Abstract
A glycerol dehydrogenase gene was selected as a multicopy suppressor rescuing the reduced hilA expression in the Salmonella enterica serovar Typhimurium cpxA mutant. A substrate of the enzyme, 1,2-propanediol, repressed hilA expression. The 1,2-propanediol-mediated repression at 150 mM, but not that at 300 mM, was abrogated by blocking the catabolism producing propionate from 1,2-propanediol.
Invasion into host epithelial cells in the intestine is the initial and essential step of pathogenesis by Salmonella. Most of the genes required for the invasion localize to a 40-kb region of the chromosome termed Salmonella pathogenicity island 1 (SPI-1) (9). Many genes within this region encode components of the Type III secretion machinery and effectors. These effectors are secreted through the functions of the type III secretion machinery. It has well been established that some of the SPI-1 region-encoded effectors, such as SipBCD, are essential mediators of invasion. The regulatory circuits of expression of the SPI-1 genes have been intensively analyzed (for reviews, see references 9, 16, and 26). One of the key steps of the circuits is regulation of the synthesis of the first global activator, HilA, since HilA is believed to be ultimately required for synthesis of both the effectors and the secretion machinery (11). Expression of the hilA gene and, consequently, the invasive phenotype is tightly regulated by a variety of environmental signals, such as oxygen concentration, osmolarity, and growth phase (13, 17, 25). Because of its important position in the circuits, many researchers have also investigated the regulatory mechanisms by which expression of the hilA gene is regulated. These works have identified many genetic factors controlling hilA expression. Briefly, a two-component system, BarA-SirA, activates hilA (3, 22). Fis, a DNA-nucleoid-associated protein (40); FadD, which is involved in uptake and degradation of fatty acids (28); and fliZ, a flagellar gene (21, 28), have been reported to be positive factors for hilA expression. Three AraC/XylS-type transcriptional factors, HilC, HilD, and RtsA, have been reported to be direct activation factors for hilA expression (12, 27, 36). The effect of CsrA-CsrB, a protein-RNA complex, on hilA expression seems be largely mediated through these three AraC/XylS-type factors (2, 3). Other two-component systems, i.e., PhoB-PhoR (28) and PhoP-PhoQ (33), have negative effects on hilA expression. Fahlen et al. screened genetic loci with negative effects on hilA and reported ams, hupB, pag, hilE, and hha as such loci (4, 14, 15).
Either, but not both, of the glycerol dehydrogenases is required for full activation of hilA expression.
In a previous paper, we characterized the cpxA mutant of Salmonella enterica serovar Typhimurium (31). The hilA expression level was lower in the cpxA mutant than in the cpxA+ strain, and this effect was much greater when culture was at pH 6.0 than when it was at pH 8.0. At pH 6.0, the expression level in the cpxA mutant was only approximately 7% of that in the cpxA+ strain (31). Surprisingly, the putative cognate response regulator, CpxR, had no effect on hilA expression (31). In order to examine the mechanism of the CpxR-independent activation of hilA by CpxA, we screened multicopy suppressors that compensated for the phenotype of the cpxA mutant from a total Salmonella DNA library constructed with the pBR322 (7) vector. The cpxA mutant harboring pSN849 (Table 1) (31), which is white on pH 6.0-adjusted X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) agar (31), was transformed with the library. Dark blue colonies on the agar were screened, and the plasmids they contained were analyzed. We found that these suppressors fell into one of the following three genes: hilC, rtsA, or STM3529 (encoding a putative NAD-dependent glycerol dehydrogenase) (29). The effects of the hilC and rtsA genes on hilA expression have already been described in several reports (12, 27, 36). We were interested in the function of STM3529 (called glhA hereafter, for glycerol dehydrogenase, hypothetical or alternative). We found that the multicopy effect of the cloned glhA on hilA expression was not necessarily complete but was partial. Indeed, at pH 6.0, introduction of the plasmid containing glhA into the cpxA mutant restored hilA expression to only 50% of the level in the cpxA+ strain (data not shown). This suggested that the phenotype of the cpxA mutant could not be explained by the involvement of glhA only, and we judged that glhA was a factor regulating hilA independently of the cpxA pathway. We further investigated the role of glhA in this regulation.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Source (reference) |
