Abstract
Premature termination (nonsense) codons trigger rapid mRNA decay by the nonsense-mediated mRNA decay (NMD) pathway. Two conserved proteins essential for NMD, UPF1 and UPF2, are phosphorylated in higher eukaryotes. The phosphorylation and dephosphorylation of UPF1 appear to be crucial for NMD, as blockade of either event in Caenorhabditis elegans and mammals largely prevents NMD. The universality of this phosphorylation/dephosphorylation cycle pathway has been questioned, however, because the well-studied Saccharomyces cerevisiae NMD pathway has not been shown to be regulated by phosphorylation. Here, we used in vitro and in vivo biochemical techniques to show that both S. cerevisiae Upf1p and Upf2p are phosphoproteins. We provide evidence that the phosphorylation of the N-terminal region of Upf2p is crucial for its interaction with Hrp1p, an RNA-binding protein that we previously showed is essential for NMD. We identify specific amino acids in Upf2p's N-terminal domain, including phosphorylated serines, which dictate both its interaction with Hrp1p and its ability to elicit NMD. Our results indicate that phosphorylation of UPF1 and UPF2 is a conserved event in eukaryotes and for the first time provide evidence that Upf2p phosphorylation is crucial for NMD.
Cells have evolved many quality control mechanisms to eliminate aberrant proteins and mRNAs that interfere with normal cellular functions. One such mechanism is the nonsense-mediated mRNA decay (NMD) pathway, which eliminates mRNAs that contain premature termination (nonsense) codons within the protein coding region, thereby preventing the synthesis of truncated proteins with dominant-negative and deleterious gain-of-function activities (3, 7, 14, 16, 24, 42, 47). The importance of this surveillance mechanism is underscored by its conservation in a wide variety of organisms, including Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila melanogaster, plants, and mammals (12, 13, 16, 25, 27, 34, 38).
Several genes essential for NMD have been identified in yeast, most notably UPF1, UPF2, and UPF3, all of which destabilize nonsense codon-containing mRNAs without affecting the decay rate of most wild-type mRNAs (16, 25). The yeast UPF1 gene encodes the protein Upf1p, which has RNA-binding and RNA-dependent ATPase/helicase activities (8, 9, 45). Yeast UPF3 encodes the basic protein Upf3p, which harbors several nuclear localization and nuclear export signals that allow the protein to shuttle between the nucleus and the cytoplasm (39, 40). Yeast UPF2 encodes the adaptor protein Upf2p, which forms a complex with both Upf1p and Upf3p (6, 19). Single or multiple deletions of each of these three UPF genes produce similar effects on mRNA decay, consistent with the notion that the Upf proteins function as a molecular complex in a single pathway (19).
The NMD pathway is elicited by recognition of a nonsense codon, but the precise features distinguishing a premature termination codon from a bona fide stop codon remain unknown. In S. cerevisiae, one model for NMD suggests that the degradation machinery discriminates between a premature and a normal termination codon through a loosely conserved downstream sequence element (DSE) located 3′ of the premature termination codon (36, 50). In this model, transcripts that contain a premature termination codon but lack a DSE are not degraded by the NMD pathway (36). We previously identified the RNA-binding protein Hrp1p as a factor that specifically binds to the DSE in the PGK1 transcript in yeast (15). We found that Hrp1p also interacts with Upf1p and that mutant forms of Hrp1p that are unable to specifically bind the downstream sequence element or Upf1p are unable to promote rapid degradation of nonsense codon-containing PGK1 mRNAs (15). Hrp1p also appears to have a general role in NMD, as it was recently shown to also direct the decay of premature termination codon-bearing PPR1 transcripts (22).
In higher eukaryotes, phosphorylation of UPF1 is crucial for NMD. This was initially revealed by genetic and biochemical studies in C. elegans suggesting that its UPF1 orthologue, SMG-2, must undergo cycles of phosphorylation and dephosphorylation to function in NMD (32). These cycles are maintained by proteins that promote either its phosphorylation (SMG-1, SMG-3, and SMG-4) or its dephosphorylation (SMG-5, SMG-6, and SMG-7) (2, 32). A recent study showed that SMG-1 is a phosphatidylinositol-related kinase that directly phosphorylates SMG-2 in vitro and is essential for NMD in vivo (17). Biochemical and in vivo RNA interference depletion studies suggest that mammalian NMD also depends on a cycle of UPF1 phosphorylation and dephosphorylation regulated by other NMD proteins (10, 31, 33, 41, 46, 49).
While phosphorylation has been shown to be crucial for NMD in worms and mammalian cells, nothing is known about the role of phosphorylation in NMD in S. cerevisiae, a widely used model organism to study this surveillance pathway. No study has definitively identified yeast Upf1p as a phosphoprotein, and there have been no reports identifying S. cerevisiae SMG-1, SMG-5, SMG-6, or SMG-7 orthologues that could phosphorylate or dephosphorylate yeast Upf1p. This has led to the hypothesis that, unlike higher organisms, S. cerevisiae does not require a cycle of UPF1 phosphorylation and dephosphorylation to elicit NMD (12). Another unaddressed question is whether phosphorylation of NMD proteins other than UPF1 is essential to elicit NMD. Human UPF2 was recently shown to be a phosphoprotein (5), but the functional relevance of its phosphorylation has not yet been determined.
In this study, we addressed both of these issues. Using in vitro and in vivo biochemical experiments, we found that yeast Upf1p and Upf2p are both phosphoproteins. We provide several lines of evidence that Upf2p's phosphorylation status regulates both its function and its ability to interact with Hrp1p, an RNA-binding protein essential for S. cerevisiae NMD. Our study supports the notion that NMD is a conserved, phosphorylation-regulated response, and it provides evidence for a specific role of UPF2 phosphorylation in NMD.
MATERIALS AND METHODS
Strains, growth conditions, and plasmids.
S. cerevisiae strain KC2 (MATα ura3-52 trp1 leu2-2 tyr7-1 his4-38 met14) was used as the wild-type strain. This strain and the KC2 strains harboring a deletion of the chromosomal copy of either UPF1 or UPF2 have been described previously (44).
The yeast 2μm plasmid pG-1 was used as the vector (44). Yeast transformations were performed by the lithium acetate method (37). The FLAG-UPF2 allele has been described previously (44). The upf2 alleles used in this study were constructed by site-directed mutagenesis (15) using the FLAG-UPF2 allele as the template. All constructs were confirmed by DNA sequencing.
Immunoprecipitation experiments.
Cells were grown in 15-ml cultures to an optical density (600 nm) of 0.7 to 0.8 and lysed with glass beads in lysis buffer (20 mM HEPES, pH 7.5, 100 mM KOAc, 5 mM EDTA, and 20% glycerol) containing 1 mM phenylmethylsulfonyl fluoride and 2 μg/ml protease inhibitors. The extract (100 μg) was mixed with 10 μl anti-FLAG M2 monoclonal antibody (Sigma, St. Louis, Mo.) immobilized on protein A-Sepharose beads (Sigma, St. Louis, Mo.) and incubated at 4°C for 3 h with constant rocking. The beads were then washed five times with cold lysis buffer, and the associated proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and detected by Western blotting. To assess the role of RNA in the protein-protein interactions, lysates were treated with various concentrations of RNase A for 20 minutes at room temperature prior to the immunoprecipitations. Where indicated, the same blot was probed first with an anti-Hrp1p polyclonal antibody (kindly provided by Michael F. Henry), followed by anti-FLAG M2 monoclonal (Sigma), anti-Upf1p (kindly provided by Allan Jacobson), and anti-Upf2p (from the NHLBI Program for Genomic Applications-Antibody Production Facility, The University of Texas Southwestern Medical Center at Dallas) antibodies. The percentage of Hrp1p binding of each mutant Upf2 protein was calculated by comparing the amount of Hrp1p being precipitated to that obtained with vector (0%) and wild-type Upf2p (100%). The amount of precipitated Hrp1p was standardized to the amount of FLAG-Upf2p. The values shown represent averages from five independent experiments. Phosphatase treatments were performed by incubating immunopurified FLAG-Upf2p (40 ng) with 10 U of calf intestinal phosphatase for 30 min at 30°C. Reactions were electrophoresed in an 8% SDS-polyacrylamide gel and analyzed by Western blot hybridization using an anti-Upf2p polyclonal antibody.
