Abstract
Transcriptome analyses have previously revealed that a gene encoding the putative amino acid transporter CtrA (YhdG) is one of the major targets of the pleiotropic regulator CodY in Lactococcus lactis and Bacillus subtilis. The role of ctrA in L. lactis was further investigated with respect to both transport activity as well as CodY-mediated regulation. CtrA is required for optimal growth in media containing free amino acids as the only amino acid source. Amino acid transport studies showed that ctrA encodes a secondary amino acid transport system that is specific for branched-chain amino acids (BCAAs) (isoleucine, leucine, and valine) and methionine, which is in disagreement with its previously proposed function (a cationic amino acid transporter), which was assigned based on homology. We propose to rename CtrA BcaP, for branched-chain amino acid permease. BcaP is a member of a group of conserved transport systems, as homologs are widely distributed among gram-positive bacteria. Deletion of bcaP resulted in the loss of most of the BCAA uptake activity of L. lactis, indicating that BcaP is the major BCAA carrier of this organism. Deletion of bcaP together with a second (putative) BCAA permease, encoded by brnQ, further reduced the viability of the strain. DNA microarray analysis showed that deletion of bcaP predominantly affects genes belonging to the regulons of the transcriptional regulator CodY, which is involved in global nitrogen metabolism and needs BCAAs for its activation, and of CmbR, which is involved in sulfur amino acid metabolism.
CodY is a well-studied transcriptional regulator that was first identified as the nutritional repressor of the dipeptide permease operon in Bacillus subtilis (55). Functional homologs of CodY are present in several gram-positive bacteria in which the protein is involved in the regulation of a wide array of genes (44, 56). A recent study has unraveled the genome-wide effects of CodY on gene expression of Lactococcus lactis (6). In this lactic acid bacterium (LAB), the majority of the CodY-regulated genes are involved in the proteolytic system (5, 18, 19). Proteolysis is essential to L. lactis, as it allows this organism to utilize the caseins present in milk as a source of essential amino acids (32). Efficient casein utilization requires the activities of an extracellular proteinase and various peptide transporters and intracellularly located peptidases (reviewed in reference 28). In addition to the transport systems for (oligo)peptides, L. lactis contains multiple (putative) permeases that facilitate the internalization of free amino acids (4). At least 10 of these systems, differing with respect to their specificity, have been characterized biochemically in various LAB to date (29), but most of the encoding genes have not yet been identified (33).
Besides the prominent role of CodY in coordinating expression of the genes that constitute the proteolytic system of L. lactis, transcriptome analysis of a lactococcal codY mutant revealed that several transcriptional units involved in the metabolism of certain amino acids are CodY controlled as well (6). Particularly, transcript levels of the glutamate, histidine, and branched-chain amino acid (BCAA) biosynthetic operons (gltDB, his, and leu-ilv, respectively) were strongly elevated upon deletion of codY. Although L. lactis MG1363 is auxotrophic for histidine and BCAAs (49), due to frameshifts and small deletions in the coding regions of their biosynthetic operons, expression of the genes is apparently still tightly regulated.
The finding that CodY also regulates the transcription of genes involved in the metabolism of amino acids other than BCAAs was surprising, since BCAAs are solely responsible for the activation of lactococcal CodY (5, 19, 48) and thus have a central role in CodY-mediated regulation. In both L. lactis and B. subtilis, BCAAs modulate the activity of CodY by increasing the affinity of the regulator for its operator sites. In B. subtilis, an additional level of regulation of CodY activity is provided by GTP, a marker of the energy state of the cell, which stimulates CodY activity independently of BCAAs (50, 54). CodY is most active in rapidly growing L. lactis cells, when BCAAs are abundant. When intracellular levels drop, repression by CodY is relieved, which results in the derepression of the proteolytic system and a concomitant increase in the capacity to utilize milk proteins.
L. lactis preferably uses oligopeptides as a source of amino acids during growth in milk (25). Although free amino acids are rather scarce in milk (43), they probably affect intracellular amino acid pools via the activity of amino acid uptake systems. These uptake systems, therefore, could be important for CodY-mediated regulation. Indeed, one of the newly identified members of the lactococcal CodY regulon encodes a putative amino acid transporter (6). This gene, ctrA, was found to be one of the main targets of CodY in DNA microarray experiments. Transcriptional regulation of ctrA by CodY was confirmed by expression studies in which the upstream region of ctrA was fused to the reporter gene lacZ; the amount of lacZ mRNA was increased almost 10-fold in a codY deletion strain grown in nitrogen-rich medium. In addition, these experiments showed that the mRNA of the reporter was highly abundant in the mutant strain, indicating that ctrA is preceded by a strong promoter. The upstream region of ctrA contains three copies of a conserved nucleotide stretch, termed the CodY box, that has recently been shown to serve as an operator site for CodY (6, 19a). Electrophoretic mobility shift assays and DNase I footprinting analyses revealed that CodY directly interacts with this sequence in the upstream region of ctrA (6). B. subtilis contains a homolog of CtrA (i.e., YhdG). Like lactococcal ctrA, transcription of yhdG is strongly affected by CodY, as it was derepressed 116-fold in a codY deletion strain in a DNA microarray analysis (44).
The ctrA gene product (a predicted cationic amino acid transporter) was designated as such because it contains domains that are conserved between the cationic amino acid permeases LysP, PotE, and AnsP, which transport lysine, putrescine/ornithine, and γ-aminobutyrate, respectively (4, 41). Considering the tight regulation of ctrA in both L. lactis and B. subtilis and the absence of an apparent link between the metabolism of cationic amino acids and CodY regulation, we wondered which role ctrA serves in the physiology of L. lactis. Here, we report on the cloning, functional expression, and characterization of ctrA. We show that CtrA, in disagreement with its predicted function, is a transporter of BCAAs and demonstrate its importance for CodY-mediated regulation. Based on its newly identified function, we propose to rename CtrA BcaP, for branched-chain amino acid permease.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Strains and plasmids used in this study are listed in Table 1. Lactococcus lactis was grown in twofold-diluted M17 broth (61) supplemented with 0.5% glucose (GM17) at 30°C or on GM17 solidified with 1.5% agar. When appropriate, 5 μg ml−1 of erythromycin and/or chloramphenicol (both from Sigma Chemical Co., St. Louis, MO) was added. Alternatively, cells were grown in a chemically defined medium (CDM), prepared as described previously (42), supplemented with 1% Casitone (Difco Laboratories, Detroit, MI) or with specific dipeptides where indicated. Growth in 96-well plates incubated at 30°C was monitored using a GENios microtiter plate reader (Tecan, Grödig, Austria). The A595 of the cultures was measured every 30 min following 15 s of shaking.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant phenotype or genotype | Source or reference |
|---|---|---|
| L. lactis strains | ||
| MG1363 | Lac− Prt−; plasmid-free derivative of NCDO712 | 13 |
| NZ9000 | MG1363 pepN::nisRK | 31 |
| NZ9700 | Nisin-producing transconjugant of MG1363 containing Tn5276 | 30 |
| MGbrnQ | MG1363 derivate, chromosomal deletion of brnQ | This work |
| MGbcaPbrnQ | MG1363 derivate, chromosomal deletion of bcaP and brnQ | This work |
| MGbcaP | MG1363 derivate, chromosomal deletion of bcaP | This work |
| Plasmids | ||
| pNZ8048 | Cmr; expression vector carrying the nisin-inducible PnisA | 26 |
| pNZ9530 | Eryr; nisRK cloned in pIL252; constitutive expression of nisRK | 26 |
| pVE6007 | Cmr; temperature-sensitive replication derivate of pWV01 | 40 |
| pORI280 | Eryrori+ RepA−; lacZ expressed constitutively from P32 promoter | 38 |
| pNGbcaP | pNG8048 containing bcaP gene | This work |
| pNGbcaP-H6 | pNG8048 containing bcaP-his6 of L. lactis MG1363 behind PnisA | This work |
| pORIΔbcaP | Eryr LacZ+; pORI280 containing bcaP deletion construct | This work |
| pORIΔbrnQ | Eryr LacZ+; pORI280 containing brnQ deletion construct | This work |
| pORIΔbcaPbrnQ | Eryr LacZ+; pORI280 containing bcaP and brnQ deletion construct | This work |
DNA manipulation, molecular cloning, and transformation.
