strain, along with other genes involved in different sugar metabolic pathways (58).
One of the five CF-negative mutants identified in our screen corresponds to an insertion in the yedQ gene. The protein with an unknown function, YedQ, is predicted to be a membrane-associated protein that contains a GGDEF (DUF1) domain. Proteins displaying a GGDEF domain are widespread, are present in most bacterial genomes, and form a large family of proteins that have been associated with diguanylate cyclase activity and c-di-GMP production (22, 29). The discovery of c-di-GMP and the control of its intracellular level by proteins with diguanylate cyclase (GGDEF domain proteins) and/or phosphodiesterase (EAL domain proteins) activities originated from work on cellulose production in G. xylinus (52, 63). In Salmonella, the GGDEF domain protein AdrA has been shown to be involved in cellulose synthesis regulation and its c-di-GMP synthase activity has been recently demonstrated (45, 56). Although most of the GGDEF domain proteins have not yet been experimentally characterized, it is now considered that c-di-GMP, in addition to its role as an allosteric activator of cellulose synthesis, is a second messenger involved in a number of cellular and behavioral functions, such as the transition from sessility to motility, cell cycle-dependent localization of proteins, and cellulose and biofilm formation (3, 6, 22, 23, 29, 33, 42, 45, 50, 51, 56, 58).
The construction of a null yedQ mutant in strain 1094 and its plasmid complementation confirmed that, under all tested conditions, YedQ is required for cellulose synthesis and biofilm formation in this strain. A site-directed mutagenesis in the GGEEF motif of YedQ showed that this motif is essential for cellulose synthesis in E. coli 1094, as well as for YedQ1094 to complement an AdrA defect in Salmonella serovar Enteritidis. Altogether, these results suggest that, like AdrA in Salmonella, YedQ contains a functional GGDEF domain and that we have identified a new cellulose regulatory protein in E. coli 1094 that is probably involved in the regulation of the level of intracellular c-di-GMP through its GGDEF domain activity (56). During the course of this study, gcpA (STM1987), a gene encoding a GGDEF protein sharing 64.7% identity with YedQ, was shown to be critical for cellulose and biofilm formation in Salmonella serovar Enteritidis (23). However, unlike YedQ, which is required for E. coli 1094 cellulose synthesis in rich and minimal media at both 30 and 37°C, in Salmonella serovar Enteritidis, GcpA regulates cellulose production under specific culture conditions in a CsgD-independent manner. Indeed, while in Salmonella serovar Enteritidis cellulose production depends on CsgD and AdrA in LB medium at 28°C, it depends only on GcpA in ATM at 37°C (23). These differences suggest that yedQ and gcpA may not be similarly regulated in E. coli 1094 and Salmonella serovar Enteritidis, as suggested by the differences observed between the two promoters. Therefore, while the roles of YedQ/GcpA could be different in E. coli and Salmonella serovar Enteritidis, they are both involved in CsgD- and AdrA-independent regulation of cellulose production.
Our finding that an rpoS insertion mutant leads to a CF-negative phenotype in E. coli 1094 suggested that the CsgD-dependent and -independent pathways, depicted in Fig. 6, may share a common root, i.e., that like adrA, yedQ expression could also be regulated by RpoS. However, the expression of an inducible overexpressed copy of yedQ in a 1094rpoS mutant did not reverse the CF-negative phenotype of a 1094rpoS mutant on LB-CF plates (data not shown). Moreover, a pyedQ-lacZ transcriptional fusion was still expressed in a 1094rpoS background, showing that RpoS is not required for the expression of yedQ (Fig. 4; see also Fig. S2 in the supplemental material). Although we did not obtain evidence that yedQ could be subjected to any significant transcriptional regulation under our experimental conditions, we cannot exclude yedQ regulation at either a transcriptional or a posttranscriptional level. Nevertheless, RpoS is required for the CF- and CR-binding abilities of strain 1094, suggesting that in this strain, RpoS could regulate the transcription of the cellulose bcs operons or a metabolic pathway important for cellulose biosynthesis, such as UDP-glucose synthesis, depending on environmental conditions (Fig. 6).
