Abstract
To analyze the transcriptional response of Yersinia enterocolitica cells to prolonged growth at low temperature, a collection of luxCDABE transposon mutants was cultivated in parallel at optimal (30°C) and suboptimal (10°C) temperatures and screened for enhanced promoter activities during growth until entering stationary phase. Among 5,700 Y. enterocolitica mutants, 42 transcriptional units were identified with strongly enhanced or reduced promoter activity at 10°C compared to 30°C, and changes in their transcriptional levels over time were measured. Green fluorescent protein fusions to 10 promoter regions confirmed the data. The temporal order of induction of the temperature-responsive genes of Y. enterocolitica was deduced, starting with the expression of cold shock genes cspA and cspB and the elevated transcription of a glutamate-aspartate symporter. Subsequently, cold-adapted cells drastically up-regulated genes encoding environmental sensors and regulators, such as UhpABC, ArcA, and methyl-accepting chemotaxis protein I (MCPI). Among the most prominent cold-responsive elements that were transcriptionally induced during growth in early and middle exponential phase are the insecticidal toxin genes tcaA and tcaB, as well as genes involved in flagellar synthesis and chemotaxis. The expression pattern of the late-exponential- to early-stationary-growth phase is dominated by factors involved in biodegradative metabolism, namely, a histidine ammonia lyase, three enzymes responsible for uptake and utilization of glycogen, the urease complex, and a subtilisin-like protease. Double-knockout mutants and complementation studies demonstrate inhibitory effects of MCPI and UhpC on the expression of a putative hemolysin transporter. The data partially delineate the spectrum of gene expression of Y. enterocolitica at environmental temperatures, providing evidence that an as-yet-unknown insect phase is part of the life cycle of this human pathogen.
A hallmark of microorganisms is their adaptability to environmental changes. Many bacteria are likely to encounter several alterations in milieu during their life cycle and must therefore be able to sense and adequately respond to these changes (1). Major contributions to our understanding of the complex molecular mechanisms underlying the successful adaptation come from the study of pathogenic bacteria. Pathogens such as Salmonella enterica and Listeria monocytogenes have temperature-regulated genes, obviously because their life cycles involve growth in a mammalian host, as well as exposure to environmental temperatures. An association between temperature and gene regulation has long been established in the yersiniae, due to their biphasic life cycles, during which they encounter a variety of ambient conditions (31, 64). Yersinia pestis, the causative agent of plague, regulates genes encoding capsule, iron transporters, outer membrane proteins, and toxins in response to temperatures found in the flea vector or in the mammalian host (37). In Yersinia enterocolitica, the Yop proteins and YadA are only produced at 37°C (14), while the invasin gene inv is maximally expressed at 26°C and is repressed at body temperature (20). To identify genes playing a role in host temperature adaptation, DNA microarray analysis was applied to, e.g., group A Streptococcus (61), Borrelia burgdorferi (51, 62), Campylobacter jejuni (63), and Y. pestis (27).
Considerable knowledge about the response to a temperature downshift comes from studies of mesophilic microorganisms such as Escherichia coli (34) and Bacillus subtilis (25, 73), as well as of psychrotolerant microorganisms such as L. monocytogenes (5), Arthrobacter globiformis (7), Pseudomonas fragi (29), and Y. enterocolitica (45). Extended by the application of macro- and microarray techniques (6, 35, 54), these data revealed specific mechanisms necessary for cold shock adaptation (48). Those requirements include modifications of the translation apparatus and the membrane that occur within a period of approximately 3 h after a sudden temperature drop (52, 73). This acclimatization phase is characterized by an increased but transient expression of cold shock-induced proteins like CspA, NusA, GyrA, RecA, IF-2, or polynucleotide phosphorylase and by cold acclimatization proteins continuously expressed at elevated levels (26, 30, 48, 66). By selective capture of transcribed sequences, it was shown only recently that L. monocytogenes acclimatization to 10°C probably involves general microbial stress responses and alterations in amino acid metabolism, as well as degradative metabolism (39).
However, most studies of differential gene expression in response to alterations in environmental conditions have been restricted to few target loci or, applying microarray technologies, to single time points, but no systematic, real-time analyses of gene expression of psychrotolerant bacteria following cold acclimatization have been available so far. Underused reporter gene technologies based on bioluminescence are able to overcome such limitations (55, 69). The use of promoter fusions with bioluminescent luxCDABE genes provides an independent analysis method compared to hybridization experiments, as demonstrated by the investigation of E. coli gene responses to DNA damage (69). Since the luxCDABE system produces light without the addition of exogenous substrate, it allows the efficient, facile, and repeatable measurement of promoter activities at many time points. A further advantage of the luciferase system over the green fluorescent protein (GFP) reporter is its high sensitivity, which enables the detection of transcriptional signals over a large dynamic range and thus the identification of down-regulated genes (68).
To describe the long-term response of a pathogen to low temperature, we chose Y. enterocolitica, a bacterium that is widely distributed in nature in aquatic and terrestrial reservoirs, as well as in animals. The presence of this bacterium in frozen and chilled food products is of major concern, since it has been shown that unprocessed food contaminated with pathogenic Y. enterocolitica is the vector of human infections that may give rise to severe gastrointestinal illness (8, 9). Genome-wide transposon mutagenesis of strain W22703 was carried out using a plasmid-borne mini-Tn5-based promoter probe transposon that was mobilized by conjugation (74). After random insertion of luxCDABE into the chromosome, the transcription of luxCDABE depended on the activity of an exogenous promoter located upstream. We established a screening assay based on the luxCDABE reporter system to determine the transcriptional profile of Y. enterocolitica genes during growth at 30°C and 10°C. We used this approach, an alternative to microarray profiling, to deduce a long-term expression profile of Y. enterocolitica in response to temperature downshift and to identify potential regulatory circuits underlying this response. Promoters which responded to temperature changes by significantly decreased or increased transcription were identified by monitoring the luminescence of single mutants over all growth phases. Our data describe a major set of cold-responsive genes completely diverse from the well-investigated cold shock genes, providing unforeseen insights into putative environmental reservoirs of Y. enterocolitica outside its mammalian hosts.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. All cultures were grown in Luria-Bertani (LB) broth (10-g/liter tryptone, 5-g/liter yeast extract, 5-g/liter NaCl), or on LB agar (LB broth supplemented with 1.5% agar). E. coli was grown at 37°C, and Y. enterocolitica was grown at the indicated temperature. If appropriate, the media were supplemented with the following antibiotics: kanamycin (50 μg/ml), nalidixic acid (20 μg/ml), tetracycline (12 μg/ml), or chloramphenicol (20 μg/ml). Strains were frozen in 13% glycerol at −70°C for permanent collection.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli S17.1 λpir | λpir lysogen of S17.1 (Tpr Smrthi pro hsdR− M+recA RP4::2-Tc::Mu-Km::Tn7) | 60 |
| E. coli DH5αMCR | F−mcrA Δ(mcr-hsdRMS-mcrBC) φ80lacZM15 (lacZYA-argF) U169 recA1 endA1 supE44 thi-1 gyrA96 relA1 | Invitrogen, Carlsbad, CA |
| Yersinia enterocolitica W22703 | Biovar 2, serovar O:9; NaIr Res− Mod+ | 14 |
| Y. enterocolitica NCTC10460 | Biovar 3, serovar O:1; Cmr | NCTC |
| W22703-YE0480::lux | Y. enterocolitica W22703 with luxCDABE reporter inserted into YE0480 | This study |
| W22703-YE0480::lux-uhpC′ | W22703-YE0480::lux with disrupted uhpC by pKRG9 insertion at nucleotide 717 | This study |
| W22703-YE0480::lux-YE2575′ | W22703-YE0480::lux with disrupted YE2575 by pKRG9 insertion at nucleotide 830 | This study |
| Plasmids | ||
| pUT mini-Tn5 luxCDABE Km2 | Suicide vector, ori R6K, mini-Tn5 Km2 luxCDABE transposon, mob+ (RP4); Apr Kmr | 74 |
| pKRG9 | Derivative of suicide vector pGP704; ori R6K mob+ (RP4); Cmr Aps | M. Déjosez, personal communication |
| pKRG9-uhpC′ | pKRG9 with 558-bp intragenic fragment of uhpC | This study |
| pKRG9-ybcM′ | pKRG9 with 313-bp intragenic fragment of ybcM | This study |
| pKRG9-YE2575′ | pKRG9 with 808-bp intragenic fragment of YE2575 | This study |
| pACYC184 | Cloning vector, P15A origin; Cmr Tcr | 13 |
| pACYC-uhpC | pACYC184 with a 1,761-bp EcoRI fragment carrying uhpC and 0 bp of its upstream region | This study |
| pACYC-YE2575 | pACYC184 with a 2,677-bp ScaI fragment carrying YE2575 and 362 bp of its upstream region | This study |
| pPROBE-NT | Promoter probe vector, pBBR1 replicon, gfp reporter; Kmr | 42 |
| pNT-PcspB | pPROBE-NT with 441 bp of cspB upstream sequence cloned in front of the gfp reporter | This study |
| pNT-PgltP | As above with 637-bp upstream sequence of gltP | This study |
| pNT-PYE0480 | As above with 422-bp upstream sequence of YE0480 | This study |
| pNT-PYE0951 | As above with 331-bp upstream sequence of YE0951 | This study |
| pNT-PYE0960 | As above with 500-bp upstream sequence of YE0960 | This study |
| pNT-PYE2463 | As above with 701-bp upstream sequence of YE2463 | This study |
| pNT-PYE2537 | As above with 310-bp upstream sequence of YE2537 | This study |
| pNT-PYE2575 | As above with 262-bp upstream sequence of YE2575 | This study |
| pNT-PYE2848 | As above with 503-bp upstream sequence of YE2848 | This study |
| pNT-PYE2922 | As above with 292-bp upstream sequence of YE2922 | This study |
luxCDABE reporter mutant library.