Routine DNA manipulations were performed as described previously (51). Total chromosomal DNA from L. lactis was extracted as described previously (39). Minipreparations of plasmid DNA from L. Lactis were made using the High Pure plasmid isolation kit from Roche Molecular Biochemicals (Mannheim, Germany). Restriction enzymes and T4 DNA ligase were purchased from Roche Molecular Biochemicals. PCR amplifications were carried out using Pwo DNA polymerase (Roche Molecular Biochemicals) and oligonucleotides listed in Table 2. PCR products were purified with the High Pure PCR product purification kit (Roche Molecular Biochemicals). Electrotransformation of L. lactis was performed using a Bio-Rad Gene Pulser (Bio-Rad Laboratories, Richmond, CA) as described previously (22).
TABLE 2.
Oligonucleotides used in this study
| Name | Sequence (5′-3′)a |
|---|---|
| ctrA-P1 | GCTCTAGACTAGAATTAGGACATATATCAC |
| ctrA-P2 | CGCGGATCCCTCATAAATCCCATAATAAATCCTC |
| ctrA-P3 | CGCGGATCCTGGTTCCTTATTGGAATTGCG |
| ctrA-P4 | CCGGAATTCAACGGCTGGTACGATACGAAC |
| brnQ-P1 | GCTCTAGACATTAGTCCAAATGGCGATACC |
| brnQ-P2 | CGCGGATCCGATAGTCTTTACCAGCTAGTTTC |
| brnQ-P3 | CGCGGATCCGAGCTTCTAATATTTAGGAGCTC |
| brnQ-P4 | CCGGAATTCCATCTCGGTTTTAACGTCTGAAC |
| ctrA-N | CTAGACCACCATGGGATTTATGAGAAAAGCC |
| ctrA-C | CTAGTCTAGACGTCTTATTTCTTTTTGCGACG |
| ctrA-CH6 | CTAGTCTAGATTAGTGATGGTGATGGTGATGTTTCTTTTTGCGACGATTTCCATAA |
Restriction enzyme sites are underlined.
Construction of L. lactis deletion mutant strains.
DNA fragments containing approximately 600 bp of the flanking regions of the bcaP gene of L. lactis MG1363 were obtained by PCR using the oligonucleotide pairs ctrA-P1/ctrA-P2 and ctrA-P3/ctrA-P4 (Table 2), cut with the appropriate restriction endonucleases, and ligated into plasmid pORI280, a conditionally replicating vector (38). The resulting plasmid, pORIΔbcaP, was introduced into L. lactis MG1363 by electroporation together with the helper plasmid pVE6007. Following chromosomal integration of pORIΔbcaP, cells were grown for about 60 generations under nonselective conditions after which a clone, designated MGbcaP, in which bcaP was deleted was obtained. The bcaP mutation was confirmed by PCR and Southern blot analysis (51) using the ECL direct nucleic acid labeling system (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom). L. lactis MG1363 strains in which either brnQ or both brnQ and bcaP are deleted (MGbrnQ and MGbcaPbrnQ, respectively) were obtained using the same double-crossover strategy with primer combinations brnQ-P1/brnQ-P2 and brnQ-P3/brnQ-P4 for the deletion of brnQ (Table 2).
Overproduction and immunodetection of native and histidine-tagged BcaP.
Full-length bcaP of L. lactis MG1363 was amplified from the chromosome by PCR using oligonucleotides ctrA-N and ctrA-C (Table 2), which introduced 5′ NcoI and 3′ XbaI restriction enzyme sites, respectively. Alternatively, a sequence encoding a hexahistidine tag was fused to the 3′ end of bcaP using oligonucleotides ctrA-N and ctrA-CH6. The purified PCR products were digested with NcoI and XbaI and ligated downstream of the nisin-inducible promoter PnisA using the corresponding sites in pNG8048, resulting in pNGbcaP and pNGbcaP-H6, respectively. The plasmids were introduced into L. lactis strain NZ9000, MG1363, or MGbcaP, together with plasmid pNZ9530 (26) in the latter two strains, to enable nisin-induced production of BcaP or histidine-tagged BcaP (BcaP-H6), as described previously (7).
Production of BcaP-H6 was detected by Western hybridization. To this end, samples were taken from cultures growing in GM17 containing the appropriate antibiotics. After 2 h of induction with a 1,000-fold-diluted supernatant of a culture of the nisin-producing L. lactis strain NZ9700 (30) grown overnight, approximately 108 cells were harvested by centrifugation, resuspended in 100 μl Birnboim solution A (51) containing 1 mg/ml lysozyme (Merck KGaA, Darmstadt, Germany), and incubated for 15 min at 48°C. Proteins were separated on 10% polyacrylamide gels by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described previously (36) and transferred onto polyvinylidene difluoride membranes (Roche Molecular Biochemicals). BcaP-H6 was visualized with specific anti-His antibodies and peroxidase-anti-mouse conjugates (both from Amersham Biosciences) according to the supplier's instructions.
Amino acid transport assays.
L. lactis MG1363 (pNGbcaP; pNZ9530), L. lactis MG1363, or L. lactis MGbcaP, with the latter two harboring plasmids pNG8048 and pNZ9530, was grown at 30°C in 50 ml of GM17. At an optical density at 600 nm (OD600) of approximately 0.4, 50-μl volumes of supernatant of the nisin-producing strain L. lactis NZ9700 (30) were added to the cultures to induce BcaP production. At an OD600 of approximately 1.0, cells were harvested by centrifugation at 6,300 × g at 4°C. Cell pellets were washed with 25 ml of ice-cold CDM, concentrated to an OD600 of 10.0 in CDM lacking amino acids, and stored on ice until use. Cells were energized prior to transport assays by 5 min of incubation at 30°C in the presence of 0.5% glucose. Subsequently, 100 μl of cell suspension was mixed with 100 μl of CDM containing 500 μM of the unlabeled amino acid of interest and 0.05 μCi of the same amino acid in a 14C- or 35S-labeled form (Amersham Biosciences). For competition experiments, a 10-fold excess of unlabeled amino acid was added. Mixtures were incubated at 30°C for various time intervals while being stirred. The reactions were stopped by the addition of 2 ml of ice-cold 0.1 M LiCl, followed by filtration through 0.45-μm-pore-size nitrocellulose filters (Schleicher & Schuell GmbH, Dassel, Germany). Reaction tubes and filters were washed with another 2 ml of ice-cold 0.1 M LiCl. Subsequently, the filters were transferred to vials containing 2 ml of scintillation fluid (Packard BioScience, Groningen, The Netherlands), and radioactivity was determined using a Packard TriCarb 2000 CA liquid scintillation analyzer (Packard Instruments, Meriden, CT).
DNA microarray analysis.
DNA microarray experiments were performed essentially as described previously (6). Briefly, RNA was isolated from four separately grown cultures of L. lactis MG1363 and L. lactis MGbcaP. Subsequently, single-strand reverse transcription (amplification) and indirect labeling of total RNA with either Cy3 or Cy5 dye (Amersham Biosciences) were performed in duplicate. Labeled cDNA samples were hybridized onto slides containing amplicons representing 2,450 open reading frames (ORFs) of L. lactis MG1363 spotted in duplicate. After hybridization, slides were washed and scanned. Slide data were processed and normalized as described previously (62), yielding average ratios of gene expression levels of the mutant to those of the wild-type strain. Expression of a gene was considered to be significantly altered when its ratio of expression in the mutant compared to the wild type was >1.5 or <0.67 and had a CyberT Bayesian P value of <0.001. All DNA microarray data, including the slide images and raw data, obtained in this study are available online (http://molgen.biol.rug.nl/publication/bcaP_data/).
Nucleotide sequence accession numbers.
The L. lactis MG1363 bcaP and brnQ nucleotide sequences have been assigned GenBank accession numbers DQ377686 and DQ377687, respectively.