RNA isolation and Northern blot analysis.
Total RNA was isolated, and mRNA abundance was determined by Northern blotting (15). Random-primed DNA probes were prepared from a 0.6-kb EcoRI-HindIII fragment spanning a region of the CYH2 mRNA. The values shown represent averages from three independent experiments. The activity of each mutant Upf2p protein in NMD was calculated by comparing the ratio of pre-CYH2 to CYH2 in upf2Δ strains transformed with the mutant allele to those for strains transformed with either vector only (0%) or wild-type Upf2p (100%).
GST pull-down experiments.
Upf1p and Upf2p were purified as FLAG fusion proteins from yeast using anti-FLAG M2 beads (Sigma) as described previously (9, 44). Glutathione S-transferase (GST)-Hrp1p was purified from Escherichia coli as described previously (15). All purified proteins were dialyzed in storage buffer (25 mM Tris, pH 7.5, 50 mM KCl, 1 mM dithiothreitol, and 20% glycerol), aliquoted, and frozen at −70°C.
Purified GST-Hrp1p (1 μg) was immobilized on 10 μl glutathione-Sepharose 4B beads and combined with the indicated amount of purified FLAG-tagged proteins in 0.5 ml of Ipp150 buffer (10 mM Tris, pH 8.0, 150 mM NaCl, and 0.1% Nonidet P-40). The mixtures were incubated at 4°C for 1.5 h with constant rocking. For some experiments, 5 μg of RNase A was included in the incubation mixture. In other experiments, 15 μg of RNase A was added to the mixture after the 1.5-h incubation at 4°C, and then the mixture was incubated for another 30 min. The beads were then washed three times with 1 ml of cold Ipp150 buffer. The FLAG-tagged proteins that remained associated with the beads were released by boiling in the loading buffer, resolved by 12% SDS-PAGE, and transferred to a polyvinylidene fluoride membrane for Western blotting. FLAG-tagged proteins were visualized with anti-FLAG antibody.
In vivo labeling experiments.
Cultures were grown in low-phosphate medium (35) at 30°C to an optical density (600 nm) of 0.6, after which 1 mCi of [32P]orthophosphate was added to the medium for 90 min. Cells were lysed with glass beads in lysis buffer containing 1 mM phenylmethylsulfonyl fluoride, 2 μg/ml protease inhibitors, and 100 nM okadaic acid (Calbiochem, San Diego, Calif.). The extract (400 μg) was mixed with 30 μl of either anti-Upf2p or anti-FLAG antibodies immobilized on protein A-Sepharose beads. Protein A-agarose beads alone were used as control for nonspecific binding. The mixtures were incubated at 4°C for 3 h with constant rocking. The beads were then washed, and the associated proteins were resolved by SDS-PAGE and subjected to Western blotting. Increasing concentrations of FLAG peptide (0 to 0.5 μg/μl) were added to the immunoprecipitations to test the specificity of the affinity matrix against FLAG-Upf1p. The immunoprecipitated proteins were transferred to a polyvinylidene difluoride membrane, and a Western blot assay was performed using the anti-FLAG monoclonal antibody. The amount of labeling was standardized with the amount of protein being precipitated. The values shown represent averages from two independent experiments. For the Upf2 experiments, the percent phosphorylation of each upf2 mutant was calculated by comparing the adjusted labeling efficiency with that obtained with vector only (0%) and wild-type Upf2p (100%). The results of these experiments were quantitated using a Bio-Rad Molecular Imager FX (Bio-Rad, Hercules, Calif.).
RESULTS
Hrp1p interacts with Upf2p, an essential component of NMD.
Our previous studies showed that Hrp1p is essential for NMD and that it interacts with both the NMD factor Upf1p and the DSE required for NMD (15). To better understand how Hrp1p functions in NMD, we assessed whether Hrp1p also interacts with the NMD factor Upf2p. We isolated cytoplasmic extracts from a yeast strain expressing FLAG-Upf2p in place of endogenous Upf2p. The extracts were immunoprecipitated with an anti-FLAG monoclonal antibody followed by Western blot analysis with an anti-Hrp1p antibody. This analysis revealed that Upf2p efficiently coimmunoprecipitated with Hrp1p (Fig. 1A, lane 4). The interactions were specific, as no Hrp1p was immunoprecipitated when a vector containing only FLAG was expressed (lane 3). For positive and negative controls, we probed the same Western blot with anti-Upf1p and anti-Pgk1p antibodies, respectively. These experiments showed that Upf2p specifically interacts with Upf1p but fails to coimmunoprecipitate with Pgk1p (lane 4, anti-Upf1p and anti-Pgk1p).
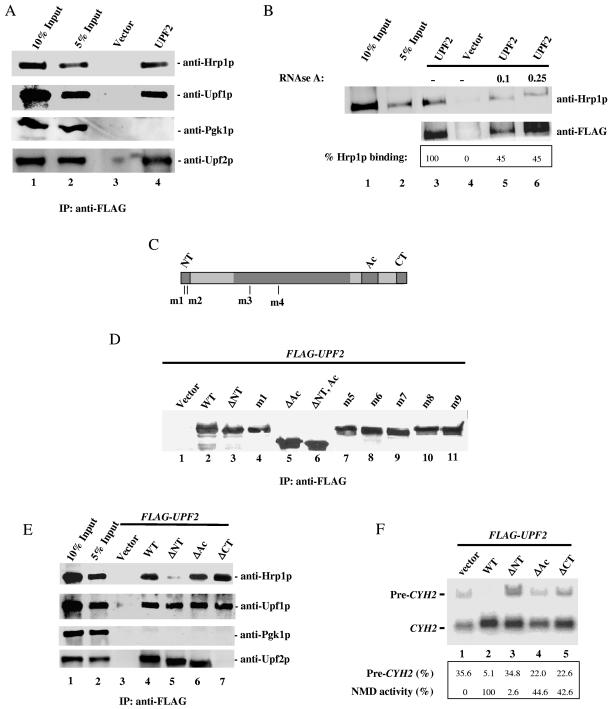
FIG. 1.