|---|---|---|
| E. coli K-12 | ||
| JM109 | endA1 recA1 gyrA96 thi hsdR17(rK− mK+) relA1 supE44 Δ(lac-proAB) [F′ traD36 proAB+lacIqZΔM15], initial host for PCR-produced DNA cloning | C. Yanisch-Perron (41) |
| MS8 | MC1061 rnh::cat Cmr, host for manipulation of pKH5002 and its derivatives | H. Ohmori (32) |
| S. enterica serovar Typhimurium | ||
| SL1344 | Standard virulent strain, hisG rpsL | SGSCa |
| KK1501 | LT2 hsdSA29 hsdB121 hsdL6 metA22 metE551 trpC2 ilv-452 rpsL120 xyl-404 galE719 FelS2, initial host of DNAs prepared in E. coli | K. Kutsukake (23) |
| SN3529 | SL1344 glhA::tetA Tetr | This study |
| SN4108 | SL1344 ΔgldA::aphT Kmr | This study |
| SN6000 | SL1344 glhA::tetA ΔgldA::aphT Tetr Kmr | This study |
| SN2036 | SL1344 ΔpocR::aphT Kmr | This study |
| SN2040 | SL1344 ΔpduCDEGH::aphT Kmr | This study |
| Plasmids | ||
| pBR322 | pMB1 replicon cloning vector, Ampr Tetr | F. Boliver (7) |
| pKH5002 | Suicide vector for gene disruption, Ampr | H. Ohmori (32) |
| pSN849 | hilA′-′lacZ translational fusion gene cloned into pHSG595, Cmr | S. Nakayama (31) |
| pGlhA | 1,706-bp Sau3AI-NruI fragment consisting almost entirely of the glhA gene cloned into the BamHI-NruI sites of pBR322, Ampr | This study |
| pPocR | PCR-generated fragment consisting of the pocR gene cloned into the SphI-SalI sites of pBR322, Ampr | This study |
SGSC, Salmonella Genetic Stock Center, Calgary, Canada.
We constructed the glhA::tetA mutant SN3529 (Table 1) according to the method of allele exchange using the suicide vector pKH5002 (32) and monitored the hilA expression level in this mutant. However, the expression level was not significantly different from that in the wild-type (WT) parent, SL1344 (Table 2), suggesting that GlhA was not necessarily required for activation of hilA. In the published genome sequence of Salmonella enterica serovar Typhimurium (29), there are two putative NAD-dependent glycerol dehydrogenase genes, gldA and glhA. We hypothesized that these two genes were redundant for activation of hilA. Hence, we constructed the ΔgldA::aphT mutant SN4108 (Table 1) and the glhA::tetA ΔgldA::aphT double mutant SN6000 (Table 1) according to the method reported by Datsenko and Wanner (10) and characterized them. hilA expression in the double mutant was decreased to approximately 60% of that in the parent, whereas in the ΔgldA::aphT single mutant, SN4108, the expression level was not significantly decreased (Table 2). These mutations, which are not linked on the chromosome, had a significant effect only when they were combined. We also confirmed that the glhA-expressing plasmid, pGlhA, which was selected as a multicopy suppressor (Table 1), fully complemented the hilA expression in the glhA::tetA ΔgldA::aphT double mutant (data not shown). On the other hand, attempts to clone the gldA gene on plasmid vectors resulted in instability of the resultant plasmids (data not shown). Overexpression of GldA, but not of GlhA, thus might have some toxic effect on the bacterial cells under our culture conditions. Thus, we could not definitely demonstrate complementation of the double mutant by the cloned gldA gene. However, these results strongly suggest that either, but not both, of the two genes is required for full activation of hilA expression. We believe this means that the total finite activity of glycerol dehydrogenase is required for full activation, although demonstration of the glycerol dehydrogenase activities of the GlhA and GldA products is needed for this to be confirmed.
TABLE 2.
hilA expression levels reported by pSN849 in glycerol dehydrogenase mutants
| Strain (relevant genotype) | β-Galactosidase activitya | % Expression compared to that in SL1344 |
|---|---|---|
| SL1344 (glhA+gldA+) | 896 ± 119 | 100 |
| SN3529 (glhA::tetA gldA+) | 942 ± 63 | 110 |
| SN4108 (glhA+ ΔgldA::aphT) | 736 ± 25 | 88 |
| SN6000 (glhA::tetA ΔgldA::aphT) | 500 ± 8 | 60 |
Activities are expressed as mean Miller units ± standard deviations. For culture conditions, see the legend to Fig. 1.
PDL represses hilA expression.