RESULTS
Characterization of the L. lactis MG1363 bcaP (ctrA) locus.
A gene encoding the putative amino acid transporter CtrA was previously identified in a genome-wide screen for CodY targets in L. lactis MG1363 (6). The results presented below show that CtrA is in fact a branched-chain amino acid permease. Therefore, we propose to rename the gene and protein bcaP and BcaP, respectively. The 1,398-nucleotide-containing bcaP ORF encodes a protein of 465 amino acid residues with a predicted molecular mass of 49.6 kDa and an isoelectric point of 9.91. The bcaP ORF starts at an AUG codon following a proper lactococcal ribosome-binding site (GAGGA). Regions that could serve as promoter elements are present upstream of the start codon. These putative −35 (TTGACA) and −10 (TAAAAT) sequences are separated by 17 bp and are likely to constitute the promoter, as DNA fragments comprising this region can readily drive transcription (6). In addition, three copies of a CodY-responsive element (CodY box) can be discerned in the regulatory region of bcaP. Downstream of bcaP, a palindromic sequence could form a stem-loop structure with a predicted free energy value of −13.8 kcal/mol that could serve as a rho-independent transcriptional terminator.
bcaP is required for optimal growth of L. lactis in media containing free amino acids as the sole amino acid source.
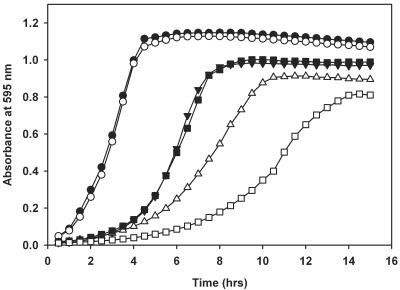
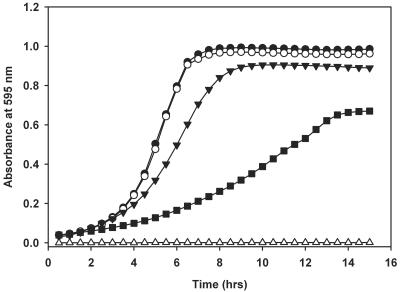
As transcription of bcaP is tightly regulated by the global regulator CodY in both L. lactis and B. subtilis and the protein is conserved among several gram-positive bacteria, we wondered which biological function it serves. To address this question, the entire bcaP gene was removed from the chromosome of L. lactis MG1363 by double-crossover recombination. The mutated strain, MGbcaP, was examined for its ability to grow in media with different amino acid or peptide contents (Fig. 1). Growth of strains MG1363 and MGbcaP was similar in CDM supplemented with Casitone. This nitrogen source consists of a tryptic digest of milk caseins and contains a wide variety of peptides of different lengths and compositions as well as free amino acids. The absence of a growth difference between the strains in this medium indicates that bcaP does not serve any apparent physiological function during growth under nitrogen-rich conditions. MGbcaP behaved differently when growth was monitored in basal CDM, which contains a mixture of all 20 amino acids in the free form in concentrations ranging from 0.2 to 5 mM but lacks peptides as amino acid source. MGbcaP reached a lower final cell density, and the maximum growth rate was significantly reduced (almost twofold) compared to that of the wild-type strain (Fig. 1). When the amino acid concentration was lowered to 75% of that in CDM (CDM75), growth of strain MGbcaP was hampered even more (Fig. 1), while the wild-type strain showed normal growth rates and final cell densities (data not shown). These results show that the presence of bcaP is required for optimal growth of L. lactis in media containing limiting amounts of amino acids.
FIG. 1.
Growth curves of L. lactis MG1363 and its bcaP deletion mutant. Growth rates of L. lactis MG1363 and MGbcaP in CDM supplemented with Casitone (closed and open circles, respectively) are shown. L. lactis MG1363, MGbcaP, and MGbcaP overproducing BcaP were grown in CDM containing free amino acids as the sole source of amino acids (closed triangles, open triangles, and closed squares, respectively). Alternatively, L. lactis MGbcaP was grown in CDM75, a medium in which the amino acid concentration was lowered to 75% of that in CDM (open squares). Cells were grown at 30°C, and the absorbance of the culture at 595 nm was measured every 30 min. Shown are the means of at least two independent experiments.
Phenotypic complementation of the bcaP gene deletion.
In order to rule out the possibility that the observed differences in growth were due to polar effects on the transcription of neighboring genes that could have been introduced through construction of the mutant, L. lactis MGbcaP was complemented by full-length bcaP. To this end, the bcaP gene was placed under the control of a nisin-inducible promoter in plasmid pNG8048 and introduced into the bcaP mutant strain. However, no protein band corresponding to BcaP could be detected by SDS-PAGE after induction of BcaP synthesis with nisin. Therefore, a histidine tag was fused to the 3′ terminus of bcaP, and proper BcaP synthesis was verified by SDS-PAGE followed by Western hybridization. A protein band specific for BcaP-H6 could indeed be detected by Western analysis upon induction of BcaP-H6 synthesis (data not shown). The size of the protein in this band corresponded to a lower molecular mass than the predicted 50.4 kDa for BcaP-H6, but it has been reported previously that membrane proteins can migrate relatively fast using SDS-PAGE (58). As can be seen in Fig. 1, normal growth was restored when native BcaP synthesis was induced in MGbcaP (pNGbcaP) grown in CDM containing the free amino acid mixture. These results show that bcaP can be provided in trans to complement MGbcaP and that polar effects of the bcaP mutation on growth are absent.
A bcaP mutation is bypassed by the addition of the BCAAs or BCAA-containing dipeptides to the growth medium.
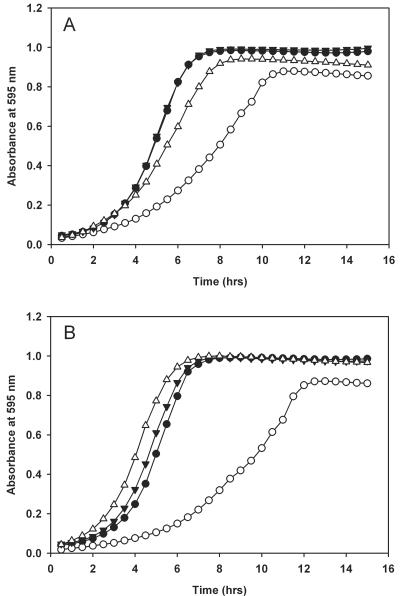
The experiments described above indicated that bcaP encodes a functional protein that is required for optimal growth in the presence of free amino acids. To find out whether BcaP facilitates amino acid transport, as assumed from BLAST results and, if so, to gain insight in its substrate specificity, growth experiments with MGbcaP were carried out in media with different amino acid concentrations. Reasoning that the absence of BcaP in the mutant strain would likely result in suboptimal intracellular levels of its substrate(s), the addition of an excess of these amino acids is expected to restore normal growth, provided that an alternative internalization pathway (e.g., another carrier) is present. Based on homology, BcaP is predicted to transport cationic amino acids, hence its original name, CtrA (4). However, no stimulatory effect on the growth of MGbcaP was observed when the medium was supplemented with lysine, arginine, putrescine, ornithine, or γ-aminobutyrate at a concentration of 5 mM of each of these compounds (data not shown). When 5 mM of the three BCAAs (leucine, isoleucine, and valine) was added together, both the maximum cell density and growth rate (almost twofold) of MGbcaP were increased significantly (Fig. 2A). The addition of 15 mM BCAAs fully restored growth (data not shown), while growth of the wild-type strain remained unchanged. In contrast, growth of MGbcaP was not enhanced when the medium was supplemented with the same amount of proline, alanine, and histidine (Fig. 2A) and all 14 other non-branched-chain amino acids (data not shown).
FIG. 2.
