Abstract
Transcription of the carAB operon encoding the unique carbamoylphosphate synthase of Escherichia coli reflects the dual function of carbamoylphosphate in the biosynthesis of arginine and pyrimidine nucleotides. The tandem pair of promoters is regulated by various mechanisms depending on the needs of both pathways and the maintenance of a pyrimidine/purine nucleotide balance. Here we focus on the linker regions that impose the distribution of target sites for DNA-binding proteins involved in pyrimidine- and purine-specific repression of the upstream promoter P1. We introduced deletions and insertions, and combinations thereof, in four linkers connecting the binding sites for integration host factor (IHF), PepA, PurR, and RNA polymerase and studied the importance of phasing and spacing of the targets and the importance of the nucleotide sequence of the linkers. The two PepA binding sites must be properly aligned and separated with respect to each other and to the promoter for both pyrimidine- and purine-mediated repression. Similarly, the phasing and spacing of the IHF and PEPA2 sites are strictly constrained but only for pyrimidine-specific repression. The IHF target is even dispensable for purine-mediated regulation. Thus, a correct localization of PepA within the higher-order nucleoprotein complex is a prerequisite for the establishment of pyrimidine-mediated repression and for the coupling between purine- and pyrimidine-dependent regulation. Our data also suggest the existence of a novel cis-acting pyrimidine-specific regulatory target located around position −60. Finally, the analysis of a P1 derivative devoid of its control region has led to a reappraisal of the effect of excess adenine on P1 and has revealed that P1 has no need for a UP element.
In Escherichia coli, carbamoylphosphate, a precursor common to the biosynthesis of arginine and the pyrimidine nucleotides, is synthesized by a unique carbamoylphosphate synthase, encoded by the carAB operon. The profile of carAB expression reflects this key position and dual metabolic function: transcription initiation is inhibited by arginine-, pyrimidine-, and purine-dependent mechanisms (for a recent survey by Charlier and Glansdorff, see reference 5). The carAB control region contains two promoters and various target sites for regulatory and architectural proteins. The arginine-specific repression of the downstream promoter P2 and the molecular details of the liganded ArgR-operator interaction are reasonably well documented (9, 25). In contrast, our knowledge of regulation of the P1 promoter is still incomplete and patchy. P1 activity is mainly repressed by excess pyrimidines and to a smaller extent by purines (Fig. 1) (2, 12, 21). P1 is also down regulated by UTP-sensitive reiterative transcription (16) and, upon nutritional stress, by stringent control (2). At least three multifunctional proteins, integration host factor (IHF), PepA (aminopeptidase A), and PyrH (UMP kinase), are involved in the predominant protein-dependent pyrimidine-specific repression of P1 (6, 8, 18), but many questions remain concerning their interplay and communication to the RNA polymerase. In this study we focus on the role of the base pair composition and length of the intervening sequences connecting the target sites for the DNA-binding proteins and define the basic requirements to bring about the regulatory responses.
FIG. 1.
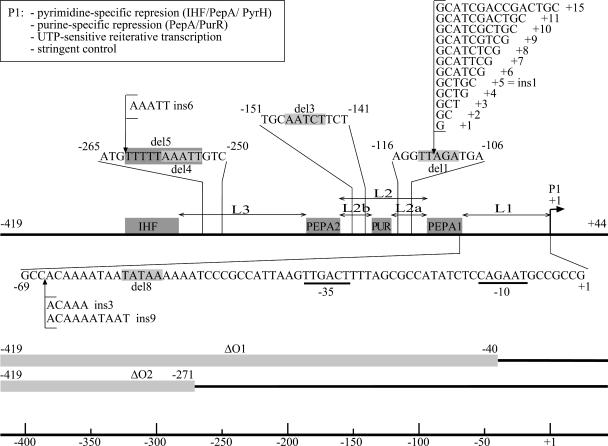
Schematic representation of the carP1 control region and of the various mutations analyzed in this work. The IHF, PepA, and PurR binding sites are indicated with gray colored boxes. Numbering is with respect to the start of P1 transcription (+1). The positions and nucleotide sequences of the various insertion mutations are indicated. The 5- and 10-bp deletions are indicated with gray-boxed sequences. The large deletions ΔO1 and ΔO2 are indicated with gray colored bars.