Transposon-bearing suicide plasmid pUT mini-Tn5 luxCDABE Km2 (17, 74) was transferred from E. coli donor strain S17.1 λpir (60) to acceptor strain Y. enterocolitica W22703 (14) by liquid mating as previously described (74) with the following modifications: donor and acceptor strains were grown to an optical density at 600 nm (OD600) of 0.5, and 100 μl of each culture was mixed and supplied with 800 μl of 10 mM MgSO4 solution. Following an incubation time of 5 min at room temperature (RT), cells were pelleted (3,000 rpm; 5 min at RT), resuspended in 100 μl LB broth, and spread onto antibiotic-free LB agar plates. The mating was incubated for 24 h at 30°C, and approximately 20% of the bacterial lawn was scraped off carefully and resuspended in 1 ml LB broth containing 13% glycerol. To avoid redundancy of the mutant library, five independent matings were performed, and aliquots of each mating were frozen in liquid N2 and stored at −70°C. Frozen aliquots were adjusted with LB medium to an OD600 of ∼0.3. Subsequently, 100 μl of this suspension was plated on LB selection plates (145-mm diameter) with kanamycin (200 μg/ml) and nalidixic acid and incubated for 24 h at 30°C. From each mating, 2,400 to 3,600 mutants were obtained, and each third colony was individually transferred to 96-well master plates filled with 800 μl LB containing antibiotics, as above, per well. Cells were cultivated for 16 h at 30°C on a shaker for microtiter plates. These overnight cultures were used for a subsequent luciferase-screening assay. In a preliminary test, Y. enterocolitica strain NCTC10460 instead of W22703 was used, leading to the identification of the cspA::luxCDABE mutant.
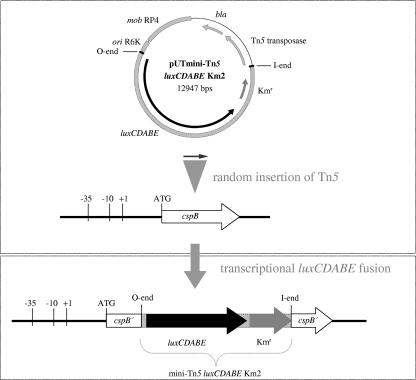
Random integration of the transposon insertions was experimentally proven by Southern blot analysis using DNA samples of 10 transposants. For that purpose, chromosomal DNA was restricted with Alw44I, gel separated, and hybridized with a 542-bp fragment from luxA, yielding bands of various length (data not shown). The resulting library of 5,700 luxCDABE transposants was used for the bioluminescence measurements. As an example, the genotype resulting from the random insertion of mini-Tn5 luxCDABE into cspB is schematically shown in Fig. 1.
FIG. 1.
Example of gene arrangement after insertion of the promoterless luxCDABE operon into Y. enterocolitica cspB. The suicide plasmid pUT mini-Tn5 luxCDABE Km2 was transferred via conjugation into Y. enterocolitica W22703, followed by random insertion of the promoterless luxCDABE operon and a kanamycin resistance gene into the Y. enterocolitica genome. The illustration depicts the transcriptional fusion of luxCDABE to the cspB promoter as a result of transposon mutagenesis. Position and orientation of the genes are indicated by arrows, and the mini-Tn5 transposon flanked by the O end and the I end is marked by dotted regions. Kmr, kanamycin resistence gene; bla, beta-lactamase gene.
Luciferase screening assay.
The screening assay to identify temperature-responsive promoters was performed with a set of 60 microtiter plates. For that purpose, 96-well plates with clear bottoms (Matrix Technologies, Hudson, N.H.) were inoculated from the master plates and shaken overnight at 500 rpm and 30°C. Fresh cultures were prepared in duplicate by the dilution of 10 μl of each overnight culture into 190 μl LB medium. One 96-well plate was continuously incubated at 30°C, and the other one was incubated at 30°C for 1 h, cooled to 10°C, and further incubated at this temperature. Of both plates, OD405 and bioluminescence (at 490 nm), the latter indicated as relative light units (RLU), were measured in parallel every hour with a Wallac VICTOR2 1420 multilabel counter (Perkin Elmer Life Sciences, Turku, Finland). To avoid interference, high-quality interference filters with an 8-nm bandwidth were used. To allow a direct comparison of 10°C and 30°C measurements, relative light units were referred to the growth phase by dividing the RLU by the respective OD405 value (RLU/OD). Following this normalization of data, we identified the start of the temperature-dependent promoter response as the OD value exhibiting the first significantly increased bioluminescence signal that was maintained or enhanced for at least a further 4 h.
The luciferase activity in cells growing at 10°C has been shown to decrease 12.3 fold in comparison to growth at 30°C (11). This correlates with other experimental data (41) and with the Arrhenius equation predicting a 1.5- to 4-fold reduction in enzyme reaction rate, following a temperature decrease of 10°C, given that the light production by the luciferase genes implies four independent catalytic steps (68). The corrected light emission at 10°C was thus determined as the ratio RLU/OD, multiplied by the factor 12.3. The response ratio of each mutant was calculated as the quotient of maximal RLU/OD at 10°C times 12.3 and maximal RLU/OD at 30°C.
DNA sequencing and characterization of transposon insertion sites.
Chromosomal DNA of each cold-induced transposon mutant was isolated and completely digested with ClaI, HindIII, SphI, SspI, or DraI (MBI Fermentas, Vilnius, Lithuania). Fragments were treated with T4 DNA ligase (Gibco, California) to allow self ligation resulting in circular molecules, and subsequent inverse PCR (50) was performed using transposon-specific primers derived from the O end or the I end of mini-Tn5 (Table 2). The PCR fragments obtained were sequenced by SequiServe (Vaterstetten, Germany) and MWG (Ebersberg, Germany) with primers hybridizing within the transposon at a distance of 26 to 97 bp to the O end or the I end. Each obtained sequence was screened for ClaI, HindIII, SphI, SspI, or DraI restriction sites and for the partial transposon sequence. Mapping of the mini-Tn5 luxCDABE insertions was conducted by using the Y. enterocolitica BLAST Server from the Sanger Institute (http://www.sanger.ac.uk/Projects/Y_enterocolitica/). Sequences without similarities to sequenced strain Y. enterocolitica 8081 were classified as specific for strain W22703. Promoter sequences located upstream of the transposon insertions were identified using a promoter prediction program (http://www.fruitfly.org/seq_tools/promoter.html).
TABLE 2.
Primers used in this study
| Target gene | 5′-3′ sequencee | Modification | Template |
|---|---|---|---|
| Sequencing by inverse PCR | |||
| GTGCAATCCATTAATTTTGGTG | SspIc | ||
| GAGCATGCGCCATCTTATGC | ClaIc | ||
| GCGCATCGGGCTTCCCATAC | ClaIc | ||
| GGCGGCCTATTCTTTAGTTC | HindIIIc | ||
| TGTTCCGTTGCGCTGCCCGGa | ClaIc | ||
| GTTGCGCTGCCCGGATTACAGa | HindIIIc | ||
| GAAATCACCATGAGTGACGACTG | HindIIIc | ||
| CATACGTATCCTCCAAGCCb | DraI,c SspIc | ||
| GTACCGAGCTCGAATTCGCGb | ClaIc | ||
| GTCATACGTATCCTCCAAGCb | HindIII,c SphIc | ||
| CAGATGCGATTAATTGGGCG | SphIc | ||
| GAGCAAATATTGCCTTATG | DraIc | ||
| Construction of pKRG knockout vectors | |||
| uhpC | CGATGAGCTCCCGATAATGCCA | SacId | |
| uhpC | GCGATCTAGACGATAACGTGCC | XbaId | |
| Test primer | CCACTTACCCACACTTGG | ||
| YE2575 | CGATGAGCTCATCATAGCATCAGCG | SacId | |
| YE2575 | GCGATCTAGATCACCAGCTTGC | XbaId | |
| Test primer | GCAGCAATTTCTGATGCG | ||
| ybcM | CGATGAGCTCTCAAGCATACGG | SacId | |
| ybcM | GCGATCTAGAGTCATTCGCTGG | XbaId | |
| Test primer | GGTCAATTGACAGGCAGC | ||
| Test primer | CCTGTTCAGCTACTGACG | pKRG9 | |
| Test primer | GTACTAAGCTCTCATGGC | pKRG9 | |
| Construction of pACYC184 complementation vectors | |||
| Test primer | GGTATTCACTCCAGAGCG | pACYC184 | |
| Test primer | ACCAGACCGTTCAGCTGG | pACYC184 | |
| YE2575 | CCTAGTACTCCGTATCGCTAACACC | ScaId | |
| YE2575 | CCTAGTACTGGATACCCAACTGAGG | ScaId | |
| uhpC | CCTGAATTCTCAAGCTTCAGCTTGTTTC | EcoRId | |
| uhpC | CCTGAATTCGGTGTATTGCACTGGC | EcoRId | |
| Cloning of gfp promoter probe vectors | |||
| cspB | GCATGTCGACCCATGTCTGATG | SalId | |
| cspB | CCGGAATTCGCCAATAGATGG | EcoRId | |
| gltP | GCATGTCGACCGCTACGTGATT | SalId | |
| gltP | CCGGAATTCCTTTGCATCCCA | EcoRId | |
| YE2463 | GCATGTCGACGGTTATAAGACGAATACC | SalId | |
| YE2463 | CCGGAATTCTTAATAAAATTAACCCGCG | EcoRId | |
| YE2537 | GCATGTCGACCCCAGAGTGCTT | SalId | |
| YE2537 | CCGGAATTCGCCAAGCTGAAT | EcoRId | |
| YE2575 | TAATGTCGACTAATGGGGTCAG | SalId | |
| YE2575 | GTTAGAATTCCATAAGTACGTC | EcoRId | |
| YE2848 | GCATGTCGACGCATGCACTTACCCG | SalId | |
| YE2848 | CCGGAATTCGGTGAAAGATTGTGCTG | EcoRId | |
| YE2922 | GAAAGTCGACTAATCTCCTCTTG | SalId | |
| YE2922 | TTAAGAATTCGAATGTTTTTCTTTGC | EcoRId | |
| YE0480 | CAACGTCGACAGGTGGTTCTAATGGC | SalId | |
| YE0480 | GCGTGAATTCGCGCACATTAAATTATAATTTTA | EcoRId | |
| YE0951 | GCATGTCGACGCGATGATAAAAACAGGG | SalId | |
| YE0951 | CCGGAATTCCACAAGAGCACTCACC | EcoRId | |
| YE0960 | GCATGTCGACGACTTCTGAGTGCTGG | SalId | |
| YE0960 | CCGGAATTCTGTGGCTGCACATGG | EcoRId | |
| Test primer | TCCGTATGTTGCATCACC | pPROBE-NT | |
| Test primer | GATCGGAAGCTTGCATGC | pPROBE-NT |
I-end sequencing.
