Abstract
Type B histone acetyltransferases are thought to catalyze the acetylation of the NH2-terminal tails of newly synthesized histones. Although Hat1p has been implicated in cellular processes, such as telomeric silencing and DNA damage repair, the underlying molecular mechanisms by which it functions remain elusive. In an effort to understand how Hat1p is involved in the process of DNA double-strand break (DSB) repair, we examined whether Hat1p is directly recruited to sites of DNA damage. Following induction of the endonuclease HO, which generates a single DNA DSB at the MAT locus, we found that Hat1p becomes associated with chromatin near the site of DNA damage. The nuclear Hat1p-associated histone chaperone Hif1p is also recruited to an HO-induced DSB with a similar distribution. In addition, while the acetylation of all four histone H4 NH2-terminal tail domain lysine residues is increased following DSB formation, only the acetylation of H4 lysine 12, the primary target of Hat1p activity, is dependent on the presence of Hat1p. Kinetic analysis of Hat1p localization indicates that it is recruited after the phosphorylation of histone H2A S129 and concomitant with the recombinational-repair factor Rad52p. Surprisingly, Hat1p is still recruited to chromatin in strains that cannot repair an HO-induced double-strand break. These results indicate that Hat1p plays a direct role in DNA damage repair and is responsible for specific changes in histone modification that occur during the course of recombinational DNA repair.
Newly synthesized histones H3 and H4 are rapidly acetylated in the cytoplasm on their NH2-terminal tail domains. This is an evolutionarily conserved modification that is removed once the histones are packaged into nucleosomes, suggesting a role for this modification in chromatin assembly (5). The acetylation of newly synthesized histones is thought to be catalyzed by type B histone acetyltransferases (9). The lone type B histone acetyltransferase identified to date is Hat1p (22, 30). When isolated from a variety of eukaryotic organisms, Hat1p is found in a complex with a WD40 repeat-containing regulatory subunit (Hat2p in Saccharomyces cerevisiae or Rbap46 in mammalian cells) (20, 25, 36, 50). Consistent with its classification as a type B histone acetyltransferase, Hat1p is highly specific for histones that are free in solution with no demonstrable activity when chromatin-associated histones are used as a substrate. In addition, the enzyme is specific for histone H4 lysine residues 5 and 12, which are the sites of acetylation found on the newly synthesized pool of histone H4 in a wide variety of eukaryotic organisms (11, 13, 20, 22, 30, 34, 42, 50). Hence, Hat1p has the requisite biochemical activities to function in the acetylation of newly synthesized histone H4. However, evidence directly linking Hat1p to the acetylation of histones in vivo is lacking (32).
The acetylation of newly synthesized histones by type B histone acetyltransferases is presumed to play a role in histone deposition. Circumstantial evidence in support of this idea was provided by in vivo investigations into the function of Hat1p in S. cerevisiae that demonstrated that, while not essential for viability, Hat1p is involved in telomeric silencing and DNA double-strand break (DSB) repair (21, 33). Defects in heterochromatin structure and DNA repair are commonly seen in yeast containing mutations in chromatin assembly factors, where functional redundancy has complicated the analysis of histone deposition in the organism (1a). Evidence that these enzymes may play a more direct role in chromatin assembly comes from the observation that Hat1p is not strictly a cytoplasmic enzyme (4, 20, 25, 32, 50). Indeed, characterization of yeast Hat1p isolated from nuclei showed that the Hat1p-Hat2p complex is joined by Hif1p, a histone H3/H4 chaperone with chromatin assembly activity (4, 32).
To shed light on the mechanisms by which Hat1p functions, we have explored in more detail the role of Hat1p in DNA double-strand break repair. While S. cerevisiae with a hat1Δ mutation is not DNA damage sensitive, combining a hat1Δ mutation with a mutation in the histone H3 tail in which lysine residues 9, 18, and 27 are changed to arginine (H3 K9,18,27R) results in sensitivity to methyl methanesulfonate. These mutants also display reduced repair of HO-induced DSBs, suggesting that they are defective in the process of recombinational repair (33).
The repair of damaged DNA occurs in a chromatin context, and this packaging clearly influences the repair process (31). This is evidenced by recent studies that have implicated a large number of chromatin-modifying activities in the DNA repair process (7, 10, 14, 19, 23, 28, 33, 40, 46, 49). The chromatin structure has the potential to impact multiple steps in the DNA repair process. These include the initiation and propagation of DNA damage checkpoint signals, creating accessible templates for the DNA repair machinery and the reassembly of proper chromatin structure following the completion of the repair process.
The S. cerevisiae mating-type switching system has proven to be a valuable tool for the study of DNA DSB repair (16). Switching of the mating-type-specific genes at the MAT locus is initiated by the induction of the HO endonuclease, which specifically cuts only at the MAT locus. Sequences homologous to those surrounding the HO cut site are present at the silent mating loci, HML and HMR, which can used by the recombinational repair machinery to effect a gene conversion event at MAT with a subsequent change of mating type (17). Placing the HO gene under the control of an inducible promoter provides for the generation of a defined lesion (at the MAT locus) at which to use chromatin immunoprecipitation (ChIP) to monitor the recruitment of factors to the site of a DSB (44, 53). In addition to repair factors, such as Rad51p, Rad52p, Rad54p, Rad55p, and Rad57p, a number of proteins with chromatin-modifying activities have been shown to be specifically recruited to the site of an HO-induced DSB. These include the INO80, SWI/SNF, and RSC chromatin-remodeling complexes and Esa1p, Gcn5p, Sir2p, Rpd3, and Hst1p histone-modifying activities (10, 14, 28, 46, 49).
We have employed an inducible HO endonuclease system to demonstrate that Hat1p, as well as its associated histone chaperone, Hif1p, is recruited to chromatin at DSBs and localizes to a relatively small domain surrounding the break. In addition, Hat1p influences chromatin structure at a DSB, as the increased levels of histone H4 lysine 12 acetylation that occur during recombinational repair are dependent on the presence of HAT1. Surprisingly, Hat1p recruitment can occur in the absence of repair, and Hat1p accumulates at a break with kinetics similar to those of the recombinational repair factor Rad52p. The presence of a type B histone acetyltransferase on chromatin at the site of a DSB suggests that these enzymes may play a more direct role in nuclear events than previously anticipated.
