Abstract
Regulation of the Escherichia coli stationary-phase sigma factor RpoS is complex and occurs at multiple levels in response to different environmental stresses. One protein that reduces RpoS levels is the transcription factor LrhA, a global regulator of flagellar synthesis. Here we clarify the mechanism of this repression and provide insight into the signaling pathways that feed into this regulation. We show that LrhA represses RpoS at the level of translation in a manner that is dependent on the small RNA (sRNA) chaperone Hfq. Although LrhA also represses the transcription of the sRNA RprA, its regulation of RpoS mainly occurs independently of RprA. To better understand the physiological signals affecting this pathway, a transposon mutagenesis screen was carried out to find factors affecting LrhA activity levels. The RcsCDB phosphorelay system, a cell envelope stress-sensing pathway, was found to repress lrhA synthesis. In addition, mutations in the gene encoding the DNA motor protein FtsK induce lrhA synthesis, which may explain why such strains fail to accumulate RpoS in stationary phase.
Bacteria such as Escherichia coli respond to environmental changes by mounting coordinated responses, sometimes involving more than one global regulator. One example is the transcription factor LrhA, which represses expression of several other global regulators, including the stationary-phase sigma factor RpoS and the master regulator of flagellar biosynthesis FlhDC (10, 18). It also influences biofilm development and adherence by repressing type 1 fimbrial expression (1, 18). Motility has been shown to be a key aspect of bacterial virulence in plants, and homologs of LrhA, such as HexA in Erwinia carotovora and PecT in Erwinia chrysanthemi, have been shown to play critical roles in the pathogenic response (4, 5, 12, 25). In addition to its effect on motility, HexA represses genes involved in pathogenicity, such as those encoding exoenzymes and factors involved in prodigiosin and antibiotic production (12). In these systems, HexA is also thought to modulate several global regulators, including RpoS, N-(3-oxohexanoyl)-l-homoserine lactone, and the regulatory small RNA (sRNA) RsmB (25).
In E. coli, LrhA expression decreases the levels of RpoS, thus preventing the accumulation of the sigma factor and the transcription of hundreds of stationary-phase genes (10). There are multiple factors that affect RpoS expression, and they work at every possible level of control, including transcription, translation, protein stability, and activity (26). Many environmental conditions influence rpoS translation, and most of these require the sRNA chaperone Hfq. It has been suggested that LrhA affects RpoS stability by stimulating the activity of SprE (RssB), the adaptor protein for RpoS degradation (6, 10). This was intriguing, since it was one of only a few factors affecting SprE activity, whose regulation is the sole mechanism for RpoS accumulation upon carbon starvation (24, 27, 36). Similar models involving LrhA and SprE homologs in E. carotovora and Photorhabdus temperata have been invoked but never tested (14, 25).
Another global regulator of RpoS and flagellar synthesis is the RcsCDB system. This phosphorelay consists of a membrane-bound sensor kinase (RcsC), a phosphorelay protein (RcsD), and a response regulator (RcsB) with an auxiliary protein (RcsA) (22). Although it was first characterized for its role in capsule synthesis, many additional genes in its regulon overlap those found in the RpoS regulon, including katE, osmC, osmB, and ftsZ (11, 22). Moreover, this regulatory system activates RpoS translation in a manner that partially requires the sRNA (noncoding RNA) RprA (20, 23). Most of the genes known to be controlled by Rcs are activated by this system, the only case of repression being the flhDC master regulator (9). The actual signals triggering RcsC are not known, but disruptions in the cell surface, such as loss of lipopolysaccharides or periplasmic glucans, activate the system (22).
In this paper, we show that LrhA represses rpoS translation by a mechanism that requires the sRNA chaperone Hfq. In addition, we identify a signaling mechanism that controls lrhA expression. In particular, we demonstrate that lrhA synthesis is repressed by the RcsCDB phosphorelay system.
MATERIALS AND METHODS
Bacterial strains, bacteriophages, and lacZ reporter fusions.
Strains are listed in Table 1. Standard microbiological techniques were used for strain construction (31). CNP265, the rcsB::kan rcsD::cam double mutant, was constructed by P1 transduction of the rcsD::cam allele into CNP264 with selection on chloramphenicol agar. The transductants were then screened for the presence of both mutations by resistance to both kanamycin and chloramphenicol. The Tntet and Tncam minitransposons used for mutagenesis were obtained from the vectors NK1323 and NK1324 (16). The position where the minitransposon inserted in the chromosome was identified by DNA sequencing by the Princeton University Department of Molecular Biology Synthesis and Sequencing Facility, as previously described (29).
TABLE 1.
