Abstract
β-Site β-amyloid precursor protein (APP)-cleaving enzyme 1 (BACE1) is the β-secretase in vivo for processing APP to generate amyloid β protein (Aβ). Aβ deposition in the brain is the hallmark of Alzheimer's disease (AD) neuropathology. Inhibition of BACE1 activity has major pharmaceutical potential for AD treatment. The expression of the BACE1 gene is relatively low in vivo. The control of BACE1 expression has not been well defined. There are six upstream AUGs (uAUGs) in the 5′ leader sequence of the human BACE1 mRNA. We investigated the role of the promoter and the uATGs in the 5′ untranslated region (UTR) of the human BACE1 gene in BACE1 gene transcription and translation initiation. Our results show that the first and second uATGs are the integral part of the core minimal promoter of the human BACE1 gene, while the third uAUG is skipped over by ribosomal scanning. The fourth uAUG can function as a translation initiation codon, and deletion or mutation of this uAUG increases downstream gene expression. The fourth uAUG of the BACE1 5′UTR is responsible for inhibiting the expression of BACE1. Translation initiation by the BACE1 uAUGs and physiological AUG requires intact eIF4G. Our results demonstrate that during human BACE1 gene expression, ribosomes skipped some uAUGs by leaky scanning and translated an upstream open reading frame, initiated efficiently at the fourth uAUG, and subsequently reinitiated BACE1 translation at the physiological AUG site. Such leaky scanning and reinitiation resulted in weak expression of BACE1 under normal conditions. Alterations of the leaky scanning and reinitiation in BACE1 gene expression could play an important role in AD pathogenesis.
Neuritic plaques and neurofibrillary tangles in the brain are the major pathological features of Alzheimer's disease (AD). Amyloid β protein (Aβ) is the major component in amyloid plaques. Aβ deposition in the brain is believed to play an essential role in AD pathogenesis. Aβ is generated from β-amyloid precursor protein (APP), a type 1 single-transmembrane-domain protein. APP is first cleaved by β-secretase to generate a 99-amino-acid peptide, C99. The C99 fragment is subsequently cleaved by γ-secretase to generate Aβ. β-Secretase and γ-secretase play an important role in AD pathogenesis, and reduction of Aβ by inhibition of either β-secretase or γ-secretase activity is considered to have major pharmaceutical potential for AD therapy.
BACE1 is a type 1 membrane-associated aspartyl protease of 501 amino acids (12, 40, 43, 49). BACE1 is a β-secretase in vivo and cleaves APP to generate Aβ (1, 24, 35). Recently, BACE1 was also found to cleave other proteins, including the low-density lipoprotein receptor-related protein (LRP) (45), β amyloid precursor-like proteins 1 (APLP1) (21) and 2 (APLP2) (28), a Golgi complex-resident sialyltransferase, ST6Gal I (17), and the cell adhesion protein P-selectin glycoprotein ligand 1 (23).
The BACE1 gene has a tissue-specific expression pattern. BACE1 mRNA has been found mainly in neuronal cells and in the pancreas (25, 43, 49). Although genetic analysis has failed to uncover any BACE1 coding sequence mutations in patients with familial AD (3, 27), increased β-secretase activity has been reported in the brains of some patients with familial AD (39) and greater expression levels of BACE1 were found in the cortex of sporadic AD patients than in age-matched controls (6, 7, 11, 50). BACE1 protein and activity levels increase with aging and in brain regions affected by amyloid deposition and remain increased despite significant neuronal and synaptic loss in AD (6, 7). These studies indicate that upregulated BACE1 gene expression at the level of transcription or translation could contribute to AD pathogenesis in some sporadic cases.
In eukaryotes, regulation of protein levels is controlled at various stages, such as transcription, posttranscriptional processing and modification, mRNA stability, translation initiation, posttranslational modifications, and protein clearance. A mature mRNA consists of three parts: the 5′ untranslated region (UTR) with an m7G cap, the main open reading frame (ORF), and the 3′UTR with a poly(A) tail. Protein translation in eukaryotes is predominantly initiated by a cap-dependent scanning mechanism in which 40s ribosomal subunits are recruited to the 5′ cap structure, scan in a 5′-to-3′ direction, and initiate translation at the first AUG (19). Two other less common mechanisms are internal entry on an internal ribosome entry site (IRES) and ribosome shunting (14). With respect to the 5′UTR, structured regions, the upstream ORF (uORF), and internal ribosome entry segments play important roles in translation control (46).
BACE1 gene expression is tightly regulated at the level of transcription (2, 9, 22). Between the transcription initiation site and the physiological translation initiation codon of the human BACE1 gene, there are six upstream ATGs (uATGs) that could serve as the translation initiation codons for five potential uORFs (Fig. 1). Recent studies indicate that the upstream AUGs in the 5′ untranslated region of BACE1 mRNA might have negative effects on BACE1 gene expression at the level of protein translation (4, 20, 36). However, previous reports regarding this issue were largely based on experiments using the 5′ leader sequences of BACE1 mRNA without consideration of its promoter's effect on BACE1 gene expression.
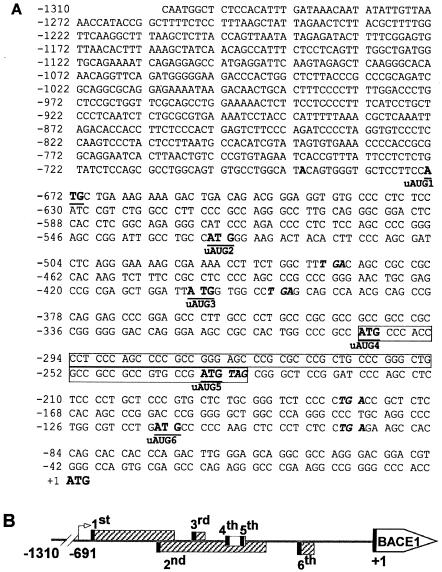
FIG. 1.
Sequence features of the human BACE1 gene minimal promoter and 5′UTR. (A) Nucleotide sequence of the minimal promoter and 5′UTR of the human BACE1 gene. The minimal promoter region corresponds to bp −1310 to −400 upstream of the physiological ATG of the human BACE1 gene, with the A designated as position +1 (2). The adenine in bold at −691 represents the transcription start site. The uATGs are indicated with both underlining and bold, and their corresponding stop codons are in italics. The fourth and fifth uATGs are in the same reading frame as the physiological ATG and their uORFs are shown in a box. The uAUGs of the corresponding upstream ORFs are also indicated. (B) Schematic diagram of the BACE1 gene uORFs. The arrow represents the transcription initiation site at −691, and +1 is the position of the adenine of the physiological start codon of the BACE1 gene ORF. Six uORFs are indicated by rectangular boxes, with the uAUGs represented by the black bars. The first, second, third, and sixth uORFs do not contain the uAUG in the same reading frame as the physiological start codon of BACE1 and are indicated by the hatched box. The uAUGs of the fourth and fifth uORFs are in the same reading frame as the physiological start codon of BACE1, and the clear box represents these uORFs.
The molecular mechanism by which the 5′UTR regulates BACE1 gene expression and its interaction with the BACE1 promoter remains elusive. In this report, we found that part of the BACE1 5′UTR has negative effects on the expression of the downstream gene under control of its own BACE1 promoter. We also found that the fourth uAUG functioned efficiently as a translation initiation codon and the rest of the uAUGs were skipped over as translation initiation codons by leaky ribosomal scanning. In addition, fourth-uAUG-initiated uORF expression had negative effects on BACE1 expression. This study suggests that leaky scanning in the 5′UTR and reinitiation at the physiological BACE1 AUG site could be responsible for the low level of expression of BACE1 under normal conditions.
MATERIALS AND METHODS
Cell culture and transfection.
Human embryonic kidney 293 (HEK293) cells and mouse neuroblastoma Neuro-2a (N2a) cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin at 37°C in a humidified incubator containing 5% CO2. Cells were grown to approximately 70 to 90% confluence and transfected with 3.8 μg of plasmid DNA on a 35-mm-diameter plate for Western blotting and RNA extractions or 500 ng of plasmid DNA per well on a 24-well plate for luciferase assays. Transfections were carried out using Lipofectamine 2000 reagent (Invitrogen) according to the supplier's protocol.
Plasmids.
pBACE1-mycHis is a mammalian expression plasmid containing BACE1 cDNA in the pcDNA3.1mycHis vector (33). Plasmid pCMV2A expresses the poliovirus protease 2A(pro). The human BACE1 minimum promoter and 5′UTR fragments corresponding to −1310 to −669, −1310 to −527, −1310 to −403, and −1310 to −401 bp upstream of the BACE1 physiological translation start codon ATG were amplified using pB1P-B (2) as a template, B-1310F (5′-CCGCTCGAGCAATGGCTCTCCACATTTG) as a forward primer, and either B-669R (5′-GGAAGCTTGCATGGAAGGAGCACCCACTG), B-527R (5′-GGAAGCTTCCATGGCAGGCAATCCGGCTC), B-403R (5′-GGAAGCTTCCATAATCCAGCTCGCGGCTC), or B-401R (5′-GGAAGCTTCACCATAATCCAGCTCGCGGCTC) as a reverse primer. The fragments were cloned into the promoterless/enhancerless luciferase reporter vector pGL3-Basic (Promega) at the XhoI and HindIII sites. The newly constructed plasmids were named pB1P-669, pB1P-527, pB1P-403, and pB1P-401, respectively.