The genetic data described above suggest that both gldA and glhA are involved in hilA activation. GldA and GlhA are predicted to have glycerol dehydrogenase activity (29). Therefore, we focused on how the glycerol dehydrogenases could be involved in gene regulation. We postulated that some organic compound, which could be a substrate of these enzymes, might influence hilA expression. A preliminary investigation indicated that glycerol, a representative substrate of the enzymes, had no effect on hilA expression when the concentration was not more than 3% (vol/vol) (about 326 mM) (data not shown). In Escherichia coli, 1,2-propanediol (PDL) is also established as a substrate of GldA, the E. coli glycerol dehydrogenase (1). Because the deduced amino acid sequences of Salmonella GldA and GlhA have significantly high similarity to that of E. coli GldA (1, 29), we examined the effect of PDL on hilA expression. We observed that addition of R-(−)-PDL into the medium at a concentration of 150 mM (at the initial point of growth and throughout the experiments described in this study) reduced hilA expression to 55.82% of that in the medium without PDL. In this initial experiment, we chose the R-(−) enantiomer of PDL because only this enantiomer is established as a substrate for the E. coli GldA (1).
This observation and the results described in the previous section raise two possibilities: first, that the glycerol dehydrogenases have a positive effect on hilA expression and R-(−)-PDL inhibits the effect through sequestration of the enzyme, and second, that R-(−)-PDL has a negative effect on hilA expression and the glycerol dehydrogenases suppress the effect through transformation of R-(−)-PDL into the dehydrogenated product, which rationally is hydroxyacetone. To distinguish these possibilities, we tested whether R-(−)-PDL further influenced hilA expression in the glhA::tetA ΔgldA::aphT double mutant, SN6000. In this experiment, we found that R-(−)-PDL further repressed hilA expression in the mutant, although the degree of repression was slightly lower than that in the parent, SL1344 (Table 3). This result strongly supports the latter possibility described above, i.e., that R-(−)-PDL has a negative effect on hilA expression and the glycerol dehydrogenases partially suppress this effect, although this needs to be confirmed by direct demonstration that the GlhA and GldA products can catabolize PDL. However, the results of the preliminary experiment described in the fifth section of this report support the interpretation that GlhA and GldA catabolize PDL (see below).
TABLE 3.
Effect of R-(−)-1,2-propanediol on hilA expression in strains SL1344 and SN6000
| Strain (relevant genotype) | β-Galactosidase activitya at PDL concn (mM):
|
% Repression by 150 mM PDLb | |
|---|---|---|---|
| 0 | 150 | ||
| SL1344 (glhA+gldA+) | 816 ± 78 | 455 ± 28 | 55.82 |
| SN6000 (glhA::tetA ΔgldA::aphT) | 491 ± 21 | 313 ± 17 | 63.74 |
Activities are expressed as mean Miller units ± standard deviations. For culture conditions, see the legend to Fig. 1.
Percent activity at 150 mM PDL compared to that at 0 mM PDL.
Dose dependency of 1,2-propanediol-mediated repression.
In the previous section, we described that addition of 150 mM R-(−)-PDL reduced hilA expression to 55.82% of that in medium without R-(−)-PDL. To better understand this repression, we investigated the dose dependency of this effect. At the same time, we examined whether the R-(−) and S-(+) enantiomers had different effects on hilA expression. The R-(−) and S-(+) enantiomers of PDL did not show a significant difference in their repressive effect at any concentration (data not shown). The reason why both enantiomers effectively repress hilA is unknown at present.
In the analysis of the dose dependency of the repressive effect, we found a striking characteristic. Namely, within the range of 50 to 150 mM, no significant decrease in hilA expression was observed with an increase in PDL concentration. This means that throughout this range of PDL concentrations, the level of hilA expression was maintained at around 55 to 60% of that in medium without PDL. However, when the concentration was further increased above this range, hilA expression again started to decrease. At 300 mM PDL, hilA expression was further reduced to 24% of that in medium without PDL. Such a unique, dual-phase nature of dose dependency implies that the PDL-mediated repression of hilA consisted of two distinct mechanisms, one functioning at lower concentrations and the other at higher concentrations. This was confirmed in the following analysis.
Involvement of pocR and the pdu operon in1,2-propanediol-mediated hilA repression.