Hrp1p and Upf2p interact. (A) Coimmunoprecipitation of Hrp1p and Upf2p. Cytoplasmic extracts from strains transformed with FLAG-UPF2 or vector only were immunoprecipitated (IP) with anti-FLAG monoclonal antibody and immunoblotted with anti-Hrp1p, anti-Upf1p (positive control), or anti-Pgk1p (negative control) antibodies. The anti-Upf2p antibody was used to demonstrate the amount of Upf2p immunoprecipitated in the assay. Lanes 1 and 2 contain 10% and 5% of the total extract used for immunoprecipitation, respectively. (B) The assay was performed as described for panel A except that cytoplasmic extracts were treated with RNase A (μg/μl concentration indicated) for 20 minutes at room temperature prior to immunoprecipitation with anti-FLAG antibody. High concentrations of RNase A were used to ensure that the RNA present in the cytoplasmic extracts was completely degraded. (C) Schematic diagram of Upf2p, showing the NT, Ac, and CT domains. The positions of the mutations in the m1 to m4 mutants are indicated. (D) Western blot analysis performed as described for panel A, demonstrating that the mutant Upf2p proteins shown are expressed at a level similar to that of wild-type Upf2p (WT). (E) Coimmunoprecipitation of mutant Upf2p proteins and Hrp1p. Cytoplasmic extracts from yeast upf2Δ strains expressing the indicated FLAG-UPF2 constructs were immunoprecipitated as described for panel A. The anti-Upf2p antibody used in these experiments recognizes a specific peptide located in the C-terminal domain of Upf2p. These amino acids are not present in the ΔCT mutant protein used in these coimmunoprecipitation studies (see lane 7, anti-Upf2p). (F) NMD activity of Upf2p mutants determined using Northern blot analysis of total cellular RNA from the upf2Δ strains in panel E. The migration of the nonsense codon-containing CYH2 pre-mRNA (pre-CYH2) and CYH2 mature mRNA (CYH2) bands is indicated. The ratio of pre-CYH2 to CYH2 mRNA gives the magnitude of NMD (quantified by phosphorimaging).
To address whether the interaction between Upf2p and Hrp1p is only because they both bind RNA, cytoplasmic extracts were treated with RNase A prior to immunoprecipitation. For these experiments, we used various concentrations of RNase A (0.1 μg/ml to 0.5 μg/ml) to ensure that the RNA present in the cytoplasmic extract was degraded to the fullest extent possible (Fig. 1B and data not shown). The interaction between Upf2p and Hrp1p was only modestly reduced by RNase treatment (compare lane 3 with lanes 5 and 6), indicating that a significant proportion of Upf2p and Hrp1p interact in vivo independently of RNA.
We also assessed the interaction between Hrp1p and Upf2p using yeast two-hybrid analysis. Full-length HRP1 and UPF2 were fused in frame to the Gal4p DNA-binding domain and activation domain, respectively. Both plasmids were transformed into a yeast strain that contained an integrated GAL1-lacZ reporter plasmid. This assay revealed a reproducible interaction between Hrp1p and Upf2p (data not shown).
The NT domain of Upf2p interacts with Hrp1p.
We next sought to identify the portion of Upf2p responsible for its interaction with Hrp1p. Upf2p contains a highly conserved N-terminal domain (NT domain; amino acids [aa] 30 to 50), an acidic domain that interacts with the release factor eRF3 (Ac domain; aa 833 to 936), and a C-terminal domain that interacts with Upf1p (CT domain; aa 937 to 1089) (Fig. 1C) (19). To determine whether any of these domains are essential for Upf2p to interact with Hrp1p, wild-type and mutant upf2 alleles lacking these domains were expressed as FLAG fusion proteins in a upf2Δ strain. These mutant proteins were expressed at levels comparable to that of wild-type Upf2p (Fig. 1D and data not shown). Cytoplasmic extracts were immunoprecipitated with an anti-FLAG antibody, and the immunoprecipitated proteins were analyzed by Western blotting with anti-Hrp1p, anti-Upf1p (positive control), anti-Pgk1p (negative control), and anti-Upf2p antibodies (Fig. 1E). We found that deletion of the NT domain significantly reduced the ability of Upf2p to interact with Hrp1p (Fig. 1E, lane 5), while deletion of the Ac or CT domain had no effect (lanes 6 and 7). Deletion of the NT domain had no effect on the ability of Upf2p to interact with Upf1p, Upf3p, or eRF3 (lane 5, anti-Upf1p, and data not shown), further demonstrating specificity.
The NT domain of Upf2p is essential for NMD.
We next assessed whether the Hrp1p interaction domain of Upf2p is essential for NMD. Northern blot analysis was conducted on total cellular RNA isolated from a upf2Δ strain transformed with wild-type or various mutant upf2 alleles. NMD was assessed by measuring the ratio of CYH2 pre-mRNA to mature mRNA, as previous studies have shown that inefficiently spliced CYH2 pre-mRNA is a natural target of the NMD pathway and that its ratio with mature CYH2 mRNA is a reliable indicator of NMD activity (19, 22). The wild-type UPF2 gene fully complemented the NMD defect in a upf2Δ strain (Fig. 1F, compare lanes 1 and 2). In contrast, the mutant lacking the NT domain was virtually inactive in complementing the defect (lane 3). This complete lack of NMD was notable because Upf2p mutants lacking the eRF3-interacting (lane 4) or Upf1p-interacting (lane 5) domains still had some NMD-promoting activity, albeit less than normal.
Upf2p-Hrp1p interactions are promoted by Upf1p and RNA.
To determine whether the interaction between Upf2p and Hrp1p is direct or instead requires other NMD components, we performed in vitro GST pull-down experiments using purified GST-Hrp1p immobilized on glutathione-Sepharose beads. Consistent with our previous findings (15), we observed binding of purified FLAG-Upf1p to immobilized GST-Hrp1p (Fig. 2A, lane 1). In contrast, FLAG-Upf2p was unable to detectably bind to GST-Hrp1p (Fig. 2A, lane 3). Thus, the interaction between Upf2p and Hrp1p is indirect, suggesting that it requires additional factors present in the cytoplasmic extracts (Fig. 1B and E). In agreement with this, we found that the addition of Upf1p allowed Upf2p to interact with Hrp1p (lane 5). This interaction was enhanced by the addition of poly(U) RNA (lane 6) but not poly(A) or poly(C) (data not shown).
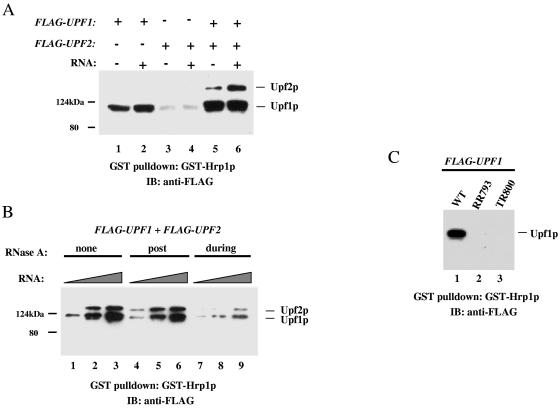
FIG. 2.
Upf2p-Hrp1p interactions promoted by Upf1p and RNA. (A) GST pull-down experiment in which immobilized GST-Hrp1p is incubated with FLAG-Upf1p and/or FLAG-Upf2p and/or poly(U) RNA. Bound proteins were detected with anti-FLAG antibody. The weak band with a size similar to that of FLAG-Upf1p in lanes 3 and 4 is GST-Hrp1p, which is weakly recognized by the anti-FLAG antibody. (B) Experiment performed as described for panel A except that RNase A (0.25 μg/μl) was added either during or after (post) complex formation (see Materials and Methods). The amount of poly(U) RNA added was 2, 20, or 200 ng. (C) Immobilized GST-Hrp1p was incubated with wild-type UPF1 or an RNA-binding-deficient UPF1 mutant (RR793 or TR800), and their interactions were examined as described for panel A.
