Abstract
Despite recent advances in characterizing the regulation of histone H3 lysine 4 (H3-K4) methylation at the GAL1 gene by the H2B-K123-specific deubiquitinase activity of Saccharomyces cerevisiae SAGA (Spt-Ada-Gcn5-acetyltransferase)-associated Ubp8p, our knowledge on the general role of Ubp8p at the SAGA-dependent genes is lacking. For this study, using a formaldehyde-based in vivo cross-linking and chromatin immunoprecipitation (ChIP) assay, we have analyzed the role of Ubp8p in the regulation of H3-K4 methylation at three other SAGA-dependent yeast genes, namely, PHO84, ADH1, and CUP1. Like that at GAL1, H3-K4 methylation is increased at the PHO84 core promoter in the UBP8 deletion mutant. We also show that H3-K4 methylation remains invariant at the PHO84 open reading frame in the Δubp8 mutant, demonstrating a highly localized role of Upb8p in regulation of H3-K4 methylation at the promoter in vivo. However, unlike that at PHO84, H3-K4 methylation at the two other SAGA-dependent genes is not controlled by Ubp8p. Interestingly, Ubp8p and H3-K4 methylation are dispensable for preinitiation complex assembly at the core promoters of these genes. Our ChIP assay further demonstrates that the association of Ubp8p with SAGA is mediated by Sgf11p, consistent with recent biochemical data. Collectively, the data show that Ubp8p differentially controls H3-K4 methylation at the SAGA-dependent promoters, revealing a complex regulatory network of histone methylation in vivo.
Transcription is a highly coordinated and orchestrated process. It takes place on the DNA template that is packaged into chromatin in the nucleus. The chromatin is an array of nucleosomes in which 146 base pairs of DNA (in each nucleosome) are wrapped around the histone octamer (27). Each octamer consists of two molecules each of the histones H2A, H2B, H3, and H4 (27). Nucleosomal arrays fold into a 30-nm fiber upon incorporation of the linker histone H1 (19, 22).
Histones are subject to posttranslational modifications such as acetylation, phosphorylation, ubiquitination, and methylation, and these modifications are known to play important roles in epigenetic regulation of transcription (33, 44). Most modifications were originally observed on the N-terminal tails of histones, with the exception of ubiquitination, which occurs on the C-terminal tails of H2A and H2B (13). However, several novel posttranslational modifications have been identified recently in the core region of the histones (45). Histone modifications alter DNA-histone interactions within and between nucleosomes, and thus they affect the higher-order chromatin structure. Alternatively, combinations of histone modifications present an interaction surface for other proteins that translate the so-called “histone code” into a gene expression pattern (42), explaining how the same chemical modification can have different functional consequences depending on the target amino acid residue.
Methylation of histone H3 at lysine 4 (K4) is associated with active chromatin in a wide range of eukaryotic organisms (5, 15, 25, 30, 32, 41) and is correlated with gene activation (38) as well as rRNA gene silencing (7, 9). Lysine methylation of histones in vivo occurs in the following three states: mono-, di-, and trimethyl. Histone H3 has been found to be dimethylated at K4 in active euchromatic regions but not in silent heterochromatic sites, whereas trimethylated H3-K4 is mainly found associated with the 5′ regions of the actively transcribing genes (28) and, thus, unlike dimethyl-H3-K4, is typically not detected at a locus prior to transcription activation (38). Interestingly, H3-K4 methylation is up-regulated in a “trans-tail” process (11, 43) by Rad6p-mediated ubiquitination of H2B at K123 (36), thus suggesting roles of H2B-K123 ubiquitination and deubiquitination in the regulation of gene activation.
A histone deubiquitinase, Ubp8p, has recently been implicated as a new component of the Saccharomyces cerevisiae SAGA complex (10, 20, 37). Ubp8p has consistently been shown to hydrolyze the ubiquitin moiety of K123-ubiquitinated H2B at the SAGA-dependent GAL1 promoter (20). Such H2B-K123 deubiquitination by Ubp8p modulates the level of H3-K4 methylation at the GAL1 promoter in a “trans-tail” process, hence altering the expression of the GAL1 gene (20). Although Ubp8p is known to regulate H3-K4 methylation at the SAGA-regulated GAL1 promoter (20), its general role at other SAGA-dependent yeast promoters is not known. To understand in greater detail the role of Ubp8p in the regulation of H3-K4 methylation and, hence, transcription, we have analyzed its contribution to regulation of H3-K4 methylation and formation of the preinitiation complex (PIC) assembly at three other SAGA-dependent yeast promoters (i.e., PHO84, ADH1, and CUP1) by a formaldehyde-based in vivo cross-linking and chromatin immunoprecipitation (ChIP) assay. Furthermore, we have analyzed the role of Sgf11p (a newly identified component of SAGA) in the association of Ubp8p with SAGA in vivo. Our results reveal differences in the regulation of H3-K4 methylation by Ubp8p. We also show that Ubp8p and Sgf11p constitute a discrete structural module within SAGA in vivo.
MATERIALS AND METHODS
Plasmids.
Plasmid SGP4 was generated by cloning a DNA fragment containing three Gal4p-binding sites into the low-copy-number plasmid pRS416 (2). The plasmids pFA6a-13Myc-KanMX6 and pFA6a-13Myc-TRP1 (26) were used for genomic Myc epitope tagging of the proteins of interest. Plasmid pRS416 was used for PCR-based gene disruption.
Yeast strains and media.
Yeast strains harboring null mutations in SPT20 (FY1097) and GCN5 (FY1370) and their isogenic wild-type equivalents, FY67 and FY1369, respectively, were obtained from Fred Winston (Harvard Medical School, Boston, MA) (34, 35). A yeast strain carrying a point mutation in histone H2B at K123 (YKH046) and its isogenic wild-type equivalent (YKH045) were obtained from Mary Ann Osley (University of New Mexico Health Science Center) (20). The Myc-tagged strains SGY1 (Spt3p-Myc, TRP1), SGY2 (Spt20p-Myc, TRP1), SGY3 (Ada2p-Myc, TRP1), and SGY4 (Gcn5p-Myc, TRP1) were generated by the insertion of multiple Myc epitope tags at the original chromosomal loci of SPT3, SPT20, ADA2, and GCN5, respectively, in strain FY631 (26). Multiple Myc epitope tags were added at the original chromosomal loci of UBP8 and SGF11 in strain W303a to generate ASY4 (Ubp8p-Myc, KAN) and ASY20 (Sgf11p-Myc, KAN), respectively. Strain ASY24 (Ubp8p-Myc, Δspt20::URA3 KAN) was generated by adding multiple Myc epitopes at the C-terminal end of Ubp8p in FY1097. Spt3p was tagged by multiple Myc epitopes in the original chromosomal locus in FY1369 and FY1370 to generate SGY97 (Spt3p-Myc, HIS3) and SGY98 (Spt3p-Myc, Δgcn5::HIS3 KAN), respectively. The strains SGY7 (Spt3p-Myc, TRP1) and SGY8 (Spt3p-Myc, Δspt20::URA3 TRP1) were generated by adding multiple Myc epitope tags at the chromosomal locus of SPT3 in FY67 and FY1097, respectively. A plasmid (SGP4) carrying three Gal4p-binding sites was transformed into ASY4 and SGY2 to generate ASY5 and SGY99, respectively. Similarly, the plasmid SGP4 was transformed into SGY27 (multiple Myc epitope tags with TRP1 at the original chromosomal locus of SPT3 in W303a) to generate SGY30. The endogenous UBP8 genes of W303a, ASY20, YKH045, and FY1370 were disrupted, using a PCR-based gene knockout method (6), to generate ASY1 (Δubp8::URA3), ASY23 (Sgf11p-Myc, Δubp8::URA3 KAN), ASY19 (Δubp8::URA3, Flag-H2B and hemagglutinin [HA]-ubiquitin), and ASY27 (Δubp8::URA3 Δgcn5::HIS3), respectively. The SGF11 gene of ASY4 was deleted to generate ASY21 (Ubp8p-Myc, Δsgf11::URA3 KAN). Similarly, the endogenous SET1 gene of W303a was knocked out to generate ASY16 (Δset1::URA3).
For studies of the GAL1 promoter in Δgcn5, Δspt20, Δsgf11, and Δubp8 mutant strains and their isogenic wild-type equivalents, cells were first grown in YPD (yeast extract-peptone plus 2% dextrose) to an optical density at 600 nm (OD600) of 0.8 and then transferred to YPG (yeast extract-peptone plus 2% galactose) for 4 h at 30°C prior to formaldehyde cross-linking. Minimal media containing either 2% dextrose, galactose, or raffinose were used for strains with the reporter plasmid SGP4. The yeast strains were grown in YPD to an OD600 of 1.0 at 30°C for studies of the ADH1, PHO84, and RPS5 genes. The CUP1 gene was induced by 1 mM CuSO4 for 25 min in synthetic complete medium (yeast nitrogen base and complete amino acid mixture plus 2% dextrose) at 30°C.
ChIP assay.
