Abstract
TSLC1/IGSF4, an immunoglobulin superfamily molecule, is predominantly expressed in the brain, lungs, and testes and plays important roles in epithelial cell adhesion, cancer invasion, and synapse formation. We generated Tslc1/Igsf4-deficient mice by disrupting exon 1 of the gene and found that Tslc1−/− mice were born with the expected Mendelian ratio but that Tslc1−/− male mice were infertile. In 11-week-old adult Tslc1−/− mice, the weight of a testis was 88% that in Tslc1+/+ mice, and the number of sperm in the semen was approximately 0.01% that in Tslc1+/+ mice. Histological analysis revealed that the round spermatids and the pachytene spermatocytes failed to attach to the Sertoli cells in the seminiferous tubules and sloughed off into the lumen with apoptosis in the Tslc1−/− mice. On the other hand, the spermatogonia and the interstitial cells, including Leydig cells, were essentially unaffected. In the Tslc1+/+ mice, TSLC1/IGSF4 expression was observed in the spermatogenic cells from the intermediate spermatogonia to the early pachytene spermatocytes and from spermatids at step 7 or later. These findings suggest that TSLC1/IGSF4 expression is indispensable for the adhesion of spermatocytes and spermatids to Sertoli cells and for their normal differentiation into mature spermatozoa.
Infertility is estimated to affect about 5% of adult human males. However, approximately 75% of these cases are diagnosed as idiopathic because the molecular mechanisms underlying these defects have not been elucidated (17, 20). Recently, male infertility has been reported as a phenotype in mice that are deficient in various single genes, providing knowledge about possible molecular targets causing male infertility in humans. So far, more than 80 genes have been identified as essential for male fertility in humans and mice (11). These genes encode a variety of proteins, including signal transduction molecules, transcription factors, metabolic enzymes, transmembrane proteins, and cytoskeletal proteins. From the pathological point of view, the causes of male infertility can be grouped into the following three categories, depending on the stage of cell differentiation affected: defects in the primordial germ cells, those in the spermatogenic cells, and those in the spermatozoa. Among these defects, abnormalities in the genes involved in the primordial germ cells and the resultant developmental defects in the reproductive organs are relatively rare, probably because such defects often cause embryonic lethality. On the other hand, spermatogenesis, a series of spermatogenic cell differentiation steps from spermatogonia to mature spermatozoa in the testes, is considered the major target of defects in male infertility. In fact, about 70% of the genes essential for male fertility are involved in this process. Another 20% of the genes have been shown to play a role in the formation, motility, or fertilizing ability of spermatozoa.
Spermatogenesis can be further divided into the following three phases: (i) the proliferative phase, in which the spermatogonia undergo rapid successive divisions; (ii) the meiotic phase, in which the spermatocytes produce cells with haploid chromosome content; and (iii) the spermiogenic phase, in which the spermatids differentiate into mature spermatozoa, which can fertilize the egg (15).
We have previously identified the TSLC1/IGSF4 gene on chromosome 11q23.2 as a tumor suppressor in sporadic lung cancer by its activity in the suppression of tumorigenicity in nude mice by a lung cancer cell line, A549 (7). TSLC1/IGSF4 is predominantly expressed in the brain, lungs, and testes and is followed by most epithelial and neuronal tissues, while the loss of its expression through promoter methylation associated with a loss of heterozygosity is observed in a variety of human tumors, including lung, esophageal, pancreatic, breast, and prostate cancers, especially in tumors with aggressive behavior (12). The TSLC1/IGSF4 protein belongs to immunoglobulin superfamily cell adhesion molecules (IgCAMs) containing three Ig-like loops in the extracellular domain and mediates cell-to-cell adhesion through homophilic and heterophilic interactions in a Ca2+- and Mg2+-independent manner (10). A mouse orthologue of the Tslc1/Igsf4 gene shows extremely high homology to human TSLC1/IGSF4, with 97% identity in the overall amino acid sequences, suggesting that TSLC1/IGSF4 plays an important role during evolution (3).
Wakayama et al. independently cloned SgIGSF, a mouse orthologue of TSLC1/IGSF4, by scanning the database of mouse expressed sequence tags and selecting a sequence homologous to the neural cell adhesion molecules (19). Expression of this molecule, SgISGF/IGSF4, was detected in the membranes of spermatogenic cells in two distinct phases, one from the intermediate spermatogonia through the early pachytene spermatocytes and the other from step 7 spermatids to step 16 residual bodies. These findings suggest that, in the testes, SgIGSF/IGSF4 may be involved in spermatogenesis (18).
To elucidate the physiological function of TSLC1/IGSF4, we generated mutant mice lacking the Tslc1/Igsf4 gene. We report in the present study that Tslc1/Igsf4-deficient mice are born without any overt abnormalities but that the males are infertile.
MATERIALS AND METHODS
Generation of mice lacking the Tslc1/Igsf4 gene.
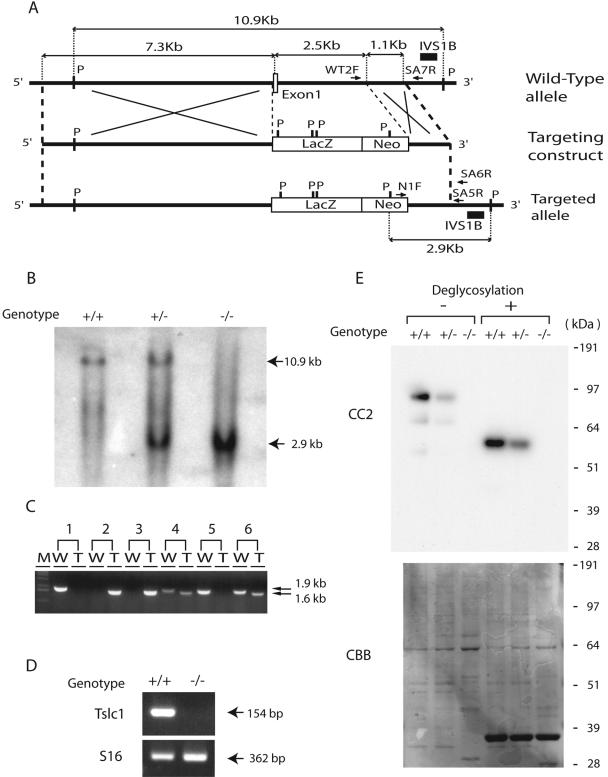
An 11-kb mouse genomic DNA fragment containing exon 1 of the Tslc1/Igsf4 gene was cloned from the mouse 129Sv/Ev bacterial artificial chromosome genomic library by hybridization with a radiolabeled probe generated from the mouse genomic sequence around exon 1 of the Tslc1/Igsf4 gene (3). From this clone, a fragment of 7.3 kb upstream of exon 1 and one of 1.1 kb within intron 1 were subcloned, and the targeting construct was generated by inserting these fragments into the 5′ and 3′ sites of the LacZ-neomycin (Neo) resistance gene cassette, respectively (Fig. 1A). The genomic fragment of 2.5 kb containing exon 1 of the Tslc1/Igsf4 gene, which was replaced by the LacZ-Neo cassette, starts with the sequence 5′-CCGACATGGCGAGTGCTGTGCTGCCGAGCGGATCCCAGTGTGCGGCGG-3′ and ends with 5′-TAGGGCTTTGCTAAGACTCTCTCTCAAACTGTATAC-3′. By homologous recombination, therefore, it was expected that all coding sequences of exon 1 and the 5′ region of intron 1 of the Tslc1/Igsf4 gene were deleted, whereas the LacZ gene as well as the neomycin resistance gene was inserted into the targeted allele so that the expression of the LacZ gene could be regulated by the endogenous promoter of the Tslc1/Igsf4 gene (Fig. 1A). Ten micrograms of the targeting vector was linearized by NotI, transfected into 129Sv/Ev iTL1 embryonic stem (ES) cells by electroporation, and selected with G418. Among the 300 neomycin-resistant cells obtained, two ES cells that had undergone homologous recombination were identified by PCR analysis using primers N1F and SA6R (Fig. 1A). The sequences of the primers are listed in Table S1 in the supplemental material. These targeted ES cells were microinjected into C57BL/6J blastocysts to generate seven male chimeras and nine female chimeras. The chimeric mice were then mated with C57BL/6J mice to obtain offspring that were heterozygous for the targeted inactivation of the Tslc1/Igsf4 gene. Germ line transmission of the targeted allele was confirmed by Southern blotting and monitored by PCR, in which the wild-type fragment was generated by primers WT2F and SA7R, while the targeted fragment was generated by primers N1F and SA5R (Fig. 1A). All animals used in this study were handled in compliance with the National Cancer Center Research Institute's guidelines for the use of animals.
FIG. 1.