In order to construct the following plasmids, we first generated plasmid pB1P-1625+36. A 541-bp section from bp −1625 to −1075 of the BACE1 promoter region was cleaved from plasmid pB1P-H (2) by XhoI and SacI. A 1,100-bp fragment corresponding to the region of BACE1 from −1074 to +36 was amplified using plasmid pB1P-E (2) as the template and B-1074F (5′-GGTGAGCTCAAGGGCACAAACAGG) and B+36R (5′-CACAAGCTTCATCCACAGCAGGAGCCAGG) as primers and subsequently cleaved by SacI and HindIII. Then, the 1,100-bp fragment and the 541-bp fragment were ligated and cloned into the pGL3-Basic vector at the XhoI and HindIII sites to generate plasmid pB1P-1625+36.
Using pB1P-1625+36 as the template, the following primer sets were used to amplify the BACE1 fragments from bp −1310 to −330, −1310 to −331, −1310 to −234, −1310 to −235, −1310 to −237, −1310 to −113, −1310 to −114, −1310 to +4, and −1310 to +6: B-1310F and B-330R (5′-GGAAGCTTGCATGGCGGGCCAGTGGCGGCTTx), B-331R (5′-GGAAGCTTCATGGCGGGCCAGTGGCGGCTT), B-234R (5′-GGAAGCTTTCATCGGCACGGCGGCGGCCAG), B-235R (5′-GGAAGCTTCATCGGCACGGCGGCGGCCAG), B-237R (5′-GGAAGCTTTCGGCACGGCGGCGGCCAG), B-113R (5′-GGAAGCTTGCATCAGGACGCCAGGGCCTG), B-114R (5′-GGAAGCTTCATCAGGACGCCAGGGCCTG), B+4R (5′-GGAAGCTTCCATGGTGGGCCCCGGCCTTCGG), and B+6R (5′-GGAAGCTTGGCCATGGTGGGCCCCGGCCTTCGG). The fragments were cloned into pGL3-Basic at the XhoI and HindIII sites to generate plasmids pB1P-330, pB1P-331, pB1P-234, pB1P-235, pB1P-237, pB1P-113, pB1P-114, pB1P+4, and pB1P+6, respectively.
Site-directed mutagenesis by PCR was performed to construct plasmids pB1P+4m and pB1P+4dm. Two sets of primers, B-1310F and B-4thmR (5′-GGTGGGAATGGCGGGCCAGTGGC) and B-4thmF (5′-CCGCCATTCCCACCCCTCCCAGCC) and B+4R, were used to amplify the BACE1 fragment from the −1310 to +4 region, resulting in the fourth uATG's being mutated from ATG to ATT at bp −235. The mutated fragments were cloned into pGL3-basic at the XhoI and HindIII sites to generate pB1P +4m.
To make the third and fourth uATG double-mutation plasmid pB1P+4dm, a mutated −1310 to +4 BACE1 fragment was amplified using pB1P+4m as the template and the following two sets of primers: B-1310F and B-3rdmR (5′-GGCCACAATAATCCAGCTCGCGGCTC) and B-3rdmF (5′-GGATTATTGTGGCCTGAGCAGCCAAC) and B+4R. The PCR fragment was cloned into pGL3-Basic at the XhoI and HindIII sites to generate pB1P+4dm with the third and fourth uATGs mutated from ATG to ATT at bp −404 and −235, respectively.
To make the fourth uATG mutation in pB1P-234 and pB1P-235, pB1P+4m was used as a template for PCR amplifications with primers B-1310F and B-234R or B-235R. The PCR fragments were cloned into pGL3-Basic for plasmids pB1P-234m and pB1P-235m, with the fourth uATG mutated to ATT. The deletion plasmid pB1P-234δm was generated by amplifying a 933-bp BACE1 fragment from pB1P+4m using B-1310F and B-234R as primers and a shorter denaturing time. The deletion fragment was cloned into the pGL3-Basic vector at the XhoI and HindIII sites. The new construct, pB1P-234δm, contains the fourth uATG mutation and lacks a 143-bp section corresponding to the third uATG-containing region from bp −451 to −309 of the human BACE1 gene.
Luciferase assay.
Cells in 24-well plates were transfected as described above with 500 ng of BACE1-luciferase reporter plasmids; 1 ng of pCMV-Rluc (Promega) expression plasmid was cotransfected into each well to normalize for transfection efficiency. Cells were harvested 24 h after transfection and lysed in 50 μl of 1× passive lysis buffer (Promega) for luciferase activity assay. The assays for firefly luciferase activity and Renilla luciferase activity were performed sequentially using one reaction tube and the luminescent signal was measured using a TD 20/20 luminometer (Turner Designs model 20/20). The relative light intensity was measured to reflect the luciferase activity. Firefly luciferase activity was normalized to Renilla luciferase activity and expressed as relative luciferase units (RLUs) to reflect promoter activity.
Western blot analysis.
Cells in a 35-mm plate were cotransfected as described above with 3.6 μg of BACE1-luciferase reporter plasmids and 0.2 μg of the pBACE1-mycHis expression plasmid in each well. Cells were harvested 48 h after transfection and lysed in 100 μl of radioimmunoprecipitation assay (RIPA)-Doc buffer (1% Triton X-100, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 0.15 M NaCl, 0.05 M Tris-HCl, pH 7.2) supplemented with protease inhibitors (Complete; Boehringer Mannheim). Cell lysates were separated on a 4 to 12% sodium dodecyl sulfate-Tris-tricine gel, and immunoblotting was performed as described previously (42). A mouse anti-firefly luciferase monoclonal antibody (anti-luciferase) (Novus Biologicals) was used to detect firefly luciferase expression. Mouse monoclonal antibody 9E10 (ATCC CRL 1729) was used to detect Myc-tagged BACE1 protein.
Quantitative reverse transcription-PCR.
Total RNA was extracted with TRI reagent (Sigma) from cells of the same well which were transfected for Western blot protein analysis. DNase I (Invitrogen) was used to treat 2 μg of the RNA sample to completely remove residual DNA. Random hexamer and ThermoScript reverse transcriptase (Invitrogen) were used to generate the first-strand cDNA from 1 μg of the total RNA sample. PCR was carried out in a 20-μl reaction mixture using Platinum PCR Supermix (Invitrogen) and 0.25 μM of each primer; 24 to 30 cycles of PCR with denaturation at 94°C for 30 s, annealing at 56°C for 30 s, and then extension at 70°C for 60 s were used to cover the linear range of the PCR amplification. The firefly luciferase gene-specific primers 5′-CTGCCTCATAGAACTGCCTGC and 5′-TGAGCCCATATCCTTGCCTG were used to amplify a 444-bp fragment of the firefly luciferase gene coding sequence. Plasmid pBACE1-MycHis-specific primers were used to amplify a 690-bp fragment of part of the BACE1 gene coding sequence with the N-terminal Myc epitope tag sequence as an internal control. The PCR products were further analyzed on a 1.2% agarose gel. Kodak Image Station 1000 software (Perkin-Elmer) was used to analyze the data.
TRAIL treatment.
HEK293 cells in 24-well plates were transfected as described above with 500 ng of plasmid pB1P+4 and 1 ng of pCMV-Rluc for 24 h and then treated with TRAIL (tumor necrosis factor-related apoptosis-inducing ligand) reagent (BIOMOL International, LP) at concentrations of 0, 0.25, 0.5, 1, or 2 μg/ml for 4 h. Cells were harvested in 1× passive lysis buffer for luciferase assays.
RESULTS
Inhibition of downstream cistron translation by the bp −403 to −234 region of the human BACE1 5′UTR.
There are six uATGs and uORFs (−673 to −473, −530 to −177, −406 to −395, −303 to −232, −237 to −232, and −116 to −93) between the transcription initiation site and the physiological translation initiation codon of the human BACE1 gene (Fig. 1). It has been reported that the 5′UTR of BACE1 mRNA decreased downstream gene expression. However, such observations were based on experiments in which the 5′UTR of BACE1 was under the control of an artificial T7 promoter (20).
Previously, we showed that the region from −1311 to −400 bp upstream of the physiological translation start codon, ATG, of the human BACE1 gene contains minimal promoter activity (2). To investigate which part of the BACE1 5′UTR is responsible for downregulation of BACE1 expression under physiological conditions, we constructed a series of BACE1 5′UTR deletion plasmids under control of the BACE1 promoter. Genomic DNA sequences containing a human BACE1 minimal promoter sequence and various lengths of the BACE1 5′UTR were cloned into the promoterless pGL3-basic vector in front of a reporter firefly luciferase gene.