Strains used in this study
| Name | Genotype | Sourcea |
|---|---|---|
| MC4100 | F−araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR | 3 |
| CNP54 | MC4100 clpX::kan | |
| CNP58 | MC4100 sprE::tet | |
| CNP109 | MC4100 lrhA49::cam | 10 |
| CNP110 | CNP109 sprE::tet | |
| CNP111 | MC4100 lrhA::spc | 10 |
| CNP112 | CNP 111 sprE::tet | |
| CNP113 | CNP 109 clpX::kan | |
| CNP114 | CNP 111 clpX::kan | |
| CNP118 | MC4100 λφ(rpoS′-lacZ+) | 29 |
| CNP119 | MC4100 λφ(rpoS750′-′lacZ) | 33 |
| CNP125 | MC4100 λφ(rpoS477′-′lacZ) | 29 |
| CNP120 | CNP118 lrhA49::cam | |
| CNP121 | CNP118 lrhA::spc | |
| CNP122 | CNP119 lrhA49::cam | |
| CNP127 | CNP125 lrhA49::cam | |
| CNP128 | CNP125 lrhA::spc | |
| CNP137 | MC4100 λφ(dsrA205p′-lacZ+) | 28 |
| CNP160 | CNP137 lrhA49::cam | |
| CNP161 | CNP137 lrhA::spc | |
| CNP166 | CNP122 rprA::kan | |
| CNP170 | MC4100 λφ(rprA′-lacZ+) | 23 |
| TC15 | MC4100 λφ(lrhA′-′lacZ) | 18 |
| TC18 | TC15 lrhA::spc | |
| TC46 | TC15 ftsK::tet | |
| TC132 | TC15 rcsD::cam | |
| CNP203 | CNP125 zed-3069::Tn10 ΔdsrA | |
| CNP205 | CNP119 rprA::kan | |
| CNP206 | CNP203 lrhA49::cam | |
| CNP207 | CNP170 lrhA49::cam | |
| CNP213 | CNP109 sprE::tet hfq::kan | 35 |
| CNP214 | CNP111 sprE::tet hfq::kan | |
| CNP215 | MC4100sprE::tet hfq::kan | |
| CNP264 | TC15 rcsB::kan | 15 |
| CNP265 | CNP264 rcsD::cam | |
| CNP266 | TC15 lon::Tn10 | |
| CNP267 | CNP170 lrhA49::cam | |
| CNP268 | CNP170 lrhA::spc | |
| CNP269 | CNP203 rprA::kan | |
| CNP270 | CNP206 rprA::kan | |
| VC4 | TC46 lrhA::spc | |
| VC16 | TC132 lrhA::spc |
Strains without a reference were constructed in this study.
rpoS transcription was monitored by the rpoS′-lacZ+ construct (29). Translation and transcription of rpoS were reported by the rpoS477′-′lacZ fusion (fusion joint at nucleotide +477 of rpoS) (29). A longer translation fusion rpoS750′-′lacZ reports transcription, translation, and protein stability of RpoS (33).
Media and growth conditions.
Lactose MacConkey agar and Luria-Bertani (LB) broth and agar were prepared as described previously (31). Bacteria were grown at 37°C unless otherwise noted.
Western blot analysis.
One-milliliter samples were prepared from exponentially growing cells (optical density at 600 nm [OD600] ∼ 0.4 to 0.5). Whole cells were pelleted and immediately resuspended in sodium dodecyl sulfate (SDS) loading buffer (31) in a volume equal to OD600/10. After boiling for 10 min, equal volumes were loaded onto 12% SDS-polyacrylamide gels (17). After electrophoresis, proteins were transferred onto nitrocellulose membranes and probed with a 1:6,000 dilution of anti-RpoS antibody (our laboratory stock). For secondary antibody, donkey anti-rabbit immunoglobulin G-horseradish peroxidase conjugate (Amersham Pharmacia Biotech) was used at a dilution of 1:6,000. The bands were detected using the ECL antibody detection kit (Amersham Pharmacia Biotech) and XAR film (Kodak).
β-Galactosidase assays.
Cultures of strains grown in LB overnight were subcultured 1:100 into fresh LB broth and grown with aeration at 37°C until the OD600 was 0.4 to 0.5. β-Galactosidase assays were performed as previously described (32) Relative β-galactosidase units were determined by the rate (in OD420 units per minute) divided by the OD600 times the volume assayed (in milliliters). Each figure depicts an individual experiment, and all experiments were repeated at least three times.
RESULTS
LrhA negatively regulates RpoS translation.
LrhA, a member of the LysR family of transcription factors, has been found to negatively affect RpoS levels (6, 10). It had been suggested that LrhA decreases RpoS stability by affecting the activity of SprE, based on results indicating that lrhA mutations did not affect RpoS levels in the absence of the RpoS degradation factors SprE and ClpX/P. Since these studies were carried out with an rpoS-lacZ fusion strain that has subsequently been shown to be unstable and unreliable because of multiple lysogens (our unpublished results), we repeated these epistasis experiments using Western blots that directly measure RpoS levels. Furthermore, we collected samples from exponentially growing cells rather than stationary-phase cells, since the mutant lrhA phenotypes are stronger during exponential growth phase.
The lrhA49::cam allele has an inserted transposon located 571 base pairs upstream from the lrhA start codon and causes lrhA transcription to be upregulated (10). This overexpression of lrhA causes RpoS levels to decrease compared to an otherwise isogenic wild-type strain (10). Conversely, a strain carrying lrhA::spc, a null allele of lrhA, has only slightly increased RpoS levels (10).