O-end sequencing.
pUTmini-Tn5 luxCDABE Km2.
Chromosomal DNA of Y. enterocolitica W22703.
Recognition sites of restriction enzymes are underlined.
Construction of insertional knockout mutants and complementing vectors.
To generate knockout mutants of uhpC, ybcM, and YE2575 by plasmid insertion via homologous recombination, short intragenic fragments from the target genes were amplified from Y. enterocolitica chromosomal DNA using the primers listed in Table 2. Fragments were digested with XbaI (MBI Fermentas, Vilnius, Lithunia) and SacI (New England Biolabs, Beverly, Mass.) and ligated into XbaI/SacI-restricted suicide plasmid pKRG9. The recombinant plasmids were transformed into E. coli S17.1 λpir by electroporation and transferred into each individual Y. enterocolitica strain by plate mating (see above). Transformants were selected on plates containing nalidixic acid, kanamycin, and chloramphenicol. To exclude illegitimate recombination, the correct insertion of the recombinant plasmid was confirmed by PCR using a gene-specific test primer and a plasmid-derived primer. Y. enterocolitica transposon mutants with and without the insertional knockouts were grown in parallel at 30°C and 10°C to measure bioluminescence and optical density as described above. To complement the double mutants W22703-YE0480::lux-YE2575′ and W22703-YE0480::lux-uhpC′, the coding sequences of YE2575 and uhpC, the first gene together with 362 bp of its upstream sequence, were amplified with the appropriate oligonucleotides listed in Table 2. The resulting fragments were cloned via ScaI (YE2575) and EcoRI (uhpC) into pACYC184. In the resulting recombinant plasmids, pACYC-YE2575 and pACYC-uhpC, the direction of transcription of both genes corresponds to that of the disrupted gene encoding chloramphenicol acetyltransferase. The constructs were verified by PCR and restriction analysis and transformed by electroporation into W22703-YE0480::lux-YE2575′ and W22703-YE0480::lux-uhpC′, respectively.
Construction of transcriptional gfp fusions and in vitro fluorescence measurements.
Promoter regions were amplified from purified Y. enterocolitica DNA by PCR using the primers listed in Table 2. PCR products were digested with EcoRI and SalI (MBI Fermentas, Vilnius, Lithunia), ligated (T4 DNA ligase; Gibco, Hudson, N.H.) into the SalI/EcoRI site of pPROBE-NT (42) and transformed into DH5αMCR. Plasmids containing the correct transcriptional gfp fusions as verified by PCR were isolated (GenElute Plasmid Mini Prep Kit; Sigma-Aldrich, Taufkirchen, Germany) and transformed into Y. enterocolitica. Bacterial cells bearing the recombinant gfp promoter probe vectors were grown in parallel at 30°C and 10°C in LB medium supplemented with kanamycin. From both cultures, 5-ml samples were harvested at an OD600 of approximately 1.8, centrifuged (8,000 rpm for 2 min at RT), shock frozen, and stored at −70°C. Pellets were washed once with 500 μl phosphate-buffered saline, centrifuged again, and resuspended in 500 μl phosphate-buffered saline. Quantitative fluorescence assays were performed using a Wallac VICTOR2 1420 multilabel counter (Perkin Elmer Life Sciences, Turku, Finland) with sterile, untreated, black 96-well microplates (Nunc, Wiesbaden, Germany). Suspensions (each, 200 μl) were measured at wavelengths of 485 nm for excitation and 520 nm for emission. Intensity readings are presented as relative fluorescence units.
RESULTS
Identification of 42 cold-responsive transcriptional units.
Out of 5,700 transposon mutants, 4,454 (78%) mini-Tn5 luxCDABE insertions into Y. enterocolitica strain W22703 showed a response ratio of <2.0 or an average light emission at background level of 200 RLUs and were classified as not differentially expressed at both temperatures. Finally, 1,238 clones were identified to be steadily upregulated at 10°C with a response ratio of at least 2.0. Although a twofold difference in relative transcript levels is a threshold commonly used for analysis and interpretation of microarray data (32), we used a threshold of 5.0 to further analyze only mutants with strongly elevated activity of the respective promoter. From the 222 mutants matching this criteria, about 40% showed light emission profiles congruent to those of mutants already identified, indicating a redundant transposon insertion, or exhibited unsteady, unreproducible profiles with strong data fluctuation. Those perturbations were especially derived from promoters with transcriptional activity slightly above background. Another 11% were withdrawn because they were false positive, due to neighboring mutants with strongly up-regulated light production in the microtiter plate. In total, we selected 109 clones for further characterization, representing approximately 1.9% of the 5,700 transposon mutants, plus 5 clones as examples for strongly inhibited luciferase activity at low temperature. The DNA of those mutants for which we observed response ratios of >5.0 or <0.6 and a steady expression pattern was digested and used for inverse PCR as described. The sequences of chromosomal fragments flanking the transposon insertion were compared to the Y. enterocolitica DNA assembly, and the results of the BLAST analysis allowed the precise localization of the transposon insertion and the orientation of the inserted reporter genes with respect to the Y. enterocolitica genome sequence, except for strain-specific fragments. This analysis revealed 42 different genes or open reading frames (ORFs) with enhanced (37 genes) or reduced (5 genes) promoter activity at 10°C compared to 30°C, 13 genes of which are probably localized within putative operon structures. This finding suggests that all proteins encoded by these putative operons are present in the cell in higher or lower copy numbers in response to low temperature. It should be noticed that the 37 genes represent all mutants characterized by a response ratio of the luciferase reporter of >5.0, whereas the five strains with down-regulated promoter activity are examples only of a larger group of mutants not further characterized. For two transposon insertions, no promoter responsible for luciferase induction could be identified, due to preliminary annotation of the respective genome region. An accumulation of independent transposon insertions was found within three ORFs or their surroundings, namely 13 insertions in both tcaB and tcaA and 10 insertions in a putative alkaline serine protease (YE2922). All transposon insertions are listed in Table 3.