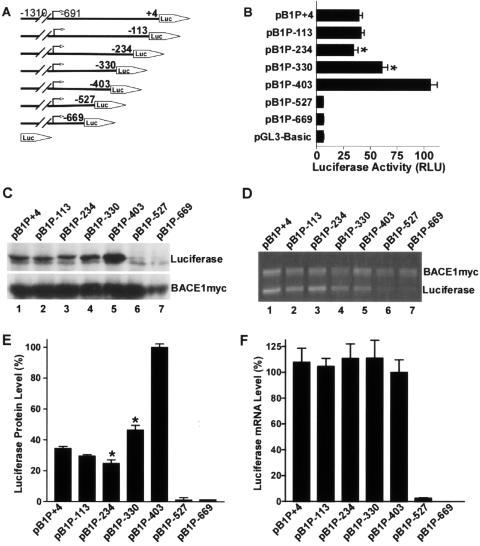
Plasmids pB1P-403, pB1P-330, pB1P-234, and pB1P+4 contain fragments covering bp −1310 to −403, −1310 to −330, −1310 to −234, and −1310 to +4 of the 5′-flanking region of the BACE1 gene, respectively, with +1 as the adenine of the physiological translation start codon ATG (Fig. 2A). As shown in Fig. 2B, the minimal BACE1 promoter plasmid pB1P-403 had significant luciferase activity. However, addition of a full-length 5′UTR fragment to the minimal promoter construct drastically reduced the luciferase activity to 39.36 ± 2.98% in pB1P+4-transfected cells relative to pB1P-403-transfected cells (P < 0.001). Plasmids containing a partial 5′UTR with deletion of 117 bp from +4 to −113 (pB1P-113) or 238 bp from +4 to −234 (pB1P-234) had a similar inhibitory effect on luciferase activity (41.21% ± 2.86% of that of pB1P+4 for pB1P-113 and 34.32% ± 4.13% of that of pB1P+4 for pB1P-234) (P > 0.05). Further deletion showed that 73 bp of the 5′UTR from −403 to −330 significantly reduced the luciferase activity to 60.52% ± 5.29% of that of pB1P-403 in pB1P-330-transfected cells (P < 0.001). These results indicate that the 169-bp fragment from −403 to −234 of the human BACE1 5′UTR downregulates downstream gene expression.
FIG. 2.
Functional deletion analysis of the human BACE1 5′UTR. (A) Schematic diagram of plasmid constructs containing the BACE1 minimal promoter and a series of truncated 5′ leader sequences. The horizontal line indicates the BACE1 minimal promoter with the 5′ leader sequence; the arrow indicates the BACE1 transcriptional start site. The box with the arrow labeled LUC represents the coding sequence of the luciferase reporter gene. The numbers indicate the endpoints of each construct, with +1 as the adenine of the physiological start codon of the BACE1 gene. (B) HEK293 cells were transiently cotransfected with the deletion plasmids and pCMV-RLuc. Firefly luciferase activity was measured 24 h after transfection, and Renilla luciferase activity was used to normalize for transfection efficiency. The values represent means ± standard error of the mean (n = 3). *, P < 0.001 by analysis of variance (ANOVA) with the posthoc Newmann-Keuls test. (C) Western blots were performed to determine the protein level of the downstream firefly luciferase cistron with antiluciferase antibody. pBACE1-mycHis was transfected with the deletion constructs, and the BACE1-Myc proteins were detected with antibody 9E10 against the Myc motif as the transfection control. (D) Quantitative RT-PCR was performed to determine the mRNA level of firefly luciferase. pBACE1-mycHis was transfected with the deletion constructs and exogenous BACE1-myc transcript was used as a transfection efficiency control. The ratio of luciferase protein to BACE1-Myc protein (E) and the ratio of luciferase mRNA to BACE1-myc mRNA (F) were quantitated by Kodak Image Analysis. Shown are the means ± standard error of the mean. *, P < 0.001 versus pB1P-403 by ANOVA with the posthoc Newmann-Keuls test.
To determine whether the decrease in luciferase activity was caused by the 5′UTR's inhibitory effect on transcription or translation, quantitative reverse transcription (RT)-PCR and Western blots were performed to measure the reporter gene luciferase protein (Fig. 2C) and mRNA (Fig. 2D) levels. Plasmid pBACE1-mycHis was cotransfected as a control. Consistent with the luciferase activity data, the luciferase protein levels in pB1P+4-, pB1P-113-, pB1P-234-, and pB1P-330-transfected cells were significantly (34.49% ± 1.29%, 29.46% ± 0.94%, 24.77% ± 2.37%, and 46.46% ± 3.02%, respectively) lower than that in pB1P-403-transfected cells (P < 0.0001) (Fig. 2E). However, the luciferase mRNA levels in pB1P+4-, pB1P-113-, pB1P-234-, and pB1P-330-transfected cells were similar to that in pB1P-403-transfected cells (P > 0.05) (Fig. 2F). These data suggest that the downregulation of the reporter luciferase gene expression by the 169-bp fragment from bp −403 to −234 of the human BACE1 5′UTR was a result not of its effect on gene transcription, but of its inhibitory effect on protein translation.
Since the minimal promoter construct pB1P-403 also contains a part of the 5′UTR sequence, we examined the role of this fragment in the regulation of BACE1 transcription and translation. Deletion of a 124-bp fragment from −403 to −527 (pB1P-527) or another 132 bp from −527 to −669 (pB1P-669) from the minimal promoter construct (pB1P-403) reduced the luciferase activity level to that of the empty vector, pGL3-Basic (Fig. 2B). There was little detectable luciferase protein (Fig. 2C and E, lanes 6 and 7) and mRNA (Fig. 2D and F, lanes 6 and 7) in pB1P-527 and pB1P-669-transfected cells. These results show that the 5′UTR fragment between bp −430 and −669, the transcription initiation site, negatively affects downstream gene transcription, suggesting that this region might be an integral part of the core promoter sequence of the human BACE1 gene.
Physiological ATG of the BACE1 gene is essential for initiation of translation of a downstream cistron.
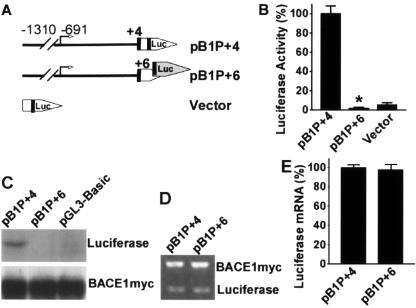
To investigate whether the uATGs affect downstream cistrons at the level of transcription or translation initiation, we first examined the role of the physiological translation start codon ATG of the human BACE1 gene using our luciferase reporter promoter system. Plasmid pB1P+4 contains the human BACE1 minimal promoter and a full-length 5′UTR in addition to the BACE1 physiological ATG codon. This BACE1 ATG codon in pB1P+4 was in the same reading frame as the ATG codon of the downstream reporter luciferase gene. There was significant luciferase activity and luciferase protein expression in cells transfected with plasmid pB1P+4 (Fig. 3B and C).
FIG. 3.
Translation initiation by the physiological BACE1 AUG codon. (A) Schematic diagram of plasmid constructs containing the BACE1 minimal promoter and the full-length 5′UTR. Plasmid pB1P+4 contains the physiological BACE1 ATG in the same reading frame as the downstream luciferase coding sequence, and pB1P+6 contains two more nucleotides at the 3′ end of the 5′UTR, which result in a frameshift in the downstream luciferase coding sequence. (B) Luciferase activity was measured 24 h after cells were cotransfected with the plasmids and pCMV-RLuc. Renilla luciferase activity was used to normalize for transfection efficiency. The values represent means ± standard error of the mean (n = 3). *, P < 0.001 by t test. The frameshift mutation abolished luciferase activity. (C) Western blotting shows that pB1P+4 has robust luciferase protein expression. However, there is no detectable level of luciferase protein in cells transfected with pB1P+6, indicating that the frameshift disrupted luciferase synthesis. pBACE1-mycHis was cotransfected with the luciferase constructs, and the BACE1-Myc proteins were used as the transfection control. (D) Quantitative RT-PCR was performed to determine the level of firefly luciferase mRNA. pBACE1-mycHis was cotransfected, and the exogenous BACE1-Myc transcript was used as the transfection efficiency control. (E) Quantitative analysis of the ratio of luciferase mRNA to BACE1-myc mRNA. The mRNA levels were quantitated by Kodak Image Analysis. Shown are the means ± standard error of the mean. There is no difference in mRNA level between the wild type and the frameshift mutant, P > 0.05 by t test.
Plasmid pB1P+6 was generated to contain two more nucleotides inserted downstream of the BACE1 physiological ATG codon, resulting in the BACE1 ATG codon's not being in the same reading frame as the ATG codon of the downstream reporter luciferase gene. The insertion of two nucleotides between the BACE1 ATG and luciferase ATG abolished the luciferase activity, and there were no detectable levels of luciferase protein in cells transfected with pB1P+6 (Fig. 3B and C). To determine if the downregulation of the reporter luciferase gene expression was due to changes in the levels of mRNA, the reporter transcript levels were analyzed. pBACE1-mycHis was cotransfected and used as a control. A pair of luciferase gene-specific primers were used to amplify reporter cDNA. There was no significant difference in reporter mRNA levels between pB1P+4- and pB1P+6-transfected cells (P > 0.05) (Fig. 3D and E). These data suggest that the physiological ATG of the human BACE1 gene in these promoter constructs does not affect downstream luciferase reporter gene transcription, but is functional and essential for the translation initiation of the BACE1 gene and other coding regions fused in frame with the physiological ATG of the human BACE1 gene.
Our results show that the luciferase reporter gene can be transcribed at similar levels whether or not the BACE1 ATG is in the same reading frame as the reporter gene ATG codon. Although pB1P+6 synthesized a transcript containing the luciferase gene coding sequence and had reporter gene mRNA levels similar to that of pB1P+4, the insertion of two nucleotides between the BACE1 ATG and the luciferase ATG resulted in a frameshift in the reporter luciferase gene coding sequence in pB1P+6. The translation units recognized the BACE1 ATG but not the downstream luciferase ATG as the translation initiation codon, and translation starting from the BACE1 ATG in pB1P+6 disrupted the ORF of the downstream luciferase cistron. Such frameshift disruption resulted in no luciferase protein synthesis and, in turn, no detectable luciferase activity. In contrast, pB1P+4 began translation at the BACE1 ATG and generated a luciferase fusion protein without disrupting the ORF of the downstream luciferase cistron. These functional fusion proteins can be detected by Western blot analysis using the firefly luciferase antibody and display luciferase enzymatic activity. These results clearly demonstrate that the physiological ATG of the human BACE1 gene is essential for its translation initiation.