MATERIALS AND METHODS
Yeast strains.
The genotypes of yeast strains used in this study are shown in Table 1. SQY392 was constructed from BY4705α by integration of a GAL-HO fragment into the ADE3 gene as described previously (6). SQY411 was then created from SQY392 by PCR-mediated gene disruption of HHF1-HHT1 and HHF2-HHT2 with URA3 and LEU2, respectively. SQY418, SQY427, and SQY450 were constructed from SQY411 by adding a 13-myc tag to the 3′ ends of the HAT1, RAD52, and HIF1 genes as previously described (37). HAT1 was also MYC tagged in strain JKM179, which lacks donor sites at HML and HMR (24). The successful epitope tagging and expression of tagged genes were confirmed by PCR analysis and Western blotting assay, respectively. SQY503 was generated from SQY411 by transformation with EcoRI- and HaeII-digested plasmid pHAT1::LYS2 to disrupt the HAT1 gene (30). HAT1 deletion was confirmed by Southern blotting analysis.
TABLE 1.
Strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| BY4705α | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 | 8 |
| SQY392 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ade3:Gal10::HO | This study |
| SQY396 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ade3:Gal10::HO hhf2-hht2::LEU2 | This study |
| SQY411 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ade3:Gal10::HO hhf2-hht2::LEU2 hhf1-hht1::URA3 (LYS2 CEN ARS)-HHF2-HHT2 | This study |
| SQY418 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ade3:Gal10::HO hhf2-hht2::LEU2 hhf1-hht1::URA3 (LYS2 CEN ARS)-HHF2-HHT2 HAT1::13myc::TRP1 | This study |
| SQY427 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ade3:Gal10::HO hhf2-hht2::LEU2 hhf1-hht1::URA3 (LYS2 CEN ARS)-HHF2-HHT2 RAD52::13myc::TRP1 | This study |
| SQY450 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ade3:Gal10::HO hhf2-hht2::LEU2 hhf1-hht1::URA3 (LYS2 CEN ARS)-HHF2-HHT2 HIF1::13myc::TRP1 | This study |
| SQY503 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ade3:Gal10::HO hhf2-hht2::LEU2 hhf1-hht1::ADE3 (TRP1 CEN ARS)-HHF2-HHT2 hat1::LYS2 | This study |
Chromatin immunoprecipitation.
Yeast cultures (500 ml) were incubated overnight at 30°C in 2% raffinose-containing YP medium (1) to an optical density at 600 nm of 0.5 to 0.8; 50 ml was removed from the culture and used as the uninduced sample for the ChIP assay. A final concentration of 2% galactose was then added to the culture to induce HO expression. Serial 50-ml samples were taken at the time points indicated in the figures and processed as described previously (43). After a 4-h incubation in galactose, cells remaining in the culture were washed at least twice with prewarmed distilled water and then reinoculated into fresh YP medium containing 2% glucose. Again, serial 50-ml samples were taken for ChIP at the time points indicated. Samples were fixed as previously described and collected by centrifugation. The cells were washed twice with 1 ml ice-cold 1× phosphate-buffered saline and then immediately frozen in liquid nitrogen. All samples were kept at −80°C until lysis. The cells were lysed, and ChIP was performed essentially as previously described with minor modifications (43). Anti-myc antibody (9E11; Covance) was used to immunoprecipitate RAD52-MYC, HAT1-MYC, and HIF1-MYC, and residue-specific acetyl lysine antibodies (Upstate) were used for precipitation of histone H4. Anti-phospho H4 serine129 was obtained from Abcam, and anti-phospho H2A serine 1 antibody was kindly provided by C. David Allis. ChIP experiments were replicated as described in the individual figure legends, and standard errors were calculated. PCR was used to quantitate the amounts of immunoprecipitated DNA fragments and input samples. Each PCR was performed in triplicate with cycling conditions as follows: 95°C for 5 min, followed by 25 to 30 cycles consisting of 95°C for 30 s, 60°C for 30 s, and 72°C for 1 min. The primer sequences used corresponded to 20 bp (5′ CTGGTTTTGGTTTTGTAGAG 3′ and 5′ CACATCAAGATCGTTTATGG 3′), 1 kb (5′ CGCCACAAGTCTTCTCTCCC 3′ and 5′ GTGACGATATTAAGTCCTCC 3′), and 5 kb (5′ GCACGATATCCGCAAGAATG 3′ and 5′ GAAGAAACTGTCCATCAAAC 3′) downstream from the HO cutting locus, as well as the promoter region of PHO5 (5′ GCCTTGCCAAGTAAGGTGAC 3′ and 5′ CGAAGGTAAAAGGTTCATAGC 3′). Optimal amplification in the linear range was empirically determined by assaying dilutions of the samples and verifying that the signals responded accordingly. For quantitation, gels were stained with Vistra Green (Amersham), and the signal was detected by scanning on a Storm phosphorimager. The intensity of DNA bands was obtained with ImageQuant 5 software. The ratio of the intensity of the sample relative to input was determined for each time point.
Physical monitoring of mating-type switch and strand invasion.
Analysis of recombinational repair was performed essentially as previously described (33). Cells were grown as described above, and 15-ml samples were harvested at the indicated time points. Analysis of strand invasion was performed as described above using the indicated primers (53).
RESULTS
Dynamics of Hat1p recruitment to sites of DNA damage.