Generation of Tslc1−/− mice. (A) Wild-type allele, targeting construct, and targeted allele of the Tslc1/Igsf4 gene. An open box and solid lines indicate an exon and introns, respectively. IVS1B is a genomic fragment used as a probe for Southern blotting. WT2F and SA7R are the primers for PCR complementary to the wild-type genomic sequence of the Tslc1/Igsf4 gene, while N1F and SA5R are the PCR primers complementary to the targeted allele. P, restriction site of PvuII. (B) Southern blot analysis of the wild-type and targeted alleles of Tslc1/Igsf4. Genomic DNA was digested with a restriction enzyme, PvuII, blotted, and hybridized with a probe, IVS1B. Fragments of 10.9 kb and 2.9 kb were derived from the wild-type and targeted alleles, respectively. (C) PCR analysis for monitoring inheritance of the targeted allele of Tslc1/Igsf4 in the progeny of the Tslc1+/− intercross. W and T indicate the wild-type allele (1.9 kb) and the targeted allele (1.6 kb), respectively. M, molecular marker. (D) RT-PCR analysis of Tslc1/Igsf4 in the testes from Tslc1+/+ and Tslc1−/− mice. A fragment of 154 bp corresponds to exons 1 to 3 of the Tslc1/Igsf4 mRNA. A ribosomal protein gene, S16, served as a control endogenous gene. (E) Western blotting of testis lysates, with or without treatment for deglycosylation, from Tslc1+/+, Tslc1+/−, and Tslc1−/− mice. The filter was hybridized with the anti-TSLC1/IGSF4 antibody CC2 (top) or stained with Coomassie brilliant blue (CBB; bottom).
Southern blotting.
Mouse genomic DNA was extracted by the phenol-chloroform extraction method. Five micrograms of genomic DNA was digested with PvuII, subjected to electrophoresis through a 0.8% agarose gel, and blotted onto a Hybond-N+ membrane (Amersham Biosciences, Buckinghamshire, United Kingdom) using the alkaline transfer method. A fragment, IVS1A, corresponding to the 1,021-bp genomic DNA in intron 1 of the murine Tslc1/Igsf4 gene, was generated by PCR using primers 16205F and 17225R (see Table S1 in the supplemental material) and cloned into a TOPO cloning vector (Invitrogen, Carlsbad, CA) to obtain pmIVS1A. Fragment IVS1B, of 436 bp, was generated by digesting pmIVS1A DNA with BstXI and was used as a probe for Southern blotting. Radiolabeling of the probe was carried out with [32P]dCTP using the Megaprime DNA labeling system (Amersham Biosciences).
Quantitative RT-PCR.
Total cellular RNAs were extracted from the testes of 16-week-old Tslc1+/+ and Tslc1−/− mice using an RNeasy Mini kit (QIAGEN, Valencia, CA). One microgram of total cellular RNA was reverse transcribed using Superscript II reverse transcriptase (RT; Invitrogen) with oligo(dT) primers, and an aliquot was amplified by real-time PCR using a Light Cycler instrument with Master SYBR green I (Roche, Mannheim, Germany). The sequences of the oligonucleotide primers used for PCR are listed in Table S1 in the supplemental material.
Antibodies.
A rabbit polyclonal antibody against 18 amino acids at the carboxyl termini of human and mouse TSLC1/IGSF4 (CC2) was generated previously (10). A rabbit polyclonal antibody against the extracellular domains of human and mouse TSLC1/IGSF4 (EC2) was gifted from H. P. Ghosh at McMaster University, Hamilton, Canada. An anti-alpha-tubulin antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Western blotting.
The testes of 25-week-old mice were removed and homogenized in a lysis buffer (1% Triton X-100, 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 1× protease inhibitor cocktail set I [Calbiochem, Darmstadt, Germany]) to obtain cell lysates. After centrifugation at 3,000 rpm at 4°C for 10 min, the supernatants were examined for protein concentration using Benchmark (Bio-Rad, Hercules, CA) and used as cell lysates. The cell lysates from the mouse testes were subjected to NuPAGE bis-Tris 4 to 12% gel electrophoresis with a morpholinepropanesulfonic acid-sodium dodecyl sulfate (MOPS-SDS) running buffer (Invitrogen) and transferred to Immobilon-P transfer membranes (Millipore Corporation, Bedford, MA). SeeBlue Plus2 (Invitrogen) was used as a marker for molecular weight. The membranes were incubated with each primary antibody at 4°C overnight and then incubated with horseradish peroxidase-linked secondary antibodies (1:5,000; Amersham Biosciences) at room temperature for 1 h after being washed with Tris-buffered saline containing 0.1% Triton X-100. The membranes were treated with Lumi-Lightplus Western blotting substrate (Roche), and the signals were detected with Hyperfilm (Amersham Biosciences). After incubation with a stripping buffer (2% SDS, 62 mM Tris-HCl, pH 6.8, 0.7% beta-mercaptoethanol) at 50°C for 30 min, the membranes were reprobed with other antibodies or stained with Coomassie brilliant blue.
Deglycosylation.
Digestion of sites of N-linked glycosylation was carried out using peptide N-glycosidase F (New England Biolabs, Beverly, MA) according to the manufacturer's instructions. Briefly, 20 μg of protein from cell lysates of the testes in 90 μl lysis buffer was denatured with 10 μl of glycoprotein denaturing buffer (5% SDS, 10% beta-mercaptoethanol) at 100°C for 10 min and then incubated with 10 μl of G7 buffer (0.5 M sodium phosphate, pH 7.5) and 10 μl of 10% NP-40 containing 1,500 U of peptide N-glycosidase F at 37°C for 5 h.
Sperm counts and motility.
After 2- to 21-week-old mice were sacrificed by cervical dislocation, their epididymides and vasa deferentia were immediately removed, cut into 2-mm-long pieces, resuspended in 1 ml of buffer containing 75 mM NaCl, 24 mM EDTA, and 0.4% bovine serum albumin, and then homogenized to dissociate somatic cells at 32°C for 10 min. The sperm cells remaining as a monodispersed suspension were counted on a hemacytometer. The motility of the sperm obtained from the epididymides and vasa deferentia was examined as reported previously (1).
Morphological examination and immunohistochemistry.
Mice 2 to 40 weeks old were necropsied for histopathological examination. The testes, epididymides, and vasa deferentia were immediately removed and fixed in Bouin's solution. The whole body was perfused with 7.4% formaldehyde solution. All organs and/or tissues were routinely processed, embedded in paraffin, and stained with hematoxylin and eosin (HE). The testes, epididymides, and vasa deferentia from Tslc1+/+, Tslc1+/−, and Tslc1−/− mice were stained with periodic acid-Schiff stain (PAS) to identify the stage of spermatid development. For immunohistochemistry, serial sections of the testes were heated to 105°C for 5 min with an antigen retrieval buffer (DakoCytomation, Glostrup, Denmark) after deparaffinization and dehydration for antigen retrieval. Nonspecific reactions were blocked with 5% normal donkey serum in phosphate-buffered saline (PBS). All sections were incubated with each primary antibody at 4°C overnight. The sections were then incubated with a horseradish peroxidase-labeled polymer (DakoCytomation) at room temperature for 1 h, rinsed with PBS, and visualized with 3,3′-diaminobenzidine (DakoCytomation). All sections were counterstained with hematoxylin.
Terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assays.
Testes from 21-week-old mice were fixed with paraformaldehyde and embedded in paraffin. After deparaffinization and dehydration, testicular sections from Tslc1+/+ mice and Tslc1−/− mice (12 sections from 3 mice in each group) were treated with 20 μg/ml proteinase K for 15 min. H2O2 solution was used for endogenous peroxidase blocking. The sections were incubated at 4°C overnight with terminal deoxynucleotidyl transferase labeling safe buffer (Takara Bio, Kyoto, Japan). After being rinsed with PBS, the sections were incubated with anti-fluorescein isothiocyanate-horseradish peroxidase conjugate (Takara Bio) at 37°C for 1 h, rinsed with PBS, and visualized with 3,3′-diaminobenzidine (DakoCytomation). All sections were counterstained with methyl green (Cab Vision, Fremont, CA).
Flow cytometry.
Testes were excised from 19-week-old Tslc1+/+ and Tslc1−/− mice (three mice in each group) and decapsulated and crushed through 20-gauge needles and 100-μm cell strainers (BD Falcon, Bedford, MA) in PBS. The cells (2 × 106) were then treated with RNase and stained with propidium iodide using a Cycle Test Plus DNA reagent kit (Becton Dickinson, San Jose, CA). All fluorescence-activated cell sorting data were analyzed using CELL Quest (version 3.3; Becton Dickinson).
Electron microscopy.
Tslc1+/+ and Tslc1−/− mice (25 weeks old) were perfused with 3% glutaraldehyde (Sigma-Aldrich, St. Louis, MO) buffered with PBS, pH 7.4, through the heart to fix all organs and tissues. The testes and epididymides were then removed, cut into 1-mm3 pieces, and stored in the same fixative at 4°C for 4 h. After being rinsed with PBS, the tissues were postfixed with 1% osmium tetroxide (Electron Microscopy Sciences, Hatfield, PA) at 4°C for 2 h. Thereafter, the samples were routinely processed, dehydrated with ethanol, and embedded in epoxy resin (TAAB, Berkshire, England). Semithin sections (0.5 μm) of the testes and epididymides were stained with toluidine blue. Ultrathin sections of the selected area were cut on a copper grid, stained with uranyl acetate and lead citrate, and examined by transmission electron microscopy in a JEM-1011 electron microscope (JEOL, Tokyo, Japan).
Oligonucleotide microarray.