Regulation of translation initiation by the BACE1 uAUGs.
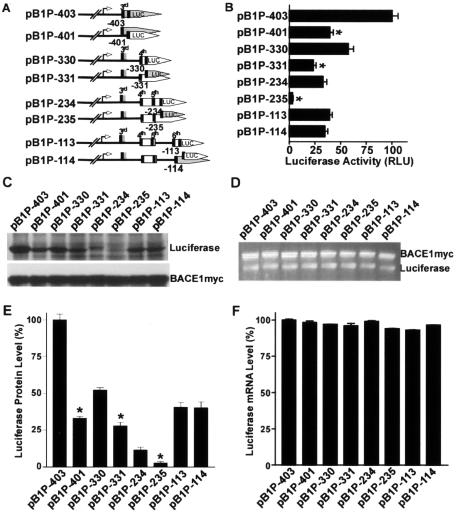
There are four uATGs within the 5′UTR between the BACE1 minimal promoter region and the physiological ATG. To examine whether these uAUGs in the 5′ leader sequence of the BACE1 mRNA could act as a functional translation initiation codon for downstream cistrons, the BACE1 minimal promoter and a series of truncated 5′ leader sequences were inserted in front of the luciferase ORF in the pGL3-basic vector and the last uATG of the BACE1 5′UTR insert was positioned either in frame or out of frame with the downstream luciferase cistron in BACE1-luciferase reporter plasmids.
If a uAUG is recognized as a translation initiation codon, it will augment or not affect the translation of an in-frame luciferase reporter carrying its own AUG, but will inhibit translation of such a reporter when out of frame. Plasmid pB1P-401 was generated to contain the third uATG in frame with the downstream luciferase ORF, and such a frameshift resulted in a significant decrease in luciferase activity to 39.51% ± 3.32% of that of pB1P-403, in which the third uATG was in the same reading frame as the downstream luciferase ORF (P < 0.001) (Fig. 4B). A similar result was also obtained by not positioning the fourth uATG in the same reading frame as the downstream luciferase ORF (pB1P-331) (Fig. 4A). Compared to pB1P-330, which contains the fourth uATG in the same reading frame as the downstream ORF, pB1P-331 showed an approximately 60% decrease in luciferase activity, from 57.54% ± 5.03% in pB1P-330 to 23.36 ± 2.75% in pB1P-331 (P < 0.001) (Fig. 4B).
FIG. 4.
Translation initiation by the uAUGs of the human BACE1 5′UTR. (A) Schematic diagram of plasmid constructs containing the BACE1 minimal promoter and the wild-type or frameshift mutant 5′UTR. The BACE1 uATG in plasmids pB1P-403, pB1P-330, pB1P-234, and pB1P-113 is in the same reading frame as the downstream luciferase coding sequence, and pB1P-401, pB1P-331, pB1P-235, and pB1P-114 were generated so that the BACE1 uATG would not be in the same reading frame as the downstream luciferase ORF, which causes a frameshift in the downstream luciferase coding sequence. The uORFs are indicated. (B) The constructs were cotransfected with pCMV-RL into cells for 24 h. Firefly luciferase activity was measured, and Renilla luciferase activity was used to normalize for transfection efficiency. The values represent means ± standard error of the mean (n = 3). *, P < 0.001 by ANOVA with the posthoc Newmann-Keuls test. (C) The luciferase reporter plasmids were also transfected with pBACE1-mycHis for Western blot analysis. Firefly luciferase was detected with antiluciferase antibody, and BACE1-Myc protein was detected with antibody 9E10 as the transfection control. (D) Quantitative RT-PCR assays of the level of firefly luciferase mRNA. pBACE1-mycHis was cotransfected, and exogenous BACE1-Myc transcript was used as the transfection efficiency control. (E) Quantitative analysis of luciferase protein levels. Values are means ± standard error of the mean (n = 3). The protein levels are normalized to the BACE1-Myc control and expressed as a percentage of the level in pB1P-403-transfected cells. *, P < 0.01 relative to the wild-type by t test. (F) Quantitative analysis of luciferase mRNA levels. Values are means ± standard error of the mean (n = 3). The luciferase mRNA levels are normalized to the BACE1-Myc control and expressed as a percentage of the level in pB1P-403-transfected cells. There is no difference in mRNA level between the wild types and the frameshift mutants, P > 0.05 by ANOVA.
The luciferase protein levels were also reduced accordingly by the frameshift mutations in plasmids pB1P-401 and pB1P-331 (Fig. 4C). Compared to the luciferase protein expression level of pB1P-403, the luciferase protein levels of pB1P-401, pB1P-330, and pB1P-331 were 32.86% ± 1.36%, 52.15% ± 1.64%, and 27.90% ± 2.43%, respectively (P < 0.001) (Fig. 4E). However, such frameshift mutations did not change the mRNA levels in the downstream luciferase cistron (P > 0.05) (Fig. 4D and F). These results suggest that the third and fourth uATGs regulate downstream luciferase gene expression at the level of translation, but not at the level of transcription.
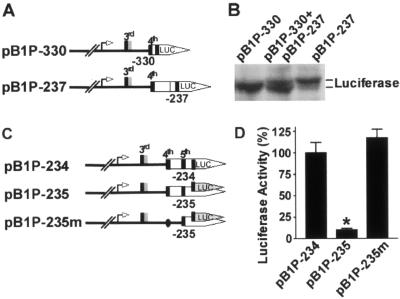
When the fifth uATG was placed in the same reading frame as the downstream luciferase ORF (pB1P-235), the luciferase activity was drastically reduced relative to that of the plasmid bearing the fifth uATG in the same reading frame as the downstream luciferase ORF (pB1P-234), from 32.64% ± 3.92% to 3.287% ± 0.56%, respectively (P < 0.001) (Fig. 2B). Positioning of the fifth uATG in the same reading frame as the downstream luciferase ORF in pB1P-234 resulted in the synthesis of a slightly higher-molecular-weight fusion luciferase protein, and the frameshift mutation in pB1P-235 inhibited luciferase protein synthesis (Fig. 4C and E, lanes 5 and 6). However, pB1P-234 and pB1P-235 had similar levels of luciferase ORF-containing transcription (P > 0.05) (Fig. 4D and F, lanes 5 and 6). This effect was very similar to that observed in previous experiments involving the engineering of the physiological ATG of the human BACE1 gene upstream of the luciferase ORF (pBIP+4 and pB1P+6; Fig. 3). This suggests that one of these three uATGs (either the third, fourth, or fifth), located upstream of the luciferase ORF in plasmid pB1P-234, functions efficiently as a translation initiation codon for the synthesis of a fusion luciferase protein with a slightly higher molecular weight than that of the native luciferase protein and that a frameshift downstream of this uAUG would completely disrupt the translation of the luciferase ORF, leading to a marked reduction in luciferase activity in pB1P-235. Since the third uATG-initiated upstream ORF has its own stop codon upstream of the ATG of the luciferase reporter gene and the fifth uATG is located at bp −234 in plasmid pB1P-234, the luciferase fusion protein was most likely expressed by translation initiated from the fourth uATG.
Similar experiments were performed to investigate the sixth uATG's effect on the downstream cistron. The luciferase activity remained the same regardless of whether or not the sixth uATG was in the same reading frame as the downstream ORF, 39.18% ± 2.72% in pB1P-113 and 34.67% ± 2.84% in the frameshift plasmid pB1P-114 (P > 0.05) relative to pBIP-403 (Fig. 4B, lane 7 and 8). Such a frameshift mutation did not affect downstream gene expression, and pB1P-114 displayed luciferase protein and mRNA levels similar to those of pB1P-113 (P > 0.05) (Fig. 4E and F, lanes 7 and 8). These results clearly demonstrate that despite being in closest proximity to the physiological ATG of the human BACE1 gene, the sixth uATG has no effect on BACE1 gene transcription and suggest that the corresponding uAUG in the BACE1 mRNA leader sequence does not function as a translation initiation codon.
Fourth uAUG functions efficiently as a translation initiation codon for downstream gene expression.
There are five uAUGs upstream of the luciferase ORF initiation codon AUG in the 5′ leader sequence of the human BACE1 mRNA transcribed from plasmid pB1P-234. The fourth and fifth uAUGs are in the same reading frame as the physiological AUG of the human BACE1 gene. The third uAUG-initiated upstream ORF has its own stop codon before the luciferase ORF initiation codon AUG. Therefore, translation beginning at the third uAUG will terminate at the stop codon and will not disrupt the translation of the downstream luciferase ORF in pB1P-234. However, since both the fourth and fifth uAUGs are in the same reading frame as the downstream ORF of luciferase transcribed from plasmid pB1P-234 and either one of these two uAUGs could function as a translation initiation codon for the downstream luciferase ORF, a frameshift mutation between these uAUGs and the AUG of the downstream ORF could result in disruption of the luciferase synthesis.