Growth of L. lactis MG1363 and MGbcaP in the presence of different amino acids and dipeptides. (A) Growth of L. lactis MG1363 and MGbcaP in CDM supplemented with amino acids P/A/H (closed and open circles, respectively) or I/L/V (closed and open triangles, respectively). (B) Growth of L. lactis MG1363 and MGbcaP in CDM supplemented with dipeptides PG/AH (closed and open circles, respectively) or LI/PV (closed and open triangles, respectively). Cells were grown at 30°C, and the optical density of the culture at 595 nm was measured every 30 min. Shown are the means of at least two independent experiments.
In accordance with these data, the addition of a mixture of BCAA-containing dipeptides (i.e., LI and PV) fully restored the growth of MGbcaP, whereas such a stimulatory effect was absent when dipeptides lacking BCAA residues were provided (Fig. 2B). The growth defect of MGbcaP was already bypassed at a concentration of 500 μM of the dipeptides, most probably because L. lactis has several highly efficient transport systems for dipeptides and contains multiple intracellular peptidases that generate free amino acids from dipeptides (28).
BcaP specifically transports branched-chain amino acids and, to a lesser extent, methionine.
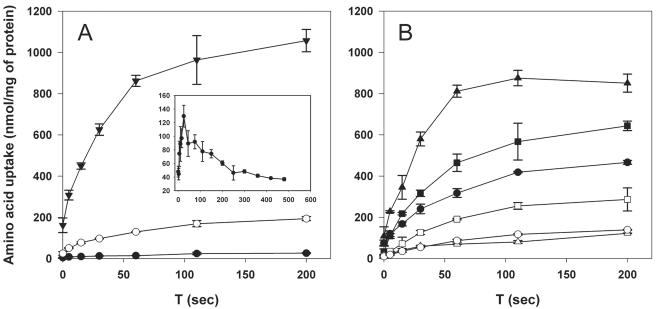
Since growth of the bcaP mutant could be restored by increasing BCAA concentrations in the medium, the lactococcal bcaP gene could encode a BCAA transporter. To test this possibility, the abilities of L. lactis MG1363 and MGbcaP to transport radioactively labeled l-isoleucine were compared (Fig. 3A). Deletion of bcaP resulted in a reduction of the initial rate of isoleucine transport by approximately ninefold, indicating that BcaP is responsible for most of the isoleucine uptake in L. lactis under these conditions. Complementation of BcaP function by overproducing BcaP in MGbcaP resulted in a strong increase (approximately 70-fold) in the uptake rate of l-[14C]isoleucine. Increasing the ratio of l-[14C]isoleucine over unlabeled isoleucine by lowering the concentration of unlabeled isoleucine in the reaction mixture from 250 to 100 μM or omitting glucose from the uptake buffer resulted in a more rapid internalization of l-[14C]isoleucine followed by a slower efflux of the isotope (Fig. 3A). Such kinetics are indicative of so-called counterflow transport and suggest that BcaP is able to facilitate bidirectional transport and most likely constitutes a secondary transporter (14, 63).
FIG. 3.
Time course of uptake of BCAAs and methionine in whole cells of L. lactis. (A) Uptake of l-isoleucine by L. lactis MG1363 (open circles), MGbcaP (closed circles), and MGbcaP (pNGbcaP) overproducing BcaP (closed triangles). l-[14C]leucine uptake in the presence of 100 μM of unlabeled substrate and in the absence of glucose in the uptake buffer is shown in the inset. (B) Uptake by L. lactis MG1363 and L. lactis MGbcaP (pNGbcaP) of l-leucine (open and closed triangles, respectively), l-valine (open and closed squares, respectively), and l-methionine (open and closed circles, respectively). Transport assays using concentrated samples of cells harvested from the exponential phase of growth were performed in the presence of 0.05 μCi 14C- or 35S-labeled amino acid and 250 μM of the corresponding unlabeled amino acid as described in Materials and Methods. Prior to the uptake assay, cells were energized by incubation for 5 min at 30°C in the presence of 0.5% glucose. T, time.
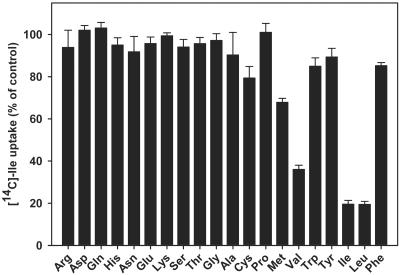
To estimate the substrate specificity of BcaP, inhibition of isoleucine transport by other amino acids was monitored in a competition experiment (Fig. 4). The addition of a 10-fold excess of isoleucine or leucine resulted in a reduction of uptake of about 80%, whereas valine was less inhibitory (64%). In addition to the BCAAs, transport of isoleucine by BcaP was inhibited by the sulfur-containing amino acids methionine (23% inhibition) and, to a lesser extent, cysteine. These results indicate that the ability of BcaP to bind amino acids increases as a function of their hydrophobicity, with the exception of those amino acids containing bulky side chains (i.e., Trp, Tyr, and Phe).
FIG. 4.
Substrate specificity of L. lactis MG1363 BcaP as determined by the inhibition of l-[14C]isoleucine uptake by l-amino acids. Shown is the percentage of l-[14C]isoleucine uptake by lactococcal cells overproducing BcaP in the presence of 250 μM unlabeled l-isoleucine (control) and a 10-fold excess of each of the 20 l-amino acids. The uptake was determined after 1 min of incubation at 30°C, and the assay was performed as described in the legend to Fig. 3. The means of three independent measurements are shown.
To test whether BcaP is capable of transporting the amino acids it recognizes, time course experiments were performed as described above for isoleucine by using radioactively labeled leucine, valine, and methionine. Consistent with the competition experiments, the transport rate of BcaP for leucine was comparable to that for isoleucine, and the rate for valine was higher than that for methionine (Fig. 3B). Uptake rates for these substrates by the bcaP mutant strain were similar to that of isoleucine (data not shown). No significant difference in transport of the nonbinding amino acid proline between the bcaP mutant and the BcaP-overproducing strain was observed (data not shown).
BcaP is conserved in several gram-positive bacteria.
The transport and growth experiments clearly show that BcaP specifically transports branched-chain amino acids and methionine, which is in disagreement with its predicted function. A BLAST search (1) with the full-length amino acid sequence against the NCBI nonredundant protein database revealed that homologs of BcaP are contained in the genomes of several gram-positive bacteria (Table 3). The amino acid sequence of lactococcal BcaP is similar to that of YhdG of B. subtilis. Like bcaP, transcription of yhdG has been shown to be strongly CodY dependent (44). Therefore, it is likely that L. lactis BcaP and B. subtilis YhdG represent functional homologs. Homologs of BcaP are present in species of Listeria, Streptococcus, Staphylococcus, and Lactobacillus, which all contain (putative) homologs of CodY (56). Notably, in the latter bacterium, a second homolog that is even more similar to lactococcal BcaP was found, although it has a C-terminal extension of about 34 amino acid residues that is not present in the L. lactis protein.
TABLE 3.
Homologs of L. lactis MG1363 BcaP in several species of gram-positive bacteria
| Species | Proteina | Protein length (amino acids) | Identity (%) | Similarity (%) | Reference or source |
|---|---|---|---|---|---|
| Lactococcus lactis MG1363 | BcaP (CtrA) | 465 | 100 | 100 | This work |
| Lactococcus lactis IL1403 | CtrA | 469 | 98 | 99 | 4 |
| Staphylococcus aureus COL | YP_187408 | 482 | 64 | 80 | 15 |
| Lactobacillus casei ATCC 334 | ZP_00384379 | 464 | 40 | 61 | b |
| Lactobacillus plantarum WCFS1 | Lp_0861 | 465 | 39 | 61 | 27 |
| Listeria innocua | Lin0648 | 463 | 36 | 61 | 16 |
| Enterococcus faecalis V583 | NP_814667 | 463 | 34 | 58 | 47 |
| Bacillus cereus G9241 | ZP_00238192 | 471 | 37 | 57 | 21 |
| Bacillus subtilis 168 | YhdG | 465 | 33 | 56 | 34 |
| Streptococcus pneumoniae R6 | YfnA | 467 | 34 | 54 | 23 |
| Streptococcus thermophilus CNRZ1066 | Str1361 | 461 | 33 | 53 | 3 |
In case no protein name was assigned, the locus tag of the encoding ORF is shown.