The functional specificity of the multiple GGDEF domain proteins producing freely diffusible c-di-GMP and present in a single bacterium is an important and currently unresolved question (29, 50). Different mechanisms have been proposed, involving either the temporal and environmental transcriptional regulation of GGDEF and EAL domain protein expression, the spatial control of the production of c-di-GMP near its cellular target proteins, or the fine tuning of the activity of GGDEF and EAL domain proteins by a specific ligand or by c-di-GMP itself (1, 29, 33, 42, 50, 59, 60). In E. coli 1094, whereas complementation experiments showed that AdrA is functional when overexpressed, transcription analysis using multicopy padrA-lacZ and pyedQ-lacZ fusions showed that adrA is weakly expressed compared to yedQ. These observations support the hypothesis that the specific use of the yedQ over the adrA cellulose pathway is not due to a defect in AdrA protein activity but may be due in part to substantial differences in gene expression. Indeed the physiological level of expression of AdrA was not sufficient to circumvent yedQ defects in E. coli 1094, whereas its expression from a high-copy-number plasmid could complement 1094yedQ. The level of protein expression has been shown to be important for the complementation of the Salmonella serovar Typhimurium adrA mutant by the Yersinia pestis GGDEF domain protein HmsT (55).
The alignment of the sequences of the adrA promoters from E. coli strains 1094 and MG1655 shows very few differences (97.7% identity) (Fig. 4C). It is not yet clear if the identified differences in the promoter sequence could explain the lack of CsgD activity on the adrA1094 promoter, thus leading to a low level of expression of adrA in E. coli 1094. Interestingly, however, CsgD can activate the adrA1094 promoter in E. coli MG1655 (Fig. 4D), suggesting that the adrA1094 promoter is functional but is independent of CsgD in E. coli 1094. It was previously suggested that CsgD could activate the csgB (curli expression) and adrA promoters by different mechanisms, since the location of the putative CsgD-binding sites are different in the two promoters (10). Our results raise the possibility that in E. coli 1094, CsgD has diverged compared to CsgDMG1655 and has lost its ability to regulate adrA while still regulating the csgB promoter. This hypothesis is under investigation.
The regulation of the level of adrA expression does not, however, formally exclude the existence of functional specificities of YedQ and AdrA, possibly related to structural differences outside of their GGDEF domains. Indeed, although YedQ and AdrA are predicted to be membrane proteins and share a GGDEF domain (with 32.9% identity and 48.8% similarity), they have distinct N-terminal domains (Fig. 2B). The N-terminal domain of YedQ is predicted by Phobius software analysis to contain two transmembrane domains and to be mainly periplasmic (32). It does not match any conserved domains in the Pfam database. On the other hand, the N-terminal domain of AdrA is a MASE2 domain, which is predicted to be an integral membrane sensory domain found in many diguanylate cyclases (39, 66). These structural differences in the N-terminal domains of the YedQ and AdrA proteins could lead to distinct localizations and interactions with potential partners and diverse abilities to sense signals for protein activation or repression, all leading to synthesis of c-di-GMP at a specific place and time.
Based on nucleotide sequence homologies, it has been proposed that the cellulose biosynthesis genes could constitute a functional module acquired by horizontal gene transfer under some environmental conditions in which the expression of the rdar morphotype could constitute a selective advantage (53). The acquisition of the capacity to activate cellulose biosynthesis through the production of c-di-GMP would have been acquired after the establishment of the cellulose genes in the genome (46). If so, owing to the large number of proteins potentially involved in the process (11 proteins with a GGDEF domain in Salmonella serovar Typhimurium and 19 in E. coli), it is possible that, depending on the strain, a GGDEF protein different from AdrA, and possibly more highly expressed due to simple activating mutations, might have fulfilled this regulatory function. In agreement with this hypothesis, E. coli strain 1094 displays a CsgD- and AdrA-independent CF-positive phenotype, which involves YedQ. Furthermore, we found that such a CsgD-independent, YedQ-dependent cellulose pathway could also be found in another commensal E. coli strain, DSM6601. Additionally, in the case of E. coli 1125, an as-yet-uncharacterized GGDEF protein, distinct from both AdrA and YedQ, may have acquired this function, as the adrA or yedQ mutants, as well as the adrA/yedQ double mutant in strain 1125, retain their CF-binding abilities (Fig. 5C). Interestingly, experimental data that do not fit the cellulose regulatory pathway schematized in Fig. 6A have been reported (see the introduction and references 23, 48, and 58). This suggests that alternative CsgD-independent pathways may also exist in these Salmonella strains and possibly in other Enterobacteriaceae. Further studies are under way to test this hypothesis. Owing to the roles of CsgD in different biological processes subject to a complex network of regulatory proteins, the appearance of CsgD-independent cellulose regulation pathways provides a way to uncouple CsgD-regulated processes (e.g., curli regulation) from cellulose synthesis. Such an uncoupling may be favored when the expression of cellulose would confer a selective advantage under conditions in which adrA is usually not expressed, such as the high temperature in the human gastrointestinal tract. This may provide a selective pressure leading to the emergence of CsgD-independent cellulose pathways involving regulated diguanylate cyclases, such as YedQ.