The ChIP assay was performed as described previously (24). Briefly, yeast cells were treated with 1% formaldehyde, collected, and resuspended in lysis buffer. Following sonication, cell lysates (800 μl lysate from 100 ml of yeast culture) were precleared by centrifugation, and then 100 μl lysate was used for each immunoprecipitation. Immunoprecipitated protein-DNA complexes were treated with proteinase K, the cross-links were reversed, and the DNA was purified. Immunoprecipitated DNA was dissolved in 20 μl TE 8.0 (10 mM Tris-HCl, pH 8.0, and 1 mM EDTA), and 1 μl of immunoprecipitated DNA was analyzed by PCR. PCR mixtures contained [α-32P]dATP (2.5 mCi for each 25-μl reaction mix), and the PCR products were detected by autoradiography after separation in a 6% polyacrylamide gel. As a control, “input” DNA was isolated from 5 μl lysate without going through the immunoprecipitation step and suspended in 100 μl TE 8.0. To compare the PCR signal arising from the immunoprecipitated DNA with that from the input DNA, 1 μl of input DNA was used for PCR analysis.
For analyses of Ubp8p recruitment and H2B-K123 ubiquitination at the PHO84, ADH1, CUP1, and RPS5 genes, we modified the above ChIP protocol as follows. An 800-μl lysate was prepared from 100 ml of yeast culture. Four hundred microliters of lysate was used for each immunoprecipitation (using 10 μl of anti-HA or anti-Myc antibody and 100 μl of protein A/G plus agarose beads from Santa Cruz Biotechnology, Inc.), and each immunoprecipitated DNA sample was dissolved in 10 μl TE 8.0, of which 1 μl was used for PCR analysis. In parallel, the PCR for “input” DNA was performed using 1 μl DNA that was prepared by dissolving purified DNA from 5 μl lysate in 100 μl TE 8.0. However, to determine whether ubiquitination occurred specifically at K123 of H2B, we performed a chromatin double-immunoprecipitation (ChDIP) assay as described previously (20). Briefly, 400 μl lysate from 50 ml yeast culture was first immunoprecipitated using anti-Flag antibody (Sigma) and protein A/G plus agarose beads. Following elution of the anti-Flag immunoprecipitate with the Flag peptide (Sigma), the eluate was immunoprecipitated using anti-HA antibody, and the immunoprecipitated DNA sample was dissolved in 10 μl TE 8.0, of which 1 μl was used for PCR analysis. The “input” DNA was isolated from 5 μl lysate and suspended in 100 μl TE 8.0, of which 1 μl was used for PCR analysis.
All ChIP experiments were repeated at least three times, and consistent results were obtained. The primer pairs used for PCR analysis were as follows: for the PHO84 upstream activation sequence (UAS), 5′-CCAGCACGTGGGGCGGAAATT-3′ and 5′-TTTAATCTAGCTAATAAGCAGGCAAAA-3′; for the PHO84 core, 5′-GATCCACTTACTATTGTGGCTCGT-3′ and 5′-GTTTGTTGTGTGCCCTGGTGATCT-3′; for PHO84 open reading frame 1 (ORF1), 5′-TAGCTGATATTGTTGGTCGTAAGAG-3′ and 5′-TACCAATACCCATGACAAAACGGTA-3′; for PHO84 ORF2, 5′-TCTGCAGACATTTTGGTCAATGGAA-3′ and 5′-AAACGTTTTTGGAACCGGCATAAC-3′; for the ADH1 UAS, 5′-GTTTCCGGGTGTACAATATGG-3′ and 5′-CTATTGTATATCTCCCCTCCGC-3′; for the ADH1 core, 5′-GGTATACGGCCTTCCTTCCAGTTAC-3′ and 5′-GAACGAGAACAATGACGAGGAAACAAAAG-3′; for ADH1 ORF1, 5′-CTGGTTACACCCACGACGGTTCTT-3′ and 5′-GCAGACTTCAAAGCCTTGTAGACG-3′; for ADH1 ORF2, 5′-CGGTAACAGAGCTGACACCAGAGA-3′ and 5′-ACGTATCTACCAACGATTTGACCC-3′; for the CUP1 UAS, 5′-GGCTGATATCTTAGCCTTGTTACT-3′ and 5′-ACAATCCATATTGCGTTGGTAGTC-3′; for the CUP1 core, 5′-TCTTCTAGAAGCAAAAAGAGCGATG-3′ and 5′-CGCTGAACATTTTATGTGATGATTG-3′; for CUP1 ORF1, 5′-ACTTCCAAAATGAAGGTCATGAGTG-3′ and 5′-AGCAGCATGACTTCTTGGTTTCTTC-3′; for the GAL4 ORF, 5′-CTTGTTCAATGCAGTCCTAGTACCC-3′ and 5′-CACAAGTCTGGATTTTAAAAGTGGCC-3′; for the GAL1 UAS, 5′-CGCTTAACTGCTCATTGCTATATTG-3′ and 5′-TTGTTCGGAGCAGTGCGGCGC-3′; for the GAL1 core, 5′-ATAGGATGATAATGCGATTAGTTTTTTAGCCTT-3′ and 5′-GAAAATGTTGAAAGTATTAGTTAAAGTGGTTATGCA-3′; and for the RPS5 UAS, 5′-AGAAACAATGAACAGCCTTGAGTTCTC-3′ and 5′-GCAGGGCCATTCTCATCTGA-3′.
The primers flanking the Gal4p-binding sites in the plasmid SGP4 were 5′-GGTGGCGGCCGCTCTAGAACTAGT-3′ and 5′-TTGACCGTAATGGGATAGGTCACG-3′.
Autoradiograms were scanned and quantitated by the NIH Image 1.62 program. The ratio of immunoprecipitated (IP) DNA to input DNA is presented as %IP. For mutant strains, the amount of immunoprecipitated DNA relative to that of the wild type (WT) is presented as %WT.
Primer extension analysis.
Primer extension analysis was performed as described previously (24). The primers used for analyses of PHO84, ADH1, and CUP1 mRNAs were as follows: for PHO84, 5′-GAAGACTTCTTTCAGCAACATG-3′; for ADH1, 5′ TATCCTTGTGTTCCAATTTACCGTGG-3′; and for CUP1, 5′-GGCATTGGCACTCATGACCTTC-3′.
RESULTS
Ubp8p is an integral component of the SAGA complex in vivo.
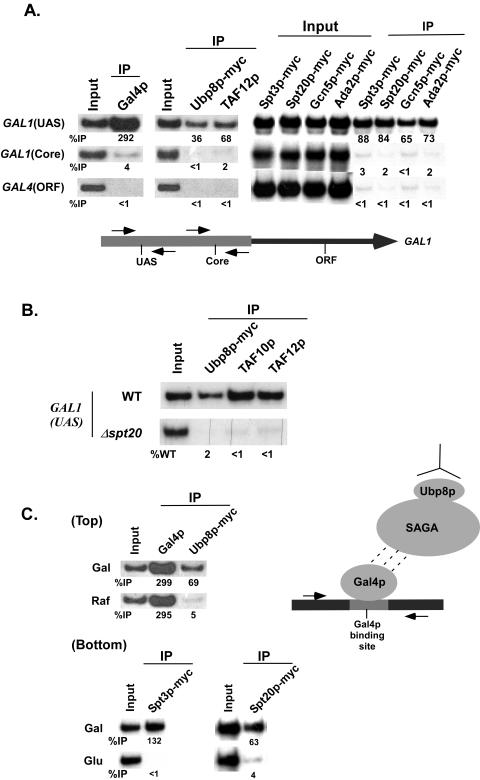
Yeast SAGA is a 1.8-MDa complex which contains at least the following 14 subunits: Ada1p, Ada2p, Ada3p, Ada5p/Spt20p, Spt3p, Spt7p, Spt8p, Gcn5p, Tra1p, TAF5p, TAF6p, TAF9p, TAF10p, and TAF12p (8, 17, 18). Recent biochemical studies (10, 20, 37) have implicated Ubp8p as a new component of the SAGA complex. However, it is not known whether Ubp8p is a SAGA component in vivo. Thus, we analyzed recruitment of Ubp8p to the GAL1 promoter by using a ChIP assay. If Ubp8p is an integral component of SAGA, it will be recruited to the GAL1 UAS along with other SAGA components, since previous studies (2-4) demonstrated recruitment of SAGA to the GAL1 UAS, but not the core promoter, by the Gal4p activation domain. Indeed, Fig. 1A shows that the SAGA subunit TAF12p, as well as Ubp8p, was present at the GAL1 UAS in galactose-containing growth medium where the Gal4p activation domain was active, consistent with the recruitment of other SAGA components (e.g., Spt3p, Spt20p, Gcn5p, and Ada2p). Similarly, Gal4p was recruited to the GAL1 UAS. Significantly, these same proteins were not associated with the GAL1 core promoter or an irrelevant DNA sequence (GAL4 ORF). Furthermore, Ubp8p and other SAGA components (TAF10p and TAF12p) were not recruited to the GAL1 UAS in a mutant with a deletion of SPT20 (Fig. 1B), which is essential for maintaining the integrity of SAGA (2, 16, 40). These results raised the possibility that Ubp8p might be a component of SAGA in vivo. However, because these experiments were performed with the intact GAL1 promoter under conditions permissive for transcription, it remained possible that other transcription components such as general transcription factors played a role in recruitment of Ubp8p to the UAS. To address this issue, we asked whether Ubp8p could be recruited by Gal4p to a plasmid bearing only Gal4p-binding sites but not other promoter elements. Figure 1C (top panel) shows that similar to the results with the GAL1 UAS, Ubp8p was recruited to a plasmid bearing Gal4p-binding sites in galactose-containing growth medium. Significantly, Ubp8p was not associated with the Gal4p-binding sites when the Gal4p activation domain was inactive in raffinose-containing growth medium, even though the levels of Gal4p recruitment were the same in both galactose- and raffinose-containing growth media. Similarly, other SAGA components such as Spt3p and Spt20p were recruited to the minimal Gal4p-binding sites in the plasmid (Fig. 1C, bottom panel) in galactose (but not in dextrose, where the Gal4p activation domain was inactive). Significantly, our previous studies demonstrated that general transcription factors such as TBP, TFIIB, mediator, and RNA polymerase II were not recruited to the minimal Gal4p-binding sites in the plasmid (2). Thus, taken together, our ChIP data provide evidence that Ubp8p exists in vivo in the same form as that defined by its biochemical copurification with SAGA (10, 20, 37).