IHF and PepA play an important architectural role in the assembly of a regulatory nucleoprotein complex. PepA plays a primordial role in the establishment of both the pyrimidine- and purine-specific repression of P1 activity (6, 7, 12). The binding of IHF to its target, situated around position −304, far upstream of the start point of P1 transcription (Fig. 1), mainly reinforces the pyrimidine-specific repression (8, 12). PyrH appears to be the sensor of the pyrimidine-specific regulatory system (18). Its mode of action and the identity of the small effector molecule(s) are still not known. A direct and specific binding of purified PyrH to the car operator could not be demonstrated in vitro, and it is unclear whether PyrH contacts the DNA or whether it is recruited solely by protein-protein contacts (7, 18).
PepA is a hexameric leucine-aminopeptidase that is also endowed with DNA-binding activity (6). PepA binds to the carAB control region and also to its own control region (6) and to the ColE1 cer site (1, 22), where it imposes in conjunction with ArgR the directionality of the site-specific resolution reaction of ColE1 multimers (15, 23). The aminopeptidase activity of the enzyme is neither required for transcriptional regulation nor for the resolution reaction (6, 20). The DNA-binding mode of PepA is not well characterized and atypical in the sense that the protein bears no classical DNA-binding motif (24). Separation-of-function pepA mutant analyses indicated the importance of the amino-terminal domain of the enzyme for DNA binding (7, 22). The same analyses revealed the existence of pepA mutants deficient for the regulatory response but not affected in the DNA-binding capacity and proficient for the resolution reaction. Therefore, binding of PepA to the P1 control region is required but not sufficient to explain its role in transcriptional repression. These observations lead us to propose the involvement of protein-protein interactions, likely with PyrH (7). Since then, the three-dimensional structures of E. coli PepA (24) and of E. coli and Pyrococcus furiosus PyrH (3, 19) have been solved. On the basis of the complementarity of charge distribution on the surfaces of PepA and PyrH, Marco-Marín and coworkers (19) recently proposed the existence of a binding platform for PyrH on the PepA hexamer, but the interaction has yet to be demonstrated.
Which operator sequences are needed for efficient binding of PepA to the carAB control region? DNase I footprinting studies have revealed two nearly 30-bp-long stretches of continuous protection and numerous regularly spaced hyperreactive sites alternating with short zones of reduced accessibility for the nuclease (6). This complex pattern suggested the existence of two areas of direct contact (PEPA1 and PEPA2 in Fig. 1) and an important remodelling of the operator upon PepA binding, possibly by wrapping a large part of the carAB control region around hexameric PepA. The contacted areas in the carAB, pepA, and cer sites exhibit poor sequence conservation, and little is known about the precise requirements for cis-acting elements in PepA binding. It is likely that PepA recognizes the local DNA structure rather than a defined array of base-specific groups.
How do the various proteins and operator DNA cooperate in modulation of promoter activity? What are the constraints imposed on the structural organization of cis-acting regulatory elements in the generation of the pyrimidine- and purine-dependent regulatory responses? Previous in vitro and in vivo data strongly suggest that P1 repression involves long-distance effects and the assembly of a specific higher-order nucleoprotein complex in which the operator DNA is profoundly remodelled (4, 6). Consequently, sequences that are not directly contacted by the various DNA-binding proteins could nevertheless play an important role in setting the regulatory signals. The linker regions dictate the helical-phase-related distribution and separation of regulatory sites for potentially interacting proteins. In conjunction with the target sites, they also determine the global configuration and conformability of the operator DNA.
In this paper we analyze the importance of the four linkers connecting the binding sites for IHF, PepA, PurR, and RNA polymerase in the establishment of pyrimidine- and purine-dependent repression of carP1 promoter activity. Previously, we have shown that a mutation altering the distance between the PEPA1 and PEPA2 regulatory sites severely affects both the pyrimidine- and purine-specific repression (12). Here we extend this analysis to all intervening stretches connecting all presently identified regulatory binding sites and the promoter. We make a distinction between effects due to changes in the nucleotide sequences of the linkers, to improper phasing of the regulatory sites, and to variations in distance with conservation of phasing. To do so, we have introduced insertion and deletion mutations of various lengths, and combinations thereof, in four selected areas of the P1 control region. Their effects on promoter activity and repressibility were assayed with a single-copy carP1-lacZ reporter gene fusion construct. We have used the electrophoretic mobility shift assay to measure the in vitro binding of PepA and IHF to mutant operators and truncated operator fragments. This enabled us to ascribe regulatory malfunctioning to defective protein-DNA binding or to interference occurring at a different stage in the development of the regulatory response, which can be either improper assembly of the higher-order complexes or aberrant cross talk with the polymerase.