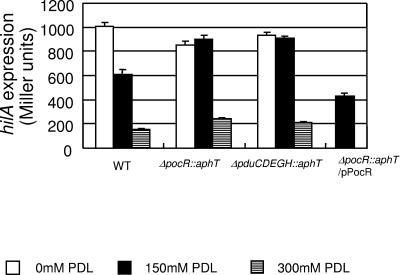
Based on the above results, we attempted to analyze how PDL repressed hilA expression. In this background, the regulatory mechanism of the pdu operon provided some hints. The pdu operon, consisting of 21 genes (pduABCDEGHJKLMNOPQSTUVWX), encodes the enzymes that degrade PDL into propionate and propanol, although the functions of some of these genes are still unknown (6, 19, 20). Expression of the operon is activated by a transcriptional factor, PocR (5, 8, 34, 35). PocR belongs to the AraC family of regulators and requires PDL as a cofactor for activation of its own DNA-binding capacity (35). Thus, pocR is a genetic factor accomplishing PDL-dependent regulation of target genes. This prompted us to investigate whether PocR is also involved in the PDL-dependent repression of hilA. Hence, we constructed the ΔpocR::aphT mutant SN2036 (Table 1) according to the method reported by Datsenko and Wanner (10) and compared its hilA expression with that of the parent WT strain, SL1344, in media containing 0 mM, 150 mM, and 300 mM PDL. As the two enantiomers of PDL had comparable effects on hilA expression (see above), we used a mixture of R-(−)- and S-(+)-PDL thereafter. In this analysis, we found that the ΔpocR::aphT mutation almost completely abrogated the repression by 150 mM PDL (Fig. 1). Introduction of the plasmid pPocR (Table 1), which expresses PocR under control of the tetA promoter on pBR322, into the mutant restored the repression (Fig. 1). Thus, the simplest interpretation of the results was that PocR, through activation of its own DNA-binding capacity, directly repressed hilA expression in the presence of PDL, the cofactor of PocR. However, at the same time, we noted another possibility. PocR is an essential activator of the pdu operon, encoding enzymes responsible for propionate production from PDL (5, 8, 34, 35), and recently Lawhon et al. have reported that propionate represses hilA expression (24). Thus, it was plausible that the observed effect of the pocR mutation was the simple outcome of a defect in propionate production in the mutant. Hence, we further constructed the ΔpduCDEGH::aphT mutant SN2040 (Table 1) according to the method reported by Datsenko and Wanner (10). (Note that pocR remains intact in this mutant, because pocR is located upstream of the pdu operon and is transcribed divergently to it.) pduCDE, the three genes in the operon, encode the subunits of diol dehydratase, which catalyzes the conversion of PDL to propionaldehyde, the initial reaction to produce propionate from PDL, whereas pduGH encodes the subunits of diol dehydratase-reactivating factor (6, 19). Thus, it was expected that in this mutant, the catabolic pathway from PDL to propionate would be completely blocked but PocR would remain active. We investigated the response of this mutant to 150 mM PDL. This clearly showed that the pattern of hilA expression in the ΔpduCDEGH::aphT mutant was indistinguishable from that in the ΔpocR::aphT mutant (Fig. 1). Because cloning of the long region corresponding to the entire pduCDEGH was unsuccessful (data not shown), we could not definitely demonstrate the complementation of this ΔpduCDEGH::aphT mutant. However, these results strongly suggest that the repression by 150 mM PDL ultimately requires the production of propionate from PDL. Furthermore, we confirmed that in the ΔpocR::aphT mutant SN2036, in the ΔpduCDEGH::aphT mutant SN2040, and in strain SL1344, hilA was repressed by propionate (24). Indeed, in all three strains, the addition of 30 mM sodium propionate reduced hilA expression to about 32% of that in medium without sodium propionate. This observation is consistent with a previous report (24) and supports our interpretation that hilA repression by 150 mM PDL is exerted through propionate produced from PDL.
FIG. 1.
Involvement of pocR and the pdu operon in hilA repression by 150 mM 1,2-propanediol. hilA expression levels reported by pSN849 in strains SL1344 (WT), SN2036 (ΔpocR::aphT), SN2040 (ΔpduCDEGH::aphT), and SN2036 harboring pPocR (ΔpocR::aphT/pPocR) were monitored through β-galactosidase assay after cultivation in media supplemented with the indicated concentrations of a mixture of R-(−)- and S-(+)-1,2-propanediol. Throughout the experiments in this study, LB supplemented with 0.1 M NaCl and 0.1 M sodium phosphate buffer (pH 7.0) was employed as the basic medium. 1,2-Propanediol or sodium propionate was added at the indicated final concentrations when used. Bacterial cultures in the indicated media were incubated and kept static at 37°C until the optical density at 600 nm reached approximately 0.7. The cultured bacteria were harvested, suspended in Z buffer (30), and used for the β-galactosidase assay. Error bars indicate standard deviations.