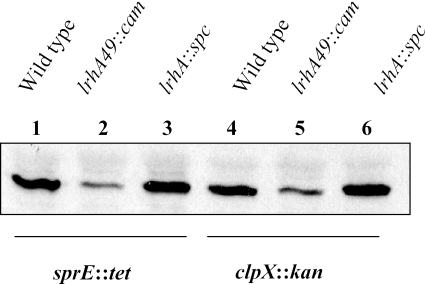
To examine whether LrhA affects RpoS levels solely by activating the degradation pathway, we tested whether there was any phenotype with these lrhA alleles in a background in which RpoS is not degraded. In the sprE::tet background, which abolishes RpoS degradation but has no effect on its translation (27), RpoS levels were decreased in the presence of the lrhA49::cam allele (Fig. 1, compare lanes 1 and 2). RpoS levels were marginally higher with the lrhA::spc allele than with lrhA+ (Fig. 1, lanes 1 and 3). Likewise, in the clpX::kan background, RpoS levels were lower in cells carrying the lrhA49::cam allele and slightly higher in lrhA::spc cells than in wild-type lrhA cells (Fig. 1, lanes 4, 5, and 6). Thus, the effect of LrhA on RpoS is not disrupted by the absence of RpoS degradation. This indicates that LrhA affects RpoS levels in a manner that is independent of protein stability.
FIG. 1.
Western blotting for RpoS shows that lrhA mutations are additive with mutations in sprE and clpX. The lrhA49::cam allele contains a transposon inserted in the chromosome upstream of the lrhA promoter, which causes LrhA levels to increase and RpoS levels to decrease (6, 10). The lrhA::spc allele is a null mutation that has been shown to slightly increase RpoS levels. Lanes 1 to 3 (CNP58, CNP110, and CNP112, respectively) show the effect of the lrhA mutations in the absence of the RpoS degradation adaptor protein SprE, and lanes 4 to 6 (CNP115, CNP113, and CNP114, respectively) show the effect in the absence of the RpoS protease chaperone ClpX. Removing either sprE or clpX has no effect on RpoS translation (29). All Western blot samples were collected from cells growing exponentially in LB medium.
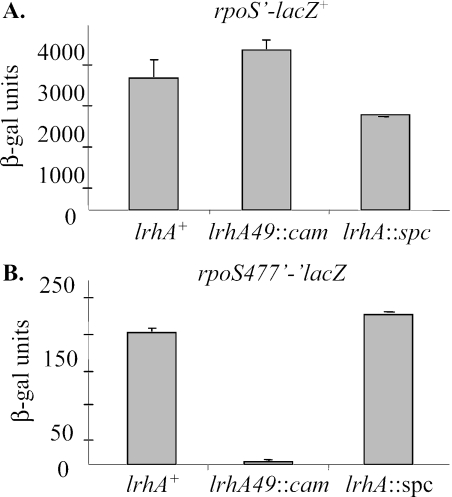
In order to determine how LrhA regulates RpoS levels, we used an rpoS′-lacZ+ operon fusion, which reports rpoS transcription, and a short protein fusion, rpoS477′-′lacZ, which reports both rpoS transcription and translation but not protein stability. SprE activity has no effect on either of these fusions (29). β-Galactosidase assays were performed on samples taken from exponentially growing cells in the wild-type background and different lrhA mutant backgrounds. Both lrhA49::cam and lrhA::spc had only minor effects on the operon fusion, less than 1.5-fold (Fig. 2A). However, the presence of lrhA49::cam dramatically decreased the LacZ activity of the rpoS477′-′lacZ fusion compared to the activity measured with lrhA+ (Fig. 2B). The lrhA::spc allele did not significantly affect the short protein fusion, similar to the minor effect it had on RpoS protein levels. These results indicate that LrhA, particularly when overexpressed, represses rpoS translation.
FIG. 2.
LrhA represses rpoS translation. (A) The rpoS′-lacZ+ operon fusion (CNP118) reports only rpoS transcription. The lrhA alleles found in CNP120 and CNP121 have a negligible influence on this transcriptional fusion. (B) The rpoS477′-′lacZ (CNP125) reports rpoS translation and transcription but does not affect RpoS stability. The lrhA49::cam allele (CNP127) strongly represses this fusion, whereas the lrhA::spc allele (CNP128) does not have a strong effect. Samples were collected from cells growing logarithmically in LB medium and processed for β-galactosidase assays as described in Materials and Methods.
The effect of LrhA on RpoS levels is dependent on Hfq.