TABLE 3.
| Insertion site | Gene | Homologue | Protein specification | Gene fusion structure | Induction (OD405) | Classification | Response ratio |
|---|---|---|---|---|---|---|---|
| Up-regulated genes | |||||||
| 3387325 | YE3114b | ymoAb | Modulator YmoA | P-luxCDABE-ymoA | 0.19 | Regulation | 123 |
| 4168711 | YE3823c | cspAc | Cold shock protein A | P-cspA′-luxCDABE | 0.20 | Regulation | 84 |
| 1066450 | YE0940 | cspB | Cold shock protein B | P-cspB′-luxCDABE | 0.28 | Regulation | 268 |
| 368636 | YE0310 | gltP | Glutamate-aspartate symport protein | P-gltP′-luxCDABE | 0.29 | Transport | 46 |
| 3033404 | YE2793 | Unknown protein | P-YE2793′-luxCDABE | 0.33 | 33 | ||
| 683797 | YE0595 | arcA | Aerobic respiration control protein ArcA | P-arcA′-luxCDABE | 0.37 | Regulation | 8 |
| 4470965 | YE4087 | uhpC | Hexose phosphate transport system regulatory protein | P-YE4090-uhpA-uhpB-uhpC′-luxCDABE | 0.41 | Regulation | 36 |
| 2732143 | YE2537 | Flagellar basal body M-ring protein | P-YE2537′-luxCDABE-fliG-fliH-fliI | 0.50 | Motility | 33 | |
| 1612069 | YE1436 | Putative transcription regulatory protein | P-YE1436′-luxCDABE | 0.52 | Regulation | 33 | |
| Strain specific | No homology | 0.53 | 6 | ||||
| 2898299 | YE2673 | Putative chemotaxis signal transduction protein | P-YE2673-luxCDABE | 0.53 | Motility/signaling | 801 | |
| 2722746 | YE2527 | ybcM | Putative AraC type regulatory protein | P-ybcM′-luxCDABE | 0.54 | Motility | 176 |
| 2649603 | YE2463 | Putative outer membrane porin | P-YE2463′-luxCDABE | 0.54 | Transport | 41 | |
| 2248595 | YE2057 | ssrAB-activated, putative virulence factor SrfA | P-YE2057′-luxCDABE-YE2058-YE2059 | 0.54 | Virulence | 25 | |
| 2259046 | YE2063 | Hypothetical membrane protein | P-YE2063′-luxCDABE | 0.55 | 73 | ||
| 2772356 | YE2577 | cheA | Chemotaxis protein | P-motA-motB-cheA′-luxCDABE-cheW | 0.57 | Regulation | 335 |
| Strain specific | No homology | 0.58 | 256 | ||||
| 2716443 | YE2521 | fleC | Flagellin FleC | P-fleA-fleB-fleC′-luxCDABE | 0.59 | Motility | 27 |
| 2769982 | YE2575 | Methyl-accepting chemotaxis protein | P-YE2575 (MCPI)′-luxCDABE-tap(MCPIV)-cheR-cheB | 0.60 | Motility/signaling | 32 | |
| 2722197 | YE2526 | fliT | Repressor of class II promoters | P-fliT′-luxCDABE | 0.60 | Motility | 22 |
| Strain specific | tcaB | Putative toxin subunit | P-tcaB′-luxCDABE | 0.65 | Virulence | 81 | |
| 2721668 | YE2525 | fliS | Putative cytoplasmic chaperone | P-luxCDABE-fliS | 0.65 | Motility | 8 |
| 4432406 | YE4063 | Hypothetical protein with EAL domain | P-YE4063′-luxCDABE | 0.66 | Signaling | 1,147 | |
| Strain specific | Hypothetical protein | 0.67 | 26 | ||||
| 1476057 | YE1324 | Hypothetical protein with EAL domain (signal transduction) | P-YE1324′-luxCDABE | 0.73 | Signaling | 98 | |
| 1074733 | YE0951b | Urea amidohydrolase | P-luxCDABE-YE0951-yeuB-ureC-ureE-ureF | 0.74 | Metabolism | 17 | |
| 2783524 | YE2586 | Mg2+ transport ATPase protein C mgtC | P-luxCDABE-YE2586 | 0.75 | Metabolism | 8 | |
| 2715679 | YE2520 | fliB | Lysin-N-methylase | P-fliB′-luxCDABE | 0.75 | Motility | 150 |
| Strain specific | tcaA | Putative toxin subunit | P-tcaA′-luxCDABE | 0.77 | Virulence | 12 | |
| 1086842 | YE0964 | Probable membrane protein | P-YE0964′-luxCDABE-YE0963 | 0.79 | 6 | ||
| 1084151 | YE0960d | Urea operon | P-YE0959-YE0960′-luxCDABE | 0.81 | Metabolism | 12 | |
| 3177216 | YE2922 | Alkaline serine protease (subtilisin) | P-YE2922′-luxCDABE | 0.83 | Metabolism | 38 | |
| 4369922 | YE4013 | 1,4-α-glucan branching enzyme GlgB | P-luxCDABE-YE4013-YE4012-YE4011 | 0.83 | Metabolism | 26 | |
| 3095861 | YE2848 | Putative chemotaxis methyl-accepting transducer | P-YE2848′-luxCDABE | 0.84 | Signaling | 95 | |
| 555157 | YE0480 | Transport of large molecules/secretion protein; Fhac homologue | P-YE0480′-luxCDABE | 0.84 | Virulence | 58 | |
| 3292901 | YE3021 | Histidine ammonia-lyase (histidase), HutH | P-YE3021′-luxCDABE-YE3020 | 0.89 | Metabolism | 13 | |
| 4037041 | YE3697 | Permease of the drug/metabolite transporter (DMT) superfamily | P-YE3697′-luxCDABE | 0.90 | Metabolism | 15 | |
| Down-regulated genes | |||||||
| 4116172 | YE3774 | Protein-Npi-phosphohistidine-sugar phosphotransferase | P-YE3774′-luxCDABE-YE3773 | 0.98 | Metabolism | 0.6 | |
| 4509439 | YE4121 | Sugar-binding periplasmic protein | P-YE4121′-luxCDABE-YE4120 (sugar transport ATP-binding protein)-YE4119 (d-xylose ABC transporter)-YE4118 (probable AraC family transcription regulatory protein) | 1.00 | Metabolism | 0.5 | |
| 4562537 | YE4164 | Heat shock protein | P-YE4164′-luxCDABE | 0.84 | 0.2 | ||
| p5086 | Not annotated | ND | 0.2 | ||||
| Strain specific | Prophage P2 OGR protein | 0.87 | 0.3 |
The first column depicts the transposon insertion with respect to the genome sequence of Y. enterocolitica 8081. Mutagenized genes are indicated by their number taken from the Y. enterocolitica annotation at the Sanger Institute homepage (column 2). Gene names and further protein specifications were obtained from BLAST analysis (columns 3 and 4). The prime symbols in column 5 mark truncated genes. ND, not determinable.
Mini-Tn5 luxCDABE genes were inserted in opposite to the putative promoters of ymoA and YE0951.
Transposon insertion was in strain NCTC10460.
As shown by a gfp fusion construct, the luxCDABE insertion is transcribed from the promoter upstream of ureD.
No genes indispensable for growth at low temperature were identified.
Given that the library is random and that the genome of Y. enterocolitica is composed of approximately 2,500 transcriptional units similar to E. coli (68), our library should allow testing of approximately 70% of all genes or operons for differential expression (38). For this calculation, we assumed that the mini-Tn5 luxCDABE insertion in 50% of the mutants was not under control of a promoter located upstream. Despite this high knockout fraction, none of the insertions affected a gene or gene complex that is dispensable for cell growth at 30°C but specifically required for growth at low temperatures. Of the tested mutants, 0.5% were growth deficient at 10°C, as well as at 30°C.
gfp reporter fusions validate gene identification.
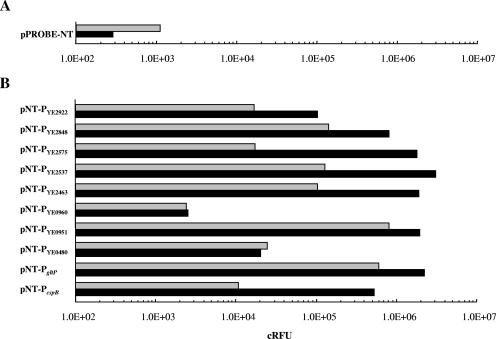
To prove the differential transcription of the identified genes, we cloned the first assigned or putative promoter regions located upstream of nine mini-Tn5 luxCDABE insertions and the promoter region of cspB as a control into vector pPROBE-NT and measured GFP expression at 30°C and 10°C. Since the autocatalytic protein modification required for fluorescent light emission by GFP proceeds slowly in bacterial cells (2), fluorescence values were taken at an OD600 value of 1.8. The background activity derived from a nonrecombinant vector in Y. enterocolitica alone was 3.9-fold (±0.3) higher at 30°C than at 10°C, and this factor was used to normalize the derived gfp reporter activities at 10°C (Fig. 2). This factor describes the reduced synthesis rate of GFP, due to the temperature decrease, and correlates with the Arrhenius equation predicting a factor of 1.5 to 4.0 (57). In all cases but one, the investigated promoters were shown to be induced 1.1 to 102 fold with an absolute range of fluorescence units from 2,536 RFU (urease operon) to 3,119,696 RFU (flagellar operon) at 10°C. An exception was observed when a gfp fusion to a putative promoter region directly upstream of YE0480 was tested. However, further annotation studies revealed that YE0480 is probably transcriptionally coupled to gene YE0479, a finding that might explain the negative result. In another cold-responsive mutant that we examined by GFP fusions, the luxCDABE genes had inserted between the ORFs YE0950 and YE0951, and a putative promoter upstream of YE0951 was found to be opposite to the direction of luxCDABE transcription. However, when the predicted promoter was amplified from this intergenic region and cloned into pPROBE-NT in correct orientation, a 2.4-fold induction of GFP at low temperature was observed (Fig. 2). This result and promoter prediction studies suggest that the selected promoter directs the polycistronic transcription of five genes (YE0951-yeuB-ureC-ureE-ureF) involved in urea metabolism. For all but one promoter probe constructs, the response ratio of gfp was significantly lower than the values deduced from the bioluminescence signal. These discrepancies are probably due to differences in both assays in copy numbers of reporter genes, half-lives, number of genes involved in light emission, reporter sensitivity, and experimental design.
FIG. 2.