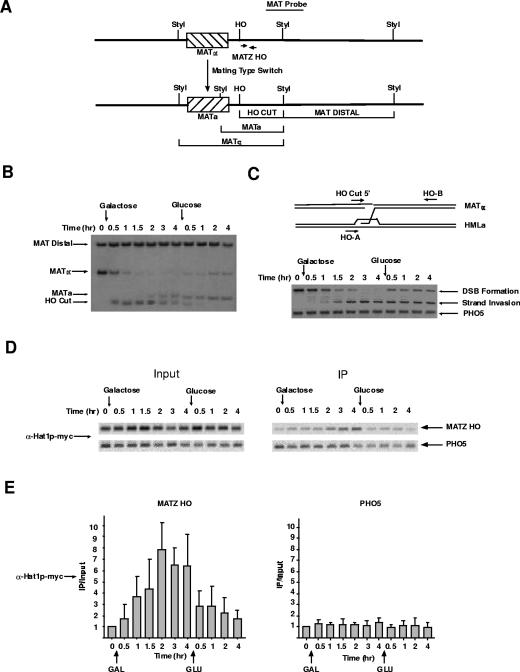
The yeast strains used in this study contain an HO gene that is under the control of a galactose-inducible promoter. Generation of a DSB at the MAT locus has been used to analyze factor recruitment in the context of both nonhomologous end joining (NHEJ) and recombinational repair. If the homologous sequences normally present at HMLα and HMRa are deleted, or if a strain lacks a factor required for recombinational repair, such as Rad52p, a DSB at MAT will be processed via NHEJ (27). As previous evidence suggested that there were no defects in NHEJ in hat1Δ/H3 K9,18,27R mutants, we chose to use strains that have intact HMLα and HMRa donor sites, which allows the HO-induced DSB to be removed through the recombinational-repair pathway (33). The formation of the strand break at MAT and subsequent events in the repair process can be physically monitored through a variety of techniques (16). This allows results obtained from ChIP to be correlated with the repair process. Figure 1A is a schematic of the MAT locus diagramming the fragments that are generated by StyI digestion of genomic DNA that allows the conversion between MATα and MATa to be distinguished on Southern blots. In all of the experiments in this study, cells were grown in the noninducing carbon source raffinose, and HO expression was induced by the addition of galactose. The cells were incubated in galactose for 4 h, after which HO expression was repressed by the addition of glucose. Figure 1B shows the kinetics of the repair events that occur during this time course. Upon HO induction, there was rapid cutting of the MATα locus (significant digestion was seen at the 30-min time point) that was complete by 2 h. The fragment representing the HO-cut MAT locus persisted as long as galactose remained in the medium and rapidly disappeared following glucose addition. Conversion of MATα to MATa (indicating recombinational repair) was first seen approximately 2 h after HO induction. This was in agreement with the results shown in Fig. 1C, in which PCR was used to detect strand invasion, showing that this early event initiated at 1 to 1.5 h post-HO induction.
FIG. 1.
The type B histone acetyltransferase Hat1p is recruited to chromatin at the site of a DNA DSB. (A) Schematic diagram of the S. cerevisiae MAT locus. The locations of endonuclease cut sites and probes used to assess the status of the locus are indicated. (B) Southern blot used to monitor the cleavage and subsequent repair of the MAT locus. The locations of the indicated fragments are detailed in panel A. The yeast strain used contains a galactose-inducible HO gene. Galactose and glucose were added to the cells at the time points indicated. (C) (Top) Schematic diagram showing the locations of PCR primers used to assess the HO-induced DSB at the MAT locus and to detect the strand invasion that occurs during recombinational repair. (Bottom) PCRs using the indicated primers, as well as primers specific to a control locus (PHO5). (D) ChIP assays were performed on samples from strain SQY418 (Hat1p-MYC) isolated at the time points shown in panels B and C. PCR fragments were generated with MATZ HO primers shown in panel A and with primers specific to the PHO5 gene using input chromatin and chromatin immunoprecipitated (IP) with an anti-myc epitope antibody as indicated. (E) PCR products, such as those shown in panel D, were quantitated on a Storm phosphorimager following staining of gels with Vistra Green. PCRs were performed in the linear range. The average of four independent ChIP assays is shown, and the IP/input values were normalized to the zero-hour time point. The error bars indicate standard errors.
Utilizing a genomic Hat1p-myc fusion, we asked whether Hat1p localized to the site of a DSB. ChIP was performed on samples taken at the same time points mentioned above. Using PCR primers that amplified a region just distal to the HO cut site, it was apparent that Hat1p was recruited to the site of DNA damage (Fig. 1D and E). Following HO induction, the association of Hat1p with the DSB steadily increased until it reached its peak level at 2 h and then plateaued. The association of Hat1p with the DSB was then rapidly lost following glucose addition. The association of Hat1p with the MAT locus was not dependent on the length of HO induction, as recruitment was also seen when HO was expressed for 1.5 h (data not shown; see Fig. 6). The chromatin association of Hat1p was specific for the site of the DSB, as there was no increase in Hat1p recruitment to a control locus on a different chromosome (PHO5) (Fig. 1D and E). In addition, a strain lacking the myc epitope showed no increased signal at the MAT locus following HO induction (data not shown). Therefore, we conclude that a type B histone acetyltransferase can interact with chromatin and that its association is closely correlated with the process of recombinational repair.
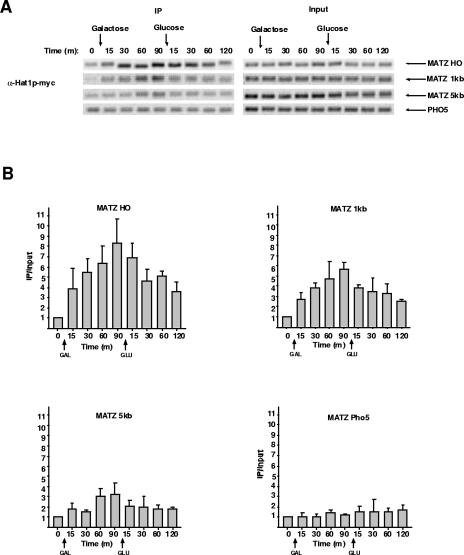
FIG. 6.
Recruitment of Hat1p to a DSB occurs in the absence of repair. ChIP assays were performed using an anti-myc antibody (HAT1-MYC) from a strain lacking the sequences at the silent mating loci that serve as donor sites for recombinational repair of an HO-induced DSB. Assays were performed as described in the legend to Fig. 1. (B) Assays shown in panel A were quantitated as described in the legend to Fig. 1. ChIP was performed twice, with multiple PCRs run on each ChIP sample.
Hat1p recruitment occurs in a localized region surrounding a DSB.