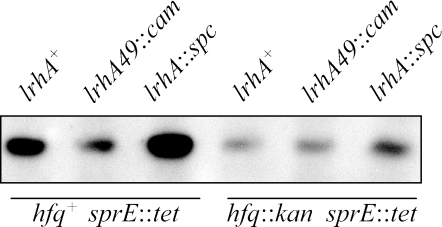
Translation of rpoS is regulated by a number of different factors, including sRNAs, HU, and H-NS (13). The pairing of the rpoS mRNA and a regulatory sRNA requires the RNA chaperone Hfq (2, 21, 23). To determine if LrhA regulation of rpoS translation requires Hfq, we measured RpoS levels by Western blotting with the different alleles of lrhA in an hfq::kan background. As expected, the hfq::kan allele dramatically reduced rpoS translation so that the RpoS protein levels were very low in an otherwise wild-type background, and it proved difficult to determine whether the lrhA49::cam allele reduced them to an even lower level. To circumvent this problem, we used a sprE::tet strain, since it has increased RpoS levels because it cannot be degraded by ClpXP. As mentioned above, with the hfq+ sprE::tet background, the strain carrying lrhA49::cam had much lower levels of RpoS while the lrhA::spc strain had higher levels (Fig. 1 and 3). However, in the hfq::kan sprE::tet background, the levels of RpoS were similar with the lrhA49::cam allele and the lrhA+ allele (Fig. 3). It should be noted that a slight increase in RpoS levels in the lrhA::spc strain occurs despite the hfq mutation. We think that this is because the Hfq requirement is not absolute. Indeed, a modest Hfq-independent effect has been documented previously in the case of other sRNAs affecting RpoS (29, 33). In summary, LrhA repression of rpoS translation is dependent on the RNA chaperone Hfq.
FIG. 3.
The sRNA chaperone Hfq is necessary for LrhA repression of rpoS translation. In the hfq::kan background, the lrhA49::cam allele (CNP213) did not reduce RpoS levels compared to those found in the lrhA+ background (CNP215), as determined by Western blotting. However, the lrhA::spc allele (CNP214) still had a positive effect on RpoS levels. A sprE::tet background was used for these experiments to increase the overall levels of RpoS, since the hfq mutation has a negative effect on RpoS synthesis and lowers its levels to below the detection threshold in wild-type cells. All samples were collected from cells growing exponentially in LB rich medium.
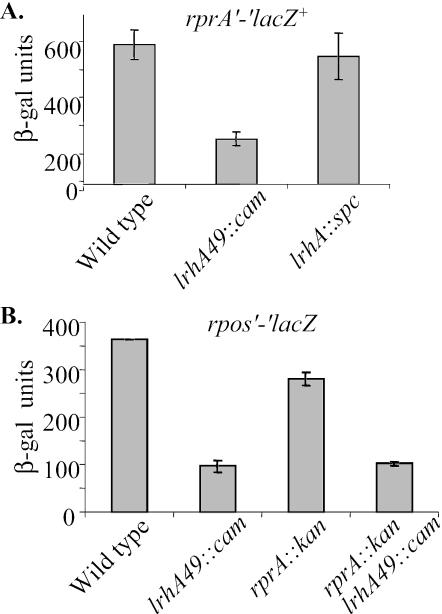
The involvement of Hfq in LrhA regulation of rpoS translation suggested that LrhA might be controlling an sRNA. To date, three sRNAs that modulate rpoS translation have been characterized: RprA, DsrA, and OxyS (28). We used a transcription fusion to dsrA and showed that LrhA did not affect its expression (data not shown). We also performed epistasis tests with both dsrA and oxyS mutants, but LrhA did not require either of these sRNAs to affect rpoS translation (data not shown). LrhA did, however, repress rprA expression. The presence of the lrhA49::cam allele caused β-galactosidase activity of the rprA′-lacZ+ fusion to drop threefold compared to activity with the wild-type lrhA allele in exponentially growing cells (Fig. 4A). As observed previously, the null lrhA allele did not have a strong phenotype and did not affect rprA expression. Thus, LrhA has a modest effect on rprA expression.
FIG. 4.
(A) LrhA represses rprA synthesis. The overexpression allele of lrhA (lrhA49::cam) in CNP267 reduces rprA′-lacZ β-galactosidase activity compared to the lrhA+ allele in CNP170. The null allele of lrhA (lrhA::spc) in CNP268 does not have a large effect on rprA′-lacZ+ β-galactosidase activity. (B) Epistasis tests with LrhA and RprA. The lrhA49::cam allele still reduces rpoS′-′lacZ activity compared to the lrhA+ allele in the rprA::kan background (CNP166 and CNP205). Thus, LrhA represses RpoS in an RprA-independent fashion. All samples were collected from exponentially growing cells in LB medium.
To determine if the LrhA effect on RpoS was completely dependent on RprA, we monitored the activity of rpoS′-′lacZ in the lrhA49::cam rprA::kan double mutant. With wild-type rprA, lrhA49::cam caused rpoS levels to decrease. In the rprA::kan background, lrhA49::cam also reduced rpoS levels by nearly the same amount (Fig. 4B). This additive effect was also seen in a ΔdsrA background (data not shown), where the RprA effect on RpoS levels is more pronounced (20). In summary, LrhA represses RprA translation threefold. Although this may contribute to the reduction of RpoS levels under certain conditions, our data indicate that LrhA mostly controls rpoS translation in an RprA-independent fashion.
RcsCDB decreases synthesis of LrhA.