Part of an ongoing genome-sequencing project, the sequence data of which are available online (http://www.doe.jgi.gov).
The amino acid sequences of the BcaP homologs are extensively conserved throughout the proteins, with the exception of the N-terminal parts. A hydropathy analysis of BcaP, using the method described previously by Kyte and Doolittle (35), predicts 12 transmembrane segments (data not shown). These transmembrane helices, with an average size of 23 amino acid residues, are linked by short hydrophilic stretches that vary in size from 3 to 40 residues. Both the N and C termini are predicted to protrude into the cytoplasm. The hydrophobicity profile of BcaP is conserved in all homologs. Among the 465 amino acid residues constituting BcaP, 326 are hydrophobic (70%), 87 are polar (18.7%), 18 are acidic (3.9%), and 34 are basic (7.3%) amino acid residues, respectively. Thus, BcaP is strongly hydrophobic, which is a typical feature of integral membrane transporters.
Role of BcaP in global gene expression in L. lactis.
As transcription of bcaP is tightly regulated by the global regulator CodY and the absence of bcaP is deleterious for the cells under amino acid-limiting conditions (Fig. 1), the effect of the removal of bcaP on global gene expression was monitored by comparing the transcriptomes of L. lactis MG1363 and L. lactis MGbcaP. Both strains were grown in CDM containing free amino acids as the sole amino acid source. RNA samples were prepared from each strain and, following cDNA synthesis and labeling, hybridized to DNA microarrays representing 2,450 genes of L. lactis MG1363. Analysis of the DNA microarray data of four biological replicates revealed that the expression of approximately 20 genes or operons was significantly up-regulated, while the expression of approximately 20 genes or operons was down-regulated upon mutation of bcaP (Table 4). The majority of the up-regulated transcriptional units are involved in (branched-chain) amino acid metabolism or peptide transport. Some of these (e.g., dpp, ilv, and his) belong to the regulon of the transcriptional regulator CodY. The extent of derepression seemed less than that observed in a strain deleted for codY (6), which might be explained, because CodY is still able to regulate its targets at low levels of BCAAs or in the absence of these cofactors (5). Interestingly, expression of cysD, cysK, cysM, and metB2 was significantly increased in MGbcaP. Expression of these genes, encoding enzymes required for methionine and cysteine biosynthesis, has recently been shown to be controlled by the transcriptional activator CmbR (57). Thus, BcaP provides a link between sulfur amino acid biosynthesis and metabolism of BCAAs in L. lactis.
TABLE 4.
Comparison of the transcriptomes of L. lactis MGbcaP and L. lactis MG1363
| Transcriptional unita | Expression ratiob | Significance (P value) | Descriptionc | Regulon |
|---|---|---|---|---|
| dppA, P, B, C, D, F | 4.5 | 10−8 | Dipeptide transport system | CodY |
| ybbE | 3.0 | 10−13 | Hypothetical protein, downstream of bcaP | |
| llmg_0328 | 2.4 | 10−6 | Hypothetical protein | |
| comX | 2.2 | 10−9 | Putative regulator | |
| gltA, citB, icD | 2.1 | 10−13 | Krebs TCA cycle enzymes | CodY |
| leuC, A | 2.0 | 10−9 | Leucine biosynthesis | CodY |
| hisC, Z, G, D, B, ymdC, hisH, A, F, I, K | 1.9 | 10−10 | Histidine biosynthesis | CodY |
| metC-cysK | 1.9 | 10−8 | Methionine/cysteine synthesis | CmbR |
| rmaG | 1.9 | 10−4 | Transcriptional regulator | |
| llmg_1066 | 1.8 | 10−6 | Putative membrane protein | |
| llmg_0472/0473 | 1.7 | 10−5 | Hypothetical proteins | |
| ilvD, B, N, C, A, aldB | 1.6 | 10−7 | BCAA biosynthesis | CodY |
| fabI | 1.6 | 10−5 | Enoyl-ACP reductase | |
| purC, H | 1.6 | 10−7 | Purine synthesis | CmbR |
| cysD | 1.6 | 10−11 | O-Acetylserine sulfhydrylase | CmbR |
| ydbE | 1.6 | 10−4 | Hypothetical protein | |
| glgD | 1.6 | 10−6 | Glucose-1-phosphate adenylyltransferase | CmbR? |
| yrbB | 1.5 | 10−10 | Hypothetical protein | |
| yriD | 1.5 | 10−6 | Hypothetical protein | CmbR |
| cysM | 1.5 | 10−7 | Cysteine synthase | CmbR |
| cpo | 0.7 | 10−6 | Hydrolase or acyltransferase | |
| yrcA | 0.7 | 10−7 | Phospho-beta-glucosidase | |
| yjaE | 0.7 | 10−4 | Hypothetical protein | |
| ynjC | 0.7 | 10−4 | Hypothetical protein | |
| argF | 0.7 | 10−6 | Ornithine carbamoyltransferase | |
| yedF | 0.6 | 10−7 | PTS II ABC component | |
| trpB | 0.6 | 10−6 | Tryptophan synthase beta chain | |
| butB | 0.6 | 10−6 | Dehydrogenase | |
| ptcA | 0.6 | 10−5 | Cellobiose-PTS component | |
| panE | 0.6 | 10−9 | Ketopantoate reductase | |
| fruA | 0.6 | 10−5 | Fructose-PTS component | |
| pgmB | 0.5 | 10−9 | Beta-phosphoglucomutase | |
| busAB | 0.5 | 10−8 | Betaine ABC transporter | |
| cstA | 0.5 | 10−4 | Carbon starvation protein | |
| bglS, ybhD | 0.5 | 10−6 | Beta-glucosidase A, hypothetical protein | |
| yecA | 0.5 | 10−5 | Transcriptional regulator | |
| chiA, yucG | 0.4 | 10−5 | Chitinase, chitin-binding protein | |
| rcfB, yxbD | 0.4 | 10−4 | Transcriptional regulator, transporter | |
| argH | 0.4 | 10−7 | Argininosuccinate lyase | |
| yjjA | 0.3 | 10−14 | Hypothetical protein |
The gene (in an operon) that shows the highest ratio of expression is underlined.
Ratio of expression in L. lactis MGbcaP over that in L. lactis MG1363.
(Putative) gene function. TCA, tricarboxylic acid; ACP, acyl carrier protein; PTS, phosphotransferase system.
BrnQ contributes to BCAA transport in L. lactis.
Although BcaP is responsible for most of the BCAA uptake in L. lactis (Fig. 3), MGbcaP can still grow in CDM containing only free amino acids (Fig. 1). Since L. lactis MG1363 is auxotrophic for BCAAs, the strain must contain at least one other transport system for BCAAs that is active under those conditions. L. lactis MG1363 contains a homolog of BrnQ, a low-affinity BCAA carrier in a number of gram-positive bacteria including Lactobacillus delbruckii (59). To investigate whether BrnQ might serve a similar purpose in L. lactis MG1363, the gene was deleted from the chromosome, and the growth characteristics of the resulting strain (MGbrnQ) were examined (Fig. 5). Unlike for MGbcaP, no significant growth difference between L. lactis MG1363 and MGbrnQ grown in CDM75 was observed. In contrast, a strain in which both brnQ and bcaP were deleted (MGbcaPbrnQ) was not viable in this medium. After a prolonged lag phase, the double-mutant strain did grow in standard CDM, but growth was significantly hampered (ninefold), and the culture reached a lower final cell density than the wild-type strain (1.5-fold). Growth of the double mutant was enhanced by the addition of BCAAs to the CDM. No differences in growth behavior between wild-type, mutant, and double-mutant strains were observed in GM17 or in CDM supplemented with Casitone (data not shown). These results indicate that brnQ most likely serves as a second, but less efficient, BCAA transport system in L. lactis MG1363.
FIG. 5.