Activitiy of gfp promoter fusions at 30°C (gray bars) and 10°C (black bars). Promoter regions of selected cold-induced genes were cloned into the gfp promoter probe vector pPROBE-NT and transformed into wild-type strain Y. enterocolitica W22703. (A) GFP expression of plasmid pPROBE-NT without recombinant promoter region at 30°C and 10°C. (B) Fluorescence measurement of 10 promoter probe constructs. The quotient of the absolute values at 10°C and 30°C obtained from the results shown in panel A was used as a correction factor. Accordingly, the absolute fluorescence values obtained from cultures grown at 10°C were multiplied by the factor 3.9 (±0.3) to give the indicated promoter activities. The data represent the means of two to five independent measurements.
Low temperature-responsive genes of Y. enterocolitica.
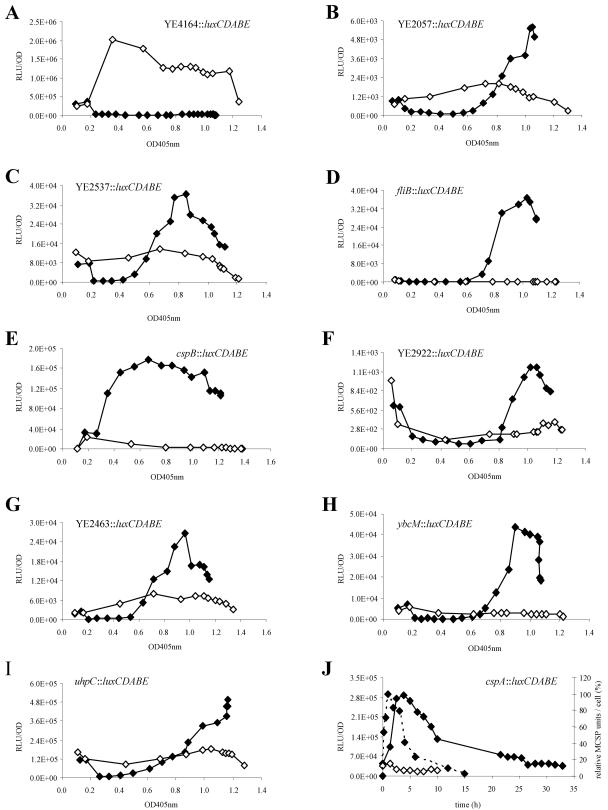
The kinetic data derived from the light emission of transposon recipients grown at 30°C and at 10°C revealed a pattern of varying gene expression profiles due to mini-Tn5 luxCDABE insertions under the control of temperature-responsive promoters (Fig. 3). The interpretation of these profiles allowed (i) determination of luciferase induction with respect to the OD405 value, (ii) quantification of the transcriptional response to low temperature by the normalized response ratios of luciferase fusions, and (iii) qualification of the induction as transient or permanent for a certain time interval. The response ratios of the mentioned genes were ranged from 6 to 1,147 (Table 3), with the maximal induction of a hypothetical protein (YE4063) that was not transcribed at 30°C and showed pronounced similarity (E value, 10−24) to conserved domains of signaling proteins. A summary of the most important genes induced during long-term response of Y. enterocolitica is provided in Fig. 4. In contrast to cold shock proteins (CSPs), several transcriptional profiles showed no clear peak but steadily increased after temperature downshift and remained at a higher expression level, e.g., the mutants YE2057::luxCDABE and uhpC::luxCDABE (Fig. 3B and I). On the other hand, the change in promoter induction was only transient in transposon mutants like YE2537::luxCDABE (Fig. 3C), indicating a maximal expression of the basal body M-ring protein at an OD405 of 0.8.
FIG.3.
Profiles of light emission were monitored during prolonged growth of Y. enterocolitica luxCDABE transposon mutants at 10°C (solid diamonds) and 30°C (open diamonds). Selected diagrams show significant decreases (A) or increases (B to J) in promoter activity in response to temperature decline. Absolute RLU values, instead of corrected light emission values considering the temperature effect, were used for the graphs. (J) The dashed line in the diagram of the cspA::luxCDABE mutant shows the relative amount of major cold shock proteins (MCSPs) following a cold shock from 30°C to 10°C, as determined by two-dimensional gel electrophoresis, with the maximum level of relative MCSP units per cell set at 100% (47).
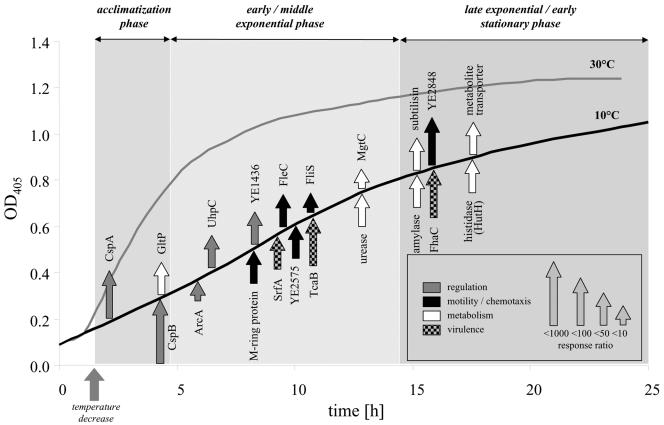
FIG. 4.
Portrait of the long-term response of Y. enterocolitica to low temperature. The temporal order and strength of gene induction are depicted. The genes were grouped into three categories: sensoring-regulation, motility-chemotaxis-virulence, and degradative metabolism. In a few cases, we used the names of homologues instead of noting the correct Y. enterocolitica nomenclature.
The genes of 37 transcriptional units significantly up-regulated during growth at 10°C were grouped into three classes according to the growth phase at which transcriptional induction was detected. At acclimatization phase (OD405 < 0.3), two transposon insertions were located within the genes encoding CspA and CspB, showing a strong normalized response ratio of 84 and 268 after cold shock. CspA was transiently expressed with a peak 4 h after temperature downshift (Fig. 3J), while light emission driven by the cspB promoter decreased only slightly during prolonged growth (Fig. 3E). The light emission profile of the cspA::luxCDABE mutant correlated with Northern blot analysis, showing inhibited cspA transcription 120 min after cold shock (46) and with the relative amount of the major CSPs as determined by two-dimensional gel electrophoresis (Fig. 3J) (45), thus demonstrating the validity of chromosomal luxCDABE promoter fusions as a tool to measure transcriptional activities. The temporal distance between the maximal amount of CspA and the corresponding luciferase activity was calculated as 180 to 195 min. The sustained luciferase activity of the cspB::luxCDABE knockout mutant might be explained by an autoregulatory effect known to occur for E. coli cspA (4). Parallel to both CSPs, mini-Tn5 luxCDABE insertions within the promoter region of ymoA, which plays a role in thermoregulation of virulence functions (15), and within the ORF of gltP, encoding a glutamate-aspartate symport protein, were shown to be upregulated within the acclimatization phase.
Early and mid-exponential growth phase (0.3 < OD405 < 0.8).
Seventy percent of the transcriptional units identified in our approach were induced at 10°C before they reached the late growth phase, most of them involved in sensoring-regulation, motility, and virulence. Besides several as-yet-uncharacterized genes, we found arcA (YE0595), which is necessary to survive prolonged starvation (59), uhpABC, controlling the expression of the hexose phosphate transporter UhpT (70), and two obviously strain-specific loci without homology to known genes. The most prominent response to temperature decrease was the enhanced transcription of motility and chemotaxis genes such as the flagellin operon or cheA. Other differentially expressed genes are YE2057, which encodes a homologue of the putative virulence factor SrfA of Salmonella, mgtC (YE2586) involved in magnesium uptake (44, 65), and YE2463 encoding an outer membrane porin. Surprisingly, we identified two mini-Tn5 luxCDABE insertions in the genes tcaA and tcaB encoding two subunits of Tca, a homologue of the insecticidal toxin complex proteins from Photorhabdus luminescens, and we measured response ratios of 12 and 81 when cultivating these mutants at 10°C (11). We also observed the induction of two putative operons involved in urease activity. One was YE0951-yeuB-ureC-ureE-ureF, encoding urea amidohydrolase, an accessory protein, and three urease subunits. The other operon was YE0959-YE960, which encodes a urease accessory protein and a urea transporter.
Late exponential and early stationary growth phase (OD > 0.8).
During late exponential growth, we observed the stimulation of the following genes: YE0480, encoding a predicted hemolysin transporter with homology to the accessory protein FhaC from Bordetella pertussis (identity, 28%; E value, 2−47); the monocistronically organized gene YE2922, encoding a homologue of subtilisin (identity 33%; E value, 6−21), which is an alkaline serine protease associated with the onset of sporulation in B. subtilis (67); two genes encoding a histidine ammonia-lyase (hutH) and a putative amylase (YE4013); and one gene (YE3697), encoding a permease of the drug/metabolite transporter (DMT) superfamily.
Down-regulated promoters.