The protocol used for sample preparation and microarray processing is available from Affymetrix (Santa Clara, CA). Briefly, 3 μg purified RNA, extracted from the testes of 16-week-old Tslc1+/+ and Tslc1−/− mice, was reverse transcribed with Superscript II reverse transcriptase (Invitrogen), using primer T7-dT24 containing a T7 RNA polymerase promoter. After a second strand of cDNA was synthesized using RNase H, Escherichia coli DNA polymerase, and E. coli DNA ligase, in vitro transcription was carried out on the cDNA to produce a biotin-labeled cRNA with a MEGAscript High Yield transcription kit (Ambion, Austin, TX), as recommended by the manufacturer. After the cRNA was linearly amplified with T7 polymerase, the biotinylated cRNA was cleaned with an RNeasy mini column (QIAGEN), fragmented to 50 to 200 nucleotides, and then hybridized to mouse genome U74A v. 2 arrays (Affymetrix). The stained microarrays were scanned with a GeneArray scanner (Affymetrix), and the signals were calculated with the Affymetrix software Microarray Suite 5.0. All of the data were scaled with the global scaling method to adjust the target intensity to 1,000.
Data analysis.
For microarray analysis, the expression value for each gene was determined by calculating the average difference (perfect match intensity minus mismatch intensity) for the probe in use for the gene. The degree of change was calculated for each sample relative to the median of the controls. Small and negative expression levels were clipped off so that they would be equal to a cutoff value arbitrarily chosen as 100. For all comparisons, statistical analysis was carried out by Student's t test, using the Stat View statistical analysis software package (version 5.0; SAS Institute, Cary, NC).
Microarray accession number.
Microarray data were deposited in the GEO database at NCBI with the accession number GSE3676.
RESULTS
Male mice deficient in the Tslc1/Igsf4 gene were infertile.
For the purpose of exploring the function of TSLC1/IGSF4 in vivo, we inactivated the Tslc1/Igsf4 gene in mouse ES cells by targeted disruption of exon 1 of the gene, thereby generating Tslc1/Igsf4-deficient mice (Fig. 1A). The targeting vector was introduced into ES cell lines, and cells that showed targeted recombination were used for the generation of mice that transmitted the disrupted gene. These mice were then mated to produce Tslc1−/− mice. The offspring of Tslc1+/− intercrosses were born at the expected Mendelian frequencies. Inheritance of the targeted gene was determined by Southern blotting (Fig. 1B) and monitored by PCR using two pairs of primers (WT2F-SA7R and N1F-SA5R) that flanked the recombination sites (Fig. 1C; see Table S1 in the supplemental material). The Tscl1/Igsf4 transcript was not detected in testes from Tslc1−/− mice by RT-PCR (Fig. 1D).
Expression of the TSLC1/IGSF4 protein was examined by Western blotting using an anti-TSLC1/IGSF4 antibody, CC2 (10). An immunoreactive signal of approximately 100 kDa, as well as a weak signal of 70 kDa, was detected in testes from Tslc1+/+ and Tslc1+/− mice, whereas no signals were detected in testes from Tslc1−/− mice (Fig. 1E). TSLC1/IGSF4 expression was also absent in other tissues from Tslc1−/− mice (data not shown), indicating that TSLC1/IGSF4 was not produced in Tslc1−/− mice. TSLC1/IGSF4 is an IgCAM carrying six potential asparagine (N)-linked glycosylation sites in its extracellular loops, and it has been shown to be modified by N glycosylation (10). Therefore, we carried out enzymatic deglycosylation of the TSLC1/IGSF4 protein by treatment with N-glycosidase F. As shown in Fig. 1E, a single signal of approximately 60 kDa was observed by Western blotting after N-glycosidase F treatment, indicating that the signals of both 100 kDa and 70 kDa were specific to the TSLC1/IGSF4 protein and were generated by distinct posttranslational modifications.
Tslc1+/− and Tslc1−/− mice did not show any overt developmental abnormalities, although significant amounts of TSLC1/IGSF4 protein were expressed in the brains and lungs, in addition to the testes, of Tslc1+/+ mice (data not shown). Intercrosses between Tslc1−/− mice, however, failed to produce any progeny. Male Tslc1−/− mice were infertile, whereas female Tslc1−/− mice as well as male and female Tslc1+/− mice were fertile.
Semen from Tslc1−/− mice contained degenerated cells.
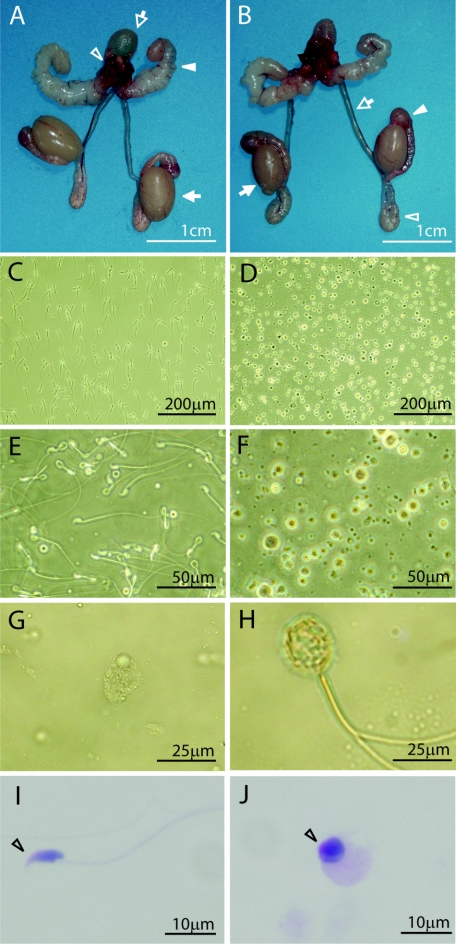
The growth of Tslc1−/− mice from birth to young adulthood was indistinguishable from that of their Tslc1+/+ littermates, except for male gonadal development. During the fetal, postnatal, and prepubertal periods, the testes of Tslc1−/− mice developed without any macroscopic abnormalities and had normal testicular descent. However, the weights of the testes in Tslc1−/− mice at 11 and 25 weeks of age were 12% (P = 0.03) and 29% (P = 0.003) lower than those in the respective Tslc1+/+ mice (Fig. 2A and B; Table 1). On the other hand, there was no significant difference either in the weights of other organs, including the seminal vesicles, epididymides, and vasa deferentia, or in the serum testosterone levels in Tslc1−/− mice and Tslc1+/+ mice (Fig. 2A and B; Table 1).
FIG. 2.
Reproductive organs and semens from Tslc1+/+ and Tslc1−/− male mice. (A and B) Morphology of reproductive organs from Tslc1+/+ (A) and Tslc1−/− (B) mice. The bladder (open arrow in panel A), prostate (open arrowhead in panel A), seminal vesicles (closed arrowhead in panel A), testes (closed arrow in panels A and B), vasa deferentia (open arrow in B), caput epididymides (closed arrowhead in panel B), and cauda epididymides (open arrowhead in panel B) are demonstrated. Note that the testes from the Tslc1−/− mice are significantly smaller than those from the Tslc1+/+ mice. (C to H) Phase-contrast microscopy of semens from Tslc1+/+ (C and E) and Tslc1−/− (D and F to H) mice. (I and J) PAS staining of semens from Tslc1+/+ (I) and Tslc1−/− mice (J). The open arrowhead indicates a possible acrosome with PAS staining.
TABLE 1.
Weights of organs, sperm parameters, and serum testosterone levels in Tslc1−/− and Tslc1+/+ mice
| Parameter | Value for Tslc1−/− mice | No. of mice analyzed | Value for Tslc1+/+ mice | No. of mice analyzed | P valueb |
|---|---|---|---|---|---|
| Total body wt (g) (25 wks) | 38.7 ± 2.8 | 4 | 37.8 ± 1.6 | 4 | NS |
| Wet wt of organs (mg)a | |||||
| Brain | 425 ± 10 | 3 | 442 ± 7.9 | 4 | NS |
| Eye | 25.5 ± 0.4 | 5 | 24.4 ± 2.1 | 3 | NS |
| Thymus | 38.7 ± 0.9 | 3 | 42.4 ± 2.2 | 3 | NS |
| Lung | 174 ± 28 | 3 | 181 ± 7.2 | 4 | NS |
| Heart | 222 ± 23 | 5 | 189 ± 19 | 4 | NS |
| Spleen | 85.5 ± 19 | 5 | 94 ± 5.0 | 4 | NS |
| Kidney | 318 ± 22 | 5 | 309 ± 22 | 4 | NS |
| Liver | 1,900 ± 80 | 5 | 1,770 ± 260 | 4 | NS |
| Stomach | 420 ± 54 | 4 | 405 ± 54 | 3 | NS |
| Intestinum tenue, pancreas, and mesenterium | 2,310 ± 130 | 5 | 2,470 ± 280 | 4 | NS |
| Intestinum crassum | 584 ± 78 | 3 | 651 ± 47 | 3 | NS |
| Seminal vesicle | 163 ± 14 | 4 | 153 ± 4.2 | 4 | NS |
| Bladder and prostate | 133 ± 18 | 4 | 143 ± 10 | 4 | NS |
| Epididymis and vas deferens | 59.4 ± 9.5 | 5 | 85.1 ± 13 | 4 | NS |
| Testis | 94.7 ± 3.0 | 5 | 134 ± 8.7 | 4 | 0.003 |
| Sperm parameters (11 wks) | |||||
| Total no. of cells (106) | 7.3 ± 1.0 | 3 | 20 ± 2.4 | 3 | 0.008 |
| No. of normal sperm (103) | 1.2 ± 0.8 | 3 | 15,000 ± 2,200 | 3 | 0.002 |
| Motile sperm/normal sperm (%) | 1.1 ± 0.4 | 3 | 83.3 ± 3.3 | 3 | <0.0001 |
| Serum testosterone level (ng/ml) | |||||
| 16 wks | 4.5 ± 4.0 | 3 | 2.5 ± 2.0 | 3 | NS |
| 25 wks | 1.7 ± 0.5 | 4 | 3.2 ± 3.0 | 4 | NS |
Brains and lungs were from 16-week-old mice. Other organs were from 25-week-old mice. For each animal, the wet weights of paired organs were averaged, and this single value was used to calculate the means±SE.