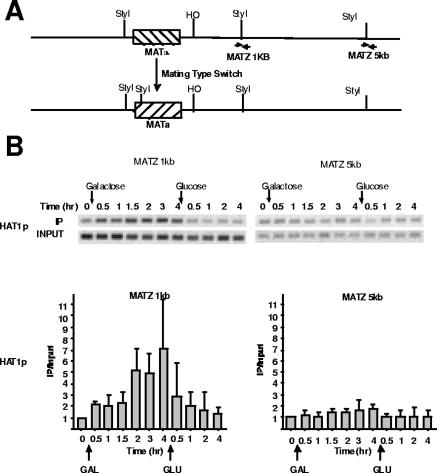
Hat1p was clearly recruited to the immediate vicinity of a DSB. To determine the extent of Hat1p localization, chromatin immunoprecipitated with Hat1p was assayed using primers located either 1 kb or 5 kb downstream of the HO cut site (depicted in Fig. 2A). At 1 kb, a similar pattern of Hat1p recruitment was seen, in which enzyme accumulation increased up to 2 h post-HO induction and then immediately dropped following glucose repression of HO expression. However, at 5 kb from the HO cut, there was little or no Hat1p accumulation (Fig. 2B). These results indicated that Hat1p localization was restricted to a relatively small domain near a DSB, extending less than 5 kb.
FIG. 2.
Hat1p localization occurs within 5 kb of a DNA double-strand break. (A) Schematic diagram of the MAT locus. The locations of PCR primers located 1 kb and 5 kb downstream of the HO DSB are indicated. (B, top) ChIP assays analyzing Hat1p association 1 kb and 5 kb downstream of the HO-induced DSB. Data are shown for four independent immunoprecipitations. PCRs were performed as described in the legend to Fig. 1. (Bottom) PCRs were quantitated and analyzed as described in the legend to Fig. 1. The error bars indicate standard errors.
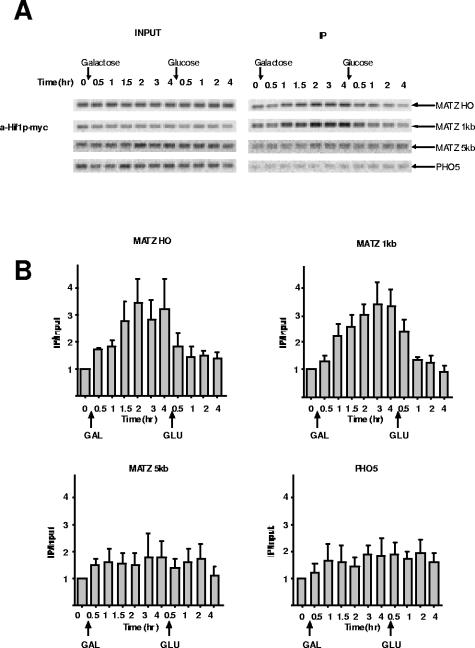
Hif1p, a component of the nuclear Hat1p-Hat2p-Hif1p complex, is recruited to a DSB.
Hat1p is found in at least two distinct complexes that differ in subcellular localization and subunit composition. While both cytoplasmic and nuclear Hat1p is associated with Hat2p, only the nuclear complex contains the histone chaperone Hif1p (4, 30, 32). To determine whether the recruitment of Hat1p to a DSB occurs in the context of the nuclear Hat1p-Hat2p-Hif1p complex, we assayed for the presence of Hif1p at the MAT locus following HO cleavage. ChIP was performed on chromatin from a strain bearing a genomic Hif1p-myc fusion that was grown under conditions identical to those mentioned above. As seen in Fig. 3A and B, Hif1p association with the MAT locus began to increase soon after HO induction and reached its maximal level after 2 h. Hif1p levels then decreased rapidly upon repression of HO. A similar pattern of Hif1p recruitment was also observed 1 kb downstream of the HO cut site, while Hif1p was nearly undetectable at 5 kb downstream. This pattern of association was highly similar to that of Hat1p, strongly suggesting that it is the nuclear Hat1p-Hat2p-Hif1p complex that is recruited to DSBs.
FIG. 3.
The Hat1p-associated histone chaperone Hif1p is recruited to a DSB. (A) ChIP was performed, in triplicate, using an anti-myc antibody with chromatin from strain SQY450 containing a HIF1-myc fusion. PCRs were performed using the indicated primers. (B) PCRs were quantitated and analyzed as described in the legend to Fig. 1. The error bars indicate standard errors.
Hat1p influences chromatin structure at a DSB.
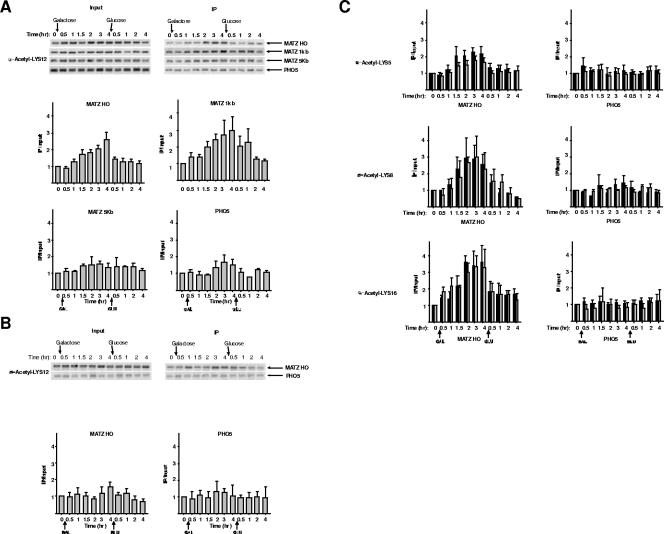
Having established the presence of Hat1p at a DSB, we sought to determine whether it was responsible for any changes in histone acetylation that occurred during the course of DNA repair. ChIP was performed with antibodies specific for the acetylation of histone H4 lysine 12, the primary in vitro target of Hat1p. Histone H4 lysine 12 acetylation levels at the MAT locus increased following DSB formation and then decreased following the addition of glucose (Fig. 4A). Similar increases in H4 lysine 12 acetylation were seen 1 kb downstream of MAT, while there were no changes in this modification 5 kb from the DSB or at a control locus (PHO5). This pattern of H4 lysine 12 acetylation changes closely mirrored the pattern of Hat1p recruitment. To test whether Hat1p was responsible for these changes in H4 lysine 12 acetylation, the ChIP analysis was repeated with chromatin from a strain containing a hat1Δ mutation (Fig. 4B). In the absence of Hat1p, creation of a DSB at the MAT locus was not accompanied by an increase in histone H4 lysine 12 acetylation. This result directly links the recruitment of Hat1p to alterations in chromatin structure that occur during the process of recombinational repair.