To investigate how poly(U) RNA enhances the interaction between Upf2p and Hrp1p, we performed RNase experiments. We found that RNase A treatment had little or no effect on complex formation when GST-Hrp1p, Upf1p, Upf2p, and/or RNA was preincubated before RNase was added (Fig. 2B, compare lanes 1 to 3 with 4 to 6). In contrast, RNase A treatment during the incubation strongly inhibited complex formation (lanes 7 to 9). This indicates that the RNA is not merely serving as a substrate that physically links these NMD proteins together. Instead, our results were consistent with poly(U) RNA serving to bind and influencing the behavior of Upf1p, as poly(U) was previously shown to be the preferred RNA substrate for Upf1p (45). To directly test this, we performed GST pull-down experiments with two upf1 mutants that are unable to bind RNA (RR793 and TR800) (45). We found that both mutants lost their ability to bind to Hrp1p in vitro (Fig. 2C) and in vivo (coimmunoprecipitation experiments; data not shown). Taken together, our results in Fig. 1 and 2 support a simple model positing that, upon RNA binding, Upf1p acquires the ability to form a stable complex with Hrp1p and Upf2p by a mechanism requiring the N-terminal region of Upf2p.
Specific N-terminal amino acids of Upf2p crucial for NMD and its interaction with Hrp1p.
Our next goal was to identify the amino acids in the N-terminal region of Upf2p required for NMD and the formation of a Upf2p/Hrp1p/Upf1p/RNA quaternary complex. Towards this end, we first replaced the three residues in the Asp-Ser-Ser (DSS) motif, which is conserved in both S. cerevisiae Upf2p and human UPF2 (Fig. 3A; aa 31 to 33 in yeast), with Ala residues to determine whether this motif dictates Upf2p's interaction with Hrp1p. This substitution (mutant m1) dramatically reduced the ability of Upf2p to interact with Hrp1p (Fig. 3B, compare lanes 4 and 5), while having no effect on the interaction between Upf2p and Upf1p (lanes 4 and 5, anti-Upf1p).
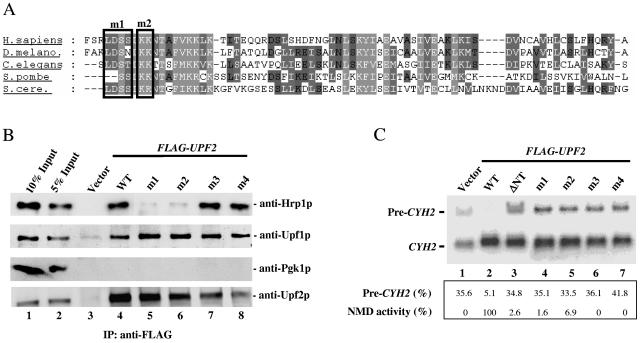
FIG. 3.
Specific N-terminal amino acids of Upf2p crucial for NMD and its interaction with Hrp1p. (A) Alignment of the N-terminal region of UPF2 homologues from S. cerevisiae, Schizosaccharomyces pombe, C. elegans, D. melanogaster, and Homo sapiens. Black boxes represent the positions of amino acid substitutions in the m1 and m2 mutants (Fig. 6A and the text indicate the substitutions made). (B) Coimmunoprecipitation of mutant Upf2p proteins and Hrp1p, performed as described for Fig. 1A. The Western blot was first probed with an anti-Hrp1p antibody, followed by anti-Upf1p (positive control) and anti-Pgk1p (negative control) antibodies. The mutant proteins were expressed at levels virtually identical to those of wild-type Upf2p, as determined by Western blot analysis (Fig. 1D and data not shown). In addition, Western blot analysis showed that the expression of Hrp1p does not vary in these different strains (data not shown). (C) NMD activity of the Upf2p mutants shown, determined using Northern blot analysis of total cellular RNA (performed as for Fig. 1F) from the upf2Δ strains used in panel B.
We next examined the role of the two conserved, positively charged Lys and Arg residues immediately downstream of the DSS motif (Fig. 3A). The m2 mutant lacking these two amino acids (KR→AA) also interacted poorly with Hrp1p (lane 4), although better than did the m1 mutant (lane 3). For comparison, we tested the effect of point mutations disrupting conserved residues in the middle region of Upf2p (mutants m3 [aa 348 and 349; FY→AA] and m4 [aa 479 and 480; KF→AA]; Fig. 1B). Neither mutation had a substantial effect on the ability of Upf2p to interact with Hrp1p or Upf1p (Fig. 3B, lanes 7 and 8, anti-Hrp1p and anti-Upf1p).
We next assessed whether the amino acid substitutions in the m1 and m2 mutants disrupted NMD. We found that neither mutant was able to restore NMD to a Upf2p-deficient strain (Fig. 3C, lanes 4 and 5). The m3 and m4 mutants were also incapable of restoring NMD (lanes 6 and 7), but since they could interact with Hrp1p (Fig. 3B), this suggests that the amino acids disrupted in the m3 and m4 mutants are responsible for interacting with an NMD protein other than Hrp1p. Taken together, our data pinpoint conserved Upf2p residues essential for NMD, including those in the N-terminal domain that mediate the interaction of Upf2p with Hrp1p.
Upf1p and Upf2p are phosphorylated in vivo.
The observation that a motif containing two Ser residues is crucial for Upf2p to support NMD and interact with Hrp1p (Fig. 3) suggested the possibility that Upf2p is a phosphoprotein. We investigated this in wild-type S. cerevisiae grown in low-phosphate medium and labeled with [32P]orthophosphate for 90 min (35). Cytoplasmic extracts were prepared in the presence of the phosphatase inhibitor okadaic acid and immunoprecipitated with an anti-Upf2p antibody. The immunoprecipitated proteins were separated by SDS-PAGE and detected by phosphorimaging. Two labeled proteins were detected; the major band migrated at the size predicted for Upf2p (Fig. 4A, lane 1). To assess whether one or both of these bands were Upf2p, the same experiment was performed with either a upf2Δ strain or a upf2Δ strain transformed with a FLAG-UPF2 allele. The upf2Δ strain yielded no bands (lane 2), whereas the strain rescued with the FLAG-UPF2 allele had the same two bands as did wild-type cells (lane 3). Incubation of purified Upf2p with calf intestine phosphatase caused a marked shift in the migration of the upper band (Fig. 4B), providing more evidence that Upf2p is a phosphoprotein. The identity of the minor band is uncertain, but it is likely to be a C-terminally truncated product of Upf2p, as a upf2Δ strain transformed with a FLAG-UPF2 gene missing the N-terminal domain lacked this band and instead expressed a smaller band (Fig. 4C, lane 3).
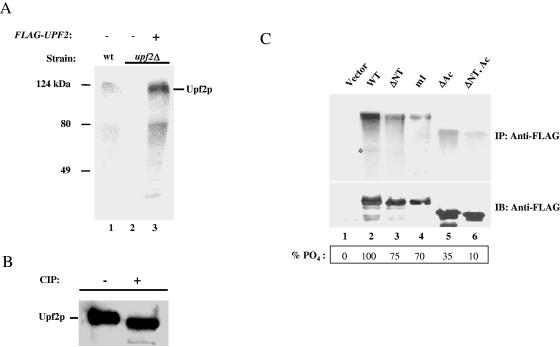
FIG. 4.
Upf2p is phosphorylated in vivo. (A) A wild-type strain (lane 1), a upf2Δ strain (lane 2), and a upf2Δ strain expressing FLAG-Upf2p (lane 3) were labeled with [32P]orthophosphate and immunoprecipitated with anti-Upf2p polyclonal antibody. Labeled proteins were detected by phosphorimaging. (B) Immunopurified FLAG-Upf2p was incubated with or without calf intestinal phosphatase (CIP) and analyzed by Western blotting with anti-Upf2p polyclonal antibody. (C) In vivo labeling experiment, performed as described for panel A, with yeast upf2Δ strains expressing the indicated upf2 constructs. The assay was performed as described for panel A, except that the cytoplasmic extracts were immunoprecipitated with anti-FLAG antibody and detected by autoradiography (upper panel) or by Western blotting with anti-FLAG antibody (lower panel). The asterisk indicates an N-terminally truncated Upf2p product.