NS, not significant.
We then examined semens from the epididymides and vasa deferentia of Tslc1−/− mice to determine the mechanisms of male infertility. Phase-contrast microscopy demonstrated that most of the semens from 16-week-old Tslc1−/− mice consisted of degenerated, uncharacterized round cells with varied morphologies (Fig. 2D, F, and G), in contrast with those from Tslc1+/+ mice (Fig. 2C and E). In addition, the number of normal sperm from Tslc1−/− mice was approximately 0.01% that from 11-week-old Tslc1+/+ mice (Table 1). Furthermore, almost all sperm from Tslc1−/− mice showed abnormal morphologies, such as large round heads (Fig. 2G), short tails, and multiple tails (Fig. 2H). In addition, the proportion of normal motile sperm in a low-viscosity buffer was only 1.1% for Tslc1−/− mice, whereas 83% of the sperm from Tslc1+/+ mice were highly mobile under the same conditions (Table 1). These findings indicate that male infertility in Tslc1−/− mice is due to quantitative and qualitative defects in mature sperm. We next stained uncharacterized round cells in semens from 16-week-old Tslc1−/− mice with PAS to detect the acrosome. As shown in Fig. 2J, the perinuclear spaces of the round cells were stained with PAS, suggesting that these cells are derived from immature spermatids.
Elongated spermatids were scarcely observed in testes from Tslc1−/− mice.
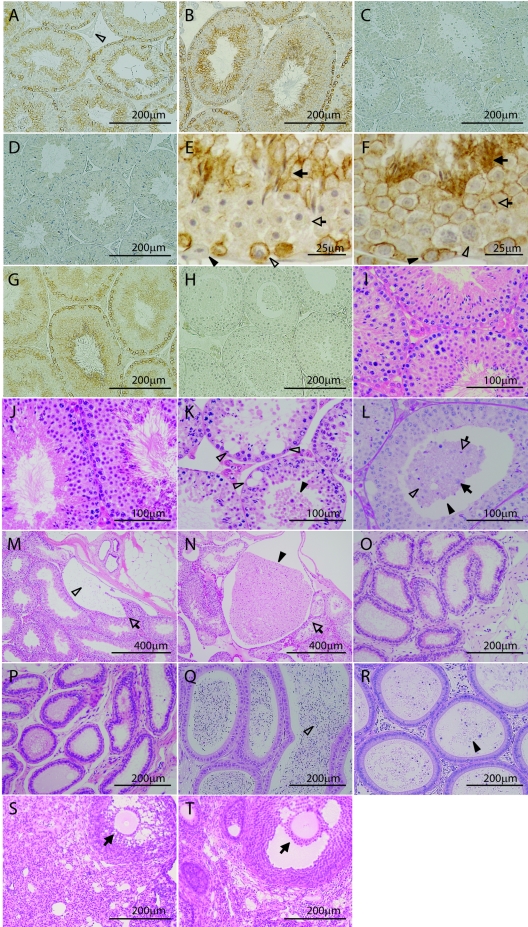
Immunohistochemical studies of Tslc1+/+ and Tslc1+/− mice with the CC2 antibody against the carboxyl-terminal end of TSLC1/IGSF4 demonstrated that the TSLC1/IGSF4 protein was present in the seminiferous tubules but not in the interstitial tissues, including the Leydig cells (Fig. 3A and B). On the other hand, no signal of the TSLC1/IGSF4 protein was detected in testes from Tslc1−/− mice (Fig. 3C). The signals observed in the testes from Tslc1+/+ mice disappeared when the CC2 antibody was preincubated with the antigenic polypeptide (Fig. 3D). Detailed analysis of the seminiferous epithelium revealed that the TSLC1/IGSF4 protein was expressed in two distinct phases of spermatogenesis: the first phase was from intermediate spermatogonia to early pachytene spermatocytes, and the second phase was from step 7 to step 16 spermatids (Fig. 3E and F). The TSLC1/IGSF4 protein was located along the membrane in these spermatogenic cells but was not present in the Sertoli cells. We also confirmed a lack of TSLC1 expression in testes from Tslc1−/− mice with the EC2 antibody against the extracellular domain of TSLC1 (Fig. 3G and H).
FIG. 3.
Immunohistochemical and histological analyses of Tslc1+/+, Tslc1+/−, and Tslc1−/− mice. (A to H) Immunohistochemical analysis of TSLC1/IGSF4 protein in testes from Tslc1+/+ (A and D to G), Tslc1+/− (B), and Tslc1−/− (C and H) mice, using the anti-TSLC1/IGSF4 antibodies CC2 (A to F) and EC2 (G and H). (A and B) The TSLC1/IGSF4 protein was detected in the seminiferous tubules but not in the interstitial compartment, including the Leydig cells (open arrowhead in panel A). (C) The TSLC1/IGSF4 protein was not detected in a testis from a Tslc1−/− mouse. (D) No signals were detected by CC2 preincubated with an excess amount of antigenic polypeptides. (E) Seminiferous epithelium at stage I. The TSLC1/IGSF4 protein was localized along the membranes of step 13 spermatids (closed arrow) and early pachytene spermatocytes (open arrowhead) but was not detected in step 1 spermatids (open arrow) or the Sertoli cells (closed arrowhead). (F) Seminiferous epithelium at stage VII. The TSLC1/IGSF4 protein was localized along the membranes of step 7 spermatids (open arrow), step 16 residual bodies (closed arrow), and preleptotene spermatocytes (closed arrowhead) but was not detected in the late pachytene spermatocytes (open arrowhead). (I to T) Histological analyses of the testes (I to N), ductuli efferentes testis (O and P), epididymides (Q and R), and ovaries (S and T) from Tslc1+/+ (I, M, O, Q, and S), Tslc1+/− (J), and Tslc1−/− (K, L, N, P, R, and T) mice by HE staining (I to K and M to T) or PAS staining (L). (K) Degenerated round cells were accumulated in the lumen (closed arrowhead), and extensive vacuolization was observed at the basal side (open arrowheads). (L) A large number of round and degenerated cells were seen in the lumen (closed arrowhead). Note that some of the cells in the lumen were stained with PAS and appeared to be derived from round spermatids (open arrowhead), elongating spermatids (open arrow), or the pachytene spermatocytes (closed arrow). (M to R) The open arrows (M and N), open arrowheads (M and Q), and closed arrowheads (N and R) indicate the rete of the testis, the spermatozoa, and the degenerated round cells, respectively. (S and T) Closed arrows indicate the secondary follicle. Mice were examined at 25 weeks of age (A to R) and 40 weeks of age (S and T).
Morphologically normal spermatogenic features were observed in the testes from Tslc1+/+ and Tslc1+/− mice, in which elongating or elongated spermatids were present in the seminiferous epithelia (Fig. 3I and J). In contrast, elongating or elongated spermatids were scarcely observed in seminiferous epithelia from Tslc1−/− mice (Fig. 3K and L). Instead, a large number of round cells with degeneration accumulated in the lumens and retia from Tslc1−/− mice (Fig. 3K, L, and N, closed arrowheads). Considerable numbers of vacuoles were also specifically observed at the basal area of the seminiferous tubules (Fig. 3K, open arrowheads). On the other hand, the basal lamina, spermatogonia, and interstitial compartments, including Leydig cells, were unaffected in testes from Tslc1−/− mice (Fig. 3K). As shown in Fig. 3L, PAS reaction revealed that the cells in the lumen were derived from the round spermatid (open arrowhead), the elongating spermatid (open arrow), or the pachytene spermatocyte (closed arrow). The ductal structure of the ductuli efferentes testis and the epididymides from Tslc1−/− mice also showed no abnormalities in comparison with those from Tslc1+/+ mice (Fig. 3O to R). Since the degenerated round cells were present in the epididymides from Tslc1−/− mice (Fig. 3R, closed arrowhead), the seminal tracts did not appear to be obstructed. In contrast to male Tslc1−/− mice, male and female Tslc1+/− mice and female Tslc1−/− mice were fertile. Histological examination of the ovaries showed no significant difference between female Tslc1+/+ and Tslc1−/− mice (Fig. 3S and T).
Sloughing and apoptosis of spermatids in Tslc1−/− mice.