The effect of glycerol dehydrogenase activity on hilA expression is due to the catabolism of PDL.
In the first section of this paper, we stated that the double mutations in glycerol dehydrogenases that are expected to catabolize PDL led to a reduction in hilA expression. Here, we found that the hilA repression by PDL at a lower concentration (150 mM) requires the function of the pdu operon. These data implied that the observed reduction of hilA expression in the glhA::tetA ΔgldA::aphT double mutant, SN6000, might also require the function of the pdu operon. Hence, we further inactivated pduCDEGH in SN6000 and monitored the hilA expression level in cells grown in medium without PDL or sodium propionate. This investigation showed that in this glhA::tetA ΔgldA ΔpduCDEGH::aphT mutant, the hilA expression level reported by pSN849 was 913 ± 25 Miller units, whereas that in SL1344 was 880 ± 21 Miller units. Thus, inactivation of pduCDEGH in SN6000 almost completely suppressed the phenotype of SN6000. This clearly indicates that the observed hilA repression in SN6000 requires the function of the pdu operon. This strongly suggests that when propionate production from the limited amount of endogenous PDL in the basic medium is blocked, PDL consumption by the glycerol dehydrogenases is no longer required for full activation of hilA. Thus, it can be interpreted that the ultimate role of the glycerol dehydrogenases in hilA regulation is to suppress the effect of propionate, which is produced from PDL. Importantly, this result may also support our interpretation that the observed effect of the glycerol dehydrogenases is via PDL and that these glycerol dehydrogenases catabolize PDL (see previous sections).
hilA repression by a higher concentration of 1,2-propandiol does not require pdu operon.
In the third section, we stated the possibility that the mechanism of PDL-dependent repression at higher concentrations is different from that at lower concentrations. Accordingly, the levels of hilA expression in the WT strain SL1344, the ΔpocR::aphT mutant SN2036, and the ΔpduCDEGH::aphT mutant SN2040 at 300 mM PDL were also monitored. Rather surprisingly, all three strains showed almost the same level of hilA expression at this concentration of PDL (Fig. 1). The results of this analysis clearly indicate that repression by 300 mM PDL does not require pocR or pduCDEGH and, consequently, the production of propionate.
The simplest explanation for these results may be that PDL itself has no effect on hilA expression when the concentration is not more than 150 mM and that it has a repressive effect only at a concentration of 300 mM or higher. However, this simple interpretation is rather unlikely, as the repression by 300 mM PDL was as strong as fourfold (Fig. 1). Thus, it is rather hard to believe that PDL has no effect at all when the concentration is 150 mM. This issue should be clarified in the course of mechanistic studies of the pathways of repression by PDL in the future.
Conclusions and perspectives.
PDL was shown to repress hilA expression. The mechanism of the PDL-dependent repression of hilA consists of two distinct pathways, one dependent on and the other independent of propionate production from PDL. These repression pathways could be weakened through consumption of PDL by the two NAD-dependent glycerol dehydrogenases of Salmonella. The specific mechanisms, as well as the biological significance, of the two repression pathways should be elucidated in the future.
PDL has been established as a safe compound (18, 38, 39) and is used as a solvent for many kinds of industrial products, including foods and cosmetics. Therefore, it can be anticipated that ingestion of PDL could prevent, at least partially, penetration of Salmonella into the intestinal epithelium through inhibition of hilA expression, without toxic effects in animals.
Acknowledgments
This work was supported by grants from the Ministry of Health, Labor, and Welfare of Japan.
We thank Akira Kushiro (Yakult Central Institute for Microbiological Research, Tokyo, Japan) for helpful discussion and advice.