To investigate which one of these two uAUGs functions as a translation initiation codon, we constructed plasmid pB1P-237, in which the fifth uATG codon and the stop codon for both the fourth and fifth uATGs of BACE1 were deleted from plasmid pB1P-234. If the fourth uAUG could serve as a translation initiation codon, a new fusion firefly luciferase protein containing extra amino acids from the BACE1 5′ leader would be expressed from plasmids pB1P-237 and pB1P-330 (Fig. 5A). Since the ORF of pB1P-237 specifies 21 extra amino acids relative to the ORF of pB1P-330, the fusion firefly luciferase protein expressed by pB1P-237 would be a little larger than the protein expressed by pB1P-330. Western blotting analysis clearly showed that the cells transfected with pB1P-237 expressed a luciferase fusion protein with a higher molecular weight than did the cells with pB1P-330 (Fig. 5B). This result clearly shows that the fourth uAUG can serve as a physiologically functional translation initiation codon.
FIG. 5.
Fourth uAUG functions efficiently as a translation initiation codon. (A) Plasmid pB1P-237 was constructed with a deletion of the fifth uATG codon and the stop codon for both the fourth and fifth uATGs of BACE1 from plasmid pB1P-234. Plasmid pB1P-237 contains 63 bp of extra nucleotides in the BACE1 5′UTR relative to pB1P-330. (B) HEK293 cells were transfected with either pB1P-330, pB1P-237, or a combination of pB1P-330 and pB1P-237. Luciferase fusion proteins were detected with antiluciferase antibody. Plasmid pB1P-237 expressed a fusion luciferase with a higher molecular weight than that of pB1P-330. The middle lane clearly shows that there are two distinct luciferase protein bands from lysates of cells transfected with plasmid pBI-330 or pBI-237. (C) Plasmid pB1P-235 contains an extra nucleotide downstream of position −234, resulting in a frameshift mutation in the luciferase ORF. Plasmid pB1P-235m was generated to have the fourth uATG codon of the BACE1 gene in plasmid pB1P-235 mutated to ATT. (D) Cells were transfected with pB1P-234, pB1P-235, or pB1P-235m and pCMV-Rluc for 24 h. Luciferase activity was measured 24 h after transfection, and Renilla luciferase activity was used to normalize for transfection efficiency. The values are means ± standard error of the mean (n = 3) and are expressed as normalized luciferase activity relative to that of the wild-type pB1P-234 control. *, P < 0.001 by ANOVA with the posthoc Newmann-Keuls test.
Next, we performed experiments to determine the role of the fifth uATG in translation initiation. Plasmid pB1P-235m was generated to contain the fourth uATG codon of the BACE1 gene in plasmid pB1P-235 mutated to ATT (Fig. 5C). The frameshift mutation in pB1P-235 caused a significant decrease in luciferase activity to 9.94% ± 1.68% of that of pB1P-234 (P < 0.001) (Fig. 5D). The reduction in luciferase activity could be caused by the effect of the frameshift mutation on translation initiation from either the fourth or fifth uAUG. If the fifth uAUG could also function as a translation initiation codon, like the fourth uAUG, the combination of the mutation in the fourth uAUG to ATT and the frameshift mutation downstream of the fifth uAUG in pB1P-235m would disrupt the downstream cistron of luciferase and inhibit luciferase activity. Instead, the mutations in pB1P-235m rescued the deficiency in luciferase activity resulting from the frameshift mutation in pB1P-235, and it displayed luciferase activity similar to that of pB1P-234, 115.50% ± 10.12% of the pB1P-234 value (P > 0.05) (Fig. 5D), indicating that the frameshift mutation in pB1P-235 affected translation initiation from the fourth uAUG, but not from the fifth uAUG. This result clearly demonstrates that the fifth uAUG might not function as a genuine translation initiation codon.
Inhibition of BACE1 translation by the fourth uAUG in the BACE1 5′UTR.
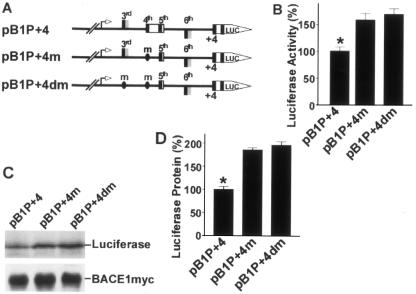
Previous experiments showed that the third and fourth uAUGs of the BACE1 mRNA leader sequence could act as translation initiation codons. These results were observed in deletion plasmids with truncated 5′UTRs (Fig. 5). To examine their role in more native conditions, site-directed mutagenesis was performed to generate plasmids pBIP+4m and pBIP+4dm. The fourth uATG in the BACE1 5′UTR was mutated to ATT in plasmid pBIP+4m and both the third and fourth uATGs were mutated to ATT in plasmid pBIP+4dm. Both plasmids contain the minimal promoter region and the full 5′UTR of the human BACE1 gene (Fig. 6A). The new constructs were transfected into cells. The luciferase assay and Western blots were performed to evaluate the effects of the mutations on the expression of the downstream cistron.
FIG. 6.
Inhibition of BACE1 translation by the fourth uAUG. (A) Plasmids pBIP+4m and pBIP+4dm were constructed by site-directed mutagenesis using wild-type full-length 5′UTR plasmid pB1P+4 as a template. pBIP+4m contains the fourth uATG mutated to ATT, and pBIP+4dm has mutations of the third and fourth uATGs to ATT. (B) The new constructs were transfected into cells for luciferase assays. Firefly luciferase activity was measured at 24 h, and Renilla luciferase activity was used to normalize for transfection efficiency. The values represent means ± standard error of the mean (n = 3). *, P < 0.001 by ANOVA with the posthoc Newmann-Keuls test. (C) Western blotting was used to detect luciferase protein expression in cells transfected with the wild-type and mutant plasmids. BACE1-Myc protein in cells cotransfected with pBACE1-mycHis was detected with anti-Myc antibody 9E10 and used as the control. (D) Quantitative analysis of luciferase protein levels. Values are means ± standard error of the mean (n = 3). The protein levels are expressed as a percentage of the level in the wild-type pB1P+4 control. *, P < 0.001 by ANOVA with the posthoc Newmann-Keuls test. The mutation of the fourth uAUG in the full-length 5′UTR constructs increased downstream gene expression.
As shown in Fig. 6B, mutation of the fourth uATG significantly increased luciferase activity to 159.4% ± 12.13% of that of the wild type (P < 0.01). There was no significant difference in luciferase activity between the single and double mutations (170.0% ± 10.48%) (P > 0.05) (Fig. 6B). Western blots revealed that protein levels were also similar between the single and double mutations, 185.6% ± 5.07% and 195.9% ± 7.76%, respectively (P > 0.05) (Fig. 6C and D). These experiments show that mutation of the fourth uAUG in the full-length 5′UTR constructs can enhance the expression of downstream cistrons but mutation of the third uAUG cannot. These results demonstrate that the fourth uAUG of the BACE1 5′UTR is responsible for inhibiting the expression of BACE1 under normal conditions.
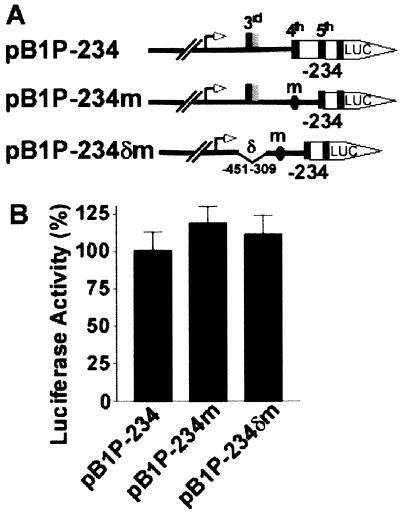
Region from −451 to −309 in the 5′UTR of BACE1 mRNA does not function as an inhibitory element.
To determine if the secondary structure of the 5′UTR of the human BACE1 gene is involved in inhibiting downstream translation initiation by the physiological ATG of the BACE1 gene, plasmids pB1P-234m and pB1P-234δm were constructed. The fourth uATG of the BACE1 5′UTR in pB1P-234 was mutated to ATT to generate plasmid pB1P-234m, which abolished translation initiation of the downstream luciferase ORF from the fourth uATG, as in pB1P-234. The third uATG-containing region from −451 to −309 was further deleted from pB1P-234m, and the resulting plasmid, pB1P-234δm, lacked the third uATG and had the fourth uATG mutated to ATT (Fig. 7A). The plasmids were transfected into N2a cells, and the luciferase activity was measured.
FIG. 7.
The −451 to −309 region of the BACE1 5′UTR contains no inhibitory elements. (A) Plasmid pB1P-234m was constructed to contain the fourth uATG mutated to ATT, and pB1P-234δm contains an additional deletion of the third uATG-containing region from −451 to −309 of pB1P-234m. (B) Cells were cotransfected with pB1P-234, pB1P-234m, or pB1P-234δm and pCMV-Rluc. Firefly luciferase activity was measured at 24 h, and Renilla luciferase activity was used to normalize for transfection efficiency. The values represent means ± standard error of the mean (n = 3). There is no difference in luciferase activity between the mutants and the wild-type control, P > 0.05 by ANOVA.