FIG. 4.
Hat1p is specifically responsible for the acetylation of histone H4 lysine 12 that occurs in response to a DNA double-strand break. (A) ChIP assays using antibodies that recognize histone H4 lysine 12 acetylation were performed on chromatin isolated from a HAT1 strain (SQY411). The primers and methods used are described in the legends to Fig. 1 and Fig. 3. (B) ChIP assays were performed as described for panel A using chromatin isolated from a hat1Δ strain (SQY503). The data shown are from three independent ChIP reactions. (C) ChIP assays using antibodies that specifically recognize histone H4 acetylated at lysine 5, 8, or 16 were performed on chromatin isolated from HAT1 (black bars) or hat1Δ (gray bars) strains. For H4 lysines 5 and 8, ChIP was performed once, and the data shown are the averages of four duplicate PCRs. For H4 lysine 16, ChIP was performed twice, and the data are the average of multiple PCRs performed with each immunoprecipitated DNA sample.
In addition to H4 lysine 12, it was recently reported that acetylation of H4 lysines 5, 8, and 16 also increases at a DSB (46). To determine whether Hat1p is responsible for these changes in histone acetylation as well, ChIP using antibodies specifically recognizing these sites of modification was performed on chromatin isolated from HAT1 and hat1Δ strains. As seen in Fig. 4C, the absence of Hat1p has no effect on the acetylation of H4 lysine 8 and lysine 16 and only marginal effects on H4 lysine 5. These results indicate that the effects of Hat1p on chromatin structure at a DSB are highly specific and are correlated with the enzyme's well-characterized in vitro activity.
Recruitment of Hat1p to a DSB is independent of recombinational repair.
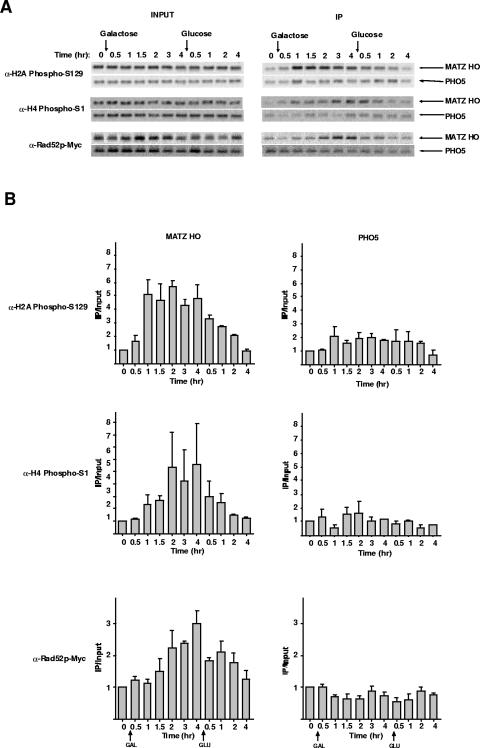
The involvement of type B HATs in the acetylation of newly synthesized histones during de novo chromatin assembly suggests a model in which the most likely role for Hat1p is in the repackaging of DNA into chromatin following the recombinational-repair event. We tested this idea in two ways. First, we compared the kinetics of Hat1p association with a DSB to other factors in the repair process. Second, we tested whether completion of recombinational repair was necessary for the association of Hat1p with the site of a DSB. Recruitment of components of the DSB repair machinery has been shown to follow a specific temporal sequence (44, 52, 53). Phosphorylation of histone H2A serine 129 is an early response to a DSB. Consistent with previous reports, in our system, H2A serine 129 phosphorylation occurred very rapidly following HO induction, reaching maximal levels within 1 h (Fig. 5A and B) (14, 41, 49). This modification was then gradually lost following shutoff of the HO gene. The phosphorylation of histone H4 (on serine 1) has also been shown to occur in the vicinity of a DSB. In agreement with recent results, this modification accumulated more slowly than H2A phosphorylation and reached a plateau approximately 2 h after HO induction (12). Levels of H4 serine 1 phosphorylation also gradually decreased after glucose addition. As a marker for the recombinational-repair machinery, we analyzed Rad52p. As seen in Fig. 5, association/dissociation of Rad52p occurred on a similar time scale as H4 serine 1 phosphorylation. In comparison, the recruitment of Hat1p to a DSB clearly occurred after H2A phosphorylation and with kinetics similar to that of Rad52p and H4 serine 1 phosphorylation. However, following DSB repair, Hat1p appeared to dissociate from chromatin more quickly than these factors.
FIG. 5.
Recruitment of Hat1p relative to other markers of DNA double-strand break repair. (A) ChIP assays using antibodies that specifically recognize histone H2A serine 129 phosphorylation, histone H4 serine 1 phosphorylation, and Rad52p (RAD52-myc fusion; SQY427) were performed as described in the legend to Fig. 1. Immunoprecipitations for histone H2A serine 129 phosphorylation, histone H4 serine 1 phosphorylation, and Rad52p were replicated three, two, and three times, respectively. (B) PCRs were quantitated and analyzed as described in the legend to Fig. 1.
The link between Hat1p recruitment and the repair event was explored further using a strain in which the silent mating loci have been deleted, preventing the recombinational repair of an HO-induced DSB. Surprisingly, in the absence of any recombinational repair (confirmed by Southern blot analysis [data not shown]), there was rapid and pronounced recruitment of Hat1p to the HO locus following induction of HO expression (Fig. 6). In the absence of recombinational repair, elevated levels of Hat1p then persisted for an extended period following the repression of HO. The spatial pattern of Hat1p recruitment was similar to that seen in strains that are repair competent, with the protein easily detectable 1 kb from the DSB and with Hat1p levels markedly reduced by 5 kb from the break. Hence, the association of Hat1p with chromatin at the site of a DSB is not dependent on the completion of the recombinational-repair process. Taken together with the timing of Hat1p association, these results suggest that the role of Hat1p at a DSB may not be strictly limited to postrepair chromatin assembly.