REFERENCES
- 1.Altaras, N. E., and D. C. Cameron. 1999. Metabolic engineering of a 1,2-propanediol pathway in Escherichia coli. Appl. Environ. Microbiol. 65:1180-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altier, C., M. Suyemoto, and S. D. Lowhon. 2000. Regulation of Salmonella enterica serovar Typhimurium invasion genes by csrA. Infect. Immun. 68:6790-6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altier, C., M. Suyemoto, A. I. Ruiz, K. D. Burnham, and R. Maurer. 2000. Characterization of two novel regulatory genes affecting Salmonella invasion gene expression. Mol. Microbiol. 35:635-646. [DOI] [PubMed] [Google Scholar]
- 4.Baxter, M. A., T. F. Fahlen, R. L. Wilson, and B. D. Jones. 2003. HilE interacts with HilD and negatively regulates hilA transcription and expression of the Salmonella enterica serovar Typhimurium invasive phenotype. Infect. Immun. 71:1295-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bobik, T. A., M. Ailion, and J. R. Roth. 1992. A single regulatory gene integrates control of vitamin B12 synthesis and propanediol degradation. J. Bacteriol. 174:2253-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bobik, T. A., G. D. Havemann, R. J. Busch, D. S. Williams, and H. C. Aldrich. 1999. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B12-dependent 1,2-propanediol degradation. J. Bacteriol. 181:5967-5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolivar, F., R. L. Rodriguez, P. J. Greene, M. C. Betlach, H. L. Hyneker, and H. W. Boyer. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2:95-113. [PubMed] [Google Scholar]
- 8.Chen, P., M. Ailion, T. A. Bobik, G. Stormo, and J. Roth. 1995. Five promoters integrate control of the cob/pdu regulon in Salmonella typhimurium. J. Bacteriol. 177:5401-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darwin, K. H., and V. L. Miller. 1999. Molecular basis of the interaction of Salmonella with the intestinal mucosa. Clin. Microbiol. Rev. 12:405-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eichelberg, K., and J. E. Galan. 1999. Differential regulation of Salmonella typhimurium type III secreted proteins by pathogenicity island 1 (SPI-1)-encoded transcriptional activators InvF and HilA. Infect. Immun. 67:4099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellermeier, C. D., and J. M. Slauch. 2003. RtsA and RtsB coordinately regulate expression of the invasion and flagellar genes in Salmonella enterica serovar Typhimurium. J. Bacteriol. 185:5096-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ernst, R. K., D. M. Dombroski, and J. M. Merrick. 1990. Anaerobiosis, type 1 fimbriae, and growth phase are factors that affect invasion of HEp-2 cells by Salmonella typhimurium. Infect. Immun. 58:2014-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fahlen, T. F., N. Mathur, and B. D. Jones. 2000. Identification and characterization of mutants with increased expression of hilA, the invasion gene transcriptional activator of Salmonella typhimurium. FEMS Immunol. Med. Microbiol. 28:25-35. [DOI] [PubMed] [Google Scholar]
- 15.Fahlen, T. F., R. L. Wilson, J. D. Boddicker, and B. D. Jones. 2001. Hha is a negative modulator of transcription of hilA, the Salmonella enterica serovar Typhimurium invasion gene transcriptional activator. J. Bacteriol. 183:6620-6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galan, J. E. 1996. Molecular genetic bases of Salmonella entry into host cells. Mol. Microbiol. 20:263-271. [DOI] [PubMed] [Google Scholar]
- 17.Galan, J. E., and R. Curtiss III. 1990. Expression of Salmonella typhimurium genes required for invasion is regulated by changes in DNA supercoiling. Infect. Immun. 58:1879-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaunt, I. F., F. M. Carpanini, P. Grasso, and A. B. Lansdown. 1972. Long-term toxicity of propylene glycol in rats. Food Cosmet. Toxicol. 10:151-162. [DOI] [PubMed] [Google Scholar]
- 19.Havemann, G. D., and T. A. Bobik. 2003. Protein content of polyhedral organelles involved in coenzyme B12-dependent degradation of 1,2-propanediol in Salmonella enterica serovar Typhimurium LT2. J. Bacteriol. 185:5086-5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Havemann, G. D., E. M. Sampson, and T. A. Bobik. 2002. PduA is a shell protein of polyhedral organelles involved in coenzyme B12-dependent degradation of 1,2-propanediol in Salmonella enterica serovar Typhimurium LT2. J. Bacteriol. 184:1253-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iyoda, S., T. Kamidoi, K. Hirose, K. Kutsukake, and H. Watanabe. 2001. A flagellar gene fliZ regulates the expression of invasion genes and virulence phenotype in Salmonella enterica serovar Typhimurium. Microb. Pathog. 30:81-90. [DOI] [PubMed] [Google Scholar]