The luciferase activity in cells transfected with pB1P-234m or pB1P-234δm increased slightly, to 118.6% ± 11.12% and 111.2% ± 12.96%, respectively, of that of the wild-type control pB1P-234 (P > 0.05) (Fig. 7B). These data indicate that neither the mutation of the fourth uATG nor the combination of the mutation and deletion of the third-uATG-containing region in constructs without the proximal region of the 5′UTR has any significant effect on luciferase activity and that the bp −451 to −309 region of the 5′UTR of human BACE1 mRNA has no inhibitory effect on human BACE1 gene expression.
Poliovirus 2A(pro) and TRAIL inhibit the translation initiated by the uATG and the physiological ATG of BACE1.
The 5′ cap (m7GpppN) of mRNA and ribosomal scanning of the 5′UTR for AUG are essential for initiation of translation in the majority of eukaryotic cells. This cap-dependent translation initiation first requires activation of the mRNA by binding of the cap-binding complex eIF4F, comprised of the initiation factors eIF4E (the 5′-cap binding protein), eIF4G, and eIF4A (the mRNA helicase) (30, 31). eIF4A interacts with eIF4G, and eIF4E binds to the eIF4G N terminus. Subsequently, eIF4F binds to the 43S complex, consisting of the 40S subunit, Met-tRNAi/eIF2/GTP, eIF3 and eIF1A, for ribosomal scanning and translation start codon (AUG) recognition.
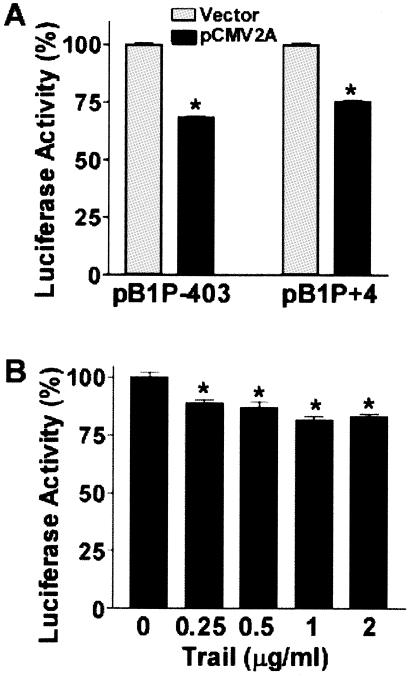
To investigate the effect of the initiation factors on BACE1 gene expression, the minimal promoter plasmid pB1P-403 or the full-length 5′UTR construct pB1P+4 was cotransfected with either control vector or plasmid pCMV2A into N2a cells. Plasmid pCMV2A expresses poliovirus protease 2A(pro), which inhibits cap-dependent translation initiation by cleaving eIF4G factor (44, 47). Overexpression of 2A(pro) markedly reduced luciferase activity in both pB1P-403- and pB1P+4-transfected cells, to 68.43% ± 0.43% and 75.15% ± 0.89%, respectively, of that of the controls (P < 0.0001) (Fig. 8A). To further confirm eIF4G's effect on BACE1 translation initiation, HEK293 cells were transfected with plasmid pB1P+4 and then treated with TRAIL. TRAIL inhibits protein synthesis, and the inhibition of protein synthesis correlates with the cleavage of eIF4G (41). Treatment of HEK293 cells with different doses of the recombinant human TRAIL ligand significantly inhibited luciferase activity in pB1P+4-transfected cells (P < 0.001) (Fig. 8B). Our data suggest that translation initiation by the BACE1 uAUGs and physiological AUG requires intact eIF4G.
FIG. 8.
Cleavage of eIF4G inhibits BACE1 uATG- and physiological ATG-initiated translation. (A) Plasmid pB1P+4 or pB1P-403 was cotransfected into N2a cells with the empty vector or pCMV2A and pCMV-Rluc. Cells were lysed in 1× reporter buffer, and the luciferase assay was performed. Luciferase activity was expressed as a percentage of that of the vector control. The values represent means ± standard error of the mean (n = 3), P < 0.001 by t test. (B) HEK293 cells were transfected with 500 ng of plasmid pB1P+4 and 1 ng of pCMV-Rluc for 24 h and then treated with TRAIL. Cells were harvested in 1× passive lysis buffer for the luciferase assay. The values represent means ± standard error of the mean (n = 3), P < 0.001 ANOVA with the posthoc Newmann-Keuls test. TRAIL treatment significantly reduced luciferase activity.
DISCUSSION
BACE1 is the essential enzyme for cleaving APP at the β-secretase site to generate Aβ during AD pathogenesis. We have previously shown that transcription of the human BACE1 gene is tightly controlled and relatively low expression of the BACE1 gene and Aβ production in vivo are partially due to its weak gene promoter (22). It has been reported that β-secretase activity is increased in the cortex of sporadic AD patients relative to that in age-matched controls (6, 7, 11, 50). BACE1 protein and activity levels increase with aging and in brain regions affected by amyloid deposition and remain increased despite significant neuronal and synaptic loss in AD (6, 7).
Since there are no significant differences in BACE1 mRNA levels between the brains of some AD patients and controls, abnormal translational regulation or protein degradation of BACE1 could be responsible for the BACE1 protein increase in sporadic AD cases (33). There is no evidence to support the idea that the human BACE1 5′UTR acts as an IRES element in cap-independent initiation of BACE1 translation (4, 36).
Ribosome shunting has been implicated as a mechanism of translation regulation in BACE1 mRNA (36). During translation initiation, ribosome shunting enables ribosomes to bind to the mRNA in a cap-dependent manner and scan through the 5′-proximal part of mRNA in the usual way but then skip over large regions of the mRNA containing stable secondary RNA structure and/or a uORF to land upstream of the initiator AUG. Ribosome shunting was initially demonstrated in the translation of the plant viral RNA of cauliflower mosaic virus (8) and for late adenoviral mRNAs (51). The shunting mechanism involves ribosomes translating a small uORF at the base of the stem-loop and then bypassing the structured regions. Base-pairing between mRNA sequences in the 5′UTR and a stem-loop structure at the 3′ end of the 18S rRNA may mediate this shunting mechanism (52).
Ribosomal shunting is rare. Our data show that the fourth uAUG of the human BACE1 5′UTR can function efficiently as a translation initiation codon and has an inhibitory effect on downstream cistrons. Furthermore, deletion of the −451 to −309 region does not affect translation efficiency. Taken together, the results of our study suggest that the shunting mechanisms might not be involved in human BACE1 translation in normal conditions.
Secondary structure in some 5′UTR regions plays an important role in regulating the initiation of translation. These regions are generally GC rich and have the potential to form stable stem-loops which would inhibit ribosomal scanning (46). RNA hairpin structure located close to the cap was thought to block scanning significantly (18, 32). The iron-responsive element (IRE) is one such stem-loop structure. Iron shortage induces binding of iron-regulatory protein to an IRE in the 5′UTR of the mRNA, which specifically suppresses translation initiation. The IRE is generally located within 50 nucleotides of the cap structure (18, 32). The APP gene was found to have an IRE of type II (37).
Since the BACE1 5′UTR has a high GC content of 75% and its free energy is −215.9 to −226.7 kcal/mol, as predicted using mFold 3.2 (http://www.bioinfo.rpi.edu/∼zukerm/rna/), it is likely to form a stable stem-loop structure (20). However, deletion of the −451 to −309 region of the 5′UTR, which disrupted the stem-loop structure in the 5′UTR and reduced the free energy to approximately −60 kcal/mol, had no effect on luciferase activity, indicating that the secondary structure in this region might not contribute to the weak translation of BACE1 initiated from its physiological initiation codon.
A majority of mRNAs in eukaryotes do not contain uAUGs, and the length of their 5′UTR is relatively short, with the average somewhere between 90 and 210 nucleotides for vertebrate mRNAs. The scanning model fits in with the translation of those mRNAs. However, 10 to 28% of mRNAs contain one or more uAUGs, some of which can affect translation by leaky scanning and ribosome reinitiation (16, 38). Initiation usually occurs at the AUG codon that is proximal to the 5′ end of an mRNA. The proximity of the AUG to the cap and its surrounding nucleotides affects translation initiation site recognition during the scanning process. If the AUGs reside in a nonoptimal context, the scanning ribosomal complex may bypass the potential starting AUGs by leaky scanning, which enables a single mRNA to encode several proteins with different N termini. In the BACE1 5′UTR, mutation of the third uAUG had no affect on translation, and this uAUG was probably skipped by the scanning ribosomal complex.
The role of uORFs in protein translational control has not been fully elucidated. Most uORFs are believed to inhibit the translation of a downstream cistron (13, 53). However, some uORFs could enhance translation of the main ORF (48). Reinitiation is a process by which the ribosome remains connected to the mRNA after translation of a uORF, continues scanning for a start site, and initiates at a new site (26). It was initially described after analysis of translational control of GCN4 mRNA in Saccharomyces cerevisiae (10). In general, it is an inefficient process and only occurs after termination of short uORFs. It has been reported that a uORF may be up to 105 nucleotides long and still successfully mediate reinitiation (34). The fourth uORF of the human BACE1 mRNA contains 69 nucleotides. Our results show that the fourth uAUG resides in an excellent context and functions efficiently as an initiation site. Mutation of the fourth uAUG reduced the inhibition of the 5′UTR on the downstream cistron. This is consistent with the reinitiation process.