DISCUSSION
Analogous to the multiple mechanisms that exist to coordinate the interactions between the transcription machinery and in vivo DNA templates, many of the factors that play roles in the repair of damaged DNA exert their effects through the manipulation of chromatin structure (31). As expected, these factors include both ATP-dependent chromatin remodeling and histone-modifying activities. In addition, chromatin-modifying activities appear to be involved at multiple stages of the DNA repair process (47, 51). The inability of the type B histone acetyltransferase Hat1p to acetylate nucleosomal substrates and its likely role in the acetylation of newly synthesized histones suggested that the enzyme likely influenced processes such as DNA repair indirectly through downstream chromatin assembly pathways (29, 30). The identification of a nuclear complex that contains Hat1p and Hat2p in association with the histone chaperone Hif1p opened up the possibility that Hat1p may play a more direct role in nuclear events (4, 32). The goal of this study was to determine if Hat1p becomes physically associated with chromatin in the context of a DNA DSB. The unexpected observation that a type B histone acetyltransferase can be recruited to chromatin, and that this recruitment can occur early in the repair process, significantly alters our view of the potential points at which Hat1p may participate in the well-orchestrated series of events that leads to the repair of damaged DNA.
The earliest events in the process of DNA repair involve damage sensing and the activation of the DNA damage checkpoint. These steps are clearly impacted by chromatin structure. The phosphorylation of histone H2AX (or the major H2A variant in yeast), catalyzed by the checkpoint kinases ATM and ATR (Mec1p and Tel1p in S. cerevisiae), is one of the earliest responses to a DNA DSB (35, 51). The presence of this modification at a DSB is important for subsequent changes in chromatin structure, as it is specifically recognized by components of the NuA4 histone acetyltransferase and INO80 chromatin-remodeling complexes (14, 28, 49). In addition, while not induced by DNA damage, methylation of histone H3 K79 (or histone H4 K20 in Schizosaccharomyces pombe) is also involved in the DNA damage checkpoint through the recruitment of the checkpoint proteins 53BP1/Rad9p/Crb2 (15, 18, 38). A number of results indicate that it is unlikely that Hat1p influences these early events. First, in the absence of Hat1p, the S-phase DNA damage checkpoint operates normally (33). Second, the recruitment of Hat1p to a DSB occurs well after the appearance of phosphorylated histone H2A.
Following activation of the DNA damage checkpoint, a series of chromatin-modifying activities are then recruited to a DSB. NuA4 and the RSC chromatin remodeling complexes are recruited within minutes of DSB induction (10, 14). While the precise roles of these factors in DNA repair are not known, their presence at sites of damage prior to the arrival of repair proteins suggests that they are involved in making the chromatin structure accessible to the repair machinery. Following NuA4 and RSC, recruitment of a number of chromatin-modifying activities progresses more gradually, reaching peak values 2 to 4 h post-HO induction. In addition to Hat1p, this group includes the INO80 and SWI/SNF complexes and the histone acetyltransferase Gcn5p (10, 28, 46, 49). Precisely ordering the recruitment of these proteins is difficult, given the level of resolution provided by current techniques. Important components of the DNA repair machinery, such as the RAD52 epistasis group proteins, are also recruited to DSBs during this period (44, 52, 53). The concurrent localizations of numerous chromatin-modifying activities with the DNA repair machinery suggests that they act to facilitate multiple steps in the DNA repair pathway. Indeed, mutations in components of the INO80 complex cause defects in the resection of DNA at the HO cut site to form the single-stranded DNA that is required for the strand invasion step of recombinational repair (49). In addition, SWI/SNF appears to play an important role in the synapsis between the HO cut site and homologous sequences at the silent mating loci (10).
The timing of Hat1p localization at a DSB is similar to that of Rad52p, and the extent of the domain occupied by Hat1p is comparable to that occupied by components of the recombinational-repair machinery (41, 44, 52). In addition, repair of a DSB is not required for the recruitment of Hat1p to chromatin. Therefore, these observations open up the possibility that Hat1p may be acting to directly facilitate the DNA repair process rather than (or in addition to) functioning in the restoration of chromatin structure that must follow the completion of recombinational repair. In this respect, the Hat1p-dependent acetylation of histone H4 lysine 12 may be important for generating a chromatin structure that is competent for interaction with the recombinational-repair machinery.
The inability of Hat1p to acetylate histones in a chromatin context makes it unlikely that this acetylation is the result of the enzyme directly modifying histones that were associated with chromatin prior to the introduction of the DSB (30). Rather, an alternative model involves the participation of Hat1p in a histone exchange process that would be analogous to the histone exchange events that are responsible for the replication-coupled assembly of histone H3.3 in regions of active chromatin in higher eukaryotes (2, 3, 26, 39, 48). This exchange would be predicted to be specific for the substitution of histone H3/H4 tetramers, given the physical association of Hat1p with these histones, as well the observation that histone H2A phosphorylation levels remain high for extended periods at the site of a DSB. This model is supported by several lines of evidence. First, histone exchange factors clearly play a role in DNA repair, and the coupling of histone acetylation with histone exchange at sites of DNA damage has been observed with the Drosophila Tip60 complex (23). Second, human homologs of all three components of the Hat1p-Hat2p-Hif1p complex copurify with both replication-dependent and replication-independent chromatin assembly complexes (45). Third, in addition to acetylated NH2-terminal tails, histone H4 that copurifies with nuclear Hat1p can also be acetylated on lysine 91 in the core domain. This modification can function to destabilize the histone octamer structure and, therefore, may help in creating a chromatin structure that is accessible to the repair factors (54). A more precise determination of the time at which Hat1p is recruited to a DSB relative to the arrival of recombinational-repair factors will be necessary before the mechanism(s) by which Hat1p influences DNA repair is fully elucidated.
The results presented here provide important new details about the activity of Hat1p. While the in vitro activity of Hat1p is highly specific for histone H4 lysine 12, and to a lesser extent H4 lysine 5, deletion of the HAT1 gene has no effect on the steady-state levels of histone H4 acetylation (32). Indirect evidence that histone H4 lysine 12 is the in vivo target of Hat1p was provided by the observation that mutating H4 lysine 12 to arginine phenocopies a hat1Δ in the context of telomeric silencing (21). The demonstration that the increased levels of H4 lysine 12 acetylation that occur at the site of a DNA DSB are dependent on the presence of Hat1p is the first direct confirmation of the in vivo substrate specificity of this enzyme. In addition, the fact that Hat1p can physically associate with chromatin indicates that, rather than simply modifying the histones that are used in downstream chromatin assembly pathways, type B histone acetyltransferases can remain associated with histones throughout the entire histone deposition process. While the precise functions of these enzymes remain unknown, it is clear that they are a more integral component of the chromatin assembly process than previously envisioned.