Growth of L. lactis MG1363, MGbrnQ, and MGbcaPbrnQ in different media. Growth levels of L. lactis MG1363 (closed circles), MGbrnQ (open circles), and MGbcaPbrnQ (open triangles) in CDM75, MGbcaPbrnQ (closed triangles) in CDM75 supplemented with amino acids I/L/V, and MGbcaPbrnQ (squares) in standard CDM are shown. Cells were grown at 30°C, and the optical density of the culture at 595 nm was measured every 30 min. Shown are the means of at least two independent experiments.
DISCUSSION
During the past decades, various studies have led to the identification of transport systems responsible for the uptake of amino acids in LAB (29). Although many of the properties of these carriers were revealed, most of the genetic elements involved remained elusive. In the current study, a tightly regulated and conserved gene encoding a putative cationic amino transporter, CtrA (BcaP), was cloned, and its protein product was functionally characterized. For several reasons we conclude that the encoded protein indeed constitutes an amino acid transporter that, in contrast to its predicted function, specifically transports BCAAs and, to a lesser extent, methionine. Firstly, the growth defect brought about by the removal of bcaP from the chromosome was complemented by the addition of BCAAs or BCAA-containing dipeptides to the medium and not by those containing other amino acids. Secondly, the bcaP deletion strain showed a strongly reduced uptake activity of BCAAs and methionine compared to that of its parental strain. Finally, overexpression of plasmid-encoded CtrA in MGbcaP resulted in a significant increase in the uptake of BCAA and methionine. In view of its newly identified function, we have renamed CtrA BcaP, for branched-chain amino acid permease.
We show that the presence of bcaP is beneficial to L. lactis when it is grown in CDM containing free amino acids as the sole amino acid source and that this advantage is lost when a complex mixture of peptides (in the form of Casitone) is added to the medium (Fig. 1). When peptides are available, BcaP is probably superfluous, as intracellular BCAA pools can be maintained through the uptake and intracellular cleavage of peptides into free amino acids. Complementation of growth of the bcaP mutant was already achieved upon the addition of 500 μM of BCAA-containing dipeptides, which are substrates for the highly efficient transport systems encoded by dpp (52, 53) and dtpT (12, 20), whereas concentrations of the corresponding free amino acids in CDM need to be over 10-fold higher for full complementation. It remains to be established which physiological role BcaP serves for L. lactis growing in milk, as amino acid concentrations in this environment are low and oligopeptides are the main source of nitrogen (25). It may be that BcaP facilitates the excretion of redundant BCAAs under these conditions.
The absence of an apparent physiological function of BcaP during growth under peptide-rich conditions could explain why its transcription is tightly regulated by CodY, the master regulator of the genes constituting the proteolytic system of L. lactis (6). The bcaP gene is one of the main targets of the repressor, and it has been shown that bcaP transcription is directly regulated by CodY (6). CodY regulates its targets through a so-called CodY box, a DNA motif that serves as a high-affinity binding site for CodY. Upstream of bcaP, no less than three copies of the CodY box are present, which might reflect the stringent regulation of bcaP by CodY. As CodY activity itself is modulated by BCAAs (5, 48, 54), the regulator is expected to be fully active when BCAAs or peptides containing BCAAs are abundant. However, it appears that bcaP is not fully repressed in peptide-rich medium, as a difference in isoleucine uptake between the isogenic wild-type and bcaP mutant strains could still be discerned when both were grown in GM17 (Fig. 3). Preliminary experiments indicate that in a strain in which codY is deleted, some peptide-dependent regulation of bcaP transcription still occurs (data not shown). This suggests the presence of a second, CodY-independent mechanism of repression of bcaP transcription.
As for many LAB, L. lactis MG1363 is auxotrophic for a number of amino acids (BCAAs, Met, and His). Since cells of MGbcaP are viable in media containing free amino acids as the only amino acid sources, an additional transport system that retrieves BCAAs from the growth medium must be active under these conditions. A homolog of the well-conserved BCAA permease BrnQ (45, 59, 60) is specified by the chromosome of L. lactis MG1363, and our data suggest that the product of this gene is responsible for part of the BCAA uptake in MGbcaP. A region of BcaP is homologous to a stretch of amino acids of BrnQ, which might be an indication that this region contains a substrate (BCAA) recognition domain. Future studies will be required to detail structure-function relationships of these conserved transport systems.
Previously, a lactococcal amino acid carrier with specificity for BCAAs was characterized biochemically using membrane isolates, but no gene encoding this carrier was identified (8-10). Transport of BCAAs by the carrier was inhibited by l-amino acids containing branched or aliphatic side chains with at least three methyl groups. As amino acid uptake by BcaP is strongly inhibited by BCAAs and methionine, the latter of which can be regarded as an aliphatic amino acid since the S atom in the side chain is largely nonreactive, the data presented in the current study indicate that bcaP might be the structural gene encoding this unknown carrier described previously by Driessen and coworkers (8-10). Moreover, those authors have shown that substrates of the transport system are translocated in symport with one proton and that transport can be bidirectional, which fits well with our observations that BcaP constitutes a secondary transport system that enables the counterflow of its substrates. However, part of the BCAA uptake by L. lactis, as observed previously by Driessen and coworkers, might be due to BrnQ activity. BrnQ was characterized as a low-affinity BCAA carrier in Lactobacillus delbruckii (59). In L. lactis, BrnQ activity is probably also responsible for only a minor fraction of total BCAA uptake, at least under the conditions examined in this work, as the functional removal of bcaP resulted in a loss of most of the isoleucine transport capacity (Fig. 3) and deletion of brnQ alone did not have any effect on growth (Fig. 5). The redundancy of BCAA transport systems in L. lactis stresses the importance of BCAAs for the cell. BCAAs serve as precursors for branched-chain fatty acids and are the most abundant amino acids in membrane proteins. Transcription of L. lactis brnQ was recently shown to be affected by environmental stress (65). In Salmonella enterica serovar Typhimurium and Corynebacterium glutamicum, transcription of brnQ is modulated by BCAAs (37, 46), but the mechanisms remain unknown. Future work will have to show whether expression of brnQ in L. lactis is regulated.
Surprisingly, the strain in which both brnQ and bcaP were deleted was still viable in CDM containing only free amino acids, although growth started only after a prolonged lag phase and the culture reached a much lower final cell density (Fig. 5) than the wild-type or single-mutant strains. These results suggest the presence of at least one more system that enables BCAA transport in L. lactis. Alternatively, the cells might acquire the necessary BCAAs through passive diffusion via the membrane. To identify candidates that could be responsible for the remaining BCAA influx, we searched the lactococcal genome for other putative BCAA uptake system genes. The product of ydgC shares homology with Aap, a general amino acid permease of Rhizobium leguminosarum (64). Transcription of this gene, like bcaP, is dependent on CodY (6) and might therefore be important for BCAA metabolism. In addition, L. lactis MG1363 contains a copy of azl, an operon suggested to be involved in BCAA transport in B. subtilis (2). Future work will aim to establish whether lactococcal ydcG and azl encode functional proteins and whether they are involved in BCAA transport.
A number of genes encoding enzymes involved in sulfur amino acid metabolism were differentially expressed upon removal of bcaP (Table 4). In L. lactis, most of these genes are controlled by CmbR (11, 57). The activity of this LysR-type transcriptional activator is modulated by O-acetyl serine (17), a precursor in cysteine biosynthesis. In addition, it has been suggested that other regulatory mechanisms might be important for coordinating sulfur metabolism (57). It could be that the changes in the expression of sulfur metabolism genes, as a consequence of the bcaP mutation, are the result of changes in the intracellular pool of methionine. Similarly, up-regulation of many target genes of the pleiotropic regulator CodY in MGbcaP might be explained by lowered intracellular BCAA levels in this strain, leading to the inactivation of CodY and the concomitant derepression of genes belonging to the CodY regulon. Most of these genes encode transporters and amino acid biosynthetic enzymes that could help to counteract the effects of the deletion of bcaP. Since BcaP is responsible for most of the uptake of BCAAs and (at least) part of the methionine, this transporter provides a link between BCAA and sulfur amino acid metabolism.