Alternatively, spliced forms of the 5′UTR of mRNA have also been reported to control protein translation. mRNAs containing short transcripts of the 5′UTR among multiple transcripts in the ribonucleotide reductase M2 gene (5) and argininosuccinate synthase gene (29) exhibit high translation efficiency. A short 5′UTR transcript of the BACE1 mRNA containing only 364 nucleotides has been reported (4). It is worth examining whether this short transcript is altered in the brains of AD patients and what might be its role in AD pathogenesis. Some peptides translated from uORFs have been shown to inhibit translation (15). Future study to investigate if the four ORF-translated peptides have any effect on BACE1 translation is warranted.
In conclusion, our results clearly demonstrate that the fourth uAUG in the 5′UTR of the human BACE1 mRNA can function efficiently as a translation initiation site and that the fourth uORF further contributes to the weak translation of BACE1. Leaky scanning and reinitiation are involved in inhibition of physiological AUG-initiated BACE1 translation. Such leaky scanning and reinitiation result in weak expression of BACE1 under normal conditions. Alterations of leaky scanning and reinitiation in BACE1 gene expression could play an important role in AD pathogenesis. Future studies will determine what factors might affect this leaky scanning and reinitiation process in some sporadic AD cases. This approach will further define the molecular mechanism of BACE1 translational regulation in AD pathogenesis and its therapeutic potential for AD therapy.
Acknowledgments
We thank Xiulian Sun, Hong Qing, Odysseus Zis, and Diane Parsons for their technical assistance and helpful comments. We thank Martin Holcik and Robert G. Korneluk for providing plasmid pCMV2A.
This work was supported by the Canadian Institutes of Health Research (CIHR), the Jack Brown and Family Alzheimer's Research Foundation, and the Michael Smith Foundation for Health Research (to W.S.). W.S. is the holder of the Canada Research Chair in Alzheimer's Disease. W.Z. was the recipient of an Arthur & June Willms Fellowship.
REFERENCES
- 1.Cai, H., Y. Wang, D. McCarthy, H. Wen, D. R. Borchelt, D. L. Price, and P. C. Wong. 2001. BACE1 is the major beta-secretase for generation of Abeta peptides by neurons. Nat. Neurosci. 4:233-234. [DOI] [PubMed] [Google Scholar]
- 2.Christensen, M. A., W. Zhou, H. Qing, A. Lehman, S. Philipsen, and W. Song. 2004. Transcriptional regulation of BACE1, the β-amyloid precursor protein β-secretase, by Sp1. Mol. Cell. Biol. 24:865-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cruts, M., B. Dermaut, R. Rademakers, G. Roks, M. Van den Broeck, G. Munteanu, C. M. van Duijn, and C. Van Broeckhoven. 2001. Amyloid beta secretase gene (BACE) is neither mutated in nor associated with early-onset Alzheimer's disease. Neurosci. Lett. 313:105-107. [DOI] [PubMed] [Google Scholar]
- 4.De Pietri Tonelli, D., M. Mihailovich, A. Di Cesare, F. Codazzi, F. Grohovaz, and D. Zacchetti. 2004. Translational regulation of BACE-1 expression in neuronal and non-neuronal cells. Nucleic Acids Res. 32:1808-1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong, Z., Y. Liu, and J. T. Zhang. 2005. Regulation of ribonucleotide reductase M2 expression by the upstream AUGs. Nucleic Acids Res. 33:2715-2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukumoto, H., B. S. Cheung, B. T. Hyman, and M. C. Irizarry. 2002. Beta-secretase protein and activity are increased in the neocortex in Alzheimer disease. Arch. Neurol. 59:1381-1389. [DOI] [PubMed] [Google Scholar]
- 7.Fukumoto, H., D. L. Rosene, M. B. Moss, S. Raju, B. T. Hyman, and M. C. Irizarry. 2004. Beta-secretase activity increases with aging in human, monkey, and mouse brain. Am. J. Pathol. 164:719-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Futterer, J., Z. Kiss-Laszlo, and T. Hohn. 1993. Nonlinear ribosome migration on cauliflower mosaic virus 35S RNA. Cell 73:789-802. [DOI] [PubMed] [Google Scholar]
- 9.Ge, Y. W., B. Maloney, K. Sambamurti, and D. K. Lahiri. 2004. Functional characterization of the 5′ flanking region of the BACE gene: identification of a 91 bp fragment involved in basal level of BACE promoter expression. FASEB J. 18:1037-1039. [DOI] [PubMed] [Google Scholar]
- 10.Hinnebusch, A. G. 1993. Gene-specific translational control of the yeast GCN4 gene by phosphorylation of eukaryotic initiation factor 2. Mol. Microbiol. 10:215-223. [DOI] [PubMed] [Google Scholar]
- 11.Holsinger, R. M., C. A. McLean, K. Beyreuther, C. L. Masters, and G. Evin. 2002. Increased expression of the amyloid precursor beta-secretase in Alzheimer's disease. Ann. Neurol. 51:783-786. [DOI] [PubMed] [Google Scholar]
- 12.Hussain, I., D. Powell, D. R. Howlett, D. G. Tew, T. D. Meek, C. Chapman, I. S. Gloger, K. E. Murphy, C. D. Southan, D. M. Ryan, T. S. Smith, D. L. Simmons, F. S. Walsh, C. Dingwall, and G. Christie. 1999. Identification of a novel aspartic protease (Asp 2) as beta-secretase. Mol. Cell Neurosci. 14:419-427. [DOI] [PubMed] [Google Scholar]
- 13.Iacono, M., F. Mignone, and G. Pesole. 2005. uAUG and uORFs in human and rodent 5′untranslated mRNAs. Gene 349:97-105. [DOI] [PubMed] [Google Scholar]
- 14.Jackson, R. J. 2000. A comparative view of initiation site selection mechanisms. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Jousse, C., A. Bruhat, V. Carraro, F. Urano, M. Ferrara, D. Ron, and P. Fafournoux. 2001. Inhibition of CHOP translation by a peptide encoded by an open reading frame localized in the chop 5′UTR. Nucleic Acids Res. 29:4341-4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawaguchi, R., and J. Bailey-Serres. 2005. mRNA sequence features that contribute to translational regulation in Arabidopsis. Nucleic Acids Res. 33:955-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kitazume, S., Y. Tachida, R. Oka, K. Shirotani, T. C. Saido, and Y. Hashimoto. 2001. Alzheimer's beta-secretase, beta-site amyloid precursor protein-cleaving enzyme, is responsible for cleavage secretion of a Golgi-resident sialyltransferase. Proc. Natl. Acad. Sci. USA 98:13554-13559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozak, M. 1989. Circumstances and mechanisms of inhibition of translation by secondary structure in eukaryotic mRNAs. Mol. Cell. Biol. 9:5134-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozak, M. 1999. Initiation of translation in prokaryotes and eukaryotes. Gene 234:187-208. [DOI] [PubMed] [Google Scholar]
- 20.Lammich, S., S. Schobel, A. K. Zimmer, S. F. Lichtenthaler, and C. Haass. 2004. Expression of the Alzheimer protease BACE1 is suppressed via its 5′-untranslated region. EMBO Rep. 5:620-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, Q., and T. C. Sudhof. 2004. Cleavage of amyloid-beta precursor protein and amyloid-beta precursor-like protein by BACE 1. J. Biol. Chem. 279:10542-10550. [DOI] [PubMed] [Google Scholar]
- 22.Li, Y., W. Zhou, Y. Tong, G. He, and W. Song. 2006. Control of APP processing and Aβ generation level by BACE1 enzymatic activity and transcription. FASEB J. 20:285-292. [DOI] [PubMed] [Google Scholar]
- 23.Lichtenthaler, S. F., D. I. Dominguez, G. G. Westmeyer, K. Reiss, C. Haass, P. Saftig, B. De Strooper, and B. Seed. 2003. The cell adhesion protein P-selectin glycoprotein ligand-1 is a substrate for the aspartyl protease BACE1. J. Biol. Chem. 278:48713-48719. [DOI] [PubMed] [Google Scholar]
- 24.Luo, Y., B. Bolon, S. Kahn, B. D. Bennett, S. Babu-Khan, P. Denis, W. Fan, H. Kha, J. Zhang, Y. Gong, L. Martin, J. C. Louis, Q. Yan, W. G. Richards, M. Citron, and R. Vassar. 2001. Mice deficient in BACE1, the Alzheimer's beta-secretase, have normal phenotype and abolished beta-amyloid generation. Nat. Neurosci. 4:231-232. [DOI] [PubMed] [Google Scholar]
- 25.Marcinkiewicz, M., and N. G. Seidah. 2000. Coordinated expression of beta-amyloid precursor protein and the putative beta-secretase BACE and alpha-secretase ADAM10 in mouse and human brain. J. Neurochem. 75:2133-2143. [DOI] [PubMed] [Google Scholar]