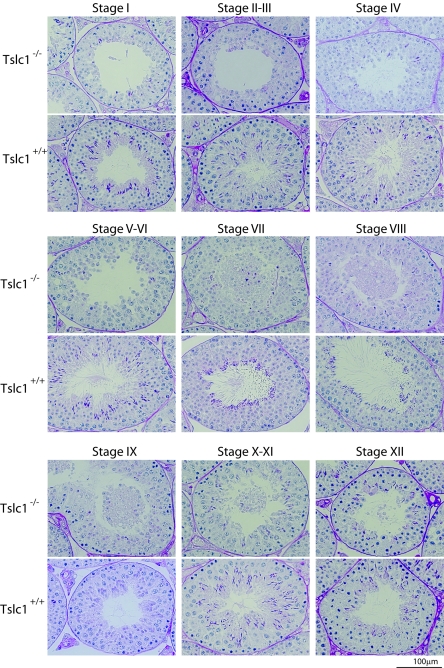
To further characterize the defect in Tslc1−/− mice, detailed staging analysis of spermatogenesis was carried out. In Tslc1−/− mice, sloughed cells were observed mainly in stages VII to IX (Fig. 4). The numbers of spermatids in steps 10 to 16 were markedly decreased, in contrast to those from Tslc1+/+ mice. However, synchronous spermatogenesis in each tubule was essentially not affected. To unveil which phase of spermatogenic cells sloughed off into the lumen, we classified the types of sloughed cells and counted the numbers of cells. These analyses characterized only 10 to 15% of the round cells in the lumen, as it was not possible to determine the phases of the remaining cells due to severe degeneration. As shown in Table 2, among the characterized sloughed cells, 98% were determined to be spermatids, and the remaining 2% were spermatocytes in the pachytene phase, while no spermatogonia were observed. Notably, about 60% of the characterized cells were spermatids in steps 7 to 9, suggesting that sloughing from the seminiferous epithelia occurred mainly in the spermatids in steps 7 to 9 in the tubules in stages VII to IX. The absence of spermatids in step 10 and later in the tubules in stages X to XII and I to VIII suggests that the maturation of the majority of the spermatids was arrested around step 10.
FIG. 4.
Staging analyses of the testes from 21-week-old Tslc1+/+ and Tslc1−/− mice by PAS staining.
TABLE 2.
Sloughed cells from seminiferous epithelium in testes from Tslc1−/− micea
| Type of cells | No. of cells (%) |
|---|---|
| Spermatogonia | 0 (0.0) |
| Spermatocytes | 12 (2.5) |
| Spermatids | 474 (97.5) |
| Steps 1-3 | 16 (3.3) |
| Steps 4-6 | 84 (17.3) |
| Steps 7-9 | 298 (61.3) |
| Steps 10-13 | 38 (7.8) |
| Steps 14-16 | 38 (7.8) |
| Total | 486 (100) |
Twelve testicular sections from three 25-week-old Tslc1−/− mice were examined.
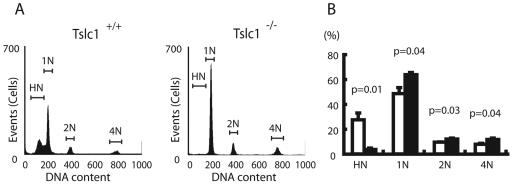
We next examined the DNA contents of testicular cells from Tslc1+/+ and Tslc1−/− mice, using flow cytometry (Fig. 5). Tslc1−/− mice showed markedly decreased HN cell fractions (representing elongated spermatids [steps 10 to 16]), increased 1N cell fractions (round spermatids [steps 1 to 9]), and slightly increased 2N and 4N cell fractions (spermatogonia and spermatocytes) compared to Tslc1+/+ mice. This finding is consistent with the histological observation of a maturation arrest of spermatogenesis at the stages between round spermatids and elongated spermatids. In addition, the presence of 1N cell fractions (haploid cells) suggests that meiosis was not affected in Tslc1−/− testes.
FIG. 5.
Flow cytometric analyses of cells isolated from the testes of Tslc1+/+ and Tslc1−/− mice. (A) The flow cytograms demonstrate four discrete peaks: an HN (haploid) peak representing elongated spermatids, a 1N (haploid) peak representing round spermatids, a 2N (diploid) peak representing G1-phase spermatogonia, and a 4N (tetraploid) peak representing pachytene spermatocytes and G2-phase spermatogonia. (B) Relative amounts of four cell populations in the testes. Open and closed boxes indicate cells from Tslc1+/+ and Tslc1−/− mice, respectively.
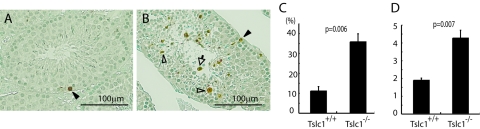
To examine the possible involvement of apoptosis in the mature arrest of the spermatids, we carried out a TUNEL assay on the testes from Tslc1+/+ and Tslc1−/− mice. A considerable number of spermatocytes and spermatids in Tslc1−/− mice were positive in the TUNEL assay (Fig. 6B). In contrast, a small number of spermatocytes, but not spermatids, in Tslc1+/+ mice were TUNEL positive (Fig. 6A). As shown in Fig. 6C and D, Tslc1−/− testes showed significantly increased numbers of TUNEL-positive tubules and TUNEL-positive cells compared with Tslc1+/+ testes. These results suggest that the primary role of TSLC1/IGSF4 is suppression of apoptosis, although we cannot exclude the possibility that TSLC1/IGSF4 directly functions as a survival factor in sperm development.
FIG. 6.
Detection of apoptosis by TUNEL assays. (A and B) Histochemistry of the testes from Tslc1+/+ (A) and Tslc1−/− (B) mice by TUNEL assay. Cells stained brown are TUNEL-positive cells. Nuclei were counterstained with methyl green (green). Closed arrowheads and open arrowheads indicate spermatocytes and spermatids, respectively. The open arrow indicates the sloughed cell. (C) Ratios of TUNEL-positive tubules to total tubules. (D) Average numbers of TUNEL-positive cells in TUNEL-positive tubules.
Delayed maturation of spermatocytes in developing Tslc1−/− mice.
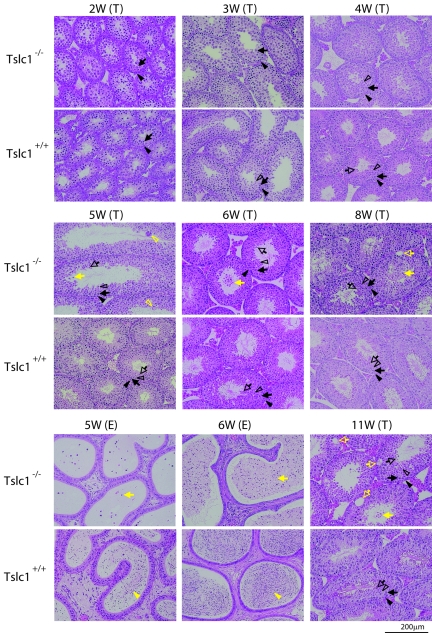
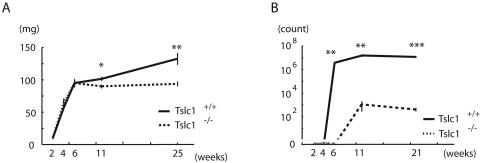
To understand detailed features of the maturation defect of spermatogenesis in Tslc1−/− mice, the postnatal development of the testes was compared in Tslc1+/+ and Tslc1−/− mice (Fig. 7). The spermatogonia and Sertoli cells of 2-week-old mice were observed at the basal sites of the seminiferous tubules. In addition, the primary spermatocytes appeared as round cells with large nuclei at the luminal sites in both Tslc1+/+ and Tslc1−/− mice. The round spermatids appeared at the luminal sites in Tslc1+/+ mice by 3 weeks of age. In Tslc1−/− mice, however, the round spermatids were not seen until 4 weeks of age. Furthermore, elongated spermatids were scarcely observed at all time points in Tslc1−/− mice. Therefore, it is speculated that germ cell differentiation is delayed when the spermatocytes proceed to round spermatids and is finally arrested when the round spermatids proceed to elongated spermatids in Tslc1−/− mice. In Tslc1+/+ mice, elongated spermatids and spermatozoa appeared at 4 weeks and 5 weeks of age, respectively. The epididymides from Tslc1+/+ mice were filled with spermatozoa by 6 weeks of age. In contrast, the round spermatids began to slough into the lumen and multinucleated giant cells appeared at 5 weeks of age in Tslc1−/− mice. Sloughing of the spermatids was also seen in mice 5 weeks of age or older. Consistent with these findings, the epididymides were filled with degenerated round cells by the time the mice were 6 weeks of age. Then, several weeks after the spermatids began to slough, vacuoles appeared at the basal portions of the seminiferous tubules in the Tslc1−/− testes. This occurred in mice around 8 weeks of age and developed more prominently by the time the mice were 11 weeks of age. Testicular weight showed a significant reduction in Tslc1−/− mice compared with that in Tslc1+/+ mice at 11 weeks of age (Fig. 8A). It should be noted, however, that very few elongated spermatids and even spermatozoa could be found among the degenerated round cells in the Tslc1−/− testes of 5-week-old mice and in the epididymides of 11-week-old mice, respectively. Therefore, maturation arrest of the spermatids caused by a TSLC1/IGSF4 deficiency did not seem to be complete (Fig. 8B).
FIG. 7.