Acknowledgments
We are grateful to Eric Geertsma, Hein Trip, and Arjen Nauta for helpful suggestions and critical review of the manuscript. We thank Koen Cornelissen for valuable contributions to this study.
This work was supported by Friesland Foods Corporate Research and the Dutch Ministry of Economic Affairs (SENTER).
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Belitsky, B. R., M. C. Gustafsson, A. L. Sonenshein, and W. C. Von. 1997. An lrp-like gene of Bacillus subtilis involved in branched-chain amino acid transport. J. Bacteriol. 179:5448-5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolotin, A., B. Quinquis, P. Renault, A. Sorokin, S. D. Ehrlich, S. Kulakauskas, A. Lapidus, E. Goltsman, M. Mazur, G. D. Pusch, M. Fonstein, R. Overbeek, N. Kyprides, B. Purnelle, D. Prozzi, K. Ngui, D. Masuy, F. Hancy, S. Burteau, M. Boutry, J. Delcour, A. Goffeau, and P. Hols. 2004. Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat. Biotechnol. 22:1554-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.den Hengst, C. D., P. Curley, R. Larsen, G. Buist, D. van Sinderen, O. P. Kuipers, and J. Kok. 2005. Probing direct interactions between CodY and the oppD promoter of Lactococcus lactis. J. Bacteriol. 187:512-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.den Hengst, C. D., S. A. F. T. van Hijum, J. M. Geurts, A. Nauta, J. Kok, and O. P. Kuipers. 2005. The Lactococcus lactis CodY regulon: identification of a conserved cis-regulatory element. J. Biol. Chem. 280:34332-34342. [DOI] [PubMed] [Google Scholar]
- 7.de Ruyter, P. G., O. P. Kuipers, and W. M. de Vos. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Driessen, A. J., S. de Jong, and W. N. Konings. 1987. Transport of branched-chain amino acids in membrane vesicles of Streptococcus cremoris. J. Bacteriol. 169:5193-5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Driessen, A. J., K. J. Hellingwerf, and W. N. Konings. 1987. Mechanism of energy coupling to entry and exit of neutral and branched chain amino acids in membrane vesicles of Streptococcus cremoris. J. Biol. Chem. 262:12438-12443. [PubMed] [Google Scholar]
- 10.Driessen, A. J., J. Kodde, S. de Jong, and W. N. Konings. 1987. Neutral amino acid transport by membrane vesicles of Streptococcus cremoris is subject to regulation by internal pH. J. Bacteriol. 169:2748-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernandez, M., M. Kleerebezem, O. P. Kuipers, R. J. Siezen, and R. van Kranenburg. 2002. Regulation of the metC-cysK operon, involved in sulfur metabolism in Lactococcus lactis. J. Bacteriol. 184:82-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foucaud, C., E. R. Kunji, A. Hagting, J. Richard, W. N. Konings, M. Desmazeaud, and B. Poolman. 1995. Specificity of peptide transport systems in Lactococcus lactis: evidence for a third system which transports hydrophobic di- and tripeptides. J. Bacteriol. 177:4652-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geertsma, E. R., R. H. Duurkens, and B. Poolman. 2005. The activity of the lactose transporter from Streptococcus thermophilus is increased by phosphorylated IIA and the action of beta-galactosidase. Biochemistry 44:15889-15897. [DOI] [PubMed] [Google Scholar]
- 15.Gill, S. R., D. E. Fouts, G. L. Archer, E. F. Mongodin, R. T. Deboy, J. Ravel, I. T. Paulsen, J. F. Kolonay, L. Brinkac, M. Beanan, R. J. Dodson, S. C. Daugherty, R. Madupu, S. V. Angiuoli, A. S. Durkin, D. H. Haft, J. Vamathevan, H. Khouri, T. Utterback, C. Lee, G. Dimitrov, L. Jiang, H. Qin, J. Weidman, K. Tran, K. Kang, I. R. Hance, K. E. Nelson, and C. M. Fraser. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187:2426-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. Garcia-del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 17.Golic, N., M. Schliekelmann, M. Fernandez, M. Kleerebezem, and R. van Kranenburg. 2005. Molecular characterization of the CmbR activator-binding site in the metC-cysK promoter region in Lactococcus lactis. Microbiology 151:439-446. [DOI] [PubMed] [Google Scholar]
- 18.Guédon, E., P. Renault, S. D. Ehrlich, and C. Delorme. 2001. Transcriptional pattern of genes coding for the proteolytic system of Lactococcus lactis and evidence for coordinated regulation of key enzymes by peptide supply. J. Bacteriol. 183:3614-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guédon, E., P. Serror, S. D. Ehrlich, P. Renault, and C. Delorme. 2001. Pleiotropic transcriptional repressor CodY senses the intracellular pool of branched-chain amino acids in Lactococcus lactis. Mol. Microbiol. 40:1227-1239. [DOI] [PubMed] [Google Scholar]
- 19a.Guédon, E., B. Sperandio, N. Pons, S. D. Ehrlich, and P. Renault. 2005. Overall control of nitrogen metabolism in Lactococcus lactis by CodY, and possible models for CodY regulation in Firmicutes. Microbiology 151:3895-3909. [DOI] [PubMed] [Google Scholar]
- 20.Hagting, A., E. R. Kunji, K. J. Leenhouts, B. Poolman, and W. N. Konings. 1994. The di- and tripeptide transport protein of Lactococcus lactis. A new type of bacterial peptide transporter. J. Biol. Chem. 269:11391-11399. [PubMed] [Google Scholar]
- 21.Hoffmaster, A. R., J. Ravel, D. A. Rasko, G. D. Chapman, M. D. Chute, C. K. Marston, B. K. De, C. T. Sacchi, C. Fitzgerald, L. W. Mayer, M. C. Maiden, F. G. Priest, M. Barker, L. Jiang, R. Z. Cer, J. Rilstone, S. N. Peterson, R. S. Weyant, D. R. Galloway, T. D. Read, T. Popovic, and C. M. Fraser. 2004. Identification of anthrax toxin genes in a Bacillus cereus associated with an illness resembling inhalation anthrax. Proc. Natl. Acad. Sci. USA 101:8449-8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holo, H., and I. F. Nes. 1995. Transformation of Lactococcus by electroporation. Methods Mol. Biol. 47:195-199. [DOI] [PubMed] [Google Scholar]
- 23.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D. J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P. M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183:5709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reference deleted.
- 25.Juillard, V., D. Le Bars, E. R. Kunji, W. N. Konings, J. C. Gripon, and J. Richard. 1995. Oligopeptides are the main source of nitrogen for Lactococcus lactis during growth in milk. Appl. Environ. Microbiol. 61:3024-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kleerebezem, M., M. M. Beerthuyzen, E. E. Vaughan, W. M. de Vos, and O. P. Kuipers. 1997. Controlled gene expression systems for lactic acid bacteria: transferable nisin-inducible expression cassettes for Lactococcus, Leuconostoc, and Lactobacillus spp. Appl. Environ. Microbiol. 63:4581-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. Fiers, W. Stiekema, R. M. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kok, J., and G. Buist. 2003. Genetics of proteolysis in Lactococcus lactis, p. 189-224. In B. J. B. Wood and W. M. de Vos (ed.), Genetics of lactic acid bacteria. Kluwer Academics/Plenum Publishers, New York, N.Y.