- 26.Morris, D. R., and A. P. Geballe. 2000. Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol. 20:8635-8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicolaou, M., Y. Q. Song, C. A. Sato, A. Orlacchio, T. Kawarai, H. Medeiros, Y. Liang, S. Sorbi, E. Richard, E. I. Rogaev, Y. Moliaka, A. C. Bruni, R. Jorge, M. Percy, R. Duara, L. A. Farrer, P. St Georg-Hyslop, and E. A. Rogaeva. 2001. Mutations in the open reading frame of the beta-site APP cleaving enzyme (BACE) locus are not a common cause of Alzheimer's disease. Neurogenetics 3:203-206. [DOI] [PubMed] [Google Scholar]
- 28.Pastorino, L., A. F. Ikin, S. Lamprianou, N. Vacaresse, J. P. Revelli, K. Platt, P. Paganetti, P. M. Mathews, S. Harroch, and J. D. Buxbaum. 2004. BACE (beta-secretase) modulates the processing of APLP2 in vivo. Mol. Cell. Neurosci. 25:642-649. [DOI] [PubMed] [Google Scholar]
- 29.Pendleton, L. C., B. L. Goodwin, L. P. Solomonson, and D. C. Eichler. 2005. Regulation of endothelial argininosuccinate synthase expression and NO production by an upstream open reading frame. J. Biol. Chem. 280:24252-24260. [DOI] [PubMed] [Google Scholar]
- 30.Pestova, T. V., S. I. Borukhov, and C. U. Hellen. 1998. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature 394:854-859. [DOI] [PubMed] [Google Scholar]
- 31.Pestova, T. V., and C. U. Hellen. 2000. The structure and function of initiation factors in eukaryotic protein synthesis. Cell. Mol. Life Sci. 57:651-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pickering, B. M., and A. E. Willis. 2005. The implications of structured 5′ untranslated regions on translation and disease. Semin. Cell. Dev. Biol. 16:39-47. [DOI] [PubMed] [Google Scholar]
- 33.Qing, H., W. Zhou, M. A. Christensen, X. Sun, Y. Tong, and W. Song. 2004. Degradation of BACE by the ubiquitin-proteasome pathway. FASEB J. 18:1571-1573. [DOI] [PubMed] [Google Scholar]
- 34.Rajkowitsch, L., C. Vilela, K. Berthelot, C. V. Ramirez, and J. E. McCarthy. 2004. Reinitiation and recycling are distinct processes occurring downstream of translation termination in yeast. J. Mol. Biol. 335:71-85. [DOI] [PubMed] [Google Scholar]
- 35.Roberds, S. L., J. Anderson, G. Basi, M. J. Bienkowski, D. G. Branstetter, K. S. Chen, S. Freedman, N. L. Frigon, D. Games, K. Hu, K. Johnson-Wood, K. E. Kappenman, T. T. Kawabe, I. Kola, R. Kuehn, M. Lee, W. Liu, R. Motter, N. F. Nichols, M. Power, D. W. Robertson, D. Schenk, M. Schoor, G. M. Shopp, M. E. Shuck, S. Sinha, K. A. Svensson, G. Tatsuno, H. Tintrup, J. Wijsman, S. Wright, and L. McConlogue. 2001. BACE knockout mice are healthy despite lacking the primary β-secretase activity in brain: implications for Alzheimer's disease therapeutics. Hum. Mol. Genet. 10:1317-1324. [DOI] [PubMed] [Google Scholar]
- 36.Rogers, G. W., Jr., G. M. Edelman, and V. P. Mauro. 2004. Differential utilization of upstream AUGs in the beta-secretase mRNA suggests that a shunting mechanism regulates translation. Proc. Natl. Acad. Sci. USA 101:2794-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rogers, J. T., J. D. Randall, C. M. Cahill, P. S. Eder, X. Huang, H. Gunshin, L. Leiter, J. McPhee, S. S. Sarang, T. Utsuki, N. H. Greig, D. K. Lahiri, R. E. Tanzi, A. I. Bush, T. Giordano, and S. R. Gullans. 2002. An iron-responsive element type II in the 5′-untranslated region of the Alzheimer's amyloid precursor protein transcript. J. Biol. Chem. 277:45518-45528. [DOI] [PubMed] [Google Scholar]
- 38.Rogozin, I. B., A. V. Kochetov, F. A. Kondrashov, E. V. Koonin, and L. Milanesi. 2001. Presence of ATG triplets in 5′ untranslated regions of eukaryotic cDNAs correlates with a ‘weak’ context of the start codon. Bioinformatics 17:890-900. [DOI] [PubMed] [Google Scholar]
- 39.Russo, C., G. Schettini, T. C. Saido, C. Hulette, C. Lippa, L. Lannfelt, B. Ghetti, P. Gambetti, M. Tabaton, and J. K. Teller. 2000. Presenilin-1 mutations in Alzheimer's disease. Nature 405:531-532. [DOI] [PubMed] [Google Scholar]
- 40.Sinha, S., J. P. Anderson, R. Barbour, G. S. Basi, R. Caccavello, D. Davis, M. Doan, H. F. Dovey, N. Frigon, J. Hong, K. Jacobson-Croak, N. Jewett, P. Keim, J. Knops, I. Lieberburg, M. Power, H. Tan, G. Tatsuno, J. Tung, D. Schenk, P. Seubert, S. M. Suomensaari, S. Wang, D. Walker, V. John, et al. 1999. Purification and cloning of amyloid precursor protein beta-secretase from human brain. Nature 402:537-540. [DOI] [PubMed] [Google Scholar]
- 41.Stoneley, M., S. A. Chappell, C. L. Jopling, M. Dickens, M. MacFarlane, and A. E. Willis. 2000. c-Myc protein synthesis is initiated from the internal ribosome entry segment during apoptosis. Mol. Cell. Biol. 20:1162-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun, X., Y. Wang, H. Qing, M. A. Christensen, Y. Liu, W. Zhou, Y. Tong, C. Xiao, Y. Huang, S. Zhang, X. Liu, and W. Song. 2005. Distinct transcriptional regulation and function of the human BACE2 and BACE1 genes. FASEB J. 19:739-749. [DOI] [PubMed] [Google Scholar]
- 43.Vassar, R., B. D. Bennett, S. Babu-Khan, S. Kahn, E. A. Mendiaz, P. Denis, D. B. Teplow, S. Ross, P. Amarante, R. Loeloff, Y. Luo, S. Fisher, J. Fuller, S. Edenson, J. Lile, M. A. Jarosinski, A. L. Biere, E. Curran, T. Burgess, J. C. Louis, F. Collins, J. Treanor, G. Rogers, and M. Citron. 1999. Beta-secretase cleavage of Alzheimer's amyloid precursor protein by the transmembrane aspartic protease BACE. Science 286:735-741. [DOI] [PubMed] [Google Scholar]
- 44.Ventoso, I., A. Barco, and L. Carrasco. 1998. Mutational analysis of poliovirus 2Apro: distinct inhibitory functions of 2Apro on translation and transcription. J. Biol. Chem. 273:27960-27967. [DOI] [PubMed] [Google Scholar]
- 45.von Arnim, C. A. F., A. Kinoshita, I. D. Peltan, M. M. Tangredi, L. Herl, B. M. Lee, R. Spoelgen, T. T. Hshieh, S. Ranganathan, F. D. Battey, C.-X. Liu, B. J. Bacskai, S. Sever, M. C. Irizarry, D. K. Strickland, and B. T. Hyman. 2005. The low density lipoprotein receptor-related protein (LRP) is a novel β-secretase (BACE1) substrate. J. Biol. Chem. 280:17777-17785. [DOI] [PubMed] [Google Scholar]
- 46.Willis, A. E. 1999. Translational control of growth factor and proto-oncogene expression. Int. J. Biochem. Cell. Biol. 31:73-86. [DOI] [PubMed] [Google Scholar]
- 47.Wyckoff, E., J. Hershey, and E. Ehrenfeld. 1990. Eukaryotic initiation factor 3 is required for poliovirus 2A protease-induced cleavage of the p220 component of eukaryotic initiation factor 4F. Proc. Natl. Acad. Sci. USA 87:9529-9533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yaman, I., J. Fernandez, H. Liu, M. Caprara, A. A. Komar, A. E. Koromilas, L. Zhou, M. D. Snider, D. Scheuner, R. J. Kaufman, and M. Hatzoglou. 2003. The zipper model of translational control: a small upstream ORF is the switch that controls structural remodeling of an mRNA leader. Cell 113:519-531. [DOI] [PubMed] [Google Scholar]
- 49.Yan, R., M. J. Bienkowski, M. E. Shuck, H. Miao, M. C. Tory, A. M. Pauley, J. R. Brashier, N. C. Stratman, W. R. Mathews, A. E. Buhl, D. B. Carter, A. G. Tomasselli, L. A. Parodi, R. L. Heinrikson, and M. E. Gurney. 1999. Membrane-anchored aspartyl protease with Alzheimer's disease beta-secretase activity. Nature 402:533-537. [DOI] [PubMed] [Google Scholar]
- 50.Yang, L. B., K. Lindholm, R. Yan, M. Citron, W. Xia, X. L. Yang, T. Beach, L. Sue, P. Wong, D. Price, R. Li, and Y. Shen. 2003. Elevated beta-secretase expression and enzymatic activity detected in sporadic Alzheimer disease. Nat. Med. 9:3-4. [DOI] [PubMed] [Google Scholar]
- 51.Yueh, A., and R. J. Schneider. 1996. Selective translation initiation by ribosome jumping in adenovirus-infected and heat-shocked cells. Genes Dev. 10:1557-1567. [DOI] [PubMed] [Google Scholar]
- 52.Yueh, A., and R. J. Schneider. 2000. Translation by ribosome shunting on adenovirus and hsp70 mRNAs facilitated by complementarity to 18S rRNA. Genes Dev. 14:414-421. [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, Z., and F. S. Dietrich. 2005. Identification and characterization of upstream open reading frames (uORF) in the 5′ untranslated regions (UTR) of genes in Saccharomyces cerevisiae. Curr. Genet. 48:77-87. [DOI] [PubMed] [Google Scholar]