Acknowledgments
We thank C. David Allis for providing anti-phospho-histone H4 serine 1 antibody, James Haber for providing plasmids, and Amy Knapp for critical reading of the manuscript.
This work was supported by the National Institutes of Health (R01 GM62970 to M.R.P.).
REFERENCES
- 1.Adams, A., D. E. Gottschling, C. A. Kaiser, and T. Stearns. 1997. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 1a.Adams, C. R., and R. T. Kamakaka. 1999. Chromatin assembly: biochemical identities and genetic redundancy. Curr. Opin. Genet. Dev. 9:185-190. [DOI] [PubMed] [Google Scholar]
- 2.Ahmad, K., and S. Henikoff. 2002. Histone H3 variants specify modes of chromatin assembly. Proc. Natl. Acad. Sci. USA 99(Suppl. 4:16477-16484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmad, K., and S. Henikoff. 2002. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9:1191-1200. [DOI] [PubMed] [Google Scholar]
- 4.Ai, X., and M. R. Parthun. 2004. The nuclear Hat1p/Hat2p complex: a molecular link between type B histone acetyltransferases and chromatin assembly. Mol. Cell 14:195-205. [DOI] [PubMed] [Google Scholar]
- 5.Annunziato, A. T., and J. C. Hansen. 2000. Role of histone acetylation in the assembly and modulation of chromatin structures. Gene Expr. 9:37-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bahler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943-951. [DOI] [PubMed] [Google Scholar]
- 7.Bird, A. W., D. Y. Yu, M. G. Pray-Grant, Q. Qiu, K. E. Harmon, P. C. Megee, P. A. Grant, M. M. Smith, and M. F. Christman. 2002. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419:411-415. [DOI] [PubMed] [Google Scholar]
- 8.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 9.Brownell, J. E., and C. D. Allis. 1996. Special HATs for special occasions: linking histone acetylation to chromatin assembly and gene activation. Curr. Opin. Genet. Dev. 6:176-184. [DOI] [PubMed] [Google Scholar]
- 10.Chai, B., J. Huang, B. R. Cairns, and B. C. Laurent. 2005. Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 19:1656-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang, L., S. S. Loranger, C. Mizzen, S. G. Ernst, C. D. Allis, and A. T. Annunziato. 1997. Histones in transit: cytosolic histone complexes and diacetylation of H4 during nucleosome assembly in human cells. Biochemistry 36:469-480. [DOI] [PubMed] [Google Scholar]
- 12.Cheung, W. L., F. B. Turner, T. Krishnamoorthy, B. Wolner, S. H. Ahn, M. Foley, J. A. Dorsey, C. L. Peterson, S. L. Berger, and C. D. Allis. 2005. Phosphorylation of histone H4 serine 1 during DNA damage requires casein kinase II in S. cerevisiae. Curr. Biol. 15:656-660. [DOI] [PubMed] [Google Scholar]
- 13.Chicoine, L. G., I. G. Schulman, R. Richman, R. G. Cook, and C. D. Allis. 1986. Nonrandom utilization of acetylation sites in histones isolated from Tetrahymena. Evidence for functionally distinct H4 acetylation sites. J. Biol. Chem. 261:1071-1076. [PubMed] [Google Scholar]
- 14.Downs, J. A., S. Allard, O. Jobin-Robitaille, A. Javaheri, A. Auger, N. Bouchard, S. J. Kron, S. P. Jackson, and J. Cote. 2004. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol. Cell 16:979-990. [DOI] [PubMed] [Google Scholar]
- 15.Giannattasio, M., F. Lazzaro, P. Plevani, and M. Muzi-Falconi. 2005. The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J. Biol. Chem. 280:9879-9886. [DOI] [PubMed] [Google Scholar]
- 16.Haber, J. E. 1995. In vivo biochemistry: physical monitoring of recombination induced by site-specific endonucleases. Bioessays 17:609-620. [DOI] [PubMed] [Google Scholar]
- 17.Haber, J. E. 1998. Mating-type gene switching in Saccharomyces cerevisiae. Annu. Rev. Genet. 32:561-599. [DOI] [PubMed] [Google Scholar]
- 18.Huyen, Y., O. Zgheib, R. A. Ditullio, Jr., V. G. Gorgoulis, P. Zacharatos, T. J. Petty, E. A. Sheston, H. S. Mellert, E. S. Stavridi, and T. D. Halazonetis. 2004. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 432:406-411. [DOI] [PubMed] [Google Scholar]
- 19.Ikura, T., V. V. Ogryzko, M. Grigoriev, R. Groisman, J. Wang, M. Horikoshi, R. Scully, J. Qin, and Y. Nakatani. 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102:463-473. [DOI] [PubMed] [Google Scholar]
- 20.Imhof, A., and A. P. Wolffe. 1999. Purification and properties of the Xenopus hat1 acetyltransferase: association with the 14-3-3 proteins in the oocyte nucleus. Biochemistry 38:13085-13093. [DOI] [PubMed] [Google Scholar]
- 21.Kelly, T. J., S. Qin, D. E. Gottschling, and M. R. Parthun. 2000. Type B histone acetyltransferase Hat1p participates in telomeric silencing. Mol. Cell. Biol. 20:7051-7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kleff, S., E. D. Andrulis, C. W. Anderson, and R. Sternglanz. 1995. Identification of a gene encoding a yeast histone H4 acetyltransferase. J. Biol. Chem. 270:24674-24677. [DOI] [PubMed] [Google Scholar]
- 23.Kusch, T., L. Florens, W. H. Macdonald, S. K. Swanson, R. L. Glaser, J. R. Yates III, S. M. Abmayr, M. P. Washburn, and J. L. Workman. 2004. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science 306:2084-2087. [DOI] [PubMed] [Google Scholar]
- 24.Lee, S. E., J. K. Moore, A. Holmes, K. Umezu, R. D. Kolodner, and J. E. Haber. 1998. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94:399-409. [DOI] [PubMed] [Google Scholar]