In conclusion, the regulation of enterobacterial cellulose production is more complex than previously recognized and, in E. coli, can depend on alternative, and possibly widespread, cellulose regulatory pathways. The characterization of an alternative pathway for the regulation of cellulose production in E. coli illustrates the diversity of the regulatory arsenal used by bacteria to control the synthesis of polysaccharidic components of the extracellular matrix.
Supplementary Material
Acknowledgments
We thank Patricia Latour-Lambert for her technical assistance. We are grateful to Pierre Beguin for the gift of purified endoglucanase. We thank C. Beloin, I. Lasa, B. Lakowski, P. Beguin, C. Latasa, J. Valle, B. Le Quéré, T. Msadek, and C. Guet for critical reading of the manuscript. We thank Chantal Le Bouguenec, Uli Dobrindt, and Inigo Lasa for the gifts of the E. coli and Salmonella strains used in this study. We thank C. Guet and F. Norel for the gifts of pZE12-gfp and pSTC14, respectively.
J.-M.G. is supported by Institut Pasteur, CNRS URA 2172, and Fondation BNP PARIBAS grants. S.D.R. is supported by Sanofi-Pasteur.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org.
REFERENCES
- 1.Aldridge, P., R. Paul, P. Goymer, P. Rainey, and U. Jenal. 2003. Role of the GGDEF regulator PleD in polar development of Caulobacter crescentus. Mol. Microbiol. 47:1695-1708. [DOI] [PubMed] [Google Scholar]
- 2.Amikam, D., and M. Y. Galperin. 2005. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22:3-6. [DOI] [PubMed] [Google Scholar]
- 3.Ausmees, N., R. Mayer, H. Weinhouse, G. Volman, D. Amikam, M. Benziman, and M. Lindberg. 2001. Genetic data indicate that proteins containing the GGDEF domain possess diguanylate cyclase activity. FEMS Microbiol. Lett. 204:163-167. [DOI] [PubMed] [Google Scholar]
- 4.Beloin, C., S. Da Re, and J. M. Ghigo. August 2005, posting date. Chapter 8.3.1.3, Colonization of abiotic surfaces. In R. Curtiss III, A. Böck, J. L. Ingraham, J. B. Kaper, F. C. Neidhardt, M. Riley, and C. L. Squires (ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, D.C. [Online.] http://www.ecosal.org.