Morphological analysis of germ cells from Tslc1+/+ and Tslc1−/− mice during postnatal development. Testes (T) and epididymides (E) of juvenile mice from 2 to 11 weeks of age were examined by HE staining. Closed arrowheads, closed arrows, open arrowheads, and open arrows in black indicate spermatogonia, spermatocytes, round spermatids, and elongated spermatids, respectively. Closed arrowheads, closed arrows, open arrowheads, and open arrows in yellow indicate spermatozoa, sloughed cells, multinucleated giant cells, and vacuoles, respectively.
FIG. 8.
Weights of testes and numbers of normal sperm during postnatal development of Tslc1+/+ and Tslc1−/− mice. (A) Weights of testes. (B) Numbers of normal sperm. *, P < 0.05; **, P < 0.005; ***, P < 0.0001.
Electron microscopic examination of spermatogenic cells and Sertoli cells.
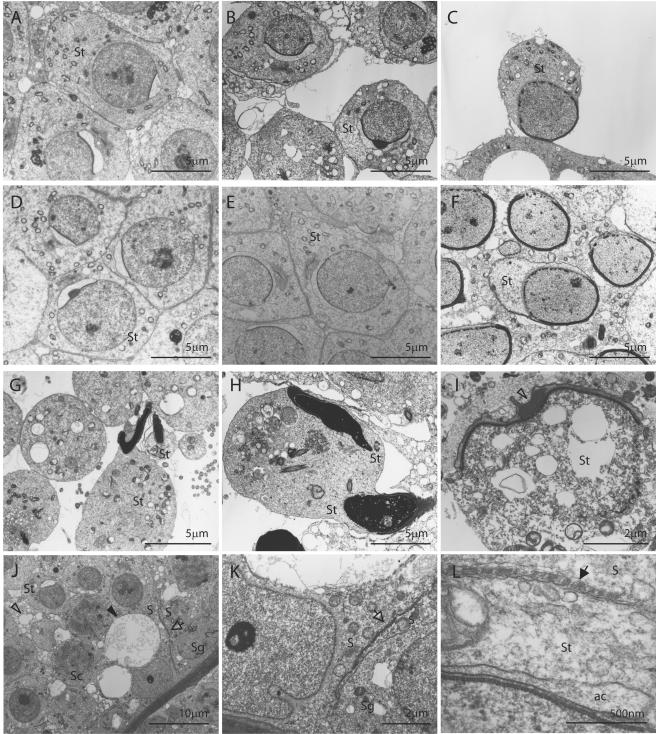
To further elucidate the pathological defect, electron microscopic analysis was carried out using Tslc1−/− mice as well as Tslc1+/+ mice. In the testes, the morphology of the spermatids in step 5 was essentially unaffected (Fig. 9A and D). On the other hand, sloughing of the spermatids from the Sertoli cells of Tslc1−/− mice was clearly seen in steps 7 and 8. Vacuoles in the cytoplasm were preferentially observed in these sloughing spermatids compared with those in Tslc1+/+ testes (Fig. 9B, C, E, and F). Furthermore, spermatids in step 10 and later were very rare and, when present, showed more drastic morphological changes, including morphologically abnormal nuclei and a large amount of residual cytoplasm (Fig. 9G). Intercellular boundaries were not clear in some spermatids (Fig. 9H). Compared with those in the testes, sloughed spermatids observed in the epididymides were more severely degenerated, with numerous vacuoles, even in step 7 or 8 (Fig. 9I). On the other hand, Sertoli cells from Tslc1−/− mice contained numerous phagocytes and a considerable number of large vacuoles in the cytoplasm (Fig. 9J). However, the Sertoli cell-Sertoli cell junction, which separates the spermatogonia and the spermatocytes from the luminal side up to the preleptotene phase, was unaffected in Tslc1−/− mice (Fig. 9K). In addition, ectoplasmic specialization, a junctional apparatus between the spermatids and the Sertoli cells, was unaffected as long as the spermatids were present and attached to the Sertoli cells (Fig. 9L).
FIG. 9.
Electron microscopic analysis of spermatogenic cells and Sertoli cells from Tslc1+/+ and Tslc1−/− mice. (A to H) Spermatids from Tslc1−/− mice (A to C, G, and H) and Tslc1+/+ mice (D to F). Spermatids in step 5 (A and D), step 7 (B and E), step 8 (C and F), and step 10 or later (G and H) are demonstrated. (I) A degenerated cell in the epididymis from a Tslc1−/− mouse. Densely staining materials corresponding to the acrosome (open arrowhead), as well as numerous degenerated vacuoles, were observed. (J to L) Numerous figures of phagocytosis (open arrowhead in panel J) and vacuolization (closed arrowhead in panel J) were observed within the Sertoli cells. Note that the Sertoli cell-Sertoli cell junction (open arrows in panels J and K) and ectoplasmic specialization (closed arrow in panel L) were unaffected in the testes from Tslc1−/− mice. S, Sertoli cell; Sg, spermatogonium; Sc, spermatocyte; St, spermatid; ac, acrosome.
Expression profile of testes from Tslc1−/− mice.
To elucidate the molecular mechanisms underlying the TSLC1/IGSF4 deficiency and the defect in spermatogenesis, we carried out expression profiling of testes from Tslc1−/− mice and Tslc1+/+ mice, using an oligonucleotide microarray (MG-U74A v.2; Affymetrix) containing a total of 18,400 transcripts. A search for genes whose expression in the testes from Tslc1−/− mice showed a >1.3-fold increase or decrease compared with the testes from Tslc1+/+ mice in two independent experiments yielded 33 and 130 genes, respectively. It should be noted that some transcripts of the Tslc1/Igsf4 gene were listed as the most down-regulated genes, indicating that the experimental system works well (Table 3).
TABLE 3.
Genes up- and down-regulated in testes from Tslc1−/− mice
| Gene product | Accession no. | Microarray analysis results
|
Quantitative RT-PCR analysis resultsb
|
P valuec | |||
|---|---|---|---|---|---|---|---|
| Signal intensitya
|
Ratio of Tslc1−/− value/Tslc1+/+ value | Tslc1−/− value | Tslc1+/+ value | ||||
| Tslc1−/− | Tslc1+/+ | ||||||
| Up-regulated genes in Tslc1−/− testes | |||||||
| Phospholipase A2, group XIIA (Pla2g12a) | AI845798 | 13,102 | 771 | 17.0 | 11 ± 2.1 | 4.1 ± 0.8 | 0.03 |
| Purine-nucleoside phosphorylase (Pnp) | U35374 | 1,629 | 410 | 4.0 | 6.1 ± 0.7 | 2.5 ± 0.6 | 0.02 |
| Mus musculus cDNA 5′ end clone | AA874329 | 706 | 272 | 2.6 | |||
| FUS interacting protein 1 (Fusip1) | AF060490 | 695 | 282 | 2.5 | |||
| Hemoglobin, beta adult major chain (Hbb-b1) | J00413 | 1,446 | 636 | 2.3 | |||
| DnaJ homolog, subfamily A, member 2 (Dnaja2) | AA763945 | 1,833 | 860 | 2.1 | 470 ± 160 | 300 ± 120 | NS |
| Vascular cell adhesion molecule 1 (Vcam1) | U12884 | 2,321 | 1,546 | 1.5 | 110 ± 5.8 | 38 ± 2.5 | 0.0002 |
| Guanylate kinase 1 (Guk1) | U53514 | 3,225 | 2,294 | 1.4 | 23 ± 12 | 4.4 ± 0.1 | NS |
| Angiopoietin-like 4 (Angpt14) | AF110520 | 6,143 | 4,823 | 1.3 | 3.1 ± 0.2 | 1.2 ± 0.2 | 0.002 |
| Down-regulated genes in Tslc1−/− testes | |||||||
| Immunoglobulin superfamily 4 (Igsf4/Tslc1) | AF061260 | 444 | 3,943 | 0.11 | 0 ± 0 | 100 ± 18 | 0.001 |
| Immunoglobulin superfamily 4 (Igsf4/Tslc1) | AB021966 | 690 | 3,739 | 0.18 | |||
| Platelet/endothelial cell adhesion molecule 1 (Pecam1) | L06039 | 181 | 799 | 0.23 | 1.6 ± 0.2 | 1.2 ± 0.3 | NS |
| Mus musculus cDNA 3′ end clone | AW047207 | 241 | 895 | 0.27 | |||
| Peroxiredoxin 2 (Prdx2) | U20611 | 281 | 892 | 0.31 | 16 ± 3.0 | 11 ± 0.6 | NS |
| Dehydrogenase/reductase X chromosome (Dhrsx) | AI846822 | 4,509 | 11,760 | 0.38 | 11 ± 1.6 | 12 ± 2.1 | NS |
| Ornithine decarboxylase antizyme 3 (Oaz3) | AB016275 | 17,735 | 44,962 | 0.39 | 94 ± 6.8 | 150 ± 9.2 | 0.007 |
| Splicing factor, arginine/serine-rich 16 (Sfrs16) | AF042799 | 430 | 1,030 | 0.42 | |||
| Protein phosphatase 2, regulatory subunit B, beta isoform (Ppp2r2b) | AW048155 | 6,134 | 14,358 | 0.43 | 9.2 ± 1.9 | 23 ± 1.5 | 0.004 |
| Mouse endogenous murine leukemia virus modified polytopic provirus DNA | M17327 | 789 | 1,769 | 0.45 | |||
| Deleted in polyposis 1-like 1 (Dplll) | AA755260 | 14,224 | 29,836 | 0.48 | 340 ± 30 | 970 ± 130 | 0.008 |
| Lysophospholipase 1 (Lypla1) | U89352 | 6,971 | 14,600 | 0.48 | 7.8 ± 0.4 | 5.6 ± 1.6 | NS |
| Suppressor of K+ transport defect 3 (Skd3) | U09874 | 7,709 | 15,612 | 0.49 | |||
| S100 calcium binding protein A13 (S100a13) | X99921 | 773 | 1,430 | 0.54 | |||
| Protamine 1 (Prm1) | Z47352 | 18,600 | 31,700 | 0.58 | 12,000 ± 2,200 | 16,000 ± 1,700 | NS |
| RAN GTPase activating protein 1 (Rangap1) | U20857 | 4,960 | 8,480 | 0.58 | 72 ± 33 | 110 ± 47 | NS |
| Growth arrest-specific protein 6 (Gas6) | X59846 | 1,120 | 1,820 | 0.61 | 80 ± 8.2 | 170 ± 35 | 0.04 |
| Bcl2-associated athanogene 1 (Bag1) | AF022223 | 3,412 | 5,546 | 0.61 | 0.1 ± 0.1 | 1.1 ± 0.4 | NS |
| Cyclin-dependent kinase inhibitor 1C (Cdkn1c) | U22399 | 1,090 | 1,650 | 0.66 | 2.3 ± 0.2 | 1.8 ± 0.1 | NS |
| Mothers against decapentaplegic homolog 6 (Smad6) | AF010133 | 2,070 | 2,930 | 0.70 | 79 ± 11 | 94 ± 8.4 | NS |
| Sperm mitochondrion-associated cysteine-rich protein (Smcp) | M88463 | 29,700 | 38,900 | 0.76 | 430 ± 43 | 930 ± 81 | 0.002 |
Average value of two independent experiments.