Due to its high dynamic range, the luciferase reporter also allows the assessment of decreased transcriptional activity in long-term response to temperature changes (75). We mapped five mini-Tn5 luxCDABE insertion mutants responding to growth at 10°C with strong reductions in luciferase activity. One of these insertions was found to be located within gene YE4164, encoding a protein identical to heat shock proteins of the IbpA family, probably cotranscribed with YE4163, which encodes an IbpB-like heat shock protein. At 10°C, very low luciferase activity corresponding to the background level was observed for the putative promoters in front of gene YE4121, which encodes a sugar-binding periplasmic protein, and upstream of YE3774, which encodes a putative phosphotrehalase as part of a phosphotransferase system. This is in line with the observed elevated transcription levels of cheA at low temperature, resulting in a tumbling behavior of bacterial cells. The activity of the phosphotransferase system, however, that transports carbohydrate attractants into the cell is known to extend swimming runs (40). Interestingly, the sequence obtained from another transposon insertion was identical to an as-yet-unannotated region of the Yersinia virulence plasmid pYVe8081 immediately downstream of yopT/sycT, and the maximal light emission of 20,489 RLU/OD obtained for the respective mutant at 30°C was reduced by 1 order of magnitude to 3,865 RLU/OD (Table 3).
Inhibitory effect of UhpC and MCPI on the expression of YE0480.
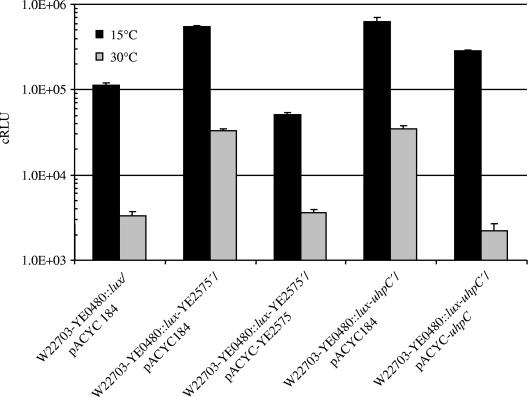
The above-described determination of induction time points of gene expression should allow potential interdependencies between the cold-responsive genes found to be uncovered. To examine the effect of knockouts of three regulatory genes from Table 3 on selected transposon mutants, insertional duplication mutagenesis of the three regulatory genes uhpC, ybcM, and YE2575 was performed with a set of 24 temperature-responsive luxCDABE mutants, resulting in a total of 72 double mutants. None of the insertion mutants showed deficient growth at 10°C or 30°C, indicating that the double knockout did not affect cell viability (data not shown). The promoter activities of the double mutants were measured at 10°C and 30°C, and the data were compared to the bioluminescence values of the corresponding mutants without additional insertional knockout mutations. In most cases, this analysis did not reveal significant changes in promoter activities of the double knockout mutants in comparison to the single luxCDABE insertion mutants. However, two exceptions were observed: the activity of the lux reporter inserted in YE0480, encoding an FhaC-like transporter, was enhanced by the factor 4.8 and 5.6 at 15°C as a result of the insertional knockouts of YE2575 and uhpC, respectively, and by a factor of approximately 10 at 30°C (Fig. 5). The cloned uhpC and YE2575 genes complemented the mutations; in the case of uhpC, the light signal of the complemented strains exceeded that of W22703-YE0480::lux, probably due to the lack of the native uhpC promoter. These results indicate that the two-component system UhpABC, as well as methyl-accepting chemotaxis protein I (MCPI) involved in chemotaxis, negatively regulates the expression of the putative hemolysin transporter encoded by YE0480.
FIG. 5.
Inhibitory effect of the insertional knockout of uhpC and YE2575 encoding MCPI on the transcription of YE0480::luxCDABE at 30°C and 15°C. Luciferase activities are shown in corrected RLU. The data represent the means and standard errors of means for three independent measurements.
DISCUSSION
Cold acclimatization versus long-term adaptation.
In many cases, gene induction has been studied immediately after sudden changes, e.g., cold shock, acid shock, or heat shock, while much less effort has been dedicated to analyzing gene expression during prolonged growth under a specific environmental stress. It is a significant result of this work that there is no overlap of the well-described cold acclimatization response directly following cold shock and the genes induced during exponential growth at low temperature. This is supported by our negative results of an in silico search for consensus regulatory regions within promoter regions or translational start regions of cold shock genes, e.g., the putative downstream box or the cold box (22, 43). Similar observations have been reported for the UV-B shock response and subsequent growth under elevated UV-B of the cyanobacterium Nostoc commune (19), and for L. monocytogenes grown to mid-exponential phase (39). The pronounced separation between shock response and long-term adaptation may be a general feature of bacterial gene expression induced by environmental stress.
Y. enterocolitica virulence factors at environmental temperature.
At least two virulence factors of Y. enterocolitica have been known to be maximally expressed at ambient temperatures, the heat-stable enterotoxin Yst (10) and the outer-membrane protein Inv (53). One of the most important aspects of differential gene expression in terms of the Y. enterocolitica temperature response that emerged from our study is the enhanced transcription of another four genes associated with virulence functions. At low temperature, we observed the upregulation of tcaA and tcaB encoding homologues of the insecticidal toxin subunits from P. luminescens that is found in the gut of entomopathogenic nematodes (72). Extracts of Y. enterocolitica grown at 10°C but not 30°C were shown to be lethal for insect larvae, and tcaA was demonstrated to be essential for this insecticidal activity. This finding gave rise to speculations about an as-yet-undisclosed niche of Y. enterocolitica in insects (11). The transcription of another putative virulence factor, a homologue of the SsrAB-regulated factor SrfA (76), was also shown to be low-temperature responsive. This is in line with the observation that srfA transcription is repressed fivefold during intracellular growth in macrophages at 37°C (21). Interestingly, microarray data of Salmonella typhimurium motility only recently revealed the surface-dependent regulation of several virulence genes, among them srfABC, thus demonstrating the association between swarming behavior and virulence, as observed with many bacteria (28, 71). YE0480 is possibly involved in hemolysin secretion, and hemolysins are predicted determinants of Photorhabdus pathogenicity against insects (12). It is remarkable that neither tcaA, tcaB, srfA, nor YE0480 showed significant transcriptional activity at 37°C (data not shown). The correlation of bacterial pathogenicity with exponential phase is reminiscent of a molecular model that describes factors involved in the association of Photorhabdus with nematodes and insects (24). Taken together, these data suggest a role of these virulence factors outside mammalian hosts, possibly in invertebrates.
Environmental sensing and motility.
The specific expression profile at low temperature discussed here strongly supports the view of temperature as an important stimulus for the long-term adaptation of Y. enterocolitica to environmental changes, mediated by sensory proteins and regulatory networks. In our approach, we observed the induction of several regulatory systems of known (ArcA and UhpABC) and predicted (YE1436, ybcM, YE4063, and YE1324) function, many of them involved in signal transduction (Table 3). The ArcAB system controls the expression of at least 40 operons involved in catabolic gene expression (3, 59), and arcA is suggested to control the rate of utilization of endogenous reserves (49). None of our results points to the presence of a cold regulon during prolonged growth at 10°C. However, YE0480, a gene encoding a homologue of the accessory protein FhaC from B. pertussis, is strongly expressed at 10°C but not at 37°C, and its transcription is negatively influenced by MCPI and UhpC, suggesting that YE0480 expression is connected with nutrient acquisition and motility regulons. Interestingly, a similar regulatory circuit was reported recently for expression and activity of a Xenorhabdus nematophila hemolysin that is required for full virulence against insects (16). These interdependencies confirm the linkage between motility and virulence properties at the level of regulation (18). The induction of flagellae and chemotaxis genes at low temperature is a well-characterized phenomenon that is in line with the finding that loss of motility at 37°C does not affect pathogenesis of yersiniae in humans (33), indicating a role of motility in the colonization of new habitats. The master flagellum regulatory operon, flhDC, is assumed to be required for full Xenorhabdus virulence against insects. Moreover, the flagellar sigma factor of Y. enterocolitica, σ28, controls the phospholipase gene yplA, whose product is exported by the flagellum secretion apparatus (58).
Degradative metabolism: further evidence for an insect stage during the Y. enterocolitica life cycle.
A predominant feature of the derived expression profile at environmental temperatures is the prevalence of genes involved in biodegradative metabolism during late growth and stationary phase. We found the significant upregulation of two urea operons and of a histidine ammonia-lyase (YE3021) with homology to the HutH histidase, all of them involved in nitrogen utilization. In addition, low temperature results in elevated expression of the α-glucan branching enzyme GlgB (YE4013) that, together with the α-amylase GlgX and GlgC, regulates the glycogen metabolism (56). Interestingly, the degradative hut operon, as well as an α-amylase, have been shown to be upregulated at low temperature in several antarctic psychrotrophic bacteria (23, 36). A role of the Y. enterocolitica alkaline serine protease, a hot spot of mini-Tn5 luxCDABE insertions and a member of the extracellular subtilisin family (67), has not yet been described. Proteases, however, represent exoenzymes that might help to degrade tissues and macromolecules of a eukaryotic host. None of the genes mentioned as being involved in degradative metabolism is expressed at 37°C (data not shown), again suggesting a role of these enzymes outside a mammalian host. It is therefore tempting to speculate that these genes play a role in bioconversion of an insect cadaver, providing a nutrient base for bacterial growth. Taken together, the long-term portrait of cold-responsive genes as derived from the luciferase assay is a promising starting point for further analysis of the association of Y. enterocolitica with invertebrates.
Acknowledgments
We are grateful to Samir Velagic and Nicole Büchl for technical assistance, to Monica Dommel for critical reading of the manuscript, to Kevin Francis for the gift of pUT mini-Tn5 luxCDABE Km2 and continuous encouragement, and to Stanley Lindow for the gift of the pPROBE-NT vector.