To better understand the signals to which LrhA responds, we sought to identify genes that affect its synthesis and activity. To this end, we performed a minitransposon mutagenesis screen in a strain carrying an lrhA′-′lacZ fusion. LysR regulators, such as LrhA, often bind ligands which regulate their activity (30). Since LrhA regulates itself (10, 18), our transposon mutagenesis screen in the lrhA′-′lacZ fusion strain could identify genes that regulated either LrhA activity or its expression, and knowledge of such factors could provide insight into the nature of the signals sensed by LrhA. Over 26,000 transposon-mutated colonies were screened for changes in LacZ activity on both X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) LB agar and lactose MacConkey agar, and linkage to the transposon was confirmed using standard techniques.
An insertion in the rcsD (yojN) gene that decreased the LacZ activity of the lrhA′-′lacZ fusion was identified. RcsD is a member of the Rcs system which responds to cell envelope stress. It is a phosphorelay protein, and upon activation of the system, it phosphorylates the response regulator RcsB, which, in turn, directs synthesis of capsule and other gene products (22). RcsD also has a phosphatase activity; therefore, rcsD null mutants have increased levels of phosphorylated RcsB, since this protein can be phosphorylated by small-molecule donors (22).
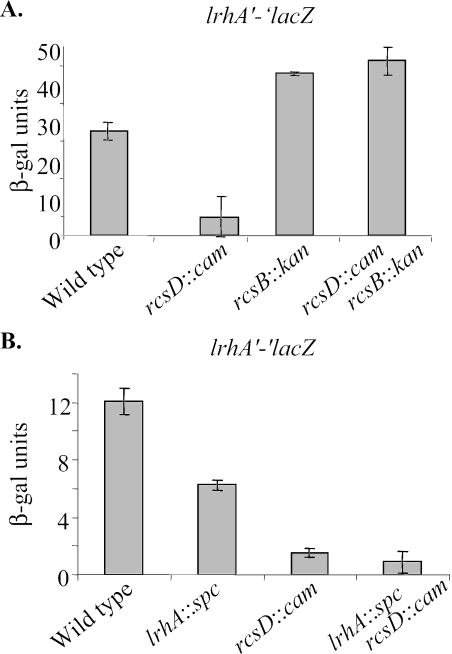
β-Galactosidase assays were performed on samples taken from exponentially growing cultures of the wild type and the rcsD::cam mutant. The rcsD::cam insertion decreased the lrhA′-′lacZ fusion activity fivefold (Fig. 5A). To confirm that the rcsD mutation affected lrhA synthesis through the Rcs pathway, a mutation was introduced in rcsB. Without RcsB, rcsD::cam did not repress lrhA expression (Fig. 5A). Both the double mutant and an rcsB::kan single mutant had only a very modest and insignificant increase in lrhA synthesis compared to wild-type strains (Fig. 5A). Thus, we predict that the phosphorylated form of RcsB, which accumulates in the rcsD mutant, negatively regulates the lrhA′-′lacZ fusion.
FIG. 5.
The Rcs phosphorelay represses lrhA synthesis. (A) β-Galactosidase assays show that the rcsD::cam mutant found in our screen decreases lrhA′-′lacZ expression fivefold (TC15 and TC132). This repression is abolished in an rcsB rcsD double mutant (CNP265), while the single mutation rcsB (CNP264) does not have a strong phenotype. (B) RcsD represses LrhA synthesis rather than affecting LrhA activity. In the absence of wild-type LrhA (TC18), synthesis of lrhA′-′lacZ is still affected by a mutation in rcsD (VC16). The lower levels of lrhA′-′lacZ activity in the lrhA::spc strain (TC18 compared to TC15) confirm that LrhA positively autoregulates itself (18). All samples were taken from exponentially growing cells and processed as described in Materials and Methods.
RcsA is an additional transcription factor that been shown to coregulate a number of promoters with RcsB. The protein is usually found at extremely low levels, since it is degraded by the Lon protease. A mutation in lon causes RcsA levels to increase approximately 100-fold, which leads to 100-fold induction of the cps genes (8, 34). This mutation was used to see if RcsA is involved in regulating lrhA synthesis. β-Galactosidase assays showed that the presence of lon::Tn10 does not cause lrhA′-′lacZ repression; in fact, in this mutant there is a slight increase (∼15%) in lrhA synthesis (data not shown). Thus, RcsA does not contribute to repression of lrhA.
To see if the rcsD mutation affects LrhA activity or synthesis, we tested if rcsD::cam still had an effect on the activity of the lrhA′-′lacZ fusion in the absence of LrhA. If it affected LrhA activity, then it would have no influence on the fusion in cells lacking LrhA. Replacing wild-type lrhA with the null allele caused lrhA′-′lacZ fusion activity to decrease (Fig. 5B). There have been conflicting reports on the nature of the LrhA autoregulation (10, 18), and these results confirm that LrhA a has positive feedback loop. The rcsD mutant affected lrhA′-′lacZ in both the wild-type and the lrhA::spc backgrounds. Therefore, the Rcs pathway represses lrhA synthesis rather than LrhA activity.