- 25.Lusser, A., A. Eberharter, A. Loidl, M. Goralik-Schramel, M. Horngacher, H. Haas, and P. Loidl. 1999. Analysis of the histone acetyltransferase B complex of maize embryos. Nucleic Acids Res. 27:4427-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKittrick, E., P. R. Gafken, K. Ahmad, and S. Henikoff. 2004. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc. Natl. Acad. Sci. USA 101:1525-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore, J. K., and J. E. Haber. 1996. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:2164-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morrison, A. J., J. Highland, N. J. Krogan, A. Arbel-Eden, J. F. Greenblatt, J. E. Haber, and X. Shen. 2004. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 119:767-775. [DOI] [PubMed] [Google Scholar]
- 29.Mosammaparast, N., Y. Guo, J. Shabanowitz, D. F. Hunt, and L. F. Pemberton. 2002. Pathways mediating the nuclear import of histones H3 and H4 in yeast. J. Biol. Chem. 277:862-868. [DOI] [PubMed] [Google Scholar]
- 30.Parthun, M. R., J. Widom, and D. E. Gottschling. 1996. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell 87:85-94. [DOI] [PubMed] [Google Scholar]
- 31.Peterson, C. L., and J. Cote. 2004. Cellular machineries for chromosomal DNA repair. Genes Dev. 18:602-616. [DOI] [PubMed] [Google Scholar]
- 32.Poveda, A., M. Pamblanco, S. Tafrov, V. Tordera, R. Sternglanz, and R. Sendra. 2004. Hif1 is a component of yeast histone acetyltransferase B, a complex mainly localized in the nucleus. J. Biol. Chem. 279:16033-16043. [DOI] [PubMed] [Google Scholar]
- 33.Qin, S., and M. R. Parthun. 2002. Histone H3 and the histone acetyltransferase Hat1p contribute to DNA double-strand break repair. Mol. Cell. Biol. 22:8353-8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richman, R., L. G. Chicoine, M. P. Collini, R. G. Cook, and C. D. Allis. 1988. Micronuclei and the cytoplasm of growing Tetrahymena contain a histone acetylase activity which is highly specific for free histone H4. J. Cell Biol. 106:1017-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rogakou, E. P., D. R. Pilch, A. H. Orr, V. S. Ivanova, and W. M. Bonner. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273:5858-5868. [DOI] [PubMed] [Google Scholar]
- 36.Ruiz-Garcia, A. B., R. Sendra, M. Galiana, M. Pamblanco, J. E. Perez-Ortin, and V. Tordera. 1998. HAT1 and HAT2 proteins are components of a yeast nuclear histone acetyltransferase enzyme specific for free histone H4. J. Biol. Chem. 273:12599-12605. [DOI] [PubMed] [Google Scholar]
- 37.Sandell, L. L., and V. A. Zakian. 1993. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell 75:729-739. [DOI] [PubMed] [Google Scholar]
- 38.Sanders, S. L., M. Portoso, J. Mata, J. Bahler, R. C. Allshire, and T. Kouzarides. 2004. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 119:603-614. [DOI] [PubMed] [Google Scholar]
- 39.Schwartz, B. E., and K. Ahmad. 2005. Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev. 19:804-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen, X., G. Mizuguchi, A. Hamiche, and C. Wu. 2000. A chromatin remodelling complex involved in transcription and DNA processing. Nature 406:541-544. [DOI] [PubMed] [Google Scholar]
- 41.Shroff, R., A. Arbel-Eden, D. Pilch, G. Ira, W. M. Bonner, J. H. Petrini, J. E. Haber, and M. Lichten. 2004. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr. Biol. 14:1703-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sobel, R. E., R. G. Cook, C. A. Perry, A. T. Annunziato, and C. D. Allis. 1995. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc. Natl. Acad. Sci. USA 92:1237-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strahl-Bolsinger, S., A. Hecht, K. Luo, and M. Grunstein. 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11:83-93. [DOI] [PubMed] [Google Scholar]
- 44.Sugawara, N., X. Wang, and J. E. Haber. 2003. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol. Cell 12:209-219. [DOI] [PubMed] [Google Scholar]
- 45.Tagami, H., D. Ray-Gallet, G. Almouzni, and Y. Nakatani. 2004. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116:51-61. [DOI] [PubMed] [Google Scholar]
- 46.Tamburini, B. A., and J. K. Tyler. 2005. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol. Cell. Biol. 25:4903-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thiriet, C., and J. J. Hayes. 2005. Chromatin in need of a fix: phosphorylation of H2AX connects chromatin to DNA repair. Mol. Cell 18:617-622. [DOI] [PubMed] [Google Scholar]
- 48.Thiriet, C., and J. J. Hayes. 2005. Replication-independent core histone dynamics at transcriptionally active loci in vivo. Genes Dev. 19:677-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Attikum, H., O. Fritsch, B. Hohn, and S. M. Gasser. 2004. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119:777-788. [DOI] [PubMed] [Google Scholar]
- 50.Verreault, A., P. D. Kaufman, R. Kobayashi, and B. Stillman. 1998. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr. Biol. 8:96-108. [DOI] [PubMed] [Google Scholar]
- 51.Vidanes, G. M., C. Y. Bonilla, and D. P. Toczyski. 2005. Complicated tails: histone modifications and the DNA damage response. Cell 121:973-976. [DOI] [PubMed] [Google Scholar]
- 52.Wang, X., and J. E. Haber. 2004. Role of Saccharomyces single-stranded DNA-binding protein RPA in the strand invasion step of double-strand break repair. PLoS Biol. 2:E21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolner, B., S. van Komen, P. Sung, and C. L. Peterson. 2003. Recruitment of the recombinational repair machinery to a DNA double-strand break in yeast. Mol. Cell 12:221-232. [DOI] [PubMed] [Google Scholar]
- 54.Ye, J., X. Ai, E. E. Eugeni, L. Zhang, L. R. Carpenter, M. A. Jelinek, M. A. Freitas, and M. R. Parthun. 2005. Histone H4 lysine 91 acetylation a core domain modification associated with chromatin assembly. Mol. Cell 18:123-130. [DOI] [PMC free article] [PubMed] [Google Scholar]