- 5.Bokranz, W., X. Wang, H. Tschape, and U. Romling. 2005. Expression of cellulose and curli fimbriae by Escherichia coli isolated from the gastrointestinal tract. J. Med. Microbiol. 54:1171-1182. [DOI] [PubMed] [Google Scholar]
- 6.Bomchil, N., P. Watnick, and R. Kolter. 2003. Identification and characterization of a Vibrio cholerae gene, mbaA, involved in maintenance of biofilm architecture. J. Bacteriol. 185:1384-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyd, A., and A. M. Chakrabarty. 1995. Pseudomonas aeruginosa biofilms: role of the alginate exopolysaccharide. J. Ind. Microbiol. 15:162-168. [DOI] [PubMed] [Google Scholar]
- 8.Branda, S. S., J. E. Gonzalez-Pastor, E. Dervyn, S. D. Ehrlich, R. Losick, and R. Kolter. 2004. Genes involved in formation of structured multicellular communities by Bacillus subtilis. J. Bacteriol. 186:3970-3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Branda, S. S., S. Vik, L. Friedman, and R. Kolter. 2005. Biofilms: the matrix revisited. Trends Microbiol. 13:20-26. [DOI] [PubMed] [Google Scholar]
- 10.Brombacher, E., C. Dorel, A. J. Zehnder, and P. Landini. 2003. The curli biosynthesis regulator CsgD co-ordinates the expression of both positive and negative determinants for biofilm formation in Escherichia coli. Microbiology 149:2847-2857. [DOI] [PubMed] [Google Scholar]
- 11.Brown, P. K., C. M. Dozois, C. A. Nickerson, A. Zuppardo, J. Terlonge, and R. Curtiss III. 2001. MlrA, a novel regulator of curli (AgF) and extracellular matrix synthesis by Escherichia coli and Salmonella enterica serovar Typhimurium. Mol. Microbiol. 41:349-363. [DOI] [PubMed] [Google Scholar]
- 12.Chaveroche, M. K., J. M. Ghigo, and C. d'Enfert. 2000. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 28:E97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiang, S. L., and E. J. Rubin. 2002. Construction of a mariner-based transposon for epitope-tagging and genomic targeting. Gene 296:179-185. [DOI] [PubMed] [Google Scholar]
- 14.Chun, K. T., H. J. Edenberg, M. R. Kelley, and M. G. Goebl. 1997. Rapid amplification of uncharacterized transposon-tagged DNA sequences from genomic DNA. Yeast 13:233-240. [DOI] [PubMed] [Google Scholar]
- 15.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Danese, P. N., L. A. Pratt, and R. Kolter. 2000. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J. Bacteriol. 182:3593-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derbise, A., B. Lesic, D. Dacheux, J. M. Ghigo, and E. Carniel. 2003. A rapid and simple method for inactivating chromosomal genes in Yersinia. FEMS Immunol. Med. Microbiol. 38:113-116. [DOI] [PubMed] [Google Scholar]
- 18.Firon, A., A. Beauvais, J. P. Latge, E. Couve, M. C. Grosjean-Cournoyer, and C. D'Enfert. 2002. Characterization of essential genes by parasexual genetics in the human fungal pathogen Aspergillus fumigatus: impact of genomic rearrangements associated with electroporation of DNA. Genetics 161:1077-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedman, L., and R. Kolter. 2004. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51:675-690. [DOI] [PubMed] [Google Scholar]
- 20.Friedman, L., and R. Kolter. 2004. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J. Bacteriol. 186:4457-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galperin, M. Y. 2004. Bacterial signal transduction network in a genomic perspective. Environ. Microbiol. 6:552-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galperin, M. Y., A. N. Nikolskaya, and E. V. Koonin. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203:11-21. [DOI] [PubMed] [Google Scholar]
- 23.Garcia, B., C. Latasa, C. Solano, F. Garcia-del Portillo, C. Gamazo, and I. Lasa. 2004. Role of the GGDEF protein family in Salmonella cellulose biosynthesis and biofilm formation. Mol. Microbiol. 54:264-277. [DOI] [PubMed] [Google Scholar]
- 24.Genevaux, P., P. Bauda, M. S. DuBow, and B. Oudega. 1999. Identification of Tn10 insertions in the rfaG, rfaP, and galU genes involved in lipopolysaccharide core biosynthesis that affect Escherichia coli adhesion. Arch. Microbiol. 172:1-8. [DOI] [PubMed] [Google Scholar]
- 25.Gerstel, U., C. Park, and U. Romling. 2003. Complex regulation of csgD promoter activity by global regulatory proteins. Mol. Microbiol. 49:639-654. [DOI] [PubMed] [Google Scholar]
- 26.Ghigo, J. M. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412:442-445. [DOI] [PubMed] [Google Scholar]
- 27.Gotz, F. 2002. Staphylococcus and biofilms. Mol. Microbiol. 43:1367-1378. [DOI] [PubMed] [Google Scholar]