FtsK reduces synthesis of lrhA.
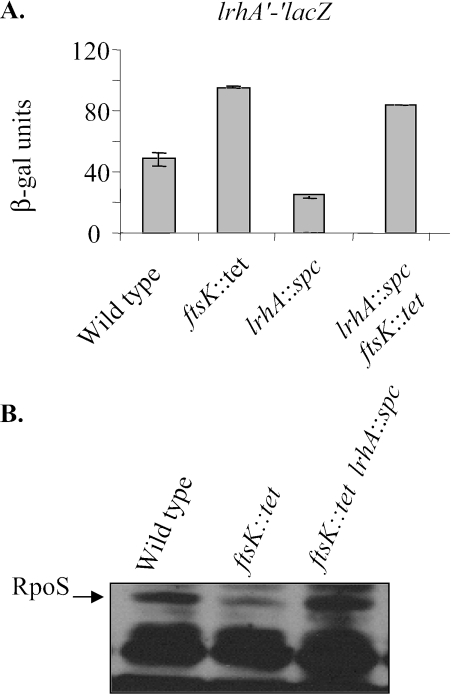
Another transposon insertion found in the screen for mutations affecting expression of the lrhA′-′lacZ fusion was in the ftsK gene, which encodes a protein involved in DNA translocation. Mutants that contained disruptions of this gene showed an increase of lrhA′-′lacZ activity compared to otherwise wild-type strains (Fig. 6A). To test if this mutation affected LrhA activity or synthesis, β-galactosidase assays were performed with a strain containing both the ftsK::tet and lrhA::spc alleles. Despite the absence of LrhA, there was still an increase in lrhA′-′lacZ levels in the ftsK mutant compared to those in the wild type. This suggests that FstK decreases LrhA levels by decreasing its synthesis.
FIG. 6.
A mutation in ftsK increases lrhA expression. (A) β-Galactosidase assays show that the ftsK::tet mutant found in our screen stimulates lrhA′-′lacZ expression (TC46 compared to TC15). This effect persists in the lrhA::spc background (VC4 compared to TC18), indicating that the mutation affects lrhA expression rather than LrhA activity. (B) The ftsK effect on RpoS levels requires wild-type LrhA. It had previously been shown that ftsK mutants reduce RpoS levels owing to posttranscription control (7). In this Western blot, the double mutant with ftsK::tet and lrhA::spc alleles (VC4) does not have any attenuation of RpoS levels in stationary phase.
Previous reports demonstrated that mutations in ftsK impede RpoS accumulation during stationary phase by acting posttranscriptionally (7). It is possible that these mutations cause an increase in the levels of LrhA, which, in turn, represses rpoS translation. To test this hypothesis, epistasis tests were carried out by using double mutations of ftsK and lrhA. Samples were taken from stationary-phase cells, and RpoS levels were assayed by Western blot analysis. As previously reported, RpoS levels were lower in stationary phase in the ftsK mutant than in the wild type. However, when the lrhA null allele was introduced into the strain, RpoS levels were no longer diminished and instead accumulated to levels seen in the wild-type strain (Fig. 6B). This is particularly striking since in other circumstances the null allele does not have a strong phenotype. Since the mutations influence RpoS levels in opposite ways, these epistasis experiments suggest that the mechanism for FtsK modulation of RpoS levels requires LrhA and presumably its regulated sRNA(s).
DISCUSSION
Several mechanisms regulate RpoS accumulation, and each one is activated under different stresses (13). In this study, we have shown LrhA decreases RpoS levels through translation control rather than protein stability. This regulation requires the sRNA chaperone Hfq and involves both an RprA-dependent and an RprA-independent mechanism. Furthermore, we have found that mutations in rcsDB and ftsK affect LrhA synthesis.
Many environmental stresses affecting RpoS do so at the level of translation. LrhA also regulates rpoS translation and appears to be involved in the cell's response to membrane perturbations through the Rcs system. Control of rpoS translation frequently includes one or more sRNAs, and we think that the LrhA pathway is no exception. LrhA represses synthesis of the sRNA RprA and most likely affects another sRNA(s) as well. It is likely that there are multiple sRNAs that regulate RpoS in addition to those already characterized. Finding the other sRNAs can be difficult in genetic screens because they are sometimes highly redundant (19) and because they are small targets for transposons. Note that in the hfq mutant the effect of lrhA overexpression was abolished but that of the lrhA null allele persisted. Accordingly, we speculate that LrhA may repress a positively regulating sRNA in addition to RprA. In the absence of LrhA, the concentration of this sRNA would increase to a level where it could affect RpoS without the aid of Hfq. There are several examples of similar sRNAs which can affect their target mRNAs without Hfq if they are overexpressed (29, 33).