REFERENCES
- 1.Abee, T., and J. A. Wouters. 1999. Microbial stress response in minimal processing. Int. J. Food Microbiol. 50:65-91. [DOI] [PubMed] [Google Scholar]
- 2.Albano, C. R., L. Randers-Eichhorn, W. E. Bentley, and G. Rao. 1998. Green fluorescent protein as a real time quantitative reporter of heterologous protein production. Biotechnol. Prog. 14:351-354. [DOI] [PubMed] [Google Scholar]
- 3.Alexeeva, S., K. J. Hellingwerf, and M. J. Teixeira de Mattos. 2003. Requirement of ArcA for redox regulation in Escherichia coli under microaerobic but not anaerobic or aerobic conditions. J. Bacteriol. 185:204-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bae, W., P. G. Jones, and M. Inouye. 1997. CspA, the major cold shock protein of Escherichia coli, negatively regulates its own gene expression. J. Bacteriol. 179:7081-7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bayles, D. O., B. A. Annous, and B. J. Wilkinson. 1996. Cold stress proteins induced in Listeria monocytogenes in response to temperature downshock and growth at low temperatures. Appl. Environ. Microbiol. 62:1116-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beckering, C. L., L. Steil, M. H. Weber, U. Volker, and M. A. Marahiel. 2002. Genomewide transcriptional analysis of the cold shock response in Bacillus subtilis. J. Bacteriol. 184:6395-6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger, F., N. Morellet, F. Menu, and P. Potier. 1996. Cold shock and cold acclimation proteins in the psychrotrophic bacterium Arthrobacter globiformis SI55. J. Bacteriol. 178:2999-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bottone, E. J. 1999. Yersinia enterocolitica: overview and epidemiologic correlates. Microbes Infect. 1:323-333. [DOI] [PubMed] [Google Scholar]
- 9.Bottone, E. J. 1997. Yersinia enterocolitica: the charisma continues. Clin. Microbiol. Rev. 10:257-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyce, J. M., E. J. Evans, Jr., D. G. Evans, and H. L. DuPont. 1979. Production of heat-stable, methanol-soluble enterotoxin by Yersinia enterocolitica. Infect. Immun. 25:532-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bresolin, G., J. A. W. Morgan, D. Ilgen, S. Scherer, and T. M. Fuchs. 2006. Low temperature-induced insecticidal activity of Yersinia enterocolitica. Mol. Microbiol. 59:503-512. [DOI] [PubMed] [Google Scholar]
- 12.Brillard, J., E. Duchaud, N. Boemare, F. Kunst, and A. Givaudan. 2002. The PhlA hemolysin from the entomopathogenic bacterium Photorhabdus luminescens belongs to the two-partner secretion family of hemolysins. J. Bacteriol. 184:3871-3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cornelis, G., and C. Colson. 1975. Restriction of DNA in Yersinia enterocolitica detected by recipient ability for a derepressed R factor from Escherichia coli. J. Gen. Microbiol. 87:285-291. [DOI] [PubMed] [Google Scholar]
- 15.Cornelis, G. R., C. Sluiters, I. Delor, D. Geib, K. Kaniga, C. Lambert de Rouvroit, M. P. Sory, J. C. Vanooteghem, and T. Michiels. 1991. ymoA, a Yersinia enterocolitica chromosomal gene modulating the expression of virulence functions. Mol. Microbiol. 5:1023-1034. [DOI] [PubMed] [Google Scholar]
- 16.Cowles, K. N., and H. Goodrich-Blair. 2005. Expression and activity of a Xenorhabdus nematophila haemolysin required for full virulence towards Manduca sexta insects. Cell. Microbiol. 7:209-219. [DOI] [PubMed] [Google Scholar]
- 17.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DiRita, V. J., N. C. Engleberg, A. Heath, A. Miller, J. A. Crawford, and R. Yu. 2000. Virulence gene regulation inside and outside. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 355:657-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ehling-Schulz, M., S. Schulz, R. Wait, A. Görg, and S. Scherer. 2002. The UV-B stimulon of the terrestrial cyanobacterium Nostoc commune comprises early shock proteins and late acclimation proteins. Mol. Microbiol. 46:827-843. [DOI] [PubMed] [Google Scholar]
- 20.Ellison, D. W., B. Young, K. Nelson, and V. L. Miller. 2003. YmoA negatively regulates expression of invasin from Yersinia enterocolitica. J. Bacteriol. 185:7153-7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eriksson, S., S. Lucchini, A. Thompson, M. Rhen, and J. C. Hinton. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47:103-118. [DOI] [PubMed] [Google Scholar]
- 22.Fang, L., Y. Hou, and M. Inouye. 1998. Role of the cold-box region in the 5′ untranslated region of the cspA mRNA in its transient expression at low temperature in Escherichia coli. J. Bacteriol. 180:90-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feller, G., T. Lonhienne, C. Deroanne, C. Libioulle, J. Van Beeumen, and C. Gerday. 1992. Purification, characterization, and nucleotide sequence of the thermolabile alpha-amylase from the antarctic psychrotroph Alteromonas haloplanctis A23. J. Biol. Chem. 267:5217-5221. [PubMed] [Google Scholar]
- 24.ffrench-Constant, R., N. Waterfield, P. Daborn, S. Joyce, H. Bennett, C. Au, A. Dowling, S. Boundy, S. Reynolds, and D. Clarke. 2003. Photorhabdus: towards a functional genomic analysis of a symbiont and pathogen. FEMS Microbiol. Rev. 26:433-456. [DOI] [PubMed] [Google Scholar]
- 25.Graumann, P., T. M. Wendrich, M. H. Weber, K. Schröder, and M. A. Marahiel. 1997. A family of cold shock proteins in Bacillus subtilis is essential for cellular growth and for efficient protein synthesis at optimal and low temperatures. Mol. Microbiol. 25:741-756. [DOI] [PubMed] [Google Scholar]
- 26.Graumann, P. L., and M. A. Marahiel. 1999. Cold shock response in Bacillus subtilis. J. Mol. Microbiol. Biotechnol. 1:203-209. [PubMed] [Google Scholar]
- 27.Han, Y., D. Zhou, X. Pang, L. Zhang, Y. Song, Z. Tong, J. Bao, E. Dai, J. Wang, Z. Guo, J. Zhai, Z. Du, X. Wang, P. Huang, and R. Yang. 2005. DNA microarray analysis of the heat- and cold-shock stimulons in Yersinia pestis. Microbes Infect. 7:335-348. [DOI] [PubMed] [Google Scholar]
- 28.Harshey, R. M. 2003. Bacterial motility on a surface: many ways to a common goal. Annu. Rev. Microbiol. 57:249-273. [DOI] [PubMed] [Google Scholar]
- 29.Hebraud, M., E. Dubois, P. Potier, and J. Labadie. 1994. Effect of growth temperatures on the protein levels in a psychrotrophic bacterium, Pseudomonas fragi. J. Bacteriol. 176:4017-4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hebraud, M., and P. Potier. 1999. Cold shock response and low temperature adaptation in psychrotrophic bacteria. J. Mol. Microbiol. Biotechnol. 1:211-219. [PubMed] [Google Scholar]
- 31.Hurme, R., and M. Rhen. 1998. Temperature sensing in bacterial gene regulation—what it all boils down to. Mol. Microbiol. 30:1-6. [DOI] [PubMed] [Google Scholar]
- 32.Ichikawa, J. K., A. Norris, M. G. Bangera, G. K. Geiss, A. B. van't Wout, R. E. Bumgarner, and S. Lory. 2000. Interaction of Pseudomonas aeruginosa with epithelial cells: identification of differentially regulated genes by expression microarray analysis of human cDNAs. Proc. Natl. Acad. Sci. USA 97:9659-9664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iriarte, M., I. Stainier, A. V. Mikulskis, and G. R. Cornelis. 1995. The fliA gene encoding σ28 in Yersinia enterocolitica. J. Bacteriol. 177:2299-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones, P. G., and M. Inouye. 1994. The cold-shock response—a hot topic. Mol. Microbiol. 11:811-818. [DOI] [PubMed] [Google Scholar]
- 35.Kaan, T., G. Homuth, U. Mader, J. Bandow, and T. Schweder. 2002. Genome-wide transcriptional profiling of the Bacillus subtilis cold-shock response. Microbiology 148:3441-3455. [DOI] [PubMed] [Google Scholar]
- 36.Kannan, K., K. L. Janiyani, S. Shivaji, and M. K. Ray. 1998. Histidine utilisation operon (hut) is upregulated at low temperature in the antarctic psychrotrophic bacterium Pseudomonas syringae. FEMS Microbiol. Lett. 161:7-14. [DOI] [PubMed] [Google Scholar]
- 37.Konkel, M. E., and K. Tilly. 2000. Temperature-regulated expression of bacterial virulence genes. Microbes Infect. 2:157-166. [DOI] [PubMed] [Google Scholar]
- 38.Lee, M. S., C. Seok, and D. A. Morrison. 1998. Insertion-duplication mutagenesis in Streptococcus pneumoniae: targeting fragment length is a critical parameter in use as a random insertion tool. Appl. Environ. Microbiol. 64:4796-4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu, S., J. E. Graham, L. Bigelow, P. D. Morse, 2nd, and B. J. Wilkinson. 2002. Identification of Listeria monocytogenes genes expressed in response to growth at low temperature. Appl. Environ. Microbiol. 68:1697-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lux, R., K. Jahreis, K. Bettenbrock, J. S. Parkinson, and J. W. Lengeler. 1995. Coupling the phosphotransferase system and the methyl-accepting chemotaxis protein-dependent chemotaxis signaling pathways of Escherichia coli. Proc. Natl. Acad. Sci. USA 92:11583-11587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maoz, A., R. Mayr, G. Bresolin, K. Neuhaus, K. P. Francis, and S. Scherer. 2002. Sensitive in situ monitoring of a recombinant bioluminescent Yersinia enterocolitica reporter mutant in real time on Camembert cheese. Appl. Environ. Microbiol. 68:5737-5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller, W. G., J. H. Leveau, and S. E. Lindow. 2000. Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol. Plant-Microbe Interact. 13:1243-1250. [DOI] [PubMed] [Google Scholar]
- 43.Mitta, M., L. Fang, and M. Inouye. 1997. Deletion analysis of cspA of Escherichia coli: requirement of the AT-rich UP element for cspA transcription and the downstream box in the coding region for its cold shock induction. Mol. Microbiol. 26:321-335. [DOI] [PubMed] [Google Scholar]
- 44.Moncrief, M. B., and M. E. Maguire. 1998. Magnesium and the role of MgtC in growth of Salmonella typhimurium. Infect. Immun. 66:3802-3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neuhaus, K. 2000. Characterization of major cold shock protein genes of the psychrotolerant food pathogens Bacillus weihenstephaniensis and Yersinia enterocolitica. Ph.D. thesis Technische Universität München, Freising, Germany.