- 28.Higgins, D. G., and P. M. Sharp. 1988. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene 73:237-244. [DOI] [PubMed] [Google Scholar]
- 29.Jenal, U. 2004. Cyclic di-guanosine-monophosphate comes of age: a novel secondary messenger involved in modulating cell surface structures in bacteria? Curr. Opin. Microbiol. 7:185-191. [DOI] [PubMed] [Google Scholar]
- 30.Joliff, G., P. Beguin, J. Millet, J. P. Aubert, P. Alzari, M. Juy, and R. J. Poljak. 1986. Crystallization and preliminary X-ray diffraction study of an endoglucanase from Clostridium thermocellum. J. Mol. Biol. 189:249-250. [DOI] [PubMed] [Google Scholar]
- 31.Jubelin, G., A. Vianney, C. Beloin, J. M. Ghigo, J. C. Lazzaroni, P. Lejeune, and C. Dorel. 2005. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J. Bacteriol. 187:2038-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kall, L., A. Krogh, and E. L. Sonnhammer. 2004. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338:1027-1036. [DOI] [PubMed] [Google Scholar]
- 33.Kirillina, O., J. D. Fetherston, A. G. Bobrov, J. Abney, and R. D. Perry. 2004. HmsP, a putative phosphodiesterase, and HmsT, a putative diguanylate cyclase, control Hms-dependent biofilm formation in Yersinia pestis. Mol. Microbiol. 54:75-88. [DOI] [PubMed] [Google Scholar]
- 34.Lutz, R., and H. Bujard. 1997. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 25:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marck, C. 1988. ‘DNA Strider’: a ‘C’ program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 16:1829-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mayer, R., P. Ross, H. Weinhouse, D. Amikam, G. Volman, P. Ohana, R. D. Calhoon, H. C. Wong, A. W. Emerick, and M. Benziman. 1991. Polypeptide composition of bacterial cyclic diguanylic acid-dependent cellulose synthase and the occurrence of immunologically crossreacting proteins in higher plants. Proc. Natl. Acad. Sci. USA 88:5472-5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Nesper, J., C. M. Lauriano, K. E. Klose, D. Kapfhammer, A. Kraiss, and J. Reidl. 2001. Characterization of Vibrio cholerae O1 El tor galU and galE mutants: influence on lipopolysaccharide structure, colonization, and biofilm formation. Infect. Immun. 69:435-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nikolskaya, A. N., A. Y. Mulkidjanian, I. B. Beech, and M. Y. Galperin. 2003. MASE1 and MASE2: two novel integral membrane sensory domains. J. Mol. Microbiol. Biotechnol. 5:11-16. [DOI] [PubMed] [Google Scholar]
- 40.Norel, F., V. Robbe-Saule, M. Y. Popoff, and C. Coynault. 1992. The putative sigma factor KatF (RpoS) is required for the transcription of the Salmonella typhimurium virulence gene spvB in Escherichia coli. FEMS Microbiol. Lett. 78:271-276. [DOI] [PubMed] [Google Scholar]
- 41.Ochman, H., and R. K. Selander. 1984. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157:690-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paul, R., S. Weiser, N. C. Amiot, C. Chan, T. Schirmer, B. Giese, and U. Jenal. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18:715-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prigent-Combaret, C., E. Brombacher, O. Vidal, A. Ambert, P. Lejeune, P. Landini, and C. Dorel. 2001. Complex regulatory network controls initial adhesion and biofilm formation in Escherichia coli via regulation of the csgD gene. J. Bacteriol. 183:7213-7223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prigent-Combaret, C., G. Prensier, T. T. Le Thi, O. Vidal, P. Lejeune, and C. Dorel. 2000. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: role of flagella, curli and colanic acid. Environ. Microbiol. 2:450-464. [DOI] [PubMed] [Google Scholar]
- 45.Romling, U. 2005. Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cell. Mol. Life Sci. 62:1234-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Romling, U. 2002. Molecular biology of cellulose production in bacteria. Res. Microbiol. 153:205-212. [DOI] [PubMed] [Google Scholar]
- 47.Römling, U., Z. Bian, M. Hammar, W. D. Sierralta, and S. Normark. 1998. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J. Bacteriol. 180:722-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Römling, U., W. Bokranz, U. Gerstel, H. Lünsdorf, M. Nimtz, W. Rabsch, H. Tschäpe, and M. W. X. Zogaj. 2003. Dissection of the genetic pathway leading to multicellular behaviour in Salmonella typhimurium and other Enterobacteriaceae, p. 231-261. In M. Wilson and D. Devine (ed.), Medical implications of biofilms. Cambridge University Press, Cambridge, England.