- 22.Johnston, C., D. A. Pegues, C. J. Hueck, C. A. Lee, and S. I. Miller. 1996. Transcriptional activation of Salmonella typhimurium invasion genes by a member of the phosphorylated response-regulator superfamily. Mol. Microbiol. 22:715-727. [DOI] [PubMed] [Google Scholar]
- 23.Kutsukake, K., S. Iyoda, K. Ohnishi, and T. Iino. 1994. Genetic and molecular analyses of the interaction between the flagellum-specific sigma and anti-sigma factors in Salmonella typhimurium. EMBO J. 13:4568-4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawhon, S. D., R. Maurer, M. Suyemoto, and C. Altier. 2002. Intestinal short-chain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA. Mol. Microbiol. 46:1451-1464. [DOI] [PubMed] [Google Scholar]
- 25.Lee, C. A., and S. Falkow. 1990. The ability of Salmonella to enter mammalian cells is affected by bacterial growth state. Proc. Natl. Acad. Sci. USA 87:4304-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lucas, R. L., and C. A. Lee. 2000. Unravelling the mysteries of virulence gene regulation in Salmonella typhimurium. Mol. Microbiol. 36:1024-1033. [DOI] [PubMed] [Google Scholar]
- 27.Lucas, R. L., and C. A. Lee. 2001. Roles of hilC and hilD in regulation of hilA expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 183:2733-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lucas, R. L., C. P. Lostroh, C. C. DiRusso, M. P. Spector, B. L. Wanner, and C. A. Lee. 2000. Multiple factors independently regulate hilA and invasion gene expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:1872-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 30.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 31.Nakayama, S., A. Kushiro, T. Asahara, R. Tanaka, L. Hu, D. J. Kopecko, and H. Watanabe. 2003. Activation of hilA expression at low pH requires the signal sensor CpxA, but not the cognate response regulator CpxR, in Salmonella enterica serovar Typhimurium. Microbiology 149:2809-2817. [DOI] [PubMed] [Google Scholar]
- 32.Ohmori, H., M. Saitoh, T. Yasuda, T. Nagata, T. Fujii, M. Wachi, and K. Nagai. 1995. The pcsA gene is identical to dinD in Escherichia coli. J. Bacteriol. 177:156-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pegues, D. A., M. J. Hantman, I. Behlau, and S. I. Miller. 1995. PhoP/PhoQ transcriptional repression of Salmonella typhimurium invasion genes: evidence for a role in protein secretion. Mol. Microbiol. 17:169-181. [DOI] [PubMed] [Google Scholar]
- 34.Rondon, M. R., and J. C. Escalante-Semerena. 1992. The poc locus is required for 1,2-propanediol-dependent transcription of the cobalamin biothynthetic (cob) and propanediol utilization (pdu) genes of Salmonella typhimurium. J. Bacteriol. 174:2267-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rondon, M. R., and J. C. Escalante-Semerena. 1996. In vitro analysis of the interactions between the PocR regulatory protein and the promoter region of the cobalamin biosynthetic (cob) operon of Salmonella typhimurium LT2. J. Bacteriol. 178:2196-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schechter, L. M., S. M. Damrauer, and C. A. Lee. 1999. Two AraC/XylS family members can independently counteract the effect of repressing sequences upstream of the hilA promoter. Mol. Microbiol. 32:629-642. [DOI] [PubMed] [Google Scholar]
- 37.Schmieger, H. 1972. Phage P22-mutants with increased or decreased transduction abilities. Mol. Gen. Genet. 119:75-88. [DOI] [PubMed] [Google Scholar]
- 38.Trancik, R. J., and H. I. Maibach. 1982. Propylene glycol: irritation or sensitization? Contact Dermatitis 8:185-189. [DOI] [PubMed] [Google Scholar]
- 39.Weil, C. S., M. D. Woodside, H. F. Jr. Smyth, and C. P. Carpenter. 1971. Results of feeding propylene glycol in the diet to dogs for two years. Food Cosmet. Toxicol. 9:479-490. [DOI] [PubMed] [Google Scholar]
- 40.Wilson, R. L., S. J. Libby, A. M. Freet, J. D. Boddicker, T. F. Fahlen, and B. D. Jones. 2001. Fis, a DNA nucleoid-associated protein, is involved in Salmonella typhimurium SPI-1 invasion gene expression. Mol. Microbiol. 39:79-88. [DOI] [PubMed] [Google Scholar]
- 41.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]