We next determined which regions of Upf2p are phosphorylated. We found that the aforementioned N-terminal-deleted mutant had a ∼30% reduction in phosphate labeling compared to wild-type Upf2p (Fig. 4C, compare lanes 3 and 2). Strikingly, the m1 mutant also had a ∼30% reduction in phosphorylation (lane 4), strongly suggesting that one or both of the Ser residues lost in this mutant are the site of N-terminal phosphorylation (Fig. 3A). However, because deletion of the N-terminal domain did not entirely abolish Upf2p phosphorylation, we predicted that the acidic domain of Upf2p is also phosphorylated, as it has three Ser residues in a context appropriate for phosphorylation by casein kinase II (18). In agreement with this, we found that deletion of the acidic domain caused a ∼65% reduction in Upf2p phosphorylation (Fig. 4C, lane 5). A double truncation of both the N-terminal and acidic domains almost completely abolished Upf2p phosphorylation (lane 6), indicating that these two regions harbor most of the phosphorylation sites in Upf2p.
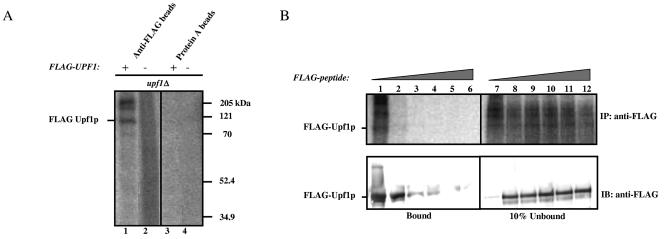
The discovery that yeast Upf2p is a phosphoprotein prompted us to assess whether the same is also true of yeast Upf1p. We found that a upf1Δ strain overexpressing FLAG-Upf1p expressed several phosphate-labeled proteins, including one that migrated at a size similar to that of Upf1p (110 kDa; Fig. 5A, lane 1). No labeled protein was immunoprecipitated with anti-FLAG antibody from the upf1Δ strain transformed with the vector alone (lane 2), nor was any protein brought down with the protein A-agarose matrix alone (lanes 3 and 4). The additional proteins immunoprecipitated with Upf1p by the anti-FLAG antibody (lane 1) are probably proteins that copurify with Upf1p. The identity of these proteins is currently under investigation.
FIG. 5.
Yeast Upf1p is phosphorylated in vivo. (A) In vivo labeling experiment performed as described for Fig. 4A using a yeast upf1Δ strain that expresses FLAG-Upf1p and a negative control that does not. (B) Increasing concentrations of FLAG peptide (0 to 400 ng/μl) were added to immunoprecipitations performed as described for panel A to test the specificity of the affinity matrix against FLAG-Upf1p (upper panel). The immunoprecipitated proteins were transferred to a polyvinylidene fluoride membrane and probed with anti-FLAG monoclonal antibody (lower panel).
To further assess specificity, we incubated the immunoprecipitated fractions with increasing concentrations of FLAG peptide. We found that this peptide specifically competed with FLAG-Upf1p for binding to the anti-FLAG antibody (Fig. 5B, upper panel). As an additional test of specificity, we transferred the immunoprecipitated proteins to a polyvinylidene fluoride membrane and performed Western blot analysis using the anti-FLAG antibody. These results confirmed that the immunoprecipitated protein band competed by the FLAG peptide was indeed Upf1p (Fig. 5B, bottom panel).
Functional role of the N-terminal Ser residues in Upf2p.
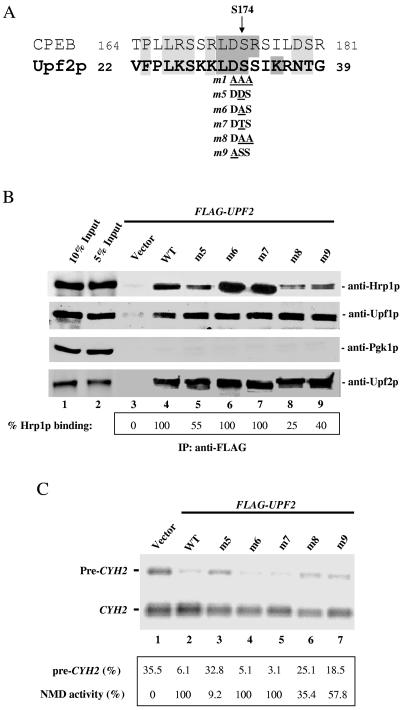
The discovery that the Upf2p m1 mutant was deficient in phosphorylation (Fig. 4C), was unable to interact with Hrp1p (Fig. 3B), and was unable to elicit NMD (Fig. 3C) suggested that one or more of the three amino acids disrupted in this mutant mediate phosphorylation-dependent interactions with Hrp1p that are essential for NMD. It seemed likely that one or both Ser residues in this motif are sites of phosphorylation, as they are in a context that highly resembles a key phosphorylated region in the Xenopus laevis cytoplasmic polyadenylation element binding (CPEB) protein (Fig. 6A) (28). To examine the role of these Ser residues, we subjected them to various mutations. Mutation of both Ser32 and Ser33 (LDSS→LDAA; mutant m8) reduced the ability of Upf2p to interact with Hrp1p by fourfold (Fig. 6B, lane 8) while having no effect on the interaction of Upf2p with Upf1p (lane 8, anti-Upf1p). In contrast, mutation of only Ser32 (LDSS→LDAS or LDTS; mutant m6 and m7, respectively) had no detectable effect (lanes 6 and 7), implying that Ser32 and Ser33 have redundant roles or that Ser33 has a secondary role. Support for the notion that Ser32 is crucial was the finding that mutation of Ser32 to Asp (LDSS→LDDS; mutant m5), which mimics a constitutively phosphorylated form of Ser, reduced the ability of Upf2p to interact with Hrp1p (lane 5). In contrast, this amino acid substitution did not impair the ability of Upf2p to interact with Upf1p (lane 5, anti-Upf1p). We also mutated the residue upstream of the two Ser residues, Asp31, as an Asp at this position was previously shown to regulate serine phosphorylation in CPEB (28). This substitution (LDSS→LASS; mutant m9) also reduced the interaction of Upf2p with Hrp1p, while having no effect on the interaction of Upf2p withUpf1p (Fig. 6B, lane 9). Collectively, these data suggest the importance of Asp31, Ser32, and Ser33 for the interaction of Upf2p with Hrp1p. However, none of the mutations affecting only one or two of these amino acids completely abolished this interaction (Fig. 6B), whereas mutation of all three amino acids did (Fig. 3B, lane 5), suggesting that the Ser and Asp residues in the Upf2p N-terminal domain can partially compensate for each other's loss.
FIG. 6.
The N-terminal domain of Upf2p harbors a phosphorylation site. (A) Sequence alignment of the phosphorylation site of Xenopus CPEB protein and the N-terminal region phosphorylated in S. cerevisiae Upf2p. (B) Coimmunoprecipitation of mutant Upf2p proteins with Hrp1p, Upf1p, and Pgk1p, performed as described for Fig. 1A. The percentage of Hrp1p interacting with the FLAG-tagged Upf2p mutants (relative to wild-type [WT]) was calculated as described in Materials and Methods. (C) NMD activity of the Upf2p mutants shown, determined using Northern blot analysis of total cellular RNA (performed as described for Fig. 1D) from the upf2Δ strains used in panel B.