- 49.Romling, U., W. Bokranz, W. Rabsch, X. Zogaj, M. Nimtz, and H. Tschape. 2003. Occurrence and regulation of the multicellular morphotype in Salmonella serovars important in human disease. Int. J. Med. Microbiol. 293:273-285. [DOI] [PubMed] [Google Scholar]
- 50.Romling, U., M. Gomelsky, and M. Y. Galperin. 2005. C-di-GMP: the dawning of a novel bacterial signalling system. Mol. Microbiol. 57:629-639. [DOI] [PubMed] [Google Scholar]
- 51.Romling, U., M. Rohde, A. Olsen, S. Normark, and J. Reinkoster. 2000. AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol. Microbiol. 36:10-23. [DOI] [PubMed] [Google Scholar]
- 52.Ross, P., H. Weinhouse, Y. Aloni, D. Michaeli, P. Weinherberg-Ohana, R. Mayer, S. Braun, E. de Vroom, G. A. van der Marel, J. H. van Boom, and M. Benziman. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325:279-281. [DOI] [PubMed] [Google Scholar]
- 53.Sakellaris, H., N. K. Hannink, K. Rajakumar, D. Bulach, M. Hunt, C. Sasakawa, and B. Adler. 2000. Curli loci of Shigella spp. Infect. Immun. 68:3780-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 55.Simm, R., J. D. Fetherston, A. Kader, U. Romling, and R. D. Perry. 2005. Phenotypic convergence mediated by GGDEF-domain-containing proteins. J. Bacteriol. 187:6816-6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Simm, R., M. Morr, A. Kader, M. Nimtz, and U. Römling. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 53:1123-1134. [DOI] [PubMed] [Google Scholar]
- 57.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 58.Solano, C., B. Garcia, J. Valle, C. Berasain, J. M. Ghigo, C. Gamazo, and I. Lasa. 2002. Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol. Microbiol. 43:793-808. [DOI] [PubMed] [Google Scholar]
- 59.Spiers, A. J., J. Bohannon, S. M. Gehrig, and P. B. Rainey. 2003. Biofilm formation at the air-liquid interface by the Pseudomonas fluorescens SBW25 wrinkly spreader requires an acetylated form of cellulose. Mol. Microbiol. 50:15-27. [DOI] [PubMed] [Google Scholar]
- 60.Spiers, A. J., S. G. Kahn, J. Bohannon, M. Travisano, and P. B. Rainey. 2002. Adaptive divergence in experimental populations of Pseudomonas fluorescens. I. Genetic and phenotypic bases of wrinkly spreader fitness. Genetics 161:33-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Starkey, M., A. K. Gray, S. I. Chang, and M. Parsek. 2004. A sticky business: the extracellular polymeric substance matrix of bacterial biofilms, p. 478. In M. Ghannoum and G. A. O'Toole (ed.), Microbial biofilms. ASM Press, Washington, D.C.
- 62.Sutherland, I. W. 2001. The biofilm matrix—an immobilized but dynamic microbial environment. Trends Microbiol. 9:222-227. [DOI] [PubMed] [Google Scholar]
- 63.Tal, R., H. C. Wong, R. Calhoon, D. Gelfand, A. L. Fear, G. Volman, R. Mayer, P. Ross, D. Amikam, H. Weinhouse, A. Cohen, S. Sapir, P. Ohana, and M. Benziman. 1998. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J. Bacteriol. 180:4416-4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang, X., J. F. Preston III, and T. Romeo. 2004. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J. Bacteriol. 186:2724-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weiner, L., and P. Model. 1994. Role of an Escherichia coli stress-response operon in stationary-phase survival. Proc. Natl. Acad. Sci. USA 91:2191-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhulin, I. B., A. N. Nikolskaya, and M. Y. Galperin. 2003. Common extracellular sensory domains in transmembrane receptors for diverse signal transduction pathways in bacteria and archaea. J. Bacteriol. 185:285-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zogaj, X., W. Bokranz, M. Nimtz, and U. Romling. 2003. Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract. Infect. Immun. 71:4151-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zogaj, X., M. Nimtz, M. Rohde, W. Bokranz, and U. Romling. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 39:1452-1463. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.