FIG. 1.
Recruitment of Ubp8p to either GAL1 UAS or the minimal Gal4p-binding sites in a plasmid. (A) Recruitment of Ubp8p and SAGA components (Spt3p, Spt20p, Gcn5p, Ada2p, and TAF12p) to the GAL1 UAS. Yeast strains expressing C-terminally Myc epitope-tagged Ubp8p or SAGA components (Spt3p, Spt20p, Gcn5p, and Ada2p) were grown at 30°C in YPG. The ChIP assay was performed as previously described (24). Primer pairs targeting the UAS or core promoter of the GAL1 gene (see Materials and Methods) were used for PCR analysis of the immunoprecipitated DNA samples. A PCR fragment corresponding to the GAL4 ORF was used as a control for background binding. Immunoprecipitation was performed using a mouse monoclonal antibody against the c-Myc epitope tag (9E10; Santa Cruz Biotechnology, Inc.) or a polyclonal antibody against TAF12p. A mouse monoclonal antibody against the DNA binding domain of Gal4p (RK5C1; Santa Cruz Biotech- nology, Inc.) was used. The primer pairs are indicated on the left. The ratio of immunoprecipitate to input is indicated below each band as %IP. (B) Recruitment of Ubp8p to GAL1 UAS in the SPT20 deletion mutant. Wild-type and SPT20 deletion mutant strains expressing Myc epitope-tagged Ubp8p were first grown in glucose-containing (YPD) medium and then shifted to galactose-containing (YPG) medium 4 h before treatment with formaldehyde. Immunoprecipitations were performed as described above. A polyclonal antibody against TAF12p was used. The percentage of DNA immunoprecipitated relative to the wild type (%WT) is indicated below each band of the mutant strain. (C) Recruitment of Ubp8p and SAGA components (Spt3p and Spt20p) to minimal Gal4p-binding sites. The plasmid containing minimal Gal4p-binding sites was transformed separately into three different yeast strains, each of which has Myc tags at the C-terminal tail of either Spt3p, Spt20p, or Ubp8p. The yeast strains thus generated were grown at 30°C in 1% yeast extract containing 2% peptone plus 2% galactose (Gal), dextrose (Glu), or raffinose (Raf). ChIP analysis was performed as described for panel A. The primers used for the PCR analysis target an area adjacent to the Gal4p-binding sites in the plasmid (see Materials and Methods). The growth media used are indicated on the left.
Ubp8p is not essential for maintaining the integrity of SAGA in vivo.
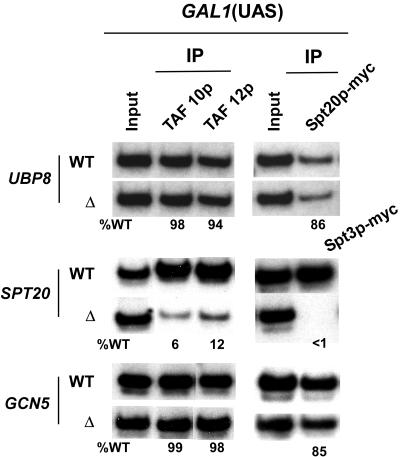
To understand in greater detail the role of Ubp8p in transcription, we first determined its contribution to maintaining the integrity of SAGA in vivo by analyzing the recruitment of SAGA to the GAL1 UAS in the Δubp8 mutant. If Ubp8p maintains SAGA's integrity, then Spt20p and other SAGA components will not be recruited to the GAL1 UAS in the Δubp8 mutant. Figure 2 shows that the deletion of UBP8 did not affect the recruitment of Spt20p and the TAF components (TAF10p and TAF12p) of SAGA. However, consistent with previous studies (2, 16, 40), SAGA was not recruited to the GAL1 UAS in yeast strains lacking SPT20 (Fig. 2), a component that integrates SAGA. On the other hand, recruitment of SAGA components such as TAF10p, TAF12p, and Spt3p was not affected in a mutant with a deletion of GCN5, which is dispensable for SAGA integrity (2, 40). Since Spt20p maintains the integrity of SAGA and its recruitment was not affected in the absence of Ubp8p, like the case with Gcn5p, Ubp8p is not required for the integrity of SAGA in vivo, consistent with recent biochemical data that demonstrated the dispensability of Ubp8p for SAGA integrity (10, 20).
FIG. 2.
Requirement of Ubp8p and SAGA subunits (Spt20p and Gcn5p) for SAGA recruitment to the GAL1 UAS. Wild-type and UBP8 and SAGA subunit (SPT20 and GCN5) deletion mutant strains were first grown in glucose-containing (YPD) medium and then shifted to galactose-containing (YPG) medium 4 h before treatment with formaldehyde. Immunoprecipitations were performed as described in the legend to Fig. 1. A primer pair targeting the GAL1 UAS was used for PCR analysis of the immunoprecipitated DNA samples.
Association of Ubp8p with SAGA is dependent on Sgf11p in vivo.
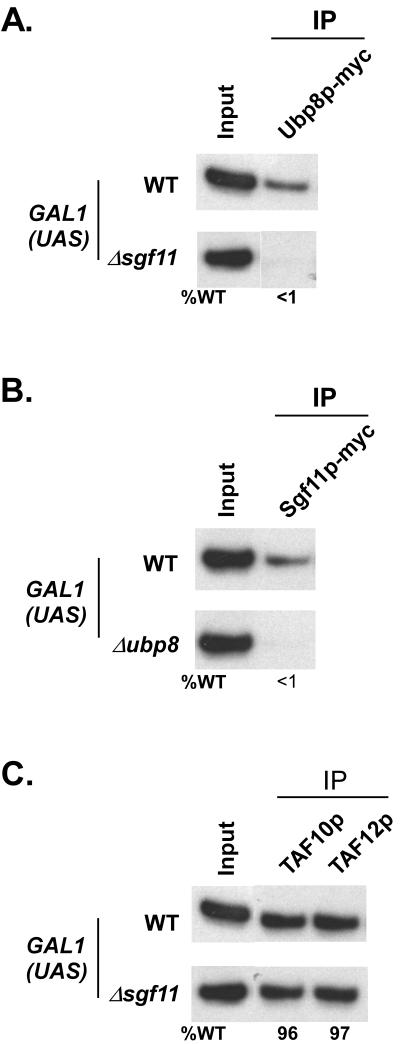
Although Ubp8p does not maintain the overall structural integrity of SAGA, it forms a structural as well as functional module within SAGA through its association with Sgf11p, as evident from recent biochemical studies (21, 23, 31). However, the existence of such a modular domain within SAGA has not been confirmed in vivo. Thus, we analyzed the recruitment of Ubp8p and Sgf11p to the GAL1 UAS in mutant strains with deletions of SGF11 and UBP8, respectively. Figure 3A shows that recruitment of Ubp8p to the GAL1 UAS was completely lost in the Δsgf11 mutant. Similarly, Sgf11p was not recruited to the GAL1 UAS in the Δubp8 mutant (Fig. 3B). Thus, Sgf11p and Ubp8p are mutually dependent on each other for recruitment in vivo. Furthermore, we show that recruitment of SAGA (TAF10p and TAF12p) to the GAL1 UAS was not altered in the Δsgf11 mutant (Fig. 3C). Thus, like Ubp8p, Sgf11p is also dispensable for the structural integrity of SAGA in vivo, consistent with recent biochemical data (21, 23, 31). Taken together, our data support the fact that Ubp8p and Sgf11p constitute a distinct modular domain within SAGA in vivo, and hence they are mutually dependent on each other for their recruitment but dispensable for the overall integrity of SAGA.
FIG. 3.