We next analyzed the ability of these various Upf2p mutants to elicit NMD. We found that only the mutants deficient in Hrp1p binding (m5, m8, and m9) had impaired ability to complement a chromosomal UPF2 deletion mutant (Fig. 6C, lanes 3, 6, and 7). In contrast, the mutants that were competent to interact with Hrp1p (m6 and m7) fully rescued NMD (Fig. 6C, lanes 4 and 5). Interestingly, the mutant that was the most deficient in NMD (m5; Fig. 6C, lane 3) displayed only a modest impairment in its interaction with Hrp1p (Fig. 6B). Because this mutant had an amino acid substitution that mimics a constitutively phosphorylated residue, this is consistent with the possibility that Upf2p must complete a cycle of phosphorylation and dephosphorylation to elicit NMD. We conclude that the DSS region in the NT domain of Upf2p is phosphorylated in vivo, is essential for Upf2p to interact with Hrp1p, and is required for NMD.
DISCUSSION
The Upf2p NT domain interacts with Hrp1p and is essential for NMD.
In this study, we demonstrated that the NMD factor Upf2p interacts with the RNA-binding protein Hrp1p (Fig. 1). We went on to show that the NT domain in Upf2p that mediates this interaction is essential for NMD (Fig. 1). Despite its high degree of conservation, the NT domain of Upf2p had not previously been assigned a function. This region harbors a nuclear localization signal consensus sequence (18), but Upf2p does not accumulate in yeast nuclei at steady state (24, 39, 40). In contrast, mammalian UPF2 has been reported to be present in the nuclear fraction of mammalian cells, but whether it is actually present in nuclei or whether it merely cofractionates with nuclei is unclear; at steady state UPF2 accumulates primarily in the region of the cytoplasm directly adjacent to nuclei (24, 26, 31).
Our site-specific mutation analysis identified highly conserved amino acids within the NT region of Upf2p (aa 30 to 33 and 35 and 36) that are crucial for both NMD and the interaction of Upf2p with Hrp1p (Fig. 3). This suggests that the ability of Upf2p and Hrp1p to interact is a functionally important step in eliciting NMD. Our in vitro studies revealed that this interaction also requires the NMD protein Upf1p and poly(U) RNA. The function of poly(U) RNA may be to activate Upf1p, as two UPF1 mutants incapable of binding RNA also lacked the ability to interact with Hrp1p in vitro and in vivo (Fig. 2). It is possible that RNA also binds and alters the properties of Upf2p, as one of the two Upf2p regions that we identified as essential for NMD is a basic patch (Lys35 and Arg36) (21).
Upf1p and Upf2p are phosphoproteins.
In this report, we demonstrated that yeast Upf2p is phosphorylated in the NT domain that mediates interactions with Hrp1p (Fig. 4). The ability of Upf2p to be phosphorylated appears to be conserved, as it was recently demonstrated that human UPF2 is also a phosphoprotein (5). Mutation of the highly conserved Ser32 and Ser33 residues in the NT domain reduced the incorporation of labeled orthophosphate into yeast Upf2p (Fig. 4C), indicating that one or both of these Ser residues are targets of phosphorylation. This phosphorylation event appears to be functionally relevant, as mutation of these Ser residues inhibited both Upf2p's interaction with Hrp1p and its ability to trigger NMD (Fig. 6). Further investigation into the phosphorylation of each of these serine residues, their effects on the interaction with Hrp1p, and their ability to elicit NMD will be required to understand their precise functional role.
The phosphorylated region of the Upf2p N-terminal domain resembles the previously characterized phosphorylated region of the Xenopus CPEB protein (35), an RNA-binding protein that stimulates the polyadenylation and translational activation of c-mos mRNA during oocyte maturation (29). The polyadenylation-inducing activity of CPEB requires that it be phosphorylated on Ser174 by the Eg2 kinase (28). This Ser residue is in a context very similar to that of the Ser32 and Ser33 residues in Upf2p (Fig. 6A).
Replacement of Ser32 in Upf2p with an amino acid that mimics a constitutively phosphorylated form of Upf2p was virtually inactive in NMD (mutant m5, Fig. 6C). This suggests the possibility that Upf2p must undergo a phosphorylation/dephosphorylation cycle to function in NMD, just as several lines of evidence suggest is the case for UPF1 (31, 46, 49). Such a phosphorylation cycle may be required for Upf2p to associate and dissociate from Hrp1p. This possibility is consistent with our finding that the DSS region that undergoes phosphorylation is absolutely required for Upf2p to interact with Hrp1p (Fig. 3B). However, we cannot rule out that the Ser-to-Asp alteration in the m5 mutant prevents NMD by a mechanism that is independent of phosphorylation. Additional studies, including an analysis of the three-dimensional structure of Upf2p in the phosphorylated and nonphosphorylated states, will be required to address this issue. In addition to the NT domain, the Ac domain of Upf2p appears to be phosphorylated, as deletion of the acidic domain reduced Upf2p phosphorylation by ∼65% (Fig. 4). We identified three Ser residues in the Ac domain that matched the criteria for a casein kinase II substrate site. Phosphorylation of these residues may regulate interactions of Upf2p with other components of the NMD pathway.
In addition to Upf2p, we found that Upf1p is a substrate for phosphorylation (Fig. 5), as was reported by another group while the manuscript was in preparation (11). This finding is significant, as the lack of previous evidence for yeast Upf1p phosphorylation had prompted some investigators to suggest that the NMD pathway in S. cerevisiae is controlled by a mechanism independent of Upf1p phosphorylation (12). Together with our Upf2p phosphorylation data, our results challenge this view and establish phosphorylation as a conserved feature of the NMD pathway in eukaryotes.
Phosphorylation of UPF1 was first demonstrated in C. elegans (32). The phosphorylation status of its UPF1 ortholog SMG-2 is altered by mutation of the essential nematode NMD genes smg-1, smg-5, smg-6, and smg-7 (2, 32). smg-1 encodes a phosphatidylinositol 3-kinase-related protein kinase that directly phosphorylates SMG-2, whereas smg-5 and smg-7 encode proteins that participate in the dephosphorylation of SMG-2 by forming a protein complex with the structural and catalytic subunits of protein phosphatase 2A (2, 17). Human UPF1 also alternates between phosphorylated and dephosphorylated states (31). It is phosphorylated by human SMG-1 (10, 49) and dephosphorylated by a mechanism mediated by the human SMG-5/7 phosphatase complex (5). Inhibition or depletion of either SMG-1 or the SMG-5/7 complex prevents NMD, suggesting that the cycle of UPF1 phosphorylation and dephosphorylation mediated by these molecules is essential for NMD (31, 46, 49). Orthologs of SMG-1, SMG-5, and SMG-7 were recently identified in D. melanogaster and shown to be essential for NMD (12). Collectively, these data provide strong evidence that UPF1 phosphorylation and dephosphorylation are conserved features of NMD in both lower and higher eukaryotes.
It will be important to identify the factors that mediate phosphorylation and dephosphorylation of S. cerevisiae Upf1p and Upf2p. Candidates include yeast protein kinases related to SMG-1 (Tor1p, Tor2p, Tel1p, and Mec1p) and a protein similar to SMG-5 (Tos5p) (2).
Models.