The Amount of Igsf4/Tslc1 in the Tslc1+/+ testis was assigned a value of 100. Data are average values ± SE of three to five independent experiments.
NS, not significant.
Among the 163 up- and down-regulated genes, we selected 6 up-regulated and 15 down-regulated genes on the basis of the amount of expression as well as their possible biological significance and then confirmed the differences in the amounts of these transcripts by quantitative RT-PCR analysis. As shown in Table 3, significant differences in gene expression were detected in four up-regulated and six down-regulated genes, including Tslc1/Igsf4. Among the gene products, Gas6 (growth arrest-specific factor 6) was shown not to be expressed in the spermatids but expressed in the spermatogonia as well as Sertoli and Leydig cells, and this protein functions to prevent apoptotic cell death in an auto- or paracrine manner with its group of receptors, Tyro3 family molecules (2). Therefore, Gas6 might play a role in enhanced apoptosis of the spermatogenic cells in Tslc1−/− mice. On the other hand, the transcripts of Oaz3 and Ppp2r2b were shown to be specifically expressed in the spermatids but not in the spermatogonia or the spermatocytes (6, 16). Therefore, a decrease in the number of these transcripts in testes from Tslc1−/− mice might be caused by a reduction in the spermatid cell population.
Similar findings were obtained by Western blotting, in which the amount of alpha-tubulin decreased dramatically in the testes from Tslc1−/− mice in comparison with those from Tslc1+/+ or Tslc1+/− mice (see Fig. S1A in the supplemental material), although no significant difference was detected in the numbers of transcripts by RT-PCR analysis (see Fig. S1B in the supplemental material). Immunohistochemical studies revealed that alpha-tubulin was preferentially expressed in mature spermatids but not in other cells in the testes (see Fig. S1C and D in the supplemental material), indicating that the marked reduction in the amount of alpha-tubulin protein was caused by a reduction in the population of mature spermatids in the testes from Tslc1−/− mice.
DISCUSSION
The present study clearly demonstrates that TSLC1/IGSF4 is essential for spermatogenesis. Although the TSLC1/IGSF4 protein is expressed in the brain, lungs, testes, and various other tissues, no overt defects other than male infertility are observed. These findings suggest that the function of TSLC1/IGSF4 is mostly compensated for by other molecules, although detailed analyses of brain function as well as disease susceptibility, including that to cancer, are being conducted. In addition, it would be valuable to compare the phenotypes of other Tslc1−/− mice, because the neo cassettes might affect the phenotype of Tslc1−/− mice in the present study. Based on the observations described above, we propose that a defect in spermatogenesis in Tslc1−/− male mice is caused by delayed maturation from spermatocytes to spermatids, the sloughing of spermatids in steps 7 to 9 from the seminiferous epithelia into the lumen, and the apoptosis and mature arrest of spermatids in step 10 and later. A small but significant number of spermatocytes were also sloughed from the seminiferous tubules. As a result, mature spermatozoa were scarcely observed in semens from Tslc1−/− mice. These findings mostly agree with the observation by immunohistochemistry that the TSLC1/IGSF4 protein was expressed in spermatogenic cells from intermediate spermatogonia to early pachytene spermatocytes and from step 7 spermatids to step 16 residual bodies. It is noteworthy, however, that no sloughing was observed in spermatogonia expressing a considerable amount of the TSLC1/IGSF4 protein. The Sertoli cell-Sertoli cell junction may prevent the spermatogonia from sloughing from the seminiferous epithelium. Indeed, the Sertoli cell-Sertoli cell junction was unaffected when examined by electron microscopic analysis. For Sertoli cells from Tslc1−/− mice, however, numerous images of phagocytosis and vacuoles were observed, although the TSLC1/IGSF4 protein was not expressed in Sertoli cells from Tslc1+/+ mice. These morphological changes in Sertoli cells were likely caused by a secondary effect in response to sloughing of the seminiferous epithelial cells because in Tslc1−/− mice, large vacuoles emerged at 8 weeks of age, whereas sloughing of the spermatids occurred at 5 weeks of age. It is also interesting that multinucleated giant cells and abnormal sperm with multiple tails were found in Tslc1−/− mice, suggesting that the intercellular cytoplasmic bridge between sister spermatids could also be disrupted by sloughing. These observations suggest that TSLC1/IGSF4 is essential for adhesion between the spermatogenic cells and between the spermatogenic cells and Sertoli cells. However, the presence of a scant amount of mature sperm implies that these functions might be compensated for by other molecules in a very small population of spermatogenic cells.
Tslc1+/− male mice were fertile and gave offspring with the expected Mendelian ratio when crossed with Tslc1+/− or Tslc1−/− female mice. These findings indicate that all haploid spermatids differentiated normally into functional spermatozoa, although half of them had lost the Tslc1/Igsf4 gene in Tslc1+/− male mice. It should be noted that neither the maturation of spermatogenic cells nor the expression pattern of the TSLC1/IGSF4 protein in these cells from Tslc1+/− mice was affected in comparison with Tslc1+/+ mice. In fact, as seen in testes from Tslc1+/+ mice, the TSLC1/IGSF4 protein was detected even in the spermatids in step 7 to the residual bodies in step 16 from Tslc1+/− mice, although the Tslc1/Igsf4 gene had been lost in one-half of these spermatids. The discrepancy between the absence of the gene and the presence of the TSLC1/IGSF4 protein in spermatids at step 7 and later can be explained if the Tslc1/Igsf4 mRNA that was transcribed in the diploid spermatogonia was transmitted into the spermatids and then translated into the protein in the spermatids at step 7 and later.
Our findings are consistent with those given in a previous report by Wakayama et al. regarding the discrepancy in the expression of SgIGSF4/IGSF4 mRNA and its protein in mouse spermatogenic cells. They found that SgIGSF/IGSF4 mRNA was detected in the spermatogonia and the early premeiotic spermatocytes that were situated adjacent to the basement membrane of the seminiferous tubules in the preleptotene to zygotene stages, whereas no mRNA signal was detected in the series of spermatids (19). On the other hand, they later observed that the SgIGSF/IGSF4 protein was expressed in spermatogenic cells from the intermediate spermatogonia to the spermatocytes and from the spermatids at step 7 to the residual bodies in step 16 (18). Thus, they speculated that the transcription of SgIGSF/IGSF4 mRNA terminates in the early steps in spermatocytes but that the translation of SgIGSF/IGSF4 restarts in the round spermatids at step 7 and later using the remaining mRNA. The SgIGSF/IGSF4 mRNA might possibly be stored in the cytoplasm as a ribonucleoprotein complex until it is recruited to the translation machinery several days later (14). Therefore, the long half-life of Tslc1/Igsf4 mRNA could prevent the spermatids lacking the Tslc1/Igsf4 gene from sloughing from the seminiferous epithelia in Tslc1+/− mice.