Ubp8p and Sgf11p are mutually dependent on each other for recruitment to the GAL1 UAS. (A) Recruitment of Ubp8p to the GAL1 UAS requires Sgf11p. Wild-type and SGF11 deletion mutant strains expressing multiple-Myc-epitope-tagged Ubp8p were first grown in glucose-containing (YPD) medium and then shifted to galactose-containing (YPG) medium 4 h before treatment with formaldehyde. Immunoprecipitations were performed as described in the legend to Fig. 1. A primer pair targeting the GAL1 UAS was used for PCR analysis of the immunoprecipitated DNA samples. (B) Recruitment of Sgf11p to the GAL1 UAS is dependent on Ubp8p. Wild-type and UBP8 deletion mutant strains expressing multiple-Myc-epitope-tagged Sgf11p were grown as described above before treatment with formaldehyde. Immunoprecipitation and PCR analysis were performed as described above. (C) Sgf11p is dispensable for recruitment of SAGA to the GAL1 UAS. Wild-type and SGF11 deletion mutant strains were grown, cross-linked, and immunoprecipitated as described above.
Ubp8p is recruited to SAGA-dependent promoters such as those of PHO84, ADH1, and CUP1.
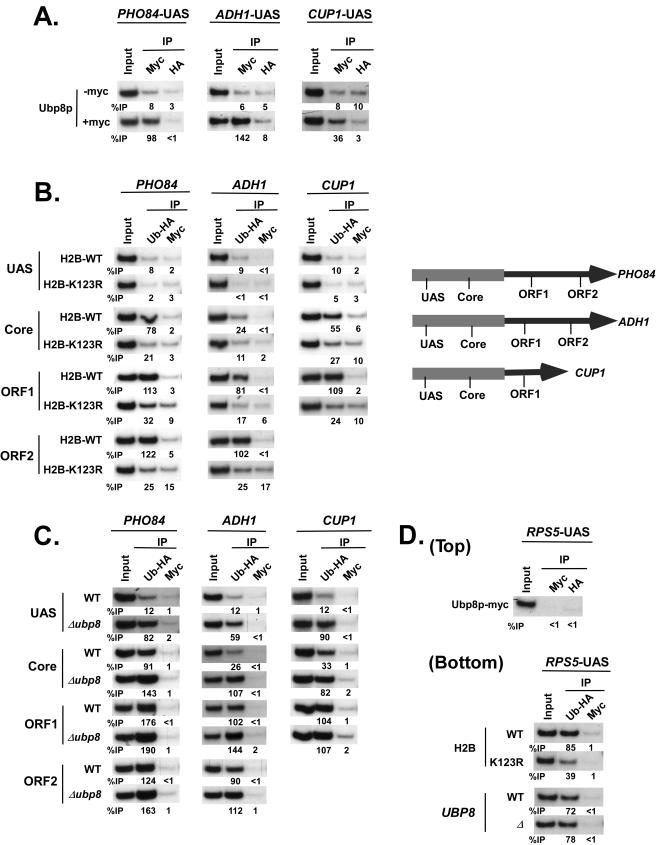
Ubp8p, being a component of SAGA, is recruited to the UAS of a SAGA-dependent gene, GAL1. To determine whether Ubp8p is associated with the UASs of other SAGA-dependent genes, we analyzed the recruitment of Ubp8p to the UASs of three representative SAGA-dependent genes, namely, PHO84, ADH1, and CUP1 (3; data not shown), using a modified ChIP assay (see Materials and Methods). Like the GAL1 UAS, the UASs of PHO84, ADH1, and CUP1 recruited Ubp8p in a yeast strain expressing Myc epitope-tagged Ubp8p (Fig. 4A). Ubp8p was not recruited to the core promoters and ORFs of these three SAGA-dependent genes (data not shown). As a control, we show that Ubp8p was not recruited to the UAS of a SAGA-independent gene, RPS5 (2) (Fig. 4D, top panel). Thus, our data demonstrate that Ubp8p is recruited to the UASs of the SAGA-dependent genes.
FIG. 4.
Analysis of Ubp8p recruitment and H2B-K123 ubiquitination at the PHO84, ADH1, and CUP1 genes, using a modified ChIP assay (see Materials and Methods). (A) Ubp8p is recruited to the UASs of the SAGA-dependent (PHO84, ADH1, and CUP1) genes. For analysis of the recruitment of Ubp8p to the PHO84 and ADH1 UASs, the yeast strains were grown in YPD to an OD600 of 1.0 at 30°C prior to formaldehyde cross-linking. The CUP1 gene was induced with 1 mM CuSO4 for 25 min in synthetic complete medium at 30°C before treatment with formaldehyde. Immunoprecipitations were performed using anti-Myc antibody. An anti-HA antibody (F-7; Santa Cruz Biotechnology, Inc.) was used to monitor the background signal. Primer pairs targeting the UASs of PHO84, ADH1, and CUP1 (see Materials and Methods) were used for PCR analysis of the immunoprecipitated DNA samples. (B) Analysis of H2B-K123 ubiquitination at PHO84, ADH1, and CUP1. Yeast cells (YKH045 [H2B wild type] and YKH046 [H2B-K123R point mutant]) were grown and cross-linked as described above. Immunoprecipitation was performed against HA-tagged ubiquitin using an anti-HA antibody. The anti-Myc antibody was used to monitor the background signal. Primer pairs targeting the UASs, core promoters, and ORFs of PHO84, ADH1, and CUP1 were used for PCR analysis of the immunoprecipitated DNA samples. ORF1 and ORF2 are located towards the 5′ and 3′ ends of the ORF, respectively. (C) The level of H2B-K123 ubiquitination is significantly elevated at the promoters but not the ORFs of PHO84, ADH1, and CUP1 in the Δubp8 mutant. The yeast strains YKH045, expressing HA-tagged ubiquitin, and ASY19 (derived by deleting UBP8 from YKH045) were grown as described above. Immunoprecipitation and PCR analysis were performed as described above. (D) Analysis of Ubp8p recruitment and H2B-K123 ubiquitination at the RPS5 UAS. (Top panel) Yeast strains expressing multiple-Myc-epitope-tagged Ubp8p were grown in YPD to an OD600 of 1.0 at 30°C, followed by formaldehyde-based cross-linking. Immunoprecipitation was carried out as described above. The primer pair targeting the RPS5 UAS was used in the PCR analysis. (Bottom panel) The yeast strains YKH045, YKH046, and ASY19 were grown and immunoprecipitated as described above.
Ubp8p regulates ubiquitination of H2B-K123 at the PHO84, ADH1, and CUP1 genes.
Although Ubp8p has recently been shown to have histone deubiquitinase activity for the removal of ubiquitin from K123-ubiquitinated H2B at a SAGA-dependent gene, GAL1 (20), its role in the regulation of H2B-K123 ubiquitination at other SAGA-dependent genes is not known. With this view, we analyzed the levels of H2B-K123 ubiquitination at three SAGA-dependent genes, PHO84, ADH1, and CUP1, in both the wild-type and Δubp8 mutant strains, using a modified ChIP assay. We used a wild-type (YKH045) or H2B-K123R (YKH046) mutant strain expressing Flag-tagged H2B and HA-tagged ubiquitin (20). Different sets of specific primer pairs (Fig. 4B) were used that could distinguish the levels of H2B-K123 ubiquitination at the UASs, core promoters, and ORFs of these genes. Two different locations in the PHO84 and ADH1 ORFs (ORF1 and ORF2 [towards the 5′ and 3′ ends of the ORFs, respectively]) were used. However, only one location in the relatively shorter CUP1 ORF (ORF1) was used.
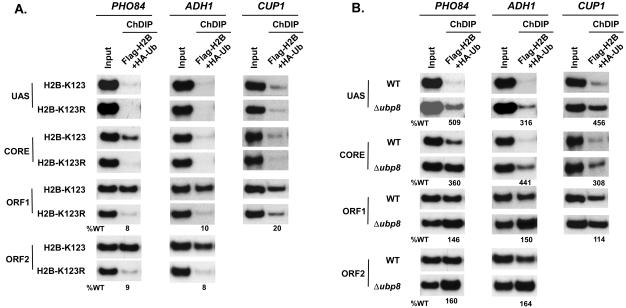
Figure 4B shows that HA-tagged ubiquitin was associated with the ORFs of PHO84, ADH1, and CUP1 in the wild-type strain. However, such results did not support H2B-K123 ubiquitination at these genes, since ubiquitination might also occur at the lysine residues of other proteins associated with these genes. To confirm that the ubiquitination occurred at K123 of H2B, we analyzed the levels of ubiquitination at these genes in the H2B-K123R point mutant strain. If K123 is involved in ubiquitination of H2B at the coding regions of these genes, then the levels of ubiquitination would be significantly reduced (or completely lost) in the H2B-K123R strain. This was indeed observed (Fig. 4B). However, it might still be possible that another ubiquitinated protein bound to these genes was immunoprecipitated in the wild-type strain (YKH045), and mutation of H2B-K123 to H2B-R123 might have altered recruitment of such a ubiquitinated protein, since the H2B-K123R mutant has been shown to affect recruitment of chromatin-associated proteins (14). Thus, to address this issue, we performed a ChDIP assay (see Materials and Methods) to specifically demonstrate the association of K123-ubiquitinated H2B with the PHO84, ADH1, and CUP1 genes. In such an assay, the first immunoprecipitation was performed using anti-Flag antibody, and the eluate was subsequently immunoprecipitated with anti-HA antibody. The immunoprecipitated DNA was analyzed by PCR, using primer pairs specific for various locations of the PHO84, ADH1, and CUP1 genes (Fig. 4B). When Flag-tagged H2B is ubiquitinated at K123, a PCR signal will be observed from the wild type but not the H2B-K123R mutant strain. Figure 5A shows the presence of PCR signals from the ORFs of PHO84, ADH1, and CUP1 in the wild type but not the H2B-K123R mutant strain, thus demonstrating the specific association of K123-ubiquitinated H2B with the coding sequences. Clearly, these results are consistent with the modified ChIP data (Fig. 4B).
FIG. 5.
Analysis of H2B-K123 ubiquitination at the PHO84, ADH1, and CUP1 genes using a ChDIP assay (see Materials and Methods). (A) ChDIP assay to analyze the levels of H2B-K123 ubiquitination in wild-type (YKH045) and H2B-K123R mutant (YKH046) strains expressing Flag-tagged H2B and HA-tagged ubiquitin. The yeast cells were grown and cross-linked as described in the legend to Fig. 4. The first immunoprecipitation was performed with anti-Flag antibody, and the precipitate was eluted with the Flag peptide. The eluate was then immunoprecipitated with anti-HA antibody. Primer pairs targeting the UASs, core promoters, and ORFs of the PHO84, ADH1, and CUP1 genes were used for PCR analysis of the immunoprecipitated DNA samples. (B) Analysis of H2B-K123 ubiquitination at PHO84, ADH1, and CUP1 in the Δubp8 mutant. The yeast strains YKH045 and ASY19 were grown as described in the legend to Fig. 4. Immunoprecipitation and PCR analysis were performed as described above.
The H2B histones at the UASs of PHO84, ADH1, and CUP1 are not ubiquitinated at K123 (Fig. 4B and 5A). Such observations raised the possibility that the targeted recruitment of Ubp8p to the UASs of these three SAGA-dependent genes might have hydrolyzed the ubiquitin moiety of K123-ubiquitinated H2B at the UASs of these genes. To test this hypothesis, we analyzed the levels of H2B-K123 ubiquitination at these genes in the Δubp8 mutant, using a modified ChIP assay. We did indeed observe the presence of H2B-K123 ubiquitination at the UASs of these three genes in the UBP8 deletion strain (Fig. 4C). As a control, we show that H2B-K123 ubiquitination at the UAS of a SAGA-independent gene, RPS5, was not altered in the Δubp8 mutant (Fig. 4D, bottom panel). We next analyzed the effect of Ubp8p on the levels of H2B-K123 ubiquitination at the core promoters and ORFs of ADH1, PHO84, and CUP1. We observed that the levels of H2B-K123 ubiquitination were increased at the core promoters of these genes in the Δubp8 mutant (Fig. 4C). However, H2B-K123 ubiquitination remained invariant at the coding sequences in the Δubp8 mutant (Fig. 4C).
To further confirm that the deletion of UBP8 specifically increased the level of H2B-K123 ubiquitination at the promoters of these three SAGA-dependent genes, we performed a ChDIP assay using wild-type and Δubp8 mutant yeast strains bearing Flag-tagged H2B and HA-tagged ubiquitin. Figure 5B shows the elevated levels of H2B-K123 ubiquitination at the UASs and core promoters, but not the ORFs, of these three SAGA-dependent genes in the Δubp8 mutant, consistent with the modified ChIP data (Fig. 4C). Thus, our results demonstrate that targeted recruitment of Ubp8p to the UASs of SAGA-dependent genes alters the levels of H2B-K123 ubiquitination at the promoters but not the ORFs in vivo. This was quite expected, since SAGA-associated Ubp8p is spatially close to both the activator binding site and TATA box, but not the ORF.
Ubp8p regulates H3-K4 methylation at the core promoter of PHO84 but not at those of ADH1 and CUP1.
The analysis of bulk nucleosomal histones revealed that the absence of H2B-K123 ubiquitination significantly lowers H3-K4 methylation (11, 43). Thus, the elevated level of H2B-K123 ubiquitination in the Δubp8 mutant would enhance H3-K4 methylation at the SAGA-dependent genes. This was indeed observed at the core promoter of the SAGA-regulated gene GAL1 (20). To verify such an observation at another SAGA-regulated gene, we analyzed H3-K4 methylation at PHO84 in the Δubp8 mutant and its isogenic wild-type equivalent.
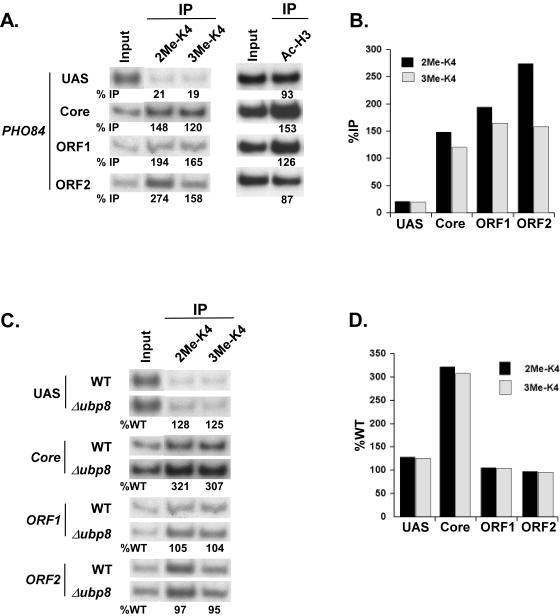
Using antibodies specific for di- and trimethylated H3-K4, we analyzed the level of H3-K4 methylation at various locations (e.g., UAS, core, and ORF) of the PHO84 gene in the wild-type yeast strain. Four sets of specific primer pairs (Fig. 4B) were used that could distinguish binding to the PHO84 UAS, core promoter, and two different locations of the ORF. Figure 6A and B show the distinct localizations of H3-K4 methylation at PHO84. The PHO84 core promoter, as well as the ORF, was dimethylated at H3-K4. Such a result is consistent with the genome-wide location analysis, which established that dimethylated H3-K4 is present in promoter and coding regions of genes, but with a bias towards the coding region (1). However, in contrast to a recent study demonstrating that H3-K4 trimethylation is concentrated at the 5′ portion of the coding regions of active genes such as PYK1 and RPS11B (28), we observed the presence of trimethyl H3-K4 throughout the coding region as well as the core promoter of the PHO84 gene (Fig. 6A and B). Neither di- nor trimethyl H3-K4 was detected at the PHO84 UAS, which was acetylated at H3-K9/14 (Fig. 6A).
FIG. 6.
Elevation of the level of H3-K4 methylation at the PHO84 core promoter in the Δubp8 mutant. (A) Analysis of H3-K9/14 acetylation and H3-K4 di- and trimethylation (2Me-K4 and 3Me-K4, respectively) at PHO84. Wild-type and UBP8 deletion mutant strains were grown and cross-linked as described in the legend to Fig. 4. Immunoprecipitations were performed using rabbit polyclonal antibodies against K9/14-diacetylated H3 (Upstate Biotechnology, Inc.), H3-dimethyl K4 (Upstate Biotechnology, Inc.), and H3-trimethyl K4 (Abcam, Inc.). Primer pairs targeting the UAS, core promoter, and two different locations of the ORF (ORF1 and ORF2) of the PHO84 gene were used for PCR analysis of the immunoprecipitated DNA samples. (B) Quantification of the H3-K4 di- and trimethylation illustrated above. The results are presented as percentages of DNA immunoprecipitated relative to the input. (C) Analysis of di- and trimethylation of H3-K4 at the PHO84 gene in the Δubp8 mutant. Immunoprecipitations were performed as described above. (D) Quantification of the experiment illustrated in panel C. The results are presented as percentages of DNA immunoprecipitated relative to the wild type.
We next analyzed the role of Ubp8p on H3-K4 methylation at the PHO84 core promoter. Both di- and trimethylation of H3-K4 at the PHO84 core promoter were elevated in the Δubp8 mutant (Fig. 6C and D). The increase in trimethylation of H3-K4 in the Δubp8 mutant was consistent with the results obtained for the GAL1 core promoter (20). However, in contrast to the fact that H3-K4 dimethylation was not affected at the GAL1 core promoter in the Δubp8 mutant (20), we observed an elevated level of dimethylated H3-K4 at the PHO84 core promoter in the Δubp8 mutant, consistent with the global role of H2B-K123 ubiquitination on the regulation of H3-K4 dimethylation (11, 43, 39). Thus, our data demonstrate that H2B-K123 deubiquitination by targeted recruitment of Ubp8p to the PHO84 promoter lowers both di- and trimethylation of H3-K4 in vivo in a “trans-tail” process. However, in contrast, Daniel et al. (10) have recently shown that H3-K4 trimethylation is significantly reduced at the GAL10 promoter in the Δubp8 mutant, indicating that Ubp8p can also up-regulate H3-K4 methylation.
To determine whether recruitment of Ubp8p at the PHO84 promoter has a long-range effect on regulation of H3-K4 methylation at the PHO84 ORF, we analyzed the levels of di- and trimethylated H3-K4 at two different locations of the PHO84 ORF in the Δubp8 mutant. Interestingly, both di- and trimethylation of H3-K4 at the PHO84 ORF were not altered in the Δubp8 mutant (Fig. 6C and D). Thus, Ubp8p has a local but not long-range effect on regulation of H3-K4 methylation in vivo.
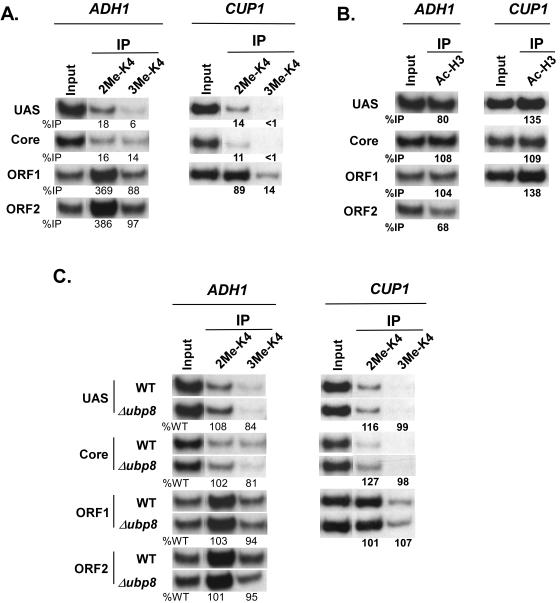
Next, we determined whether Ubp8p plays a general role in lowering H3-K4 methylation at the core promoters of two other SAGA-dependent genes, ADH1 and CUP1. Our ChIP analysis demonstrated that H3-K4 at the ORFs of CUP1 and ADH1 was di- and trimethylated (Fig. 7A). However, very low (or background) levels of H3-K4 di- and trimethylation were observed at the promoters of ADH1 and CUP1 (Fig. 7A), while these promoters were significantly acetylated at H3-K9/14 (Fig. 7B). We thus hypothesized that the elevated levels of H2B-K123 ubiquitination in the Δubp8 mutant (Fig. 4C and 5B) would enhance the levels of H3-K4 methylation at the promoters of these genes in a “trans-tail”-dependent manner. Interestingly, the levels of H3-K4 di- and trimethylation were not elevated at the core promoters or ORFs of these two genes in the Δubp8 mutant (Fig. 7C). Thus, Ubp8p does not regulate H3-K4 methylation at all SAGA-dependent genes in a “trans-tail”-dependent manner, revealing a differential regulation of H3-K4 methylation by histone deubiquitinase in vivo. This is analogous to the fact that SAGA components are differentially required for PIC assembly at the SAGA-dependent promoters (3).
FIG. 7.
Ubp8p does not regulate H3-K4 methylation at the ADH1 and CUP1 genes. (A) Analysis of di- and trimethylation of H3-K4 at the ADH1 and CUP1 genes. Wild-type and UBP8 deletion mutant strains were grown and cross-linked as described in the legend to Fig. 4. Immunoprecipitations were performed as described in the legend to Fig. 6. Primer pairs targeting the UASs, core promoters, and ORFs of ADH1 and CUP1 were used for PCR analysis of the immunoprecipitated DNA samples. (B) Analysis of H3-K9/14 acetylation at ADH1 and CUP1. Immunoprecipitations were performed as described in the legend to Fig. 6. (C) Analysis of H3-K4 di- and trimethylation at ADH1 and CUP1 in the Δubp8 mutant. Immunoprecipitations were performed as described in the legend to Fig. 6.
Ubp8p as well as H3-K4 methylation is dispensable for PIC assembly at the PHO84, ADH1, and CUP1 promoters.
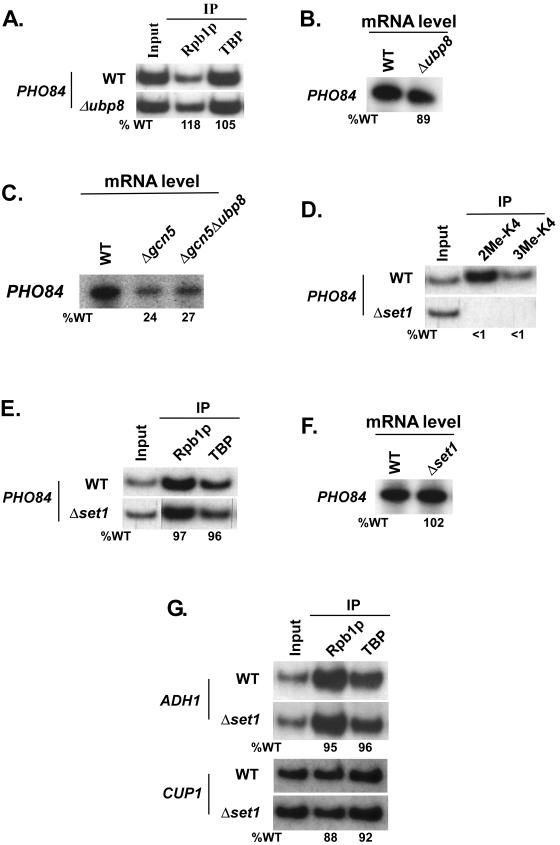
The detection of significantly high levels of di- and trimethylated H3-K4 at the promoters of several genes suggests that these modifications may be linked to transcription activation (29). Consistently, genome-wide DNA microarray analysis (38) revealed that H3-K4 methylation positively regulates transcription. Thus, we asked whether the up-regulation of H3-K4 methylation at the PHO84 core promoter in the Δubp8 mutant would stimulate the formation of the PIC assembly and, hence, transcription. To address this question, we analyzed the recruitment of TATA-binding protein (TBP) and RNA polymerase II (Rpb1), the two representative components of the PIC assembly, to the PHO84 core promoter in the Δubp8 mutant. Figure 8A shows that the increased level of H3-K4 methylation had no effect on formation of the PIC assembly at the PHO84 core promoter and, consistently, that transcription was not altered in the Δubp8 mutant (Fig. 8B).
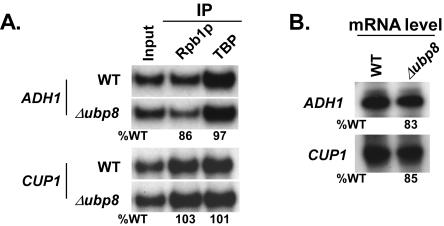
FIG. 8.
Ubp8p as well as H3-K4 methylation is not required for PIC assembly at the PHO84 core promoter. (A) Analysis of the PIC assembly at the PHO84 core promoter in the Δubp8 mutant. A primer pair targeting the PHO84 core promoter was used for PCR analysis of the immunoprecipitated DNA samples. Immunoprecipitations were performed using polyclonal antibodies against TBP and the mouse monoclonal antibody 8WG16 (Covance) against the C-terminal domain of the RNA polymerase II large subunit (Rpb1p). (B) Transcription. Total cellular RNA was prepared from the wild type or the Δubp8 mutant, and the mRNA level from the PHO84 gene was quantitated by primer extension. The percentage of transcription relative to the wild type is indicated below the gel. (C) Transcription. Total cellular RNA was prepared from the wild type and the Δgcn5 and Δgcn5 Δubp8 mutants, and the mRNA levels from the PHO84 gene were quantitated by primer extension. (D) Analysis of di- and trimethylation of H3-K4 at the PHO84 core promoter in the Δset1 strain. Immunoprecipitations were performed as described in the legend to Fig. 6. A primer pair targeting the PHO84 core promoter was used for PCR analysis of the immunoprecipitated DNA samples. (E) Analysis of the PIC assembly at the PHO84 core promoter in the Δset1 mutant. Immunoprecipitations were performed as described above. (F) Transcription. Total cellular RNA was prepared from the wild type or the Δset1 mutant, and the mRNA level from the PHO84 gene was quantitated as described above. (G) Analysis of the PIC assembly at the core promoters of ADH1 and CUP1 in the Δset1 mutant. Immunoprecipitations were performed as described above.
We next analyzed whether the UBP8 deletion mutant in combination with the Δgcn5 mutant shows a transcriptional defect of PHO84, since Henry et al. (20) demonstrated that ubp8 and gcn5 single mutants show a weak transcriptional phenotype at ADH2 but that a ubp8 gcn5 double mutant displays a synthetic one. We show that the transcription of PHO84 was significantly reduced in the Δgcn5 mutant compared with that in its isogenic wild-type equivalent (Fig. 8C). However, the Δgcn5 Δubp8 double mutant strain did not show an additional defect of PHO84 transcription (Fig. 8C). Thus, Ubp8p or an elevated level of H3-K4 methylation in the Δubp8 mutant does not seem to play a significant role in the transcription of PHO84 when combined with the Δgcn5 mutant.
To determine whether methylation of H3-K4 is at all required for PIC assembly at the PHO84 core promoter, we analyzed the recruitment of TBP and RNA polymerase II to the PHO84 core promoter in a SET1 deletion mutant. Set1p is essential for H3-K4 methylation, and thus its deletion completely removed di- and trimethylation of H3-K4 at the PHO84 core promoter (Fig. 8D). However, the deletion of SET1 did not affect formation of the PIC assembly at the PHO84 core promoter (Fig. 8E) and did not affect transcription (Fig. 8F). Thus, our data demonstrate that H3-K4 methylation is not required for formation of the PIC assembly at the PHO84 core promoter in vivo.
Next, we analyzed the role of H3-K4 methylation in formation of the PIC assembly at the core promoters of the CUP1 and ADH1 genes. A very low level of H3-K4 methylation was present at the core promoters of CUP1 and ADH1 (Fig. 7A), and thus these genes do not seem to require SET1 for formation of the PIC assembly. This was indeed observed (Fig. 8G).
Like other SAGA components, such as Spt3p and Spt8p (2, 3, 12), Ubp8p might be involved in facilitating the formation of the PIC assembly at the core promoters of ADH1 and CUP1. To test this hypothesis, we analyzed the recruitment of TBP and RNA polymerase II to the core promoters of the ADH1 and CUP1 genes and observed that Ubp8p was dispensable for formation of the PIC assembly at the core promoters of these two genes (Fig. 9A). The transcription levels from these two genes were consistently not altered in the Δubp8 mutant (Fig. 9B). Thus, unlike GAL1, GAL10, and ADH2 (10, 20), the PHO84, ADH1, and CUP1 genes do not require Ubp8p for transcription, demonstrating the differential requirement of Ubp8p for gene expression. Furthermore, Ubp8p did not show any transcriptional defect of CUP1 and ADH1, even when combined with the Δgcn5 mutant (data not shown).
FIG. 9.
Ubp8p is not required for the PIC assembly or transcription of ADH1 and CUP1. (A) Analysis of PIC assemblies at the core promoters of ADH1 and CUP1 in the Δubp8 mutant. Primer pairs targeting the core promoters of ADH1 and CUP1 were used for PCR analysis of the immunoprecipitated DNA samples. Immunoprecipitations were performed as described in the legend to Fig. 8. (B) Transcription. Total cellular RNA was prepared from the wild type or the Δubp8 mutant, and the mRNA level from the indicated gene was quantitated by primer extension.
DISCUSSION
In this report, we have analyzed in vivo the role of SAGA-associated Ubp8p in the regulation of H3-K4 methylation, PIC assembly, and transcription at three well-characterized SAGA-dependent genes, PHO84, ADH1, and CUP1. We show here that Ubp8p is associated with SAGA via Sgf11p and that its histone deubiquitinase activity lowers H3-K4 methylation at the core promoter only, not at the ORF, of the PHO84 gene. However, H3-K4 methylation at the other two SAGA-dependent genes is not regulated by Ubp8p. Interestingly, elevation of the H3-K4 methylation level at the PHO84 core promoter in the Δubp8 mutant does not stimulate formation of the PIC assembly and, hence, transcription. Finally, we demonstrate that Ubp8p as well as H3-K4 methylation is dispensable for PIC formation and transcription of these three SAGA-dependent genes in vivo.
Although Ubp8p has recently been implicated as a new component of the SAGA complex on the basis of biochemical data (10, 20, 37), it is not known whether Ubp8p is a SAGA component in vivo. If so, it would be recruited along with other SAGA components to the GAL1 UAS but not its core promoter, since SAGA is specifically recruited to the GAL1 UAS by the activator Gal4p (2-4). However, Henry et al. (20) demonstrated that Ubp8p is recruited to the GAL1 core promoter. On the other hand, Daniel et al. (10) demonstrated recruitment of Ubp8p to the GAL1 UAS. Thus, it was not clear from previous studies (10, 20) whether Ubp8p exists in vivo in the same form as that defined by its biochemical copurification with SAGA. Here we show that Ubp8p along with other SAGA components is recruited to the GAL1 UAS, but not the core promoter, or even to the minimal Gal4p-binding sites in a plasmid, supporting Ubp8p as a SAGA component in vivo. Consistently, Spt20p was also required for recruitment of Ubp8p to the GAL1 UAS. Furthermore, we show that the deletion of UBP8 has no effect on recruitment of the SAGA complex, consistent with the biochemical dispensability of Ubp8p for SAGA integrity (10, 20). Taken together, our results support the fact in vivo that Ubp8p is a SAGA component and is dispensable for the structural integrity of SAGA.
Recently, Ubp8p has been shown to form a structural module within SAGA by its interaction with Sgf11p on the basis of biochemical studies (21, 23, 31). However, evidence for formation of such a modular structure was lacking in vivo. Here we show that Ubp8p and Sgf11p are dependent on each other for recruitment to the GAL1 UAS. Furthermore, these two proteins are dispensable for the overall structural integrity of the SAGA complex. These results support the existence of a modular structural domain formed by Sgf11p and Ubp8p within SAGA in vivo.
Ubp8p is recruited to the GAL1 UAS by the Gal4p activation domain. Consistently, we also show here that Ubp8p is predominantly recruited to the UASs of three other SAGA-dependent genes, PHO84, ADH1, and CUP1. Since Ubp8p is recruited to the UAS, it is expected to play its functional role (i.e., histone H2B-K123 deubiquitination) at the promoter. Thus, H2B-K123 ubiquitination would be significantly lowered (or absent) at the UASs (and/or core promoters) of these three SAGA-dependent genes in the wild-type strain. However, the deletion of UBP8 would elevate the levels of H2B-K123 ubiquitination at the promoters of these genes. We indeed observed elevated levels of H2B-K123 ubiquitination at the UASs and core promoters but not the ORFs of PHO84, ADH1, and CUP1 in the Δubp8 mutant. Thus, UAS-localized Ubp8p has a local but not long-range functional role in the regulation of H2B-K123 ubiquitination in vivo.
Previous studies (20) demonstrated that H2B-K123 deubiquitination by Ubp8p significantly lowers the level of tri- but not dimethylated H3-K4 at the GAL1 core promoter in a “trans-tail”-dependent manner. Thus, the level of H3-K4 trimethylation is elevated at the GAL1 core promoter in the Δubp8 mutant (20). Similarly, we showed that the level of H3-K4 trimethylation at the PHO84 core promoter is increased in the Δubp8 mutant. However, in contrast to the results obtained for the GAL1 promoter (20), we observed a significant enhancement of H3-K4 dimethylation at the PHO84 core promoter in the Δubp8 mutant. Thus, our results demonstrate that H2B-K123 ubiquitination not only regulates H3-K4 trimethylation but also regulates dimethylation in vivo, consistent with mass spectrometric (39) and biochemical data (11, 43). We also showed that H3-K4 methylation at the PHO84 ORF is not altered in the Δubp8 mutant. This was expected, since H2B-K123 ubiquitination at the PHO84 ORF is not regulated by Ubp8p. Furthermore, our analysis on the levels of H3-K4 methylation at the core promoters of two other SAGA-dependent genes (ADH1 and CUP1) in the Δubp8 mutant revealed the absence of a regulatory link between H2B-K123 deubiquitination and H3-K4 methylation in vivo.
Recent studies have implicated the role of H3-K4 methylation in transcription activation (28, 38). Thus, the up-regulation of H3-K4 methylation at the PHO84 core promoter in the Δubp8 mutant might be involved in stimulating the formation of the PIC assembly and, hence, transcription. Interestingly, our results demonstrate that the elevated level as well as complete depletion of H3-K4 methylation at the PHO84 core promoter in the Δubp8 and Δset1 mutants, respectively, has no effect on PIC formation as well as transcription. In contrast, previous studies (10, 38) demonstrated the requirement of H3-K4 methylation for transcription of a subset of genes. However, it is not known currently how H3-K4 methylation can be involved in different transcriptional states. Perhaps, according to the “histone code” hypothesis (42), H3-K4 methylation in combination with other histone modifications may be playing a functional role in formation of the PIC assembly (and hence transcription activation) at the PHO84 core promoter. Thus, the loss or gain of H3-K4 methylation alone at the PHO84 core promoter does not seem to be associated with a transcriptional phenotype. However, our analysis on PIC formation at the PHO84 core promoter in the Δgcn5 Δubp8 double mutant does not show a transcriptional phenotype for Ubp8p. Probably, Ubp8p in conjunction with H3 serine 10 phosphorylation or other covalent modifications of histones has a transcriptional phenotype at PHO84. Nonetheless, this study demonstrates the lack of a regulatory link between H3-K4 methylation and PIC formation (and hence transcription) at the PHO84 core promoter.
In summary, we have shown here the differential regulation of H3-K4 methylation at the core promoters of the SAGA-dependent genes by Ubp8p. Our in vivo analysis of the function of Ubp8p in the regulation of H3-K4 methylation is remarkably consistent with the fact that the SAGA components are differentially required for gene activity (3). However, the molecular basis of the differential regulation of H3-K4 methylation at the core promoters of the SAGA-dependent genes remains to be elucidated.
Acknowledgments
We thank Michael R. Green for TBP and TAF antibodies; Fred Winston and Mary Ann Osley for yeast strains; and Praveena Krishnan, Sheila Scillufo, Stephanie Ultre, and Radiya Sojitrawala for technical assistance. We also thank Thomas Shadle and Pratibha Bajwa for critical readings of the manuscript.
This work was supported by SIU-Office of Research Development and Administration, School of Medicine and Cancer Institute.
REFERENCES
- 1.Bernstein, B. E., E. L. Humphrey, R. L. Erlich, R. Schneider, P. Bouman, J. S. Liu, T. Kouzarides, and S. L. Schreiber. 2002. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl. Acad. Sci. USA 99:8695-8697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhaumik, S. R., and M. R. Green. 2001. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 15:1935-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhaumik, S. R., and M. R. Green. 2002. Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol. Cell. Biol. 22:7365-7371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhaumik, S. R., T. Raha, D. P. Aiello, and M. R. Green. 2004. In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev. 18:333-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boggs, B. A., P. Cheung, E. Heard, D. L. Spector, A. C. Chinault, and C. D. Allis. 2002. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat. Genet. 30:73-76. [DOI] [PubMed] [Google Scholar]
- 6.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 7.Briggs, S. D., M. Bryk, B. D. Strahl, W. L. Cheung, J. K. Davie, S. Y. Dent, F. Winston, and C. D. Allis. 2001. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 15:3286-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, C. E., T. Lechner, L. Howe, and J. L. Workman. 2000. The many HATs of the transcriptional coactivators. Trends Biochem. Sci. 25:15-19. [DOI] [PubMed] [Google Scholar]
- 9.Bryk, M., S. D. Briggs, B. D. Strahl, M. J. Curcio, C. D. Allis, and F. Winston. 2002. Evidence that Set1, a factor required for methylation of histone H3, regulates rDNA silencing in S. cerevisiae by a Sir2-independent mechanism. Curr. Biol. 12:165-170. [DOI] [PubMed] [Google Scholar]
- 10.Daniel, J. A., M. S. Torok, Z. W. Sun, D. Schieltz, C. D. Allis, Y. R. Yates III, and P. A. Grant. 2004. Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J. Biol. Chem. 279:1867-1871. [DOI] [PubMed] [Google Scholar]
- 11.Dover, J., J. Schneider, M. A. Tawiah-Boateng, A. Wood, K. Dean, M. Johnston, and A. Shilatifard. 2002. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 277:28368-28371. [DOI] [PubMed] [Google Scholar]
- 12.Dudley, A. M., C. Rougeulle, and F. Winston. 1999. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev. 13:2940-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ehrenhofer-Murray, A. E. 2004. Chromatin dynamics at DNA replication, transcription and repair. Eur. J. Biochem. 271:2335-2349. [DOI] [PubMed] [Google Scholar]
- 14.Ezhkova, E., and W. P. Tansey. 2004. Proteasomal ATPases link ubiquitylation of histone H2B to methylation of histone H3. Mol. Cell 13:435-442. [DOI] [PubMed] [Google Scholar]
- 15.Gendrel, A. V., Z. Lippman, C. Yordan, V. Colot, and R. A. Martienssen. 2002. Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297:1871-1873. [DOI] [PubMed] [Google Scholar]
- 16.Grant, P. A., L. Duggan, J. Cote, S. M. Roberts, J. E. Brownell, R. Candau, R. Ohba, T. Owen-Hughes, C. D. Allis, F. Winston, S. L. Berger, and J. L. Workman. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11:1640-1650. [DOI] [PubMed] [Google Scholar]
- 17.Grant, P. A., D. E. Sterner, L. J. Duggan, J. L. Workman, and S. L. Berger. 1998. The SAGA unfolds: convergence of transcription regulators in chromatin-modifying complexes. Trends Cell Biol. 8:193-197. [DOI] [PubMed] [Google Scholar]
- 18.Hampsey, M. 1997. A SAGA of histone acetylation and gene expression. Trends Genet. 13:427-429. [DOI] [PubMed] [Google Scholar]
- 19.Hayes, J. J., and J. C. Hansen. 2001. Nucleosomes and the chromatin fiber. Curr. Opin. Genet. Dev. 11:124-129. [DOI] [PubMed] [Google Scholar]
- 20.Henry, K. W., A. Wyce, W. S. Lo, L. J. Duggan, N. C. Emre, C. F. Kao, L. Pillus, A. Shilatifard, M. A. Osley, and S. L. Berger. 2003. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 17:2648-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ingvarsdottir, K., N. J. Krogan, N. C. Emre, A. Wyce, N. J. Thompson, A. Emili, T. R. Hughes, J. F. Greenblatt, and S. L. Berger. 2005. H2B ubiquitin protease Ubp8 and Sgf11 constitute a discrete functional module within the Saccharomyces cerevisiae SAGA complex. Mol. Cell. Biol. 25:1162-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kornberg, R. D., and Y. Lorch. 1999. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98:285-294. [DOI] [PubMed] [Google Scholar]
- 23.Lee, K. K., L. Florens, S. K. Swanson, M. P. Washburn, and J. L. Workman. 2005. The deubiquitylation activity of Ubp8 is dependent upon Sgf11 and its association with the SAGA complex. Mol. Cell. Biol. 25:1173-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, X.-Y., S. R. Bhaumik, and M. R. Green. 2000. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science 288:1242-1244. [DOI] [PubMed] [Google Scholar]
- 25.Litt, M. D., M. Simpson, M. Gaszner, C. D. Allis, and G. Felsenfeld. 2001. Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science 293:2453-2455. [DOI] [PubMed] [Google Scholar]
- 26.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pingle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 27.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 28.Ng, H. H., F. Robert, R. A. Young, and K. Struhl. 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11:709-719. [DOI] [PubMed] [Google Scholar]
- 29.Noma, K., C. D. Allis, and S. I. Grewal. 2001. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science 293:1150-1155. [DOI] [PubMed] [Google Scholar]
- 30.Noma, K. I., and S. I. Grewel. 2002. Histone H3 lysine 4 methylation is mediated by Set1 and promotes maintenance of active chromatin states in fission yeast. Proc. Natl. Acad. Sci. USA 99:16438-16445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Powell, D. W., C. M. Weaver, J. L. Jennings, K. J. McAfee, Y. He, P. A. Weil, and A. J. Link. 2004. Cluster analysis of mass spectrometry data reveals a novel component of SAGA. Mol. Cell. Biol. 24:7249-7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reuben, M., and R. Lin. 2002. Germline X chromosomes exhibit contrasting patterns of histone H3 methylation in Caenorhabditis elegans. Dev. Biol. 245:71-82. [DOI] [PubMed] [Google Scholar]
- 33.Rice, J. C., and C. D. Allis. 2001. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr. Opin. Cell Biol. 13:263-273. [DOI] [PubMed] [Google Scholar]
- 34.Roberts, S. M., and F. Winston. 1996. SPT20/ADA5 encodes a novel protein functionally related to the TATA-binding protein and important for transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:3206-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberts, S. M., and F. Winston. 1997. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics 147:451-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robzyk, K., J. Recht, and M. A. Osley. 2000. Rad6-dependent ubiquitination of histone H2B in yeast. Science 287:501-504. [DOI] [PubMed] [Google Scholar]
- 37.Sanders, S. L., J. Jennings, A. Canutescu, A. J. Link, and P. A. Weil. 2002. Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol. Cell. Biol. 22:4723-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santos-Rosa, H., R. Schneider, A. J. Bannister, J. Sherriff, B. E. Bernstein, N. C. Emre, S. L. Schreiber, J. Mellor, and T. Kouzarides. 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419:407-411. [DOI] [PubMed] [Google Scholar]
- 39.Shahbazian, M. D., K. Zhang, and M. Grunstein. 2005. Histone H2B ubiquitylation controls processive methylation but not monomethylation by Dot1 and Set1. Mol. Cell 19:271-277. [DOI] [PubMed] [Google Scholar]
- 40.Sterner, D. E., P. A. Grant, S. M. Roberts, L. J. Duggan, R. Belotserkovskaya, L. A. Pacella, F. Winston, J. L. Workman, and S. L. Berger. 1999. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol. 19:86-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strahl, B. D., R. Ohba, R. G. Cook, and C. D. Allis. 1999. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc. Natl. Acad. Sci. USA 96:14967-14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 43.Sun, Z. W., and C. D. Allis. 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418:104-108. [DOI] [PubMed] [Google Scholar]
- 44.Turner, B. M. 2002. Cellular memory and the histone code. Cell 111:285-291. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, L., E. E. Eugeni, M. R. Parthun, and M. A. Freitas. 2003. Identification of novel histone post-translational modifications by peptide mass fingerprinting. Chromosoma 112:77-86. [DOI] [PubMed] [Google Scholar]