Previous studies in S. cerevisiae have led investigators to propose two primary models that explain how the NMD pathway discriminates between premature and normal translation termination events. One model posits that, for NMD to be elicited, a premature termination codon must be positioned 5′ of a downstream cis element called a DSE (see the introduction) found in most yeast mRNAs (36, 50). The DSE interacts with Hrp1p, which in turn interacts with the NMD protein Upf1p to trigger NMD (15). The central feature of this surveillance model is that NMD is elicited when the Hrp1p-bound DSE is recognized by a putative surveillance complex that assembles after the translation apparatus recognizes a stop codon (9, 43). Evidence suggests that the surveillance complex assembles and functions in the following manner (9, 44, 45). Upon recognition of a stop codon, the translation termination factors eRF1 and eRF3 are recruited to the ribosome, where they recruit Upf1p to form a complex. After hydrolysis of the peptidyl-tRNA bond, eRF1 dissociates from the complex, allowing Upf2p to join. This rearrangement of the complex results in the dissociation of eRF3, which activates the ATPase/helicase and RNA-binding activities of Upf1p and allows formation of a mature surveillance complex. This mature complex scans 3′ of the stop codon; if the stop codon is positioned upstream of a DSE, the Upf1p/Upf2p-containing surveillance complex collides with and binds to Hrp1p bound to the DSE, thereby activating NMD.
The results presented herein allow us to expand upon this surveillance model. We posit that the molecular interaction that we observed between Upf2p and Hrp1p is crucial for the final step in NMD, when the Upf2p-containing surveillance complex encounters Hrp1p bound to the DSE. We further posit that phosphorylation of the Upf2p NT domain promotes this interaction between Upf2p and Hrp1p. This is supported by our finding that the Upf2p-Hrp1p interaction was reduced as a result of mutations of Ser32 and Ser33 in the Upf2p NT domain (Fig. 6B) and completely abolished when these two amino acids and the one immediately upstream were mutated (Fig. 3B). We suggest that the phosphorylation of Upf2p occurs at an early step, such as surveillance complex assembly, to prime Upf2p for its later interaction with Hrp1p. Upf2p does not act alone to interact with Hrp1p, as we showed that RNA and Upf1p are also required for this interaction (Fig. 2). Moreover, our results suggested that the NT domain of Upf2p is required to form a stable quaternary complex between Upf1p, Upf2p, Hrp1p, and RNA and that this complex functions in an essential step of NMD. How RNA-bound Upf1p promotes this interaction and whether it also regulates Upf2p phosphorylation or dephosphorylation remain to be determined. The phosphorylation state of Upf1p may also be crucial for this molecular choreography, as recent studies indicate that the phosphorylation of human UPF1 may regulate its interaction with human UPF2 (4).
The second model proposes that NMD is triggered by the intrinsically aberrant nature of premature translation termination (20). According to this model, a premature termination codon generates an extended 3′-untranslated region (UTR) that does not drive the appropriate ribonucleoprotein remodeling events required to generate a stable mRNA. This “faux 3′-UTR” model is supported by recent studies from Jacobson and colleagues who used “toeprinting” analysis to determine the position of ribosomes on yeast transcripts containing either a normal 3′ UTR or an aberrant 3′ UTR generated by premature translation termination (1). The aberrant 3′ UTR blocked the normal release of ribosomes and promoted their aberrant migration to upstream AUG codons. If correct, we suggest that our data allow the faux 3′-UTR model to be further refined. In particular, our data support the notion that Hrp1p is crucial for normal ribonucleoprotein remodeling and that this normal function is disrupted when Hrp1p interacts with Upf2p in aberrant mRNAs harboring premature termination codons. Support for the notion that Hrp1p is involved in normal ribonucleoprotein remodeling comes from the finding that Hrp1p promotes 3′-cleavage-polyadenylation events and is linked with mRNA export (23, 30, 48).
It remains for future experiments to test these two models and to elucidate why phosphorylated UPF proteins are required to elicit NMD. Our study has shown that S. cerevisiae is a good model system to address these issues and uncover the mechanism by which phosphorylation events dictate the interactions between components of the NMD pathway to trigger rapid mRNA decay.
Acknowledgments
We are grateful to Yadiel Alameda for his help with the two-hybrid assay, Edwin García for help with immunoprecipitations, Allan Jacobson for the anti-Upf1p antibody, and Michael F. Henry for the anti-Hrp1p antibody. We also express our gratitude to the NHLBI Program for Genomic Applications-Antibody Production Facility, at The University of Texas Southwestern Medical Center at Dallas, for the anti-Upf2p antibody used in these studies. We thank Carol Wilusz for critical reading of the manuscript and helpful comments and members of the González laboratory for helpful discussions.
This work was supported by grants from the National Institutes of Health to C.I.G. (GM008102-3052, KO1 HL-04355-05, and U54 CA96297-03), M.F.W. (GM058595 and U54 CA96297-03), and S.W.P. (GM48631).
REFERENCES
- 1.Amrani, N., R. Ganesan, S. Kervestin, D. A. Mangus, S. Ghosh, and A. Jacobson. 2004. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432:112-118. [DOI] [PubMed] [Google Scholar]
- 2.Anders, K. R., A. Grimson, and P. Anderson. 2003. SMG-5, required for C. elegans nonsense-mediated mRNA decay, associates with SMG-2 and protein phosphatase 2A. EMBO J. 22:641-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, K. E., and R. Parker. 2004. Nonsense-mediated mRNA decay: terminating erroneous gene expression. Curr. Opin. Cell Biol. 16:293-299. [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharya, A., K. Czaplinski, P. Trifillis, F. He, A. Jacobson, and S. W. Peltz. 2000. Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA 6:1226-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiu, S. Y., G. Serin, O. Ohara, and L. E. Maquat. 2003. Characterization of human Smg5/7a: a protein with similarities to Caenorhabditis elegans SMG5 and SMG7 that functions in the dephosphorylation of Upf1. RNA 9:77-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui, Y., K. W. Hagan, S. Zhang, and S. W. Peltz. 1995. Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes Dev. 9:423-436. [DOI] [PubMed] [Google Scholar]
- 7.Culbertson, M. R. 1999. RNA surveillance. Unforeseen consequences for gene expression, inherited genetic disorders and cancer. Trends Genet. 15:74-80. [DOI] [PubMed] [Google Scholar]
- 8.Czaplinski, K., Y. Weng, K. W. Hagan, and S. W. Peltz. 1995. Purification and characterization of the Upf1 protein: a factor involved in translation and mRNA degradation. RNA 1:610-623. [PMC free article] [PubMed] [Google Scholar]
- 9.Czaplinski, K., M. J. Ruiz-Echevarria, S. V. Paushkin, X. Han, Y. Weng, H. A. Perlick, H. C. Dietz, M. D. Ter-Avanesyan, and S. W. Peltz. 1998. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 12:1665-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denning, G., L. Jamieson, L. E. Maquat, E. A. Thompson, and A. P. Fields. 2001. Cloning of a novel phosphatidylinositol kinase-related kinase: characterization of the human SMG-1 RNA surveillance protein. J. Biol. Chem. 276:22709-22714. [DOI] [PubMed] [Google Scholar]
- 11.de Pinto, B., R. Lippolis, R. Castaldo, and N. Altamura. 2004. Overexpression of Upf1p compensates for mitochondrial splicing deficiency independently of its role in mRNA surveillance. Mol. Microbiol. 51:1129-1142. [DOI] [PubMed] [Google Scholar]