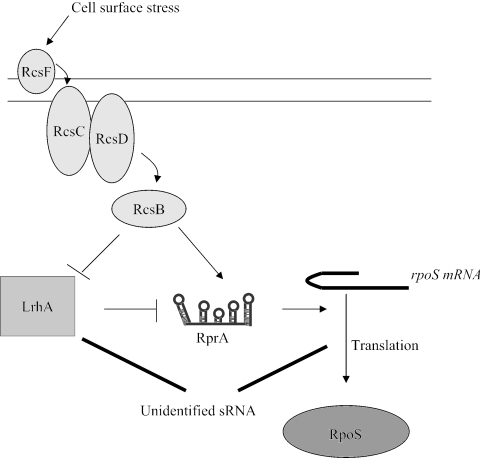
Activation of the Rcs pathway represses LrhA which, in turn, increases rpoS translation. Whether the RcsB effect on LrhA is direct or indirect is not yet apparent. It has previously been reported that stimulation of the Rcs pathway activates RpoS expression, in a manner that was partly dependent on RprA. An additional three- to fivefold induction of RpoS by the Rcs system occurred independently of RprA (23). We believe that the RprA-independent activation of RpoS by the Rcs system involves the LrhA pathway (Fig. 7).
FIG. 7.
Model of how LrhA and the Rcs phosphorelay affect RpoS translation. LrhA represses the sRNA RprA, which is an activator of RpoS translation. LrhA also regulates RpoS translation in an RprA-independent manner by an unidentified sRNA(s), which could act either positively or negatively, as indicated by heavy lines. This additional mechanism accounts for most of the effect of LrhA on RpoS translation in exponentially growing cells in LB. The Rcs system represses LrhA synthesis, and tests of epistasis demonstrate that the effect of LrhA on RpoS is independent of RcsB (data not shown). Thus, Rcs can activate RpoS translation in at least two ways: by directly stimulating RprA and by repressing LrhA synthesis.
Our screen also identified ftsK mutants that affected LrhA synthesis. We postulate that the previously reported posttranscriptional inhibitory effect of ftsK mutations on RpoS accumulation may be due to the overexpression of lrhA in these mutants (7). Our screen was not saturated, and future screens may discover additional pathways that regulate lrhA. In particular, it would be of interest to find mutations that affect LrhA activity. Since it belongs to the LysR family of proteins, there is most likely a ligand which controls its activity.
Acknowledgments
We are grateful to Natividad Ruiz for insightful discussions. We are indebted to Gottfried Unden, Susan Gottesman and Gigi Storz for kindly sending strains. We thank members of the Silhavy lab and Johan Paulsson for critical reading of the manuscript and Susan DiRenzo for her assistance in preparation of the manuscript.
T.J.S. was supported by a grant from NIGMS (GM065216).
REFERENCES
- 1.Blumer, C., A. Kleefeld, D. Lehnen, M. Heintz, U. Dobrindt, G. Nagy, K. Michaelis, L. Emody, T. Polen, R. Rachel, V. F. Wendisch, and G. Unden. 2005. Regulation of type 1 fimbriae synthesis and biofilm formation by the transcriptional regulator LrhA of Escherichia coli. Microbiology 151:3287-3298. [DOI] [PubMed] [Google Scholar]
- 2.Brown, L., and T. Elliott. 1997. Mutations that increase expression of the rpoS gene and decrease its dependence on hfq function in Salmonella typhimurium. J. Bacteriol. 179:656-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 4.Castillo, A., and S. Reverchon. 1997. Characterization of the pecT control region from Erwinia chrysanthemi 3937. J. Bacteriol. 179:4909-4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Condemine, G., A. Castillo, F. Passeri, and C. Enard. 1999. The PecT repressor coregulates synthesis of exopolysaccharides and virulence factors in Erwinia chrysanthemi. Mol. Plant-Microbe. Interact. 12:45-52. [DOI] [PubMed] [Google Scholar]
- 6.Cunning, C., and T. Elliott. 1999. RpoS synthesis is growth rate regulated in Salmonella typhimurium, but its turnover is not dependent on acetyl phosphate synthesis or PTS function. J. Bacteriol. 181:4853-4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diez, A. A., A. Tunlid, and T. Nystrom. 2002. The Escherichia coli ftsK1 mutation attenuates the induction of σS-dependent genes upon transition to stationary phase. FEMS Microbiol. Lett. 206:19-23. [DOI] [PubMed] [Google Scholar]
- 8.Ebel, W., and J. E. Trempy. 1999. Escherichia coli RcsA, a positive activator of colanic acid capsular polysaccharide synthesis, functions to activate its own expression. J. Bacteriol. 181:577-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Francez-Charlot, A., B. Laugel, A. Van Gemert, N. Dubarry, F. Wiorowski, M. P. Castanie-Cornet, C. Gutierrez, and K. Cam. 2003. RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol. Microbiol. 49:823-832. [DOI] [PubMed] [Google Scholar]
- 10.Gibson, K. E., and T. J. Silhavy. 1999. The LysR homolog LrhA promotes RpoS degradation by modulating activity of the response regulator sprE. J. Bacteriol. 181:563-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottesman, S., P. Trisler, and A. Torres-Cabassa. 1985. Regulation of capsular polysaccharide synthesis in Escherichia coli K-12: characterization of three regulatory genes. J. Bacteriol. 162:1111-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris, S. J., Y. L. Shih, S. D. Bentley, and G. P. Salmond. 1998. The hexA gene of Erwinia carotovora encodes a LysR homologue and regulates motility and the expression of multiple virulence determinants. Mol. Microbiol. 28:705-717. [DOI] [PubMed] [Google Scholar]