- 46.Neuhaus, K., K. P. Francis, S. Rapposch, A. Görg, and S. Scherer. 1999. Pathogenic Yersinia species carry a novel, cold-inducible major cold shock protein tandem gene duplication producing both bicistronic and monocistronic mRNA. J. Bacteriol. 181:6449-6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neuhaus, K., S. Rapposch, K. P. Francis, and S. Scherer. 2000. Restart of exponential growth of cold-shocked Yersinia enterocolitica occurs after down-regulation of cspA1/A2 mRNA. J. Bacteriol. 182:3285-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neuhaus, K., and S. Scherer. 2004. Life at low temperatures. In M. Dworkin et al. (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed., release 4.0. Springer-Verlag, New York, N.Y. [Online.] http://link.springer-ny.com/link/service/books/10125. Springer-Verlag, New York, N.Y.
- 49.Nyström, T., C. Larsson, and L. Gustafsson. 1996. Bacterial defense against aging: role of the Escherichia coli ArcA regulator in gene expression, readjusted energy flux and survival during stasis. EMBO J. 15:3219-3228. [PMC free article] [PubMed] [Google Scholar]
- 50.Ochman, H., A. S. Gerber, and D. L. Hartl. 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120:621-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ojaimi, C., C. Brooks, S. Casjens, P. Rosa, A. Elias, A. Barbour, A. Jasinskas, J. Benach, L. Katona, J. Radolf, M. Caimano, J. Skare, K. Swingle, D. Akins, and I. Schwartz. 2003. Profiling of temperature-induced changes in Borrelia burgdorferi gene expression by using whole genome arrays. Infect. Immun. 71:1689-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Panoff, J. M., B. Thammavongs, M. Gueguen, and P. Boutibonnes. 1998. Cold stress responses in mesophilic bacteria. Cryobiology 36:75-83. [DOI] [PubMed] [Google Scholar]
- 53.Pepe, J. C., J. L. Badger, and V. L. Miller. 1994. Growth phase and low pH affect the thermal regulation of the Yersinia enterocolitica inv gene. Mol. Microbiol. 11:123-135. [DOI] [PubMed] [Google Scholar]
- 54.Polissi, A., W. De Laurentis, S. Zangrossi, F. Briani, V. Longhi, G. Pesole, and G. Deho. 2003. Changes in Escherichia coli transcriptome during acclimatization at low temperature. Res. Microbiol. 154:573-580. [DOI] [PubMed] [Google Scholar]
- 55.Rhodius, V., T. K. Van Dyk, C. Gross, and R. A. LaRossa. 2002. Impact of genomic technologies on studies of bacterial gene expression. Annu. Rev. Microbiol. 56:599-624. [DOI] [PubMed] [Google Scholar]
- 56.Romeo, T., A. Kumar, and J. Preiss. 1988. Analysis of the Escherichia coli glycogen gene cluster suggests that catabolic enzymes are encoded among the biosynthetic genes. Gene 70:363-376. [DOI] [PubMed] [Google Scholar]
- 57.Russell, N. J. 1990. Cold adaptation of microorganisms. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 326:595-611. [DOI] [PubMed] [Google Scholar]
- 58.Schmiel, D. H., G. M. Young, and V. L. Miller. 2000. The Yersinia enterocolitica phospholipase gene yplA is part of the flagellar regulon. J. Bacteriol. 182:2314-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ševčík, M., A. Šebková, J. Wolf, and I. Rychlík. 2001. Transcription of arcA and rpoS during growth of Salmonella typhimurium under aerobic and microaerobic conditions. Microbiology 147:701-708. [DOI] [PubMed] [Google Scholar]
- 60.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology 1:784-791. [Google Scholar]
- 61.Smoot, L. M., J. C. Smoot, M. R. Graham, G. A. Somerville, D. E. Sturdevant, C. A. Migliaccio, G. L. Sylva, and J. M. Musser. 2001. Global differential gene expression in response to growth temperature alteration in group A Streptococcus. Proc. Natl. Acad. Sci. USA 98:10416-10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stevenson, B., T. G. Schwan, and P. A. Rosa. 1995. Temperature-related differential expression of antigens in the Lyme disease spirochete, Borrelia burgdorferi. Infect. Immun. 63:4535-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stintzi, A. 2003. Gene expression profile of Campylobacter jejuni in response to growth temperature variation. J. Bacteriol. 185:2009-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Straley, S. C., and R. D. Perry. 1995. Environmental modulation of gene expression and pathogenesis in Yersinia. Trends Microbiol. 3:310-317. [DOI] [PubMed] [Google Scholar]
- 65.Tao, T., P. F. Grulich, L. M. Kucharski, R. L. Smith, and M. E. Maguire. 1998. Magnesium transport in Salmonella typhimurium: biphasic magnesium and time dependence of the transcription of the mgtA and mgtCB loci. Microbiology 144:655-664. [DOI] [PubMed] [Google Scholar]
- 66.Thieringer, H. A., P. G. Jones, and M. Inouye. 1998. Cold shock and adaptation. Bioessays 20:49-57. [DOI] [PubMed] [Google Scholar]
- 67.Valbuzzi, A., E. Ferrari, and A. M. Albertini. 1999. A novel member of the subtilisin-like protease family from Bacillus subtilis. Microbiology 145:3121-3127. [DOI] [PubMed] [Google Scholar]
- 68.Van Dyk, T. K. 2002. Lighting the way: genome-wide, bioluminescent gene expression analysis. ASM News 68:222-230. [Google Scholar]
- 69.Van Dyk, T. K., Y. Wei, M. K. Hanafey, M. Dolan, M. J. Reeve, J. A. Rafalski, L. B. Rothman-Denes, and R. A. LaRossa. 2001. A genomic approach to gene fusion technology. Proc. Natl. Acad. Sci. USA 98:2555-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Verhamme, D. T., P. W. Postma, W. Crielaard, and K. J. Hellingwerf. 2002. Cooperativity in signal transfer through the Uhp system of Escherichia coli. J. Bacteriol. 184:4205-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang, Q., J. G. Frye, M. McClelland, and R. M. Harshey. 2004. Gene expression patterns during swarming in Salmonella typhimurium: genes specific to surface growth and putative new motility and pathogenicity genes. Mol. Microbiol. 52:169-187. [DOI] [PubMed] [Google Scholar]
- 72.Waterfield, N. R., D. J. Bowen, J. D. Fetherston, R. D. Perry, and R. H. ffrench-Constant. 2001. The tc genes of Photorhabdus: a growing family. Trends Microbiol. 9:185-191. [DOI] [PubMed] [Google Scholar]
- 73.Weber, M. H., and M. A. Marahiel. 2003. Bacterial cold shock responses. Sci. Prog. 86:9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Winson, M. K., S. Swift, P. J. Hill, C. M. Sims, G. Griesmayr, B. W. Bycroft, P. Williams, and G. S. Stewart. 1998. Engineering the luxCDABE genes from Photorhabdus luminescens to provide a bioluminescent reporter for constitutive and promoter probe plasmids and mini-Tn5 constructs. FEMS Microbiol. Lett. 163:193-202. [DOI] [PubMed] [Google Scholar]
- 75.Wolk, C. P., Y. Cai, and J. M. Panoff. 1991. Use of a transposon with luciferase as a reporter to identify environmentally responsive genes in a cyanobacterium. Proc. Natl. Acad. Sci. USA 88:5355-5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Worley, M. J., K. H. Ching, and F. Heffron. 2000. Salmonella SsrB activates a global regulon of horizontally acquired genes. Mol. Microbiol. 36:749-761. [DOI] [PubMed] [Google Scholar]