- 12.Gatfield, D., L. Unterholzner, F. D. Ciccarelli, P. Bork, and E. Izaurralde. 2003. Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J. 22:3960-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gatfield, D., and E. Izaurralde. 2004. Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature 429:575-578. [DOI] [PubMed] [Google Scholar]
- 14.Gehring, N. H., G. Neu-Yilik, T. Schell, M. W. Hentze, and A. E. Kulozik. 2003. Y14 and hUpf3b form an NMD-activating complex. Mol. Cell 11:939-949. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez, C. I., M. J. Ruiz-Echevarria, S. Vasudevan, M. F. Henry, and S. W. Peltz. 2000. The yeast hnRNP-like protein Hrp1/Nab4 marks a transcript for nonsense-mediated mRNA decay. Mol. Cell 5:489-499. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez, C. I., A. Bhattacharya, W. Wang, and S. W. Peltz. 2001. Nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Gene 274:15-25. [DOI] [PubMed] [Google Scholar]
- 17.Grimson, A., S. O'Connor, C. L. Newman, and P. Anderson. 2004. SMG-1 is a phosphatidylinositol kinase-related protein kinase required for nonsense-mediated mRNA decay in Caenorhabditis elegans. Mol. Cell. Biol. 24:7483-7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He, F., and A. Jacobson. 1995. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev. 9:437-454. [DOI] [PubMed] [Google Scholar]
- 19.He, F., A. H. Brown, and A. Jacobson. 1997. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol. Cell. Biol. 17:1580-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hilleren, P., and R. Parker. 1999. mRNA surveillance in eukaryotes: kinetic proofreading of proper translation termination as assessed by mRNP domain organization? RNA 5:711-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kadlec, J., E. Izaurralde, and S. Cusack. 2004. The structural basis for the interaction between nonsense-mediated mRNA decay factors UPF2 and UPF3. Nat. Struct. Mol. Biol. 11:330-337. [DOI] [PubMed] [Google Scholar]
- 22.Kebaara, B., T. Nazarenus, R. Taylor, A. Forch, and A. L. Atkin. 2003. The Upf-dependent decay of wild-type PPR1 mRNA depends on its 5′-UTR and first 92 ORF nucleotides. Nucleic Acids Res. 31:3157-3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kessler, M. M., M. F. Henry, E. Shen, J. Zhao, S. Gross, P. A. Silver, and C. L. Moore. 1997. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes Dev. 11:2545-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lykke-Andersen, J., M. D. Shu, and J. A. Steitz. 2000. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 103:1121-1131. [DOI] [PubMed] [Google Scholar]
- 25.Maquat, L. E., and G. Serin. 2001. Nonsense-mediated mRNA decay: insights into mechanism from the cellular abundance of human Upf1, Upf2, Upf3, and Upf3X proteins. Cold Spring Harbor Symp. Quant. Biol. 66:313-320. [DOI] [PubMed] [Google Scholar]
- 26.Mendell, J. T., S. M. Medghalchi, R. G. Lake, E. N. Noensie, and H. C. Dietz. 2000. Novel Upf2p orthologues suggest a functional link between translation initiation and nonsense surveillance complexes. Mol. Cell. Biol. 20:8944-8957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mendell, J. T., C. M. ap Rhys, and H. C. Dietz. 2002. Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science 298:419-422. [DOI] [PubMed] [Google Scholar]
- 28.Mendez, R., L. E. Hake, T. Andresson, L. E. Littlepage, J. V. Ruderman, and J. D. Richter. 2000. Phosphorylation of CPE binding factor by Eg2 regulates translation of c-mos mRNA. Nature 404:302-307. [DOI] [PubMed] [Google Scholar]
- 29.Mendez, R., and J. D. Richter. 2001. Translational control by CPEB: a means to the end. Nat. Rev. Mol. Cell Biol. 2:521-529. [DOI] [PubMed] [Google Scholar]
- 30.Minvielle-Sebastia, L., K. Beyer, A. M. Krecic, R. E. Hector, M. S. Swanson, and W. Keller. 1998. Control of cleavage site selection during mRNA 3′ end formation by a yeast hnRNP. EMBO J. 17:7454-7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohnishi, T., A. Yamashita, I. Kashima, T. Schell, K. R. Anders, A. Grimson, T. Hachiya, M. W. Hentze, P. Anderson, and S. Ohno. 2003. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol. Cell 12:1187-1200. [DOI] [PubMed] [Google Scholar]
- 32.Page, M. F., B. Carr, K. R. Anders, A. Grimson, and P. Anderson. 1999. SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol. Cell. Biol. 19:5943-5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pal, M., Y. Ishigaki, E. Nagy, and L. E. Maquat. 2001. Evidence that phosphorylation of human Upfl protein varies with intracellular location and is mediated by a wortmannin-sensitive and rapamycin-sensitive PI 3-kinase-related kinase signaling pathway. RNA 7:5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pulak, R., and P. Anderson. 1993. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 7:1885-1897. [DOI] [PubMed] [Google Scholar]
- 35.Rubin, G. M. 1973. The nucleotide sequence of Saccharomyces cerevisiae 5.8 S ribosomal ribonucleic acid. J. Biol. Chem. 248:3860-3875. [PubMed] [Google Scholar]
- 36.Ruiz-Echevarria, M. J., and S. W. Peltz. 1996. Utilizing the GCN4 leader region to investigate the role of the sequence determinants in nonsense-mediated mRNA decay. EMBO J. 15:2810-2819. [PMC free article] [PubMed] [Google Scholar]
- 37.Schiestl, R. H., and R. D. Gietz. 1989. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 16:339-346. [DOI] [PubMed] [Google Scholar]
- 38.Serin, G., A. Gersappe, J. D. Black, R. Aronoff, and L. E. Maquat. 2001. Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4). Mol. Cell. Biol. 21:209-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shirley, R. L., M. J. Lelivelt, L. R. Schenkman, J. N. Dahlseid, and M. R. Culbertson. 1998. A factor required for nonsense-mediated mRNA decay in yeast is exported from the nucleus to the cytoplasm by a nuclear export signal sequence. J. Cell Sci. 111:3129-3143. [DOI] [PubMed] [Google Scholar]
- 40.Shirley, R. L., A. S. Ford, M. R. Richards, M. Albertini, and M. R. Culbertson. 2002. Nuclear import of Upf3p is mediated by importin-alpha/-beta and export to the cytoplasm is required for a functional nonsense-mediated mRNA decay pathway in yeast. Genetics 161:1465-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Unterholzner, L., and E. Izaurralde. 2004. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol. Cell 16:587-596. [DOI] [PubMed] [Google Scholar]
- 42.Wagner, E., and J. Lykke-Andersen. 2002. mRNA surveillance: the perfect persist. J. Cell Sci. 115:3033-3038. [DOI] [PubMed] [Google Scholar]
- 43.Wang, J., Y. F. Chang, J. I. Hamilton, and M. F. Wilkinson. 2002. Nonsense-associated altered splicing: a frame-dependent response distinct from nonsense-mediated decay. Mol. Cell 10:951-957. [DOI] [PubMed] [Google Scholar]
- 44.Wang, W., K. Czaplinski, Y. Rao, and S. W. Peltz. 2001. The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J. 20:880-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weng, Y., K. Czaplinski, and S. W. Peltz. 1998. ATP is a cofactor of the Upf1 protein that modulates its translation termination and RNA binding activities. RNA 4:205-214. [PMC free article] [PubMed] [Google Scholar]
- 46.Wilkinson, M. F. 2003. The cycle of nonsense. Mol. Cell 12:1059-1061. [DOI] [PubMed] [Google Scholar]
- 47.Wilkinson, M. F. 2005. A new function for nonsense-mediated mRNA-decay factors. Trends Genet. 21:143-148. [DOI] [PubMed] [Google Scholar]
- 48.Xu, C., and M. F. Henry. 2004. Nuclear export of hnRNP Hrp1p and nuclear export of hnRNP Npl3p are linked and influenced by the methylation state of Npl3p. Mol. Cell. Biol. 24:10742-10756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamashita, A., T. Ohnishi, I. Kashima, Y. Taya, and S. Ohno. 2001. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 15:2215-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, S., M. J. Ruiz-Echevarria, Y. Quan, and S. W. Peltz. 1995. Identification and characterization of a sequence motif involved in nonsense-mediated mRNA decay. Mol. Cell. Biol. 15:2231-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]