- 29.Konings, W. N., B. Poolman, and A. J. Driessen. 1989. Bioenergetics and solute transport in lactococci. Crit. Rev. Microbiol. 16:419-476. [DOI] [PubMed] [Google Scholar]
- 30.Kuipers, O. P., M. M. Beerthuyzen, R. J. Siezen, and W. M. de Vos. 1993. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 216:281-291. [DOI] [PubMed] [Google Scholar]
- 31.Kuipers, O. P., P. G. de Ruyter, M. Kleerebezem, and W. M. de Vos. 1998. Quorum sensing controlled gene expression in lactic acid bacteria. Biotechnology 64:15-21. [Google Scholar]
- 32.Kunji, E. R., A. Hagting, C. J. De Vries, V. Juillard, A. J. Haandrikman, B. Poolman, and W. N. Konings. 1995. Transport of beta-casein-derived peptides by the oligopeptide transport system is a crucial step in the proteolytic pathway of Lactococcus lactis. J. Biol. Chem. 270:1569-1574. [DOI] [PubMed] [Google Scholar]
- 33.Kunji, E. R., I. Mierau, A. Hagting, B. Poolman, and W. N. Konings. 1996. The proteolytic systems of lactic acid bacteria. Antonie Leeuwenhoek 70:187-221. [DOI] [PubMed] [Google Scholar]
- 34.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, and A. Danchin. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 35.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 36.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 37.Lange, C., D. Rittmann, V. F. Wendisch, M. Bott, and H. Sahm. 2003. Global expression profiling and physiological characterization of Corynebacterium glutamicum grown in the presence of l-valine. Appl. Environ. Microbiol. 69:2521-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leenhouts, K., G. Buist, A. Bolhuis, A. ten Berge, J. Kiel, I. Mierau, M. Dabrowska, G. Venema, and J. Kok. 1996. A general system for generating unlabelled gene replacements in bacterial chromosomes. Mol. Gen. Genet. 253:217-224. [DOI] [PubMed] [Google Scholar]
- 39.Leenhouts, K. J., J. Kok, and G. Venema. 1990. Stability of integrated plasmids in the chromosome of Lactococcus lactis. Appl. Environ. Microbiol. 56:2726-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maguin, E., P. Duwat, T. Hege, D. Ehrlich, and A. Gruss. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 174:5633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marchler-Bauer, A., J. B. Anderson, P. F. Cherukuri, C. Weese-Scott, L. Y. Geer, M. Gwadz, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, G. H. Marchler, M. Mullokandov, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2005. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 33:D192-D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mierau, I., A. J. Haandrikman, O. Velterop, P. S. Tan, K. L. Leenhouts, W. N. Konings, G. Venema, and J. Kok. 1994. Tripeptidase gene (pepT) of Lactococcus lactis: molecular cloning and nucleotide sequencing of pepT and construction of a chromosomal deletion mutant. J. Bacteriol. 176:2854-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mills, O. E., and T. D. Thomas. 1981. Nitrogen sources for growth of lactic streptococci in milk. N. Z. J. Dairy Sci. Technol. 16:43-55. [Google Scholar]
- 44.Molle, V., Y. Nakaura, R. P. Shivers, H. Yamaguchi, R. Losick, Y. Fujita, and A. L. Sonenshein. 2003. Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J. Bacteriol. 185:1911-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohnishi, K., A. Hasegawa, K. Matsubara, T. Date, T. Okada, and K. Kiritani. 1988. Cloning and nucleotide sequence of the brnQ gene, the structural gene for a membrane-associated component of the LIV-II transport system for branched-chain amino acids in Salmonella typhimurium. Jpn. J. Genet. 63:343-357. [DOI] [PubMed] [Google Scholar]
- 46.Ohnishi, K., K. Matsubara, Y. Hattori, H. Sadanari, R. Yamada, and S. Fukuda. 1999. Identification of a cis-acting regulatory sequence responsible for the repression of brnQ in Salmonella typhimurium. Biochim. Biophys. Acta 1445:196-206. [DOI] [PubMed] [Google Scholar]
- 47.Paulsen, I. T., L. Banerjei, G. S. Myers, K. E. Nelson, R. Seshadri, T. D. Read, D. E. Fouts, J. A. Eisen, S. R. Gill, J. F. Heidelberg, H. Tettelin, R. J. Dodson, L. Umayam, L. Brinkac, M. Beanan, S. Daugherty, R. T. Deboy, S. Durkin, J. Kolonay, R. Madupu, W. Nelson, J. Vamathevan, B. Tran, J. Upton, T. Hansen, J. Shetty, H. Khouri, T. Utterback, D. Radune, K. A. Ketchum, B. A. Dougherty, and C. M. Fraser. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071-2074. [DOI] [PubMed] [Google Scholar]
- 48.Petranovic, D., E. Guedon, B. Sperandio, C. Delorme, D. Ehrlich, and P. Renault. 2004. Intracellular effectors regulating the activity of the Lactococcus lactis CodY pleiotropic transcription regulator. Mol. Microbiol. 53:613-621. [DOI] [PubMed] [Google Scholar]
- 49.Poolman, B., and W. N. Konings. 1988. Relation of growth of Streptococcus lactis and Streptococcus cremoris to amino acid transport. J. Bacteriol. 170:700-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ratnayake-Lecamwasam, M., P. Serror, K. W. Wong, and A. L. Sonenshein. 2001. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev. 15:1093-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 52.Sanz, Y., F. C. Lanfermeijer, P. Renault, A. Bolotin, W. N. Konings, and B. Poolman. 2001. Genetic and functional characterization of dpp genes encoding a dipeptide transport system in Lactococcus lactis. Arch. Microbiol. 175:334-343. [DOI] [PubMed] [Google Scholar]
- 53.Sanz, Y., F. Toldra, P. Renault, and B. Poolman. 2003. Specificity of the second binding protein of the peptide ABC-transporter (Dpp) of Lactococcus lactis IL1403. FEMS Microbiol. Lett. 227:33-38. [DOI] [PubMed] [Google Scholar]
- 54.Shivers, R. P., and A. L. Sonenshein. 2004. Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids. Mol. Microbiol. 53:599-611. [DOI] [PubMed] [Google Scholar]
- 55.Slack, F. J., P. Serror, E. Joyce, and A. L. Sonenshein. 1995. A gene required for nutritional repression of the Bacillus subtilis dipeptide permease operon. Mol. Microbiol. 15:689-702. [DOI] [PubMed] [Google Scholar]
- 56.Sonenshein, A. L. 2005. CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr. Opin. Microbiol. 8:203-207. [DOI] [PubMed] [Google Scholar]
- 57.Sperandio, B., P. Polard, D. S. Ehrlich, P. Renault, and E. Guedon. 2005. Sulfur amino acid metabolism and its control in Lactococcus lactis IL1403. J. Bacteriol. 187:3762-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Steffes, C., J. Ellis, J. Wu, and B. P. Rosen. 1992. The lysP gene encodes the lysine-specific permease. J. Bacteriol. 174:3242-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stucky, K., A. Hagting, J. R. Klein, H. Matern, B. Henrich, W. N. Konings, and R. Plapp. 1995. Cloning and characterization of brnQ, a gene encoding a low-affinity, branched-chain amino acid carrier in Lactobacillus delbruckii subsp. lactis DSM7290. Mol. Gen. Genet. 249:682-690. [DOI] [PubMed] [Google Scholar]
- 60.Tauch, A., T. Hermann, A. Burkovski, R. Kramer, A. Puhler, and J. Kalinowski. 1998. Isoleucine uptake in Corynebacterium glutamicum ATCC 13032 is directed by the brnQ gene product. Arch. Microbiol. 169:303-312. [DOI] [PubMed] [Google Scholar]
- 61.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Hijum, S. A., J. A. De, R. J. Baerends, H. A. Karsens, N. E. Kramer, R. Larsen, C. D. den Hengst, C. J. Albers, J. Kok, and O. P. Kuipers. 2005. A generally applicable validation scheme for the assessment of factors involved in reproducibility and quality of DNA-microarray data. BMC Genomics 6:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Veenhoff, L. M., and B. Poolman. 1999. Substrate recognition at the cytoplasmic and extracellular binding site of the lactose transport protein of Streptococcus thermophilus. J. Biol. Chem. 274:33244-33250. [DOI] [PubMed] [Google Scholar]
- 64.Walshaw, D. L., and P. S. Poole. 1996. The general L-amino acid permease of Rhizobium leguminosarum is an ABC uptake system that also influences efflux of solutes. Mol. Microbiol. 21:1239-1252. [DOI] [PubMed] [Google Scholar]
- 65.Xie, Y., L. S. Chou, A. Cutler, and B. Weimer. 2004. DNA macroarray profiling of Lactococcus lactis subsp. lactis IL1403 gene expression during environmental stresses. Appl. Environ. Microbiol. 70:6738-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]