MATERIALS AND METHODS
E. coli strains, plasmids, and growth conditions.
The genotypes and construction of E. coli strains MC1061, CSH100 (F′), and FW102 (F−); the pepA::Tn10, ihfA::Tn10, and purR::Tn10 derivatives of strain FW102; and the carP1-lacZ bearing reporter fusion constructs pET-carP1 and pFW-carP1 have been described previously (12). Complex medium (medium 853) and minimal medium (medium 132) have been described previously (14). Glucose was used at 0.5% (wt/vol), uracil and adenine at 50 μg/ml, thiamine at 0.1 μg/ml, kanamycin at 30 μg/ml, streptomycin and chloramphenicol at 20 μg/ml, and tetracycline at 7.5 μg/ml.
Site-directed mutagenesis.
Insertions and deletions were introduced in the carP1 control region using the overlap PCR method (17) using AB1Eco and AB3Bam (12) as outside primers and different pairs of complementary mutagenic oligonucleotides as inside primers. Amplification was performed with the high-fidelity Pfu DNA polymerase (Roche) and pET-carP1 plasmid DNA as the template. The amplicons were purified and digested with EcoRI and BamHI and ligated in the similarly digested and dephosphorylated vector pFW11-null (Kmr Cmr) (26). Transformation of strain MC1061 was with the CaCl2 procedure (11). Plasmid DNA was retrieved from Kmr transformants (mini plasmid extraction kit; QIAGEN), analyzed for the presence of the insert by restriction enzyme analysis, and sequenced. Recombinant plasmids were transformed into the F′ strain CSH100, and double-crossover events transferring the plasmid-borne carP1-lacZ fusion to the F′ episome were selected upon conjugation with the Smr Kms Cms F− strain FW102 or derivatives thereof, and screening for Cms transconjugants as described previously (26).
Overexpression and purification of PepA and IHF.
PepA was purified from a 2.5-liter culture of E. coli strain JM101 carrying the pKHW1 plasmid (6) grown in complex medium supplemented with ampicillin and induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at an optical density at 660 nm of 0.9 for 3 hours. Purification was performed by the method of McCulloch et al. (20) as modified by Charlier et al. (6). A final purification step by size exclusion chromatography was added. The PepA solution was concentrated to a volume of 2 ml and loaded onto a Hiload 16/60 superdex 200 preparative grade column (Amersham Biosciences). The column was equilibrated and run with the same buffer (50 mM Tris-HCl [pH 8.0], 10 mM MgCl2, 1 M NaCl, 0.1 mM dithiothreitol [DTT], and 0.01 mM MnCl2) at 0.5 ml/min.
Overproduction of IHF was achieved with E. coli strain HN880 containing a plasmid in which the transcription of ihfA and ihfB is driven by a thermo-inducible promoter (obtained from M. Faelen). Purification was performed by the method of Filutowicz et al. (13) with minor modifications. A 1-liter culture was grown at 32°C in complex medium supplemented with ampicillin and induced by increasing the temperature to 42°C at an optical density at 660 nm of 0.9 for 2 hours. The cells were harvested by centrifugation, and the bacterial pellet was resuspended in 30 ml of buffer A (25 mM Tris-HCl, pH 7.4, 1 mM EDTA, 3 mM β-mercaptoethanol, 50 mM NaCl) supplemented with 23 μg DNase I/ml. Cells were broken by sonication, and the extract was kept at 4°C for 30 min and centrifuged at 42,000 × g for 35 min to remove cell debris. The supernatant was centrifuged again for 2.5 h at 130,000 × g in an ultracentrifuge. The supernatant was recovered and subjected to a two-step ammonium sulfate precipitation. First, 0.334 g of ammonium sulfate was added to each milliliter of high-speed supernatant, and the mixture was stirred and centrifuged for 20 min at 42,000 × g. Then, 0.23 g of ammonium sulfate was added per ml of high-speed supernatant to reach the concentration of 0.564 g of ammonium sulfate per ml. After the mixture was stirred, it was centrifuged for 20 min at 42,000 × g. The precipitate was dissolved in 12 ml of buffer A and dialyzed overnight at 4°C against 2 liters of buffer A using slide-A-lyzer dialysis cassettes with a molecular weight cutoff of 3,500 (Pierce). The dialysate was filtered prior to loading onto a 5-ml HiTrap heparin HP column (Amersham Biosciences) equilibrated with buffer A. IHF was eluted over 10 column volumes with a linear gradient of 0.05 to 1.7 M NaCl in buffer A at 2.5 ml/min. IHF eluted from the column at 1.4 M NaCl.
Electrophoretic mobility shift assays.
5′-32P-single-end-labeled DNA fragments were prepared by PCR amplification with pFW-carP1 plasmid DNA or a pFW-carP1 derivative as the template. The primers were a pair of oligonucleotides, one of which was 5′ end labeled. PCR products were purified by gel electrophoresis on 6% polyacrylamide gels. Binding experiments were performed at 37°C by incubating different amounts of purified protein and a constant amount of labeled DNA (15,000 cpm) in the presence of an excess of nonspecific competitor (sonicated calf thymus DNA). PepA binding was assayed in 10 mM Tris-HCl, pH 7.9, 125 mM KCl, 10 mM MgCl2, 0.1 mM DTT, and 5% glycerol. IHF binding was assayed in 10 mM Tris-HCl, pH 7.9, 125 mM KCl, 10 mM MgCl2, 10 mM CaCl2, 0.1 mM DTT, and 25% glycerol. Separation of free and bound DNA was performed on 5% and 6% polyacrylamide gels for PepA and IHF binding, respectively. Migration was for 3 h at 8 V/cm in TEB buffer (89 mM Tris, 89 mM boric acid, 0.25 mM EDTA). Apparent dissociation equilibrium constants (KD) were determined by densitometry of the free DNA bands as a function of the protein concentration and are the mean values of at least two assays.
β-Galactosidase assays.
Specific activities were assayed in cell extracts as described previously (7). Cells were harvested by centrifugation of cultures in the exponential growth phase at a density of 4 × 108 cells/ml. The addition of adenine and/or uracil had no significant effect on the growth rate or on the F′ episome number. All assays were performed at least three times.
RESULTS
Rationale of mutant construction and analysis.
The insertion and deletion mutations analyzed in this work were introduced in the context of a single-copy F′ episome-borne reporter construct in which lacZ is fused to the carP1 promoter/operator region extending from positions −419 to +44 relative to the start of transcription (Materials and Methods). The elimination of the arginine-repressible promoter P2 rules out all possible, direct and indirect, interferences and effects of downstream RNA polymerase binding and transcription initiation at P2 (also generating transient topological effects) on P1 activity. Thus, the β-galactosidase activities of the constructs solely reflect P1 promoter activities. The making of the constructs, the methods of cultivation and harvesting of the cells in the exponential growth phase, and the enzyme assays of cell extracts have been described previously (7).
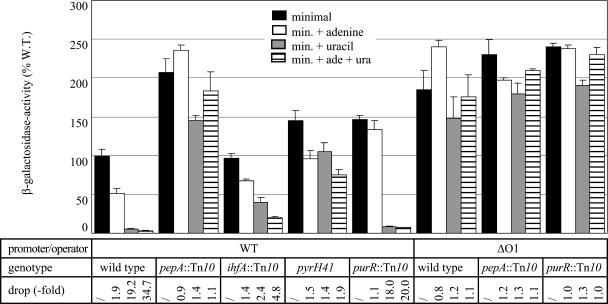
The activities of the wild-type F′-P1-lacZ construct measured in cells grown on differently supplemented media and with various genetic contexts (pepA, ihfA, pyrH, and purR) have been published before (12). These values will be used as a reference throughout this paper and are included in Fig. 2. These data clearly demonstrated that P1 activity is subject to a major repression by excess uracil (20-fold reduction) and to a minor repression by excess adenine (2-fold reduction). The pyrimidine-specific repression was shown to be PepA, IHF, and PyrH dependent, and the purine-specific repression was shown to be PurR and PepA dependent.
FIG. 2.
Histogram presentation of β-galactosidase activities measured in cell extracts of E. coli strain FW102 (wild type) or isogenic pepA::Tn10, ihfA::Tn10, pyrH41, or purR::Tn10 derivatives thereof, carrying the single-copy episome-borne carP1-lacZ fusion or its P1-ΔO1 derivative. Cells were grown on minimal medium (minimal), minimal medium supplemented with adenine (min. + adenine), uracil (min. + uracil), or both (min. + ade + ura) as indicated. One hundred percent corresponds to the activity measured in the wild-type (W. T.) strain (FW102) bearing the wild-type P1-lacZ fusion grown on minimal medium. The drop indicates the reduction in enzyme specific activity measured in cells grown in the presence of excess adenine, uracil, or both, with respect to the activity measured in the same strain grown on minimal medium. The values for the wild-type promoter construct are from Devroede et al. (12). Values are the means ± standard deviations (error bars) of at least three independent assays. /, no drop.
The P1-ΔO1 promoter construct reveals that P1 has no need for a UP element and sets apart protein-dependent repression from other direct and/or indirect pyrimidine- and purine-dependent effects.
The elimination of the whole region upstream of position −40 (P1-ΔO1 [Fig. 1]) and its replacement by a totally different and plasmid-derived stretch resulted in near-constitutive expression at a high level, indicating that this region is essential for regulation but not required for basal promoter function (Fig. 2). The level of P1 activity measured with the P1-ΔO1 construct was nearly as high as the level with the wild-type promoter in the genetically derepressed pepA::Tn10 background. Therefore, we may conclude that P1 does not require this promoter upstream A+T-rich region for high-level expression, and consequently, it is unlikely that P1 bears a functional UP element.
A close inspection of the data also revealed two minor effects. These relatively small effects go undetected in the wild-type P1 fusion construct, as they are overruled by the major pyrimidine- and purine-specific repression mechanisms. First, a small (about 1.2-fold) residual pyrimidine-dependent reduction of promoter activity was noted for the P1-ΔO1 construct (Fig. 2). A similar effect was also observed in the pepA::Tn10 background for both wild-type P1 and P1-ΔO1. This small pyrimidine-dependent reduction of promoter activity must without doubt be ascribed to a regulatory effect(s) distinct from the major protein-dependent repression. Second, the activity of the P1-ΔO1 promoter proved to be slightly but consistently stimulated (about 1.3-fold) upon the addition of adenine to both minimal and uracil-supplemented medium (Fig. 2). This effect must undoubtedly be indirect and likely reflects the cross talk between the purine and pyrimidine nucleotide biosynthetic pathways and variations in the respective nucleotide pools. Consistent with this hypothesis, in the purR::Tn10 background where the purine biosynthetic pathway is genetically derepressed, P1-ΔO1 activity was already at its maximum in unsupplemented minimal medium and was not stimulated further by excess adenine (Fig. 2). Combined, these data suggest that excess adenine exerts antagonistic effects on P1 activity—a weak indirect stimulation and a stronger PurR- and PepA-dependent repression—resulting in a net twofold down regulation.
Insertion and deletion mutations in linker regions of the P1 promoter-operator: impact of nucleotide sequence, proper phasing, and separation of regulatory sites on promoter activity and repressibility.
The positions of the zones of direct contact with PepA (PEPA1 and PEPA2) generate three main linker regions in the carP1 operator: L1 (31 bp) between the −35 promoter element and PEPA1, L2 (65 bp) comprised between PEPA1 and PEPA2, and L3 (96 bp) that connects PEPA2 to the IHF binding site (Fig. 1). As L2 contains the PUR box, the linker is compound and can be subdivided into the L2a (23-bp) and L2b (26-bp) stretches. We have introduced one-half and full-turn insertions, deletions, and combinations thereof in each one of these intervening sequences and analyzed the effects on P1 activity in the wild-type context (FW102) and in isogenic pepA, ihfA, and purR derivatives grown on different media.
One-half and full-turn insertions in the L1 linker.
The length of linker L1, between the −35 promoter element and PEPA1 was increased by 5 bp (ins3) or by 10 bp (ins9). This was done by duplication of the overlapping stretches immediately downstream of the promoter-proximal extremity of PEPA1, 5′-ACAAA- (−66 to −62) and 5′-ACAAAATAAT (−66 to −57), respectively (Fig. 1). The duplication strategy (in contrast to the insertion of unrelated sequences) minimizes the perturbation of the local sequence and therefore restricts interference with the binding of the RNA polymerase and regulatory proteins.