- 13.Hengge-Aronis, R. 2002. Signal transduction and regulatory mechanisms involved in control of the σS (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol Rev. 66:373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joyce, S. A., and D. J. Clarke. 2003. A hexA homologue from Photorhabdus regulates pathogenicity, symbiosis and phenotypic variation. Mol. Microbiol. 47:1445-1457. [DOI] [PubMed] [Google Scholar]
- 15.Kang, Y., T. Durfee, J. D. Glasner, Y. Qiu, D. Frisch, K. M. Winterberg, and F. R. Blattner. 2004. Systematic mutagenesis of the Escherichia coli genome. J. Bacteriol. 186:4921-4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kleckner, N., J. Bender, and S. Gottesman. 1991. Uses of transposons with emphasis on Tn10. Methods Enzymol. 204:139-180. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 18.Lehnen, D., C. Blumer, T. Polen, B. Wackwitz, V. F. Wendisch, and G. Unden. 2002. LrhA as a new transcriptional key regulator of flagella, motility and chemotaxis genes in Escherichia coli. Mol. Microbiol. 45:521-532. [DOI] [PubMed] [Google Scholar]
- 19.Lenz, D. H., K. C. Mok, B. N. Lilley, R. V. Kulkarni, N. S. Wingreen, and B. L. Bassler. 2004. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118:69-82. [DOI] [PubMed] [Google Scholar]
- 20.Majdalani, N., S. Chen, J. Murrow, K. St John, and S. Gottesman. 2001. Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol. Microbiol. 39:1382-1394. [DOI] [PubMed] [Google Scholar]
- 21.Majdalani, N., C. Cunning, D. Sledjeski, T. Elliott, and S. Gottesman. 1998. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. USA 95:12462-12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Majdalani, N., and S. Gottesman. 2005. The Rcs phosphorelay: a complex signal transduction system. Annu. Rev. Microbiol. [DOI] [PubMed]
- 23.Majdalani, N., D. Hernandez, and S. Gottesman. 2002. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol. Microbiol. 46:813-826. [DOI] [PubMed] [Google Scholar]
- 24.Muffler, A., D. Fischer, S. Altuvia, G. Storz, and R. Hengge-Aronis. 1996. The response regulator RssB controls stability of the σS subunit of RNA polymerase in Escherichia coli. EMBO J. 15:1333-1339. [PMC free article] [PubMed] [Google Scholar]
- 25.Mukherjee, A., Y. Cui, W. Ma, Y. Liu, and A. K. Chatterjee. 2000. hexA of Erwinia carotovora ssp. carotovora strain Ecc71 negatively regulates production of RpoS and rsmB RNA, a global regulator of extracellular proteins, plant virulence and the quorum-sensing signal, N-(3-oxohexanoyl)-l-homoserine lactone. Environ. Microbiol. 2:203-215. [DOI] [PubMed] [Google Scholar]
- 26.Peterson, C. N., M. J. Mandel, and T. J. Silhavy. 2005. Escherichia coli starvation diets: essential nutrients weigh in distinctly. J. Bacteriol. 187:7549-7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pratt, L. A., and T. J. Silhavy. 1996. The response regulator SprE controls the stability of RpoS. Proc. Natl. Acad. Sci. USA 93:2488-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Repoila, F., N. Majdalani, and S. Gottesman. 2003. Small non-coding RNAs, co-ordinators of adaptation processes in Escherichia coli: the RpoS paradigm. Mol. Microbiol. 48:855-861. [DOI] [PubMed] [Google Scholar]
- 29.Ruiz, N., and T. J. Silhavy. 2003. Constitutive activation of the Escherichia coli Pho regulon upregulates rpoS translation in an Hfq-dependent fashion. J. Bacteriol. 185:5984-5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 31.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 32.Slauch, J. M., and T. J. Silhavy. 1991. cis-acting ompF mutations that result in OmpR-dependent constitutive expression. J. Bacteriol. 173:4039-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sledjeski, D. D., A. Gupta, and S. Gottesman. 1996. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 15:3993-4000. [PMC free article] [PubMed] [Google Scholar]
- 34.Stout, V., A. Torres-Cabassa, M. R. Maurizi, D. Gutnick, and S. Gottesman. 1991. RcsA, an unstable positive regulator of capsular polysaccharide synthesis. J. Bacteriol. 173:1738-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsui, H. C., H. C. Leung, and M. E. Winkler. 1994. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol. Microbiol. 13:35-49. [DOI] [PubMed] [Google Scholar]
- 36.Zgurskaya, H. I., M. Keyhan, and A. Matin. 1997. The σS level in starving Escherichia coli cells increases solely as a result of its increased stability, despite decreased synthesis. Mol. Microbiol. 24:643-651. [DOI] [PubMed] [Google Scholar]