Finally, we examined the expression profiles of whole testes from Tslc1+/+ and Tslc1−/− mice. The number of up- and down-regulated genes in the present study was smaller than that reported in most similar studies of other genes. This result could be due to the fact that TSLC1/IGSF4 is a membrane protein and is not directly involved in the transcriptional control of other genes. Among the genes which were significantly up-regulated in the testes from Tslc1−/− mice, the Pla2g12a gene, encoding a group XIIA secretory phospholipase A2 precursor, showed a 17-fold increase in expression in the Tslc1−/− testes. The physiological function of this enzyme has not been determined yet, although other members of this gene family are known to be involved in the metabolism of arachidonic acid. On the other hand, Gas6 is a potentially interesting gene among those down-regulated in Tslc1−/− testes because it is a ligand of the Tyro3 family of receptors, consisting of Tyro3, Axl, and Mer, which are known to prevent apoptotic cell death (2, 4, 5, 8). Furthermore, male mice deficient in each of these genes (Tyro3−/−, Axl−/−, and Mer−/− mice) showed a complete loss of mature sperm owing to the progressive death of differentiating spermatogenic cells (9). The loss of TSLC1/IGSF4 function might enhance the apoptosis of spermatids by modifying the signaling of the Gas6 and Tyro3 cascade in Tslc1−/− testes. Sperm mitochondrion-associated cysteine-rich protein (Smcp) is another candidate molecule that could be implicated in the pathogenesis of Tslc1−/− testes because Smcp is directly involved in sperm motility. Moreover, Smcp homozygous mutant mice showed male infertility due to asthenozoospermia (13). Decreased expression of Smcp might be involved in the low motility of the sperm in Tslc1−/− mice. Characterization of these molecules could uncover the physiological function of the TSLC1/IGSF4 cascade, although we must consider the heterogeneity of the seminiferous tubules in terms of stages. TSLC1/IGSF4 was originally identified as a tumor suppressor gene in lung cancer. No spontaneous tumors, however, have developed in eight Tslc1−/− mice over 1 year of age. Studies of chemical carcinogenesis as well as irradiation studies in Tslc1−/− mice are being conducted in order to understand the role of TSLC1/IGSF4 in the oncogenesis of various organs.
Most of the genes that have been reported to be essential for spermatogenesis in mice also show a variety of indispensable functions in nongerm tissues. However, no obvious phenotypic abnormality, except that in the testes, was observed in Tslc1−/− mice, suggesting that the defects in TSLC1/IGSF4 function are complemented by other molecules in other tissues. This fact is especially interesting because a decreased number or morphological abnormalities of spermatozoa are often the only phenotypes recognized in most infertile human males. Further studies on the function of TSLC1/IGSF4, including carcinogenesis experiments, are required for understanding the physiological and pathological significance of TSLC1/IGSF4.
Supplementary Material
Acknowledgments
We thank Louise van der Weyden for her participation in fruitful discussions, Hara P. Ghosh for providing the EC2 antibody, Takashi Takaki for his generous help in electron microscopic analysis, and Masami Ishii for technical assistance.
Financial support consisted of a grant-in-aid for Third Term Comprehensive Control Research for Cancer from the Ministry of Health, Labor, and Welfare, Japan (Y.M.); a grant-in-aid for scientific research on priority areas for cancer (no. 17015048 for Y.M. and no. 17012003 for D.N.) from the Ministry of Education, Culture, Sports, Science, and Technology, Japan; and a grant for the promotion of fundamental studies in health sciences from the Organization for Pharmaceutical Safety and Research (OPSR) (Y.M.). D.Y., T. F., and S.K. are the recipients of research resident fellowships from the Foundation for the Promotion of Cancer Research of Japan.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Baker, J., M. P. Hardy, J. Zhou, C. Bondy, F. Lupu, A. R. Bellve, and A. Efstratiadis. 1996. Effects of an Igf1 gene null mutation on mouse reproduction. Mol. Endocrinol. 10:903-918. [DOI] [PubMed] [Google Scholar]
- 2.Chan, M. C., J. P. Mather, G. McCray, and W. M. Lee. 2000. Identification and regulation of receptor tyrosine kinases Rse and Mer and their ligand Gas6 in testicular somatic cells. J. Androl. 21:291-302. [PubMed] [Google Scholar]
- 3.Fukami, T., H. Satoh, E. Fujita, T. Maruyama, H. Fukuhara, M. Kuramochi, S. Takamoto, T. Momoi, and Y. Murakami. 2002. Identification of the Tslc1 gene, a mouse orthologue of the human tumor suppressor TSLC1 gene. Gene 295:7-12. [DOI] [PubMed] [Google Scholar]
- 4.Goruppi, S., E. Ruaro, and C. Schneider. 1996. Gas6, the ligand of Axl tyrosine kinase receptor, has mitogenic and survival activities for serum starved NIH3T3 fibroblasts. Oncogene 12:471-480. [PubMed] [Google Scholar]
- 5.Hasanbasic, I., J. Cuerquis, B. Varnum, and M. D. Blostein. 2004. Intracellular signaling pathways involved in Gas6-Axl-mediated survival of endothelial cells. Am. J. Physiol. Heart Circ. Physiol. 287:H1207-H1213. [DOI] [PubMed] [Google Scholar]
- 6.Hatano, Y., H. Shima, T. Haneji, A. B. Miura, T. Sugimura, and M. Nagao. 1993. Expression of PP2A B regulatory subunit beta isotype in rat testis. FEBS Lett. 324:71-75. [DOI] [PubMed] [Google Scholar]
- 7.Kuramochi, M., H. Fukuhara, T. Nobukuni, T. Kanbe, T. Maruyama, H. P. Ghosh, M. Pletcher, M. Isomura, M. Onizuka, T. Kitamura, T. Sekiya, R. H. Reeves, and Y. Murakami. 2001. TSLC1 is a tumor-suppressor gene in human non-small-cell lung cancer. Nat. Genet. 27:427-430. [DOI] [PubMed] [Google Scholar]
- 8.Lee, W. P., Y. Wen, B. Varnum, and M. C. Hung. 2002. Akt is required for Axl-Gas6 signaling to protect cells from E1A-mediated apoptosis. Oncogene 21:329-336. [DOI] [PubMed] [Google Scholar]
- 9.Lu, Q., M. Gore, Q. Zhang, T. Camenisch, S. Boast, F. Casagranda, C. Lai, M. K. Skinner, R. Klein, G. K. Matsushima, H. S. Earp, S. P. Goff, and G. Lemke. 1999. Tyro-3 family receptors are essential regulators of mammalian spermatogenesis. Nature 398:723-728. [DOI] [PubMed] [Google Scholar]
- 10.Masuda, M., M. Yageta, H. Fukuhara, M. Kuramochi, T. Maruyama, A. Nomoto, and Y. Murakami. 2002. The tumor suppressor protein TSLC1 is involved in cell-cell adhesion. J. Biol. Chem. 277:31014-31019. [DOI] [PubMed] [Google Scholar]
- 11.Matzuk, M. M., and D. J. Lamb. 2002. Genetic dissection of mammalian fertility pathways. Nat. Cell Biol. 4(Suppl.):S41-S49. [DOI] [PubMed] [Google Scholar]
- 12.Murakami, Y. 2005. Involvement of a cell adhesion molecule, TSLC1/IGSF4, in human oncogenesis. Cancer Sci. 96:543-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nayernia, K., I. M. Adham, E. Burkhardt-Gottges, J. Neesen, M. Rieche, S. Wolf, U. Sancken, K. Kleene, and W. Engel. 2002. Asthenozoospermia in mice with targeted deletion of the sperm mitochondrion-associated cysteine-rich protein (Smcp) gene. Mol. Cell. Biol. 22:3046-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pires-daSilva, A., K. Nayernia, W. Engel, M. Torres, A. Stoykova, K. Chowdhury, and P. Gruss. 2001. Mice deficient for spermatid perinuclear RNA-binding protein show neurologic, spermatogenic, and sperm morphological abnormalities. Dev. Biol. 233:319-328. [DOI] [PubMed] [Google Scholar]
- 15.Rusell, L. D., R. A. Ettlin, A. P. Sinha Hikim, and E. D. Clegg. 1990. Histological and histopathological evaluation of the testis. Cache River, St. Louis, Mo.
- 16.Tosaka, Y., H. Tanaka, Y. Yano, K. Masai, M. Nozaki, K. Yomogida, S. Otani, H. Nojima, and Y. Nishimune. 2000. Identification and characterization of testis specific ornithine decarboxylase antizyme (OAZ-t) gene: expression in haploid germ cells and polyamine-induced frameshifting. Genes Cells 5:265-276. [DOI] [PubMed] [Google Scholar]
- 17.Tourtellotte, W. G., R. Nagarajan, A. Auyeung, C. Mueller, and J. Milbrandt. 1999. Infertility associated with incomplete spermatogenic arrest and oligozoospermia in Egr4-deficient mice. Development 126:5061-5071. [DOI] [PubMed] [Google Scholar]
- 18.Wakayama, T., H. Koami, H. Ariga, D. Kobayashi, Y. Sai, A. Tsuji, M. Yamamoto, and S. Iseki. 2003. Expression and functional characterization of the adhesion molecule spermatogenic immunoglobulin superfamily in the mouse testis. Biol. Reprod. 68:1755-1763. [DOI] [PubMed] [Google Scholar]
- 19.Wakayama, T., K. Ohashi, K. Mizuno, and S. Iseki. 2001. Cloning and characterization of a novel mouse immunoglobulin superfamily gene expressed in early spermatogenic cells. Mol. Reprod. Dev. 60:158-164. [DOI] [PubMed] [Google Scholar]
- 20.Yan, W., L. Ma, K. H. Burns, and M. M. Matzuk. 2004. Haploinsufficiency of kelch-like protein homolog 10 causes infertility in male mice. Proc. Natl. Acad. Sci. USA 101:7793-7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.