Abstract
Tailless is an orphan nuclear receptor that controls terminal body patterning in Drosophila. Genetic analyses have revealed both positive and negative regulatory interactions of Tailless with various target genes, leading to the idea that, like many other nuclear receptors, Tailless mediates both activation and repression of transcription. In this paper, we have examined the consequences of converting Tailless into an obligate repressor and compared the activities of the resulting protein with those of wild-type Tailless. We find that this repressor form of Tailless behaves like the intact protein in gain- and loss-of-function experiments, being sufficient to support normal embryonic development and establish accurate patterns of gene expression even for positive Tailless targets such as hunchback and brachyenteron. This suggests that Tailless functions exclusively as a transcriptional repressor in the embryo and that the observed positive interactions of Tailless with specific targets are secondary effects involving repression of repressors. We provide evidence that knirps is one such repressor gene acting between Tailless and its indirect positive targets. Finally, our results indicate that Tailless exerts an active mechanism of repression via its ligand-binding domain and that this activity is largely independent of the activation function 2 (AF2) motif characteristic of most nuclear receptors.
Patterning of the early Drosophila embryo depends on a complex network of transcriptional regulation. Maternal and zygotic transcription factors cross-interact and progressively subdivide the embryo into spatial domains up to a resolution of one or two cells. By the mid-blastoderm stage, approximately 20 to 30 products are expressed in gradients, bands, and stripes that prefigure the future body plan of the adult (for a review, see reference 41). Although genetic and molecular analyses have identified many of the regulatory mechanisms involved in this system, basic questions about the function of several key regulators remain unsolved.
The Tailless (Tll) nuclear receptor controls the specification of the most anterior and posterior (terminal) embryonic structures (4, 45, 54, 57). At the mid-blastoderm stage, tll and a second terminal gene, huckebein (hkb), are activated at each pole of the embryo in response to the maternal Torso signaling pathway (2, 3, 45). Consistent with this polar expression, loss-of-function mutations in tll disrupt the development of parts of the head and the anterior brain and also delete the posterior terminal domain comprising the eighth abdominal segment, telson, and posterior gut (45, 54, 60), hence the classification of tll as a gap segmentation gene. Conversely, ectopic tll expression throughout the blastoderm embryo suppresses most of the prospective trunk and abdomen, which instead adopt a terminal fate and develop ectopic terminal elements such as the posterior filzkörper (53).
How does Tll exert its regulatory functions? Genetic analyses suggest that Tll has a dual function as both an activator and repressor of other patterning genes. For example, in the posterior region of the blastoderm embryo, Tll is required for the activation of hunchback (hb), brachyenteron (byn), and forkhead (fkh) (25, 32, 50, 57, 58), while at the same time repressing the gap genes knirps (kni), Krüppel (Kr), and giant (gt), thereby helping establish their posterior limits of expression (27, 40, 53). However, the complexity of the segmentation network makes it difficult to distinguish which of these effects are direct and which result from indirect activities via intermediary genes. Thus, although DNA-binding analyses support a direct role of Tll in activation of hb and repression of kni and Kr (19, 32, 39), definite evidence for direct versus indirect regulatory activities of Tll is in most cases lacking.
The possibility that Tll functions as both an activator and repressor is consistent with the general attributes of nuclear receptors (reviewed in references 31 and 34). Members of this family share a basic structure composed of two conserved regions: a DNA-binding domain with two zinc-binding modules and a C-terminal ligand-binding domain (LBD) that can activate or repress transcription and whose activity can be regulated by the binding of lipophilic hormones (see Fig. 1A). Nevertheless, Tll and many other nuclear receptors designated as orphan lack an identified regulatory hormone. It is therefore unclear whether Tll activity is affected in any way by the binding of a ligand. In fact, the functional properties of the Tll LBD remain uncertain: although this domain is conserved in insects and vertebrates, a sequence analysis of tll mutations did not identify an essential requirement for this region (10), raising the possibility that it is partly or completely dispensable.
FIG. 1.
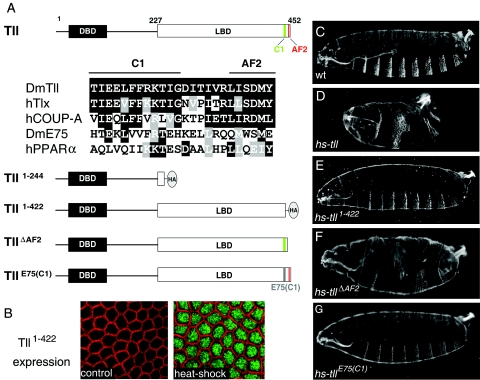
The C-terminal portion of the Tll LBD is essential for patterning activity in the early embryo. (A) Diagram of the Tll protein indicating the DNA-binding domain (DBD) and LBD and the presence of two C-terminal motifs, AF2 and C1. Numbers indicate amino acid positions. An alignment of C1 and AF2 sequences from several nuclear receptors is shown below; identical and similar residues are boxed in black and gray, respectively. The AF2 motif (also known as helix 12) conforms to the consensus ΦΦX(E/D)ΦΦ, where “Φ” denotes a hydrophobic residue and “X” is any amino acid. The C1 motif is well conserved in the human Tll ortholog, Tlx, and in the human COUP-TFA receptor, which, like Tll, belongs to subfamily 2 of nuclear receptors. In contrast, this motif has diverged in two distant receptors belonging to subfamily 1, Drosophila E75 and human peroxisome proliferator-activated receptor α. A diagram of four Tll derivatives with mutations in their LBDs is also shown. “HA” marks the positions of this epitope tag in the Tll1-244 and Tll1-422 derivatives. The E75 sequence replacing the Tll C1 motif in the TllE75(C1) construct is gray. (B) Immunodetection of Tll1-422 protein following heat shock induction using an anti-HA antibody. Detailed surface views of non-heat-shocked (control, left) and heat-shocked (right) hs-tll1-422 blastoderm embryos are shown. Tll1-422 protein (green) accumulates efficiently in the nucleus after heat shock. The cortical actin associated with the growing plasma membranes appears labeled with rhodamine-phalloidin (red). (C) Cuticle of a wild-type (wt) embryo. (D to G) Representative cuticles of heat-shocked hs-tll (D), hs-tll1-422 (E), hs-tllΔAF2 (F), and hs-tllE75(C1) (G) embryos. Note that Tll1-422 and TllE75(C1) are inactive in the assay, whereas TllΔAF2 displays considerable patterning activity.
In this paper, we have used in vivo regulatory assays to distinguish between activator and repressor functions of Tll and to examine the contribution of the Tll LBD in these activities. We find that a constitutive-repressor form of Tll behaves similarly to the endogenous protein in both gain- and loss-of-function experiments, suggesting that Tll acts primarily as a transcriptional repressor and that its positive interactions with specific targets such as hb are indirect. We also provide evidence that Tll repression involves an active mechanism mediated by an intrinsic activity of the LBD.
MATERIALS AND METHODS
DNA constructs.
Plasmid manipulations were carried out according to standard procedures. Coding segments generated by PCR were sequenced to ensure fidelity during the amplification reactions.
A 1.4-kb fragment encoding the wild-type Tll protein was generated by PCR and cloned as an EcoRI-EcoRV insert in pUCBM20 (Boehringer) and pBluescript SK(+) (Stratagene) vector plasmids. The resulting plasmids were then used as the starting material for all mutant Tll derivatives expressed under heat shock control. The Tll1-244 construct was made by replacing a HindII-EcoRV fragment encoding the Tll LBD with an EcoRV-digested PCR fragment encoding three tandem copies of the hemagglutinin (HA) tag (YPYDVPDYA). The TllEn(RD) construct was made similarly, except for using an EcoRV-digested PCR fragment encoding En amino acids 167 to 246. Tll1-422 was generated by replacing a HindIII-KpnI fragment encoding Tll amino acids 422 to 452 with a HindIII-KpnI synthetic linker containing internal SmaI and EcoRV sites and then inserting into the SmaI site the above EcoRV HA-coding fragment. For TllΔAF2, Tll coding sequences downstream of the HindIII site at position 422 were replaced with a HindIII-digested PCR fragment encoding Tll amino acids 423 to 441, thus removing amino acids 442 to 452. To make TllE75(C1), E75A coding sequences between positions 729 to 741 were joined to Tll coding sequences corresponding to amino acids 436 to 452 by recombinant PCR and the resulting fragment inserted into the HindIII site corresponding to position 422 of the Tll coding sequence. Sequences encoding wild-type and mutant Tll proteins were excised as EcoRI-EcoRV fragments and cloned into pCaSpeR-hs (see Flybase at http://flybase.bio.indiana.edu/) digested with EcoRI/StuI.
To make hb-htll(LBD), a NarI-PstI fragment including Tll LBD coding sequences and the hsp70 3′ flanking region was isolated from hs-tll and cloned into pUC57 (Fermentas). This fragment was then recovered as an EcoRV-PstI fragment (containing a 10-bp extension at the 5′ end) and triple ligated with a KpnI-NarI fragment from hb-hen (containing the hb promoter and Hairy coding sequences encoding amino acids 1 to 281) (22) and pCaSpeR4 (see Flybase) cut with KpnI and PstI. hb-htll(CTD) was constructed similarly, except that the EcoRV-PstI fragment consisted of Tll coding sequences encoding amino acids 400 to 452 joined to the hsp70 3′ flanking region.
To create tllen(RD), a 5.9-kb SalI-PstI fragment containing the tll promoter was generated by PCR and subcloned in pBluescript SK(+), resulting in plasmid pBStllp. In parallel, a 1.4-kb PstI-BamHI partially digested fragment of hs-tllen(RD), containing TllEn(RD) coding sequences encoding amino acid 38 to the C-terminal end joined to the hsp70 3′ flanking region, was subcloned in PstI/BamHI-digested pUC57. This 1.4-kb fragment was then excised as a PstI-XbaI fragment and inserted into pBStllp digested with PstI/XbaI. The resulting plasmid was then cut with PstI and ligated to a 470-bp PstI-PstI tll genomic fragment spanning from the transcription start site to codon 37 in the coding sequence, thus generating pBStllen(RD). The complete tllen(RD) sequence was then released with XhoI/XbaI and cloned into pCaSpeR4 digested with XhoI/XbaI.
Additional details on the construction of the plasmids and the sequences of the primers and linkers used in the cloning are available on request.
Fly stocks and transgenic flies.
Mutant alleles used were tlll49, tll1, and kni6 (see reference 10 and Flybase for details). For the rescue experiments shown in Fig. 4E and F, the tlll49 allele was balanced over TM3, Act-GFP and then nonfluorescent embryos or larvae were selected for cuticle analysis. For the experiments in Fig. 4G to L and 5A, tlll49 was balanced over TM3, hb-lacZ to allow identification of homozygous tlll49 embryos in the in situ hybridizations (see also below). The same balancer was used for the analysis of tll1 and tll1 kni6 mutant embryos in Fig. 5E to H. hs-kni flies were kindly provided by D. Arnosti (55).
FIG. 4.
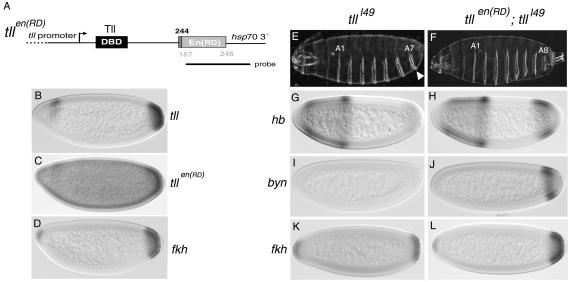
Rescue of tll mutant embryos by TllEn(RD). (A) Diagram of the tllen(RD) transgene driven by a 5.9-kb promoter region from tll. The dotted line in the promoter indicates that it is not drawn to scale. Numbers indicate amino acid positions as in Fig. 3. The probe used for the in situ hybridization shown in panel C is also depicted. DBD, DNA-binding domain. (B) Wild-type expression pattern of endogenous tll. (C) mRNA expression pattern of tllen(RD) in an otherwise-wild-type embryo. The pattern is similar to that of endogenous tll, with a visible posterior cap and a dorsal-anterior stripe. Note, however, that the transgene is also expressed at low level throughout the embryo. (D) Wild-type expression pattern of fkh. (E to L) Complementation of tlll49 mutant embryos by tllen(RD). tlll49 embryos lack structures posterior to the A7 segment (E, arrowhead) and have defective expression of hb (G), byn (I), and fkh (K) at the blastoderm stage (compare with Fig. 1C, 3I, and 3L and panel D). tlll49 embryos expressing tllen(RD) have a normal cuticle pattern (F) and restored expression of hb (H), byn (J), and fkh (L).
FIG. 5.
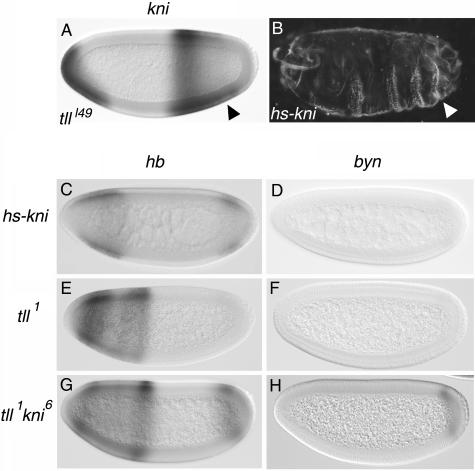
kni is partly responsible for the indirect effects of Tll on hb and byn. (A) Ectopic expression of kni in the posterior region of a tlll49 mutant embryo (arrowhead, compare with Fig. 3F). (B) Patterning defects induced by ectopic expression of kni under the control of a heat shock promoter; note the suppression of posterior terminal structures (arrowhead). (C and D) Expression patterns of hb (C) and byn (D) in heat-shocked hs-kni embryos; both genes appear significantly repressed (compare with Fig. 3I and L, respectively). (E to H) Expression of hb (E and G) and byn (F and H) in tll1 single-mutant (E and F) and tll1 kni6 double-mutant (G and H) embryos. Note that tll1 kni6 embryos exhibit enhanced posterior expression of hb and byn relative to tll1 embryos.
Germ line transformation was performed as described previously (52) by injection into y w embryos and selection for rescue of w eyes. In general, two or more lines were analyzed for each construct. For the hs constructs, homozygous viable insertions were selected for analysis. In the case of the tllen(RD) construct, the seven lines obtained had pale yellow eyes, perhaps reflecting a bias for low-expression insertions. hb-htll(LBD) lines on the X chromosome were maintained in males using a compound X chromosome [C (1)M3], whereas autosomal insertions were kept unbalanced, selecting for transformant males in each generation.
Embryo analyses.
Embryos were collected on apple juice plates at 25°C. For the heat shock experiments, 30-min egg collections were typically performed. Heat shocks were carried out by floating the apple juice plates on a water bath at 37°C for 10 min. Occasionally, 13-min heat shocks were also employed to assay relatively weak effects such as activation of byn by the wild-type Tll protein. Neither of these regimens causes significant pattern abnormalities in wild-type embryos.
Following heat shocks, embryos were allowed to continue development for cuticle analysis or dechorionated and fixed 30 to 40 min after heat shock for in situ hybridization or antibody staining. Cuticle preparations were made by dechorionating the embryos in bleach, mounting them in Hoyer's medium, and allowing them to clear overnight at 60°C. Embryos were fixed in heptane-4% formaldehyde-phosphate-buffered saline for 20 min (in situ hybridizations) or 12 min (immunostainings). In situ hybridizations were carried out using digoxigenin-UTP-labeled antisense RNA probes and antidigoxigenin antibodies conjugated to alkaline phosphatase (Roche). All the embryos shown in Fig. 3 to 5 are representative of the genotypes indicated and were obtained using side-by-side control stainings to monitor for differences in signal intensity between different experiments. For the analysis of tllen(RD) expression (Fig. 4C), a control hybridization was carried out using nontransgenic embryos to confirm the low-level ectopic expression of this transgene. The analyses of byn and fkh expression in tll single-mutant and tll kni double-mutant embryos were performed with mixed lacZ and byn or fkh probes to allow identification of homozygous mutant embryos by the lack of anterior hb-lacZ staining. For the analyses of hb expression in those mutant embryos, two-color in situ hybridizations were carried out using hb and lacZ probes labeled with digoxigenin and fluorescein, respectively (18). Immunodetection of HA-tagged proteins was performed using monoclonal antibody 12CA5 (Roche) diluted 1:100 and secondary fluorochrome- or biotin-conjugated antibodies (Molecular Probes, Amersham). In the latter case, signals were detected with the Vectastain ABC kit (Vector Laboratories). Accumulation of the TllΔAF2 and TllE75(C1) proteins following heat shock was confirmed using a guinea pig anti-Tll polyclonal antibody (kindly provided by R. Pflanz) diluted 1:150 and a secondary fluorochrome-conjugated antibody. Cortical actin was visualized with rhodamine-phalloidin (Sigma; 1:800 dilution).
FIG. 3.
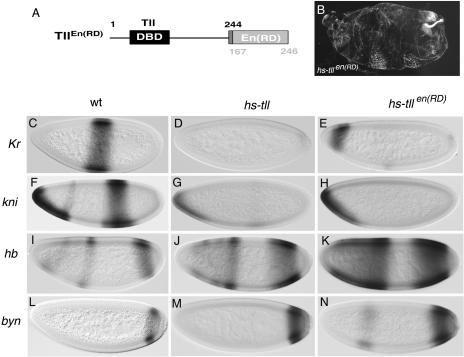
Similar effects caused by Tll and TllEn(RD) when expressed under heat shock control. (A) Diagram of the TllEn(RD) chimera; numbers indicate amino acid positions in the native Tll (above) and Engrailed (below) products. DBD, DNA-binding domain. (B) Representative cuticle phenotype induced by expression of TllEn(RD) at the blastoderm stage; note the strong suppression of central portions of the embryo, similar to that caused by ectopic Tll expression (Fig. 1D). (C to N) Expression patterns of Kr (C to E), kni (F to H), hb (I to K), and byn (L to N) in wild-type (wt) (C, F, I, and L), heat-shocked hs-tll (D, G, J, and M), and heat-shocked hs-tllen(RD) (E, H, K, and N) embryos. Tll and TllEn(RD) induce comparable levels of repression of Kr and kni and expansion of hb and byn.
RESULTS
The LBD is essential for Tll activity.
As noted in the introduction, no information is available on the function of the Tll LBD. Therefore, we tested if this domain is required for Tll activity in vivo. Previous studies showed that ubiquitous Tll expression at the blastoderm stage using a heat shock promoter induces extensive pattern deletions in the future thoracic and abdominal regions of the embryo and also leads to the formation of ectopic terminal structures such as the posterior spiracle and associated filzkörper (53). These effects result from both repression of central gap genes such as Kr and kni and ectopic activation of genes expressed at posterior terminal and subterminal regions such as hb and byn (50, 53). We used this assay to examine the activities of two Tll mutant derivatives lacking either the complete LBD or a C-terminal portion of this domain that is particularly well conserved in mammalian Tll orthologs (Tll1-244 and Tll1-422, respectively; Fig. 1A). Both constructs were tagged with the HA epitope to monitor protein expression, placed under the control of the hsp70 promoter, and then used to generate transgenic lines (see Materials and Methods).
Induction of the wild-type Tll protein with a 10-min heat shock at 37°C, administered 110 to 140 min after egg laying, causes lethal disruption of the anteroposterior pattern. More than 80% of heat-shocked hs-tll embryos show severe suppression of the segmented trunk and abdomen, resulting in phenotypes that range from deletion of three or four segments to the complete loss of metameric pattern (Fig. 1D and data not shown). We also observe the occasional formation of ectopic filzkörper structures, which often appear diffuse and disorganized (data not shown). In contrast, inducing Tll1-244 or Tll1-422 using the same heat shock conditions does not produce lethality or significant alterations in the embryonic pattern (Fig. 1B and E; data not shown). Staining of heat-shocked hs-tll1-244 embryos with an anti-HA antibody resulted in undetectable protein accumulation of the Tll1-244 derivative, suggesting that it is produced at a low level or is unstable (data not shown; see Materials and Methods). However, we observe intense nuclear accumulation of Tll1-422 following heat shock (Fig. 1B). We therefore conclude that the LBD of Tll, and in particular its C-terminal sequences, are essential for most or all regulatory activities of the protein.
We distinguish two motifs within the C-terminal portion of the Tll LBD (Fig. 1A). One consists of the final six residues corresponding to the activation function 2 (AF2) motif present at the C termini of most nuclear receptors (11) (Fig. 1A). The AF2 motif of ligand-activated receptors forms an α-helix that typically folds over the ligand and its pocket, contributing to the recruitment of transcriptional cofactors (reviewed in reference 36). We therefore examined the effects of deleting the Tll AF2-like motif in the hs-tll assay. Heat shock-induced expression of the resulting derivative, TllΔAF2, produces significant alterations of the embryonic pattern. The effects appear less severe than those caused by the intact protein, but most heat-shocked hs-tllΔAF2 embryos still die with deletions of more than three segments (Fig. 1F). In these embryos, we also observe clear repression of kni expression at the blastoderm stage (data not shown; see also below). These results suggest that the AF2-like motif makes a relatively minor contribution to Tll activity.
We also examined the effects of mutating a second motif within the Tll C-terminal domain (CTD) that shows the highest conservation when comparing the LBDs of Tll orthologs; we designate this motif C1 (Fig. 1A). The C1 motif is also conserved in other nuclear receptors such as the vertebrate COUP-TF proteins, which, like Tll, belong to subfamily 2 of nuclear receptors (38). We reasoned that a deletion of the C1 motif might alter the overall folding of the LBD, producing effects not strictly related to a specific regulatory function of this motif. In an attempt to preserve the Tll LBD structure while mutating the C1 element, we replaced this element by the corresponding sequence from a distant nuclear receptor, the Drosophila E75 protein (49). We find that expression of this mutant protein, TllE75(C1), has very little or no effect on embryonic patterning (Fig. 1G), although accumulation of TllE75(C1) is readily detected in the heat-shocked embryos (data not shown; see Materials and Methods). This indicates that the C1 motif is essential for Tll function in the early embryo.
The Tll LBD behaves as a repressor module.
We next investigated the activity of the Tll LBD when isolated from its natural context and transferred to a heterologous DNA-binding protein. For this experiment, we used an assay in which the Hairy segmentation protein interferes with expression of the Sex lethal (Sxl) gene (42). Sxl is the initial regulator of sex determination, and its expression at the blastoderm stage is normally restricted to female embryos (24, 43) (Fig. 2). In males, Sxl is repressed by the activity of several maternal factors, including the basic helix-loop-helix factor Deadpan (59). Because Deadpan is related to the Hairy segmentation protein, premature expression of Hairy driven by the hb gap gene promoter causes inappropriate repression of Sxl in the anterior halves of female embryos. This impairs dosage compensation in the female, leading to sex-specific lethality of these individuals (42). Repression of Sxl and female lethality depend on the integrity of a C-terminal repressor motif in Hairy (9). Substitution of this C-terminal motif by a heterologous repressor domain also leads to repression of Sxl (22), whereas replacement by a heterologous activation domain induces ectopic Sxl expression in males and ensuing lethality of this sex (23).
FIG. 2.
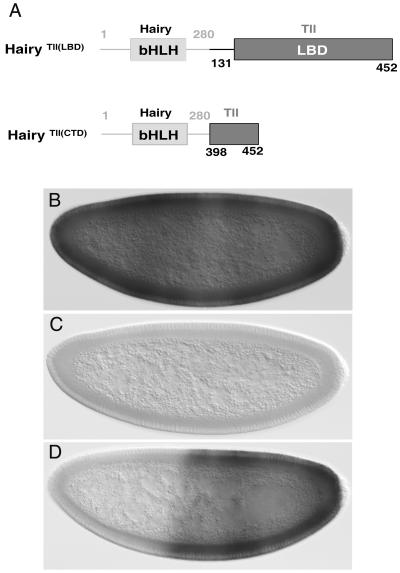
The Tll LBD has the potential to mediate transcriptional repression. (A) Diagram of Hairy-Tailless fusion proteins expressed under the control of the hb promoter. Hairy and Tll sequences are depicted with gray and black lines, respectively. Numbers indicate amino acid positions in the Hairy (above) and Tll (below) proteins. bHLH, basic helix-loop-helix. (B and C) Expression of Sxl mRNA in wild-type presumed-female (B) but not male (C) embryos. (D) Expression of Sxl in a female hb-htll(LBD) embryo; repression is observed in the anterior half. In this and subsequent figures, embryos are oriented with the anterior portion to the left and dorsal portion up.
We used the hb-h assay to express a protein chimera [HairyTll(LBD)] in which the C-terminal 57 amino acids of Hairy are replaced by a fragment of Tll (amino acids 131 to 452) that includes the complete LBD region. We find that hb-htll(LBD) efficiently represses Sxl in the anterior of female embryos (Fig. 2D). This repressor activity appears robust when compared to that of other repressor domains tested in the hb-h assay (9, 22), as it leads to 100% female lethality in all transgenic lines obtained (data not shown). These results indicate that the LBD of Tll functions as a potent repressor domain in the hb-h assay.
As mentioned above, Tll contains a C-terminal stretch of ∼25 amino acids that is well conserved in dipterans and vertebrates and that is essential for patterning activity in the embryo (Fig. 1). We asked if this CTD might suffice to mediate repression in the hb-h assay. We find that expression of a Hairy chimera containing the final 52 residues of the Tll LBD, HairyTll(CTD), does not confer female lethality (Fig. 2A and data not shown), suggesting that repression by the Tll LBD requires the integrity of this domain.
Expression of an obligate Tll repressor mimics the activity of endogenous Tll.
The preceding results indicate that the LBD is essential for Tll activity in the embryo and that this domain has the potential to mediate transcriptional repression, at least on the Sxl promoter. However, Tll has been implicated in the activation of various target genes in the early embryo (see the introduction), suggesting that it can also function as an activator of gene expression. To test this idea, we examined the effects of replacing the Tll LBD by a well-characterized repressor domain from the Engrailed protein, resulting in a protein chimera [TllEn(RD)] that should function as an obligate repressor (Fig. 3A). We reasoned that, if Tll normally exerts both activator and repressor functions, TllEn(RD) should mimic only the latter, causing opposite effects in cases where Tll acts as an activator.
We first compared the activities of Tll and TllEn(RD) by expressing both proteins under heat shock control. We find that TllEn(RD) induces cuticle phenotypes similar to those caused by the wild-type Tll protein. A 10-min heat shock during the mid-blastoderm stage causes severe segmental defects in >90% of hs-tllen(RD) embryos. These defects, as in the case of heat-shocked hs-tll embryos, consist mostly of strong suppression of abdominal and thoracic regions, as well as the occasional duplication of filzkörper structures (Fig. 3B).
To analyze in more detail the activity of TllEn(RD), we determined the responses of several genes that are affected by ectopic Tll expression. First, we examined the effects on Kr, a central gap gene that is efficiently repressed by hs-tll expression (53) (Fig. 3C and D). As expected, TllEn(RD) also causes repression of Kr in virtually all embryos; approximately 70% of them show complete elimination of Kr transcripts (Fig. 3E). We also observe similar repressor activities of Tll and TllEn(RD) on kni, another gap gene regulated by Tll (Fig. 3F to H). These results indicate that TllEn(RD) is able to mimic the normal repressor activities of Tll. They also suggest that TllEn(RD) can efficiently bind to target promoters in the absence of a functional LBD.
Next, we monitored the effects of TllEn(RD) on hb, a positive target of Tll. In stage 5 blatoderm embryos, hb expression resolves into a strong posterior stripe at about 15% egg length (EL; 0% being the posterior tip of the embryo), a central stripe at 55% EL, and a weak stripe at 75% EL (Fig. 3I). Expression of the posterior hb stripe requires endogenous tll function, and heat shock-induced expression of Tll leads to a marked expansion of this hb stripe towards the center of the embryo (3, 4, 53) (Fig. 3J). These observations, together with in vitro experiments showing binding of recombinant Tll protein to a specific enhancer in the hb promoter (32), suggest that Tll activates hb expression directly. Intriguingly, we find that TllEn(RD) also causes activation of the posterior hb stripe. In heat-shocked hs-tllen(RD) embryos, the hb stripe expands anteriorly to about 35 to 40% EL and we also see broadening of the central stripe (Fig. 3K). The activation of hb depends on the En(RD) domain, because Tll derivatives with a partially or completely deleted LBD are inactive (Fig. 1). These results suggest that activation of hb by TllEn(RD), and by Tll during normal development, is indirect and involves repression of one or more intermediary repressor genes.
We also assayed the effects of TllEn(RD) on byn, another positive target of Tll. byn is expressed as an early posterior cap that subsequently resolves into a stripe at 10% EL (Fig. 3L). Expression of byn requires endogenous Tll activity (25), and heat shock induction of Tll causes expansion of the byn stripe, reaching up to 15 to 20% EL on its anterior border (50) (Fig. 3M). Again, we see similar broadening of byn expression in response to TllEn(RD) (Fig. 3N). Thus, at least two different genes believed to be under direct positive control by Tll are activated by a constitutive repressor form of the protein, suggesting that these positive interactions represent secondary effects resulting from repression of repressors.
Expression of TllEn(RD) rescues the lack of endogenous Tll function.
We next carried out a more stringent test of TllEn(RD) function, the ability to complement tll mutant embryos. If TllEn(RD) is able to mimic Tll activities and these activities are only repressive, expression of TllEn(RD) might suffice to rescue the lack of endogenous Tll. For this experiment, the tllen(RD) coding sequences were placed under the control of a 5.9-kb promoter fragment sufficient to drive the endogenous tll expression pattern (45) (Fig. 4A). We obtained several independent lines carrying this construct, suggesting that it does not disrupt normal development. In situ hybridization experiments show that tllen(RD) is expressed in blastoderm embryos according to the endogenous tll pattern. However, expression of tllen(RD) appears weak and is also detected at low level throughout the embryo (cf. Fig. 4B and C). We speculate that this apparently ectopic expression prevents the recovery of lines producing higher levels of tllen(RD) transcripts.
We find that tllen(RD) rescues the patterning defects associated with the lack of tll function, resulting in the production of embryos with apparently normal terminal structures (Fig. 4E and F; see Materials and Methods). Many of the rescued embryos even hatch and live for 1 to 2 days, although they are unable to reach the larval LIII stage. Moreover, tll embryos carrying tllen(RD) show restored expression patterns of hb, byn, and fkh genes, considered to be normally activated by Tll (Fig. 4G to L). Taken together, our results indicate that Tll functions in the embryo exclusively as a repressor and that its positive effects on genes such as hb, byn, and fkh are the result of indirect interactions within the segmentation network.
Kni is partly responsible for the effects of Tll on hb and byn.
What might be the intermediary gene(s) between Tll and its positive target genes? Several lines of evidence support a role for kni in this context. First, kni encodes a zinc finger transcription factor with a well-characterized repressor function (1). Second, in tll mutant embryos kni expression expands towards the posterior pole, although this ectopic expression declines during stage 5 (40) (Fig. 5A). Third, ectopic expression of Kni under heat shock control causes partial or complete deletion of posterior terminal structures (47) (Fig. 5B), suggesting that Kni expression at the posterior pole blocks terminal patterning. Finally, misexpression of Kni in ventral regions of the embryo causes significant repression of the posterior hb stripe, and kni mutant embryos exhibit anteriorly expanded expression of this hb stripe (7).
We therefore tested if heat shock-induced expression of Kni represses hb, byn, or fkh. As shown in Fig. 5C, inducing Kni expression with the heat shock regimen described above leads to significant suppression of the posterior hb stripe, although residual expression persists in all embryos (compare with Fig. 3I). Likewise, Kni causes strong repression of byn (compare Fig. 3L and 5D) and, to a lesser extent, also of fkh (data not shown). These results are consistent with the hypothesis that kni is a likely intermediary gene accounting for at least some of the indirect activation functions of Tll in the blastoderm embryo.
To test this idea further, we monitored expression of hb, byn, and fkh in tll kni double-mutant embryos. If Kni-mediated repression is responsible for the reduced expression of these genes in tll mutants, expression of these genes should be restored in the double-mutant embryos. Indeed, such embryos show significant recovery of both hb and byn expression compared to tll single mutants (Fig. 5E to H). This recovery is not complete, however, indicating that other factors in addition to Kni contribute to the effects of Tll on hb and byn expression (compare Fig. 5G and H with 3I and L). In the case of fkh expression, we do not observe significant differences between tll single-mutant and tll kni double-mutant embryos (data not shown), again suggesting that Kni is not the sole factor responsible for the positive effects of Tll on endogenous target genes (see Discussion).
DISCUSSION
tll has long been known to play a key role in the establishment of unsegmented domains at the embryonic termini, and its activation in response to the Torso signaling pathway has been studied in detail. However, much less is known about the functional properties of the Tll product and how it regulates transcription of target genes. In this paper, we show that the LBD of Tll is essential for activity of the protein in the embryo and that this domain mediates strong repression in an in vivo transcriptional assay. We also find that converting the endogenous Tll protein into an obligate repressor preserves its patterning functions in the embryo. We conclude that Tll functions primarily as a transcriptional repressor and that positive genetic interactions of tll with specific targets arise from indirect regulatory events within the segmentation cascade.
Function of Tll as a dedicated repressor.
Our main evidence indicating that Tll is a dedicated repressor comes from the ability of TllEn(RD) to complement tll mutant embryos (Fig. 4). TllEn(RD) is sufficient to regulate the formation of terminal-body structures and also permits activation of secondary targets at the embryo poles. Although we have focused on events occurring at the posterior of the embryo, the ability of TllEn(RD) to support development up to larval stages suggests that Tll function in anterior regions also involves transcriptional repression. A similar conclusion applies for the role of Tll in brain development at later stages of embryogenesis (48, 60). It should be noted, however, that we cannot exclude a transcriptional-activator function of Tll during larval development or in the adult. Such an activator function would appear consistent with the inability of TllEn(RD) to rescue tll function to adulthood. Nevertheless, an alternative explanation for the latter result, which we favor, is that expression of the tllen(RD) transgene does not mimic the endogenous tll pattern exactly, eventually leading to insufficient or ectopic TllEn(RD) accumulation that blocks development (Fig. 4C).
Our results suggest that repression by Tll involves an active mechanism of action mediated by an intrinsic activity of the LBD. Thus, we find that two Tll derivatives carrying mutations in the LBD [Tll1−422 and TllE75(C1)] are largely inactive when expressed in the embryo (Fig. 1), whereas replacing the LBD by the En(RD) domain results in a fully functional protein (Fig. 3 and 4). Also, the Tll LBD mediates strong repression when fused to the DNA-binding domain of Hairy (Fig. 2). Both lines of evidence argue against a passive mechanism of Tll function whereby it simply prevents the binding of transcriptional activators to their DNA target sequences. Rather, it appears that Tll repression depends on protein-protein interactions of the LBD with other transcriptional cofactors. We do not know what these interactions are or if they involve components of the basal transcriptional apparatus or corepressor factors recruited to the promoter by the LBD. With respect to the latter possibility, genetic analyses do not support a role in Tll repression for global corepressors dCtBP and Groucho (Gro), known to associate with a variety of transcriptional repressors (reviewed in references 5 and 13). For example, dCtBP mutant embryos show normal expression of kni (37, 46), a gene that becomes derepressed in posterior regions when Tll repressor activity is absent. Also, in gro mutant embryos Tll becomes expressed in central regions of the embryo and is still able to repress Kr transcription (44). Finally, we have tested if Smrter, a demonstrated corepressor for the ecdysone nuclear receptor (EcR) (56), is involved in Tll repression and found that it is not (unpublished observations).
Given that Tll proteins represent a well-conserved subfamily of nuclear receptors, it is tempting to speculate that all members of this subfamily function exclusively as transcriptional repressors. For example, the mammalian Tlx protein, which regulates the development of brain limbic structures and the optic vesicle (35, 61), may also function as a negative regulator of gene expression. Indeed, a functional study with mice identified a role for Tlx in direct repression of the Pax2 gene during eye development (61). Also, expression of chick Tlx in Drosophila blastoderm embryos was shown to mediate efficient repression of kni similar to that caused by Tll (62). However, experiments carried out with Xenopus led to the conclusion that XTll normally mediates transcriptional activation during evagination of the eye vesicle (20). In that study, injection of mRNA encoding XTll fused to the Engrailed repressor domain blocked evagination of the optic vesicle, and this effect was reverted by coinjecting wild-type Xtll mRNA. The simplest interpretation of these results was that XTll normally has an activator function that is antagonized by the XTllEn(RD) repressor form (20). Future studies will probably identify additional roles and targets of vertebrate Tll proteins and establish a molecular basis for these regulatory interactions. Also, it will be interesting to compare the molecular activities of Tll with those of Dissatisfaction, a Drosophila Tll-related factor controlling neural development and sexual behavior (12).
Tll belongs to subfamily 2 of nuclear receptors (38). Several members of this family, including COUP-TF and TR2/TR4, regulatory proteins, appear to function predominantly as repressors (see, for example, references 6 and 29). However, another subfamily member, the mammalian HNF4 receptor, is known to activate several targets expressed in the liver and also in the kidney (reviewed in reference 51). Interestingly, COUP-TF and TR2/TR4 factors share well-conserved C1 motifs at the C termini of their LBDs, whereas this motif appears more diverged in HNF4. Because the C1 motif is essential for Tll activity, it is possible that this motif serves a specific repressor function. In contrast, the adjacent AF2 motif appears less important for Tll activity. In general, AF2 elements mediate association to coactivator factors and also control the switch between repressor and activator functions in hormone-regulated receptors (reviewed in references 34 and 36). Mutations in the AF2 motif have been shown to generate constitutive repressor forms of several receptors (17, 33, 63). Therefore, our results are consistent with the notion that Tll represents a true orphan receptor that functions primarily as a repressor.
Transcriptional repression during embryonic patterning.
Previous studies indicate that tll is required for expression of several downstream genes both in the blastoderm and at later stages of development (see the introduction) (60). Although there was little information about the mechanisms underlying these requirements, the simplest interpretation has been that they reflect direct activation by Tll. We find, however, that at least during embryonic and early larval stages Tll mediates positive interactions through the repression of repressors (Fig. 6). Specifically, our results indicate that Tll permits activation of hb and byn, at least in part, by repressing kni expression at the posterior of the embryo, and previous experiments have shown that this derepression circuit also controls activation of hairy stripe 7 (28). However, Kni appears to have a minor contribution to the regulation of fkh, because we do not observe upregulation of this gene in tll kni double-mutant embryos relative to tll single mutants (data not shown). This implies that other intermediary repressor factors mediate the positive genetic interactions of Tll on fkh. These factors could also influence hb and byn regulation, because expression of both genes is not restored to wild-type levels in tll kni double mutants (Fig. 5G and H). One such candidate repressor factor is Gt, whose expression expands posteriorly in tll mutants (27). However, this ectopic Gt expression does not reach the posterior pole of the embryo, and misexpression of Gt does not affect patterning posterior to the A6 segment (26).
FIG. 6.
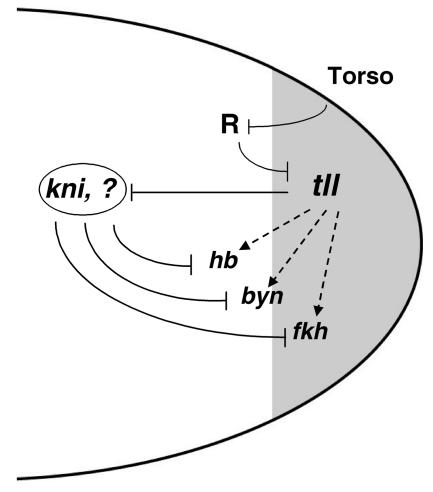
Model for the role of Tll in the regulation of gene expression at the posterior pole of the embryo. tll expression is first activated in a posterior cap (shadowed) as a result of localized Torso signaling. This activation is indirect and occurs through relief of repression: tll is silenced in middle regions of the embryo by a repressor complex (R) that is inhibited by the Torso signal at the pole, thus permitting localized tll activation by generally distributed factors. Subsequently, the Tll product acts as a repressor of central gap genes such as kni and gt and is also required for activation of secondary targets such as hb, byn, and fkh (dashed arrows). The latter function is again indirect and involves repression of kni and probably other intermediaries that mediate repression of hb, byn, and fkh. A similar model applies for the initial activation of hkb in response to Torso signaling, and the Hkb protein has been shown to act predominantly as a negative transcriptional regulator (see Discussion). Thus, repressive interactions define the regulatory network controlling terminus-specific gene expression at the posterior of the embryo.
Our results fit a growing consensus for the key role of transcriptional repression in developmental processes (8, 14, 16). Complex patterns of gene expression obviously require activation inputs, but the on/off borders of gene expression that characterize many developmental genes are often established via repression. Most transcriptional regulators that control pattern formation in the early Drosophila embryo behave as repressors. Others, including Tll and Hb, have been considered to act as both activators and repressors of gene expression, but our present results support an exclusive role of Tll in repression. This further emphasizes the crucial role of repression in embryonic terminal patterning (Fig. 6): Torso signaling mediates activation of tll and hkb via relief of repression (21, 30, 44), and both genes appear to encode repressor products. Like Tll, Hkb behaves as an activator of certain targets (e.g., fkh), but it seems likely that these positive effects are also indirect (see reference 15 and references therein). It thus appears that transcriptional repression provides a relatively simple and powerful mechanism for developing complex patterns of gene expression during evolution.
Acknowledgments
We thank Sergio González-Crespo and Ze'ev Paroush for many helpful discussions and critical comments on the manuscript. We are also grateful to Sergio Astigarraga for help and encouragement during this work and to Carlos Alonso, David Arnosti, Einat Cinnamon, Sergio González-Crespo, Marco Milán, Ze'ev Paroush, Ralf Pflanz, Paul Schedl, and the Bloomington Drosophila Stock Center for fly stocks and reagents.
This work was supported by the Spanish Ministerio de Educación y Ciencia, ICREA and the Generalitat de Catalunya. E.M. is a predoctoral fellow from the Parc Científic de Barcelona-CSIC.
REFERENCES
- 1.Arnosti, D. N., S. Gray, S. Barolo, J. Zhou, and M. Levine. 1996. The gap protein knirps mediates both quenching and direct repression in the Drosophila embryo. EMBO J. 15:3659-3666. [PMC free article] [PubMed] [Google Scholar]
- 2.Bronner, G., Q. Chu-LaGraff, C. Q. Doe, B. Cohen, D. Weigel, H. Taubert, and H. Jackle. 1994. Sp1/egr-like zinc-finger protein required for endoderm specification and germ-layer formation in Drosophila. Nature 369:664-668. [DOI] [PubMed] [Google Scholar]
- 3.Bronner, G., and H. Jackle. 1991. Control and function of terminal gap gene activity in the posterior pole region of the Drosophila embryo. Mech. Dev. 35:205-211. [DOI] [PubMed] [Google Scholar]
- 4.Casanova, J. 1990. Pattern formation under the control of the terminal system in the Drosophila embryo. Development 110:621-628. [DOI] [PubMed] [Google Scholar]
- 5.Chinnadurai, G. 2002. CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol. Cell 9:213-224. [DOI] [PubMed] [Google Scholar]
- 6.Chinpaisal, C., C. H. Lee, and L. N. Wei. 1998. Mechanisms of the mouse orphan nuclear receptor TR2-11-mediated gene suppression. J. Biol. Chem. 273:18077-18085. [DOI] [PubMed] [Google Scholar]
- 7.Clyde, D. E., M. S. Corado, X. Wu, A. Pare, D. Papatsenko, and S. Small. 2003. A self-organizing system of repressor gradients establishes segmental complexity in Drosophila. Nature 426:849-853. [DOI] [PubMed] [Google Scholar]
- 8.Courey, A. J., and S. Jia. 2001. Transcriptional repression: the long and the short of it. Genes Dev. 15:2786-2796. [DOI] [PubMed] [Google Scholar]
- 9.Dawson, S. R., D. L. Turner, H. Weintraub, and S. M. Parkhurst. 1995. Specificity for the hairy/enhancer of split basic helix-loop-helix (bHLH) proteins maps outside the bHLH domain and suggests two separable modes of transcriptional repression. Mol. Cell. Biol. 15:6923-6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diaz, R. J., R. Harbecke, J. B. Singer, F. Pignoni, W. Janning, and J. A. Lengyel. 1996. Graded effect of tailless on posterior gut development: molecular basis of an allelic series of a nuclear receptor gene. Mech. Dev. 54:119-130. [DOI] [PubMed] [Google Scholar]
- 11.Durand, B., M. Saunders, C. Gaudon, B. Roy, R. Losson, and P. Chambon. 1994. Activation function 2 (AF-2) of retinoic acid receptor and 9-cis retinoic acid receptor: presence of a conserved autonomous constitutive activating domain and influence of the nature of the response element on AF-2 activity. EMBO J. 13:5370-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finley, K. D., P. T. Edeen, M. Foss, E. Gross, N. Ghbeish, R. H. Palmer, B. J. Taylor, and M. McKeown. 1998. dissatisfaction encodes a tailless-like nuclear receptor expressed in a subset of CNS neurons controlling Drosophila sexual behavior. Neuron 21:1363-1374. [DOI] [PubMed] [Google Scholar]
- 13.Fisher, A. L., and M. Caudy. 1998. Groucho proteins: transcriptional corepressors for specific subsets of DNA-binding transcription factors in vertebrates and invertebrates. Genes Dev. 12:1931-1940. [DOI] [PubMed] [Google Scholar]
- 14.Gaston, K., and P. S. Jayaraman. 2003. Transcriptional repression in eukaryotes: repressors and repression mechanisms. Cell. Mol. Life Sci. 60:721-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldstein, R. E., G. Jiménez, O. Cook, D. Gur, and Z. Paroush. 1999. Huckebein repressor activity in Drosophila terminal patterning is mediated by Groucho. Development 126:3747-3755. [DOI] [PubMed] [Google Scholar]
- 16.Gray, S., and M. Levine. 1996. Transcriptional repression in development. Curr. Opin. Cell Biol. 8:358-364. [DOI] [PubMed] [Google Scholar]
- 17.Gurnell, M., J. M. Wentworth, M. Agostini, M. Adams, T. N. Collingwood, C. Provenzano, P. O. Browne, O. Rajanayagam, T. P. Burris, J. W. Schwabe, M. A. Lazar, and V. K. Chatterjee. 2000. A dominant-negative peroxisome proliferator-activated receptor gamma (PPARγ) mutant is a constitutive repressor and inhibits PPARγ-mediated adipogenesis. J. Biol. Chem. 275:5754-5759. [DOI] [PubMed] [Google Scholar]
- 18.Hauptmann, G., and T. Gerster. 1994. Two-color whole-mount in situ hybridization to vertebrate and Drosophila embryos. Trends Genet. 10:266. [DOI] [PubMed] [Google Scholar]
- 19.Hoch, M., N. Gerwin, H. Taubert, and H. Jackle. 1992. Competition for overlapping sites in the regulatory region of the Drosophila gene Kruppel. Science 256:94-97. [DOI] [PubMed] [Google Scholar]
- 20.Hollemann, T., E. Bellefroid, and T. Pieler. 1998. The Xenopus homologue of the Drosophila gene tailless has a function in early eye development. Development 125:2425-2432. [DOI] [PubMed] [Google Scholar]
- 21.Jiménez, G., A. Guichet, A. Ephrussi, and J. Casanova. 2000. Relief of gene repression by Torso RTK signaling: role of capicua in Drosophila terminal and dorsoventral patterning. Genes Dev. 14:224-231. [PMC free article] [PubMed] [Google Scholar]
- 22.Jiménez, G., Z. Paroush, and D. Ish-Horowicz. 1997. Groucho acts as a corepressor for a subset of negative regulators, including Hairy and Engrailed. Genes Dev. 11:3072-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiménez, G., S. M. Pinchin, and D. Ish-Horowicz. 1996. In vivo interactions of the Drosophila Hairy and Runt transcriptional repressors with target promoters. EMBO J. 15:7088-7098. [PMC free article] [PubMed] [Google Scholar]
- 24.Keyes, L. N., T. W. Cline, and P. Schedl. 1992. The primary sex determination signal of Drosophila acts at the level of transcription. Cell 68:933-943. [DOI] [PubMed] [Google Scholar]
- 25.Kispert, A., B. G. Herrmann, M. Leptin, and R. Reuter. 1994. Homologs of the mouse Brachyury gene are involved in the specification of posterior terminal structures in Drosophila, Tribolium, and Locusta. Genes Dev. 8:2137-2150. [DOI] [PubMed] [Google Scholar]
- 26.Kraut, R., and M. Levine. 1991. Mutually repressive interactions between the gap genes giant and Krüppel define middle body regions of the Drosophila embryo. Development 111:611-621. [DOI] [PubMed] [Google Scholar]
- 27.Kraut, R., and M. Levine. 1991. Spatial regulation of the gap gene giant during Drosophila development. Development 111:601-609. [DOI] [PubMed] [Google Scholar]
- 28.La Rosee, A., T. Hader, H. Taubert, R. Rivera-Pomar, and H. Jackle. 1997. Mechanism and Bicoid-dependent control of hairy stripe 7 expression in the posterior region of the Drosophila embryo. EMBO J. 16:4403-4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leng, X., A. J. Cooney, S. Y. Tsai, and M. J. Tsai. 1996. Molecular mechanisms of COUP-TF-mediated transcriptional repression: evidence for transrepression and active repression. Mol. Cell. Biol. 16:2332-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liaw, G. J., K. M. Rudolph, J. D. Huang, T. Dubnicoff, A. J. Courey, and J. A. Lengyel. 1995. The torso response element binds GAGA and NTF-1/Elf-1, and regulates tailless by relief of repression. Genes Dev. 9:3163-3176. [DOI] [PubMed] [Google Scholar]
- 31.Mangelsdorf, D. J., C. Thummel, M. Beato, P. Herrlich, G. Schutz, K. Umesono, B. Blumberg, P. Kastner, M. Mark, P. Chambon, and R. M. Evans. 1995. The nuclear receptor superfamily: the second decade. Cell 83:835-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Margolis, J. S., M. L. Borowsky, E. Steingrimsson, C. W. Shim, J. A. Lengyel, and J. W. Posakony. 1995. Posterior stripe expression of hunchback is driven from two promoters by a common enhancer element. Development 121:3067-3077. [DOI] [PubMed] [Google Scholar]
- 33.Marimuthu, A., W. Feng, T. Tagami, H. Nguyen, J. L. Jameson, R. J. Fletterick, J. D. Baxter, and B. L. West. 2002. TR surfaces and conformations required to bind nuclear receptor corepressor. Mol. Endocrinol. 16:271-286. [DOI] [PubMed] [Google Scholar]
- 34.McKenna, N. J., and B. W. O'Malley. 2002. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108:465-474. [DOI] [PubMed] [Google Scholar]
- 35.Monaghan, A. P., D. Bock, P. Gass, A. Schwager, D. P. Wolfer, H. P. Lipp, and G. Schutz. 1997. Defective limbic system in mice lacking the tailless gene. Nature 390:515-517. [DOI] [PubMed] [Google Scholar]
- 36.Nettles, K. W., and G. L. Greene. 2005. Ligand control of coregulator recruitment to nuclear receptors. Annu. Rev. Physiol. 67:309-333. [DOI] [PubMed] [Google Scholar]
- 37.Nibu, Y., H. Zhang, E. Bajor, S. Barolo, S. Small, and M. Levine. 1998. dCtBP mediates transcriptional repression by Knirps, Krüppel and Snail in the Drosophila embryo. EMBO J. 17:7009-7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nuclear Receptors Nomenclature Committee. 1999. A unified nomenclature system for the nuclear receptor superfamily. Cell 97:161-163. [DOI] [PubMed] [Google Scholar]
- 39.Pankratz, M. J., M. Busch, M. Hoch, E. Seifert, and H. Jackle. 1992. Spatial control of the gap gene knirps in the Drosophila embryo by posterior morphogen system. Science 255:986-989. [DOI] [PubMed] [Google Scholar]
- 40.Pankratz, M. J., M. Hoch, E. Seifert, and H. Jackle. 1989. Krüppel requirement for knirps enhancement reflects overlapping gap gene activities in the Drosophila embryo. Nature 341:337-340. [DOI] [PubMed] [Google Scholar]
- 41.Pankratz, M. J., and H. Jackle. 1993. Blastoderm segmentation, p. 467-516. In M. Bate and A. Martinez-Arias (ed.), The development of Drosophila melanogaster. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Parkhurst, S. M., D. Bopp, and D. Ish-Horowicz. 1990. X:A ratio, the primary sex-determining signal in Drosophila, is transduced by helix-loop-helix proteins. Cell 63:1179-1191. [DOI] [PubMed] [Google Scholar]
- 43.Parkhurst, S. M., and P. M. Meneely. 1994. Sex determination and dosage compensation: lessons from flies and worms. Science 264:924-932. [DOI] [PubMed] [Google Scholar]
- 44.Paroush, Z., S. M. Wainwright, and D. Ish-Horowicz. 1997. Torso signalling regulates terminal patterning in Drosophila by antagonising Groucho-mediated repression. Development 124:3827-3834. [DOI] [PubMed] [Google Scholar]
- 45.Pignoni, F., R. M. Baldarelli, E. Steingrimsson, R. J. Diaz, A. Patapoutian, J. R. Merriam, and J. A. Lengyel. 1990. The Drosophila gene tailless is expressed at the embryonic termini and is a member of the steroid receptor superfamily. Cell 62:151-163. [DOI] [PubMed] [Google Scholar]
- 46.Poortinga, G., M. Watanabe, and S. M. Parkhurst. 1998. Drosophila CtBP: a Hairy-interacting protein required for embryonic segmentation and hairy-mediated transcriptional repression. EMBO J. 17:2067-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rothe, M., M. Pehl, H. Taubert, and H. Jackle. 1992. Loss of gene function through rapid mitotic cycles in the Drosophila embryo. Nature 359:156-159. [DOI] [PubMed] [Google Scholar]
- 48.Rudolph, K. M., G. J. Liaw, A. Daniel, P. Green, A. J. Courey, V. Hartenstein, and J. A. Lengyel. 1997. Complex regulatory region mediating tailless expression in early embryonic patterning and brain development. Development 124:4297-4308. [DOI] [PubMed] [Google Scholar]
- 49.Segraves, W. A., and D. S. Hogness. 1990. The E75 ecdysone-inducible gene responsible for the 75B early puff in Drosophila encodes two new members of the steroid receptor superfamily. Genes Dev. 4:204-219. [DOI] [PubMed] [Google Scholar]
- 50.Singer, J. B., R. Harbecke, T. Kusch, R. Reuter, and J. A. Lengyel. 1996. Drosophila brachyenteron regulates gene activity and morphogenesis in the gut. Development 122:3707-3718. [DOI] [PubMed] [Google Scholar]
- 51.Sladek, F. M., and S. Seidel. 2001. Hepatocyte nuclear factor 4α, p. 309-361. In T. P. Burris and E. McCabe (ed.), Nuclear receptors and genetic disease. Academic Press, London, United Kingdom.
- 52.Spradling, A. C. 1986. P element-mediated transformation, p. 175-197. In D. B. Roberts (ed.), Drosophila: a practical approach. IRL Press, Oxford, United Kingdom.
- 53.Steingrímsson, E., F. Pignoni, G. J. Liaw, and J. A. Lengyel. 1991. Dual role of the Drosophila pattern gene tailless in embryonic termini. Science 254:418-421. [DOI] [PubMed] [Google Scholar]
- 54.Strecker, T. R., J. R. Merriam, and J. A. Lengyel. 1988. Graded requirement for the zygotic terminal gene, tailless, in the brain and tail region of the Drosophila embryo. Development 102:721-734. [DOI] [PubMed] [Google Scholar]
- 55.Struffi, P., M. Corado, M. Kulkarni, and D. N. Arnosti. 2004. Quantitative contributions of CtBP-dependent and -independent repression activities of Knirps. Development 131:2419-2429. [DOI] [PubMed] [Google Scholar]
- 56.Tsai, C. C., H. Y. Kao, T. P. Yao, M. McKeown, and R. M. Evans. 1999. SMRTER, a Drosophila nuclear receptor coregulator, reveals that EcR-mediated repression is critical for development. Mol. Cell 4:175-186. [DOI] [PubMed] [Google Scholar]
- 57.Weigel, D., G. Jurgens, M. Klingler, and H. Jackle. 1990. Two gap genes mediate maternal terminal pattern information in Drosophila. Science 248:495-498. [DOI] [PubMed] [Google Scholar]
- 58.Weigel, D., G. Jurgens, F. Kuttner, E. Seifert, and H. Jackle. 1989. The homeotic gene fork head encodes a nuclear protein and is expressed in the terminal regions of the Drosophila embryo. Cell 57:645-658. [DOI] [PubMed] [Google Scholar]
- 59.Younger-Shepherd, S., H. Vaessin, E. Bier, L. Y. Jan, and Y. N. Jan. 1992. deadpan, an essential pan-neural gene encoding an HLH protein, acts as a denominator in Drosophila sex determination. Cell 70:911-922. [DOI] [PubMed] [Google Scholar]
- 60.Younossi-Hartenstein, A., P. Green, G. J. Liaw, K. Rudolph, J. Lengyel, and V. Hartenstein. 1997. Control of early neurogenesis of the Drosophila brain by the head gap genes tll, otd, ems, and btd. Dev. Biol. 182:270-283. [DOI] [PubMed] [Google Scholar]
- 61.Yu, R. T., M. Y. Chiang, T. Tanabe, M. Kobayashi, K. Yasuda, R. M. Evans, and K. Umesono. 2000. The orphan nuclear receptor Tlx regulates Pax2 and is essential for vision. Proc. Natl. Acad. Sci. USA 97:2621-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu, R. T., M. McKeown, R. M. Evans, and K. Umesono. 1994. Relationship between Drosophila gap gene tailless and a vertebrate nuclear receptor Tlx. Nature 370:375-379. [DOI] [PubMed] [Google Scholar]
- 63.Zhang, J., X. Hu, and M. A. Lazar. 1999. A novel role for helix 12 of retinoid X receptor in regulating repression. Mol. Cell. Biol. 19:6448-6457. [DOI] [PMC free article] [PubMed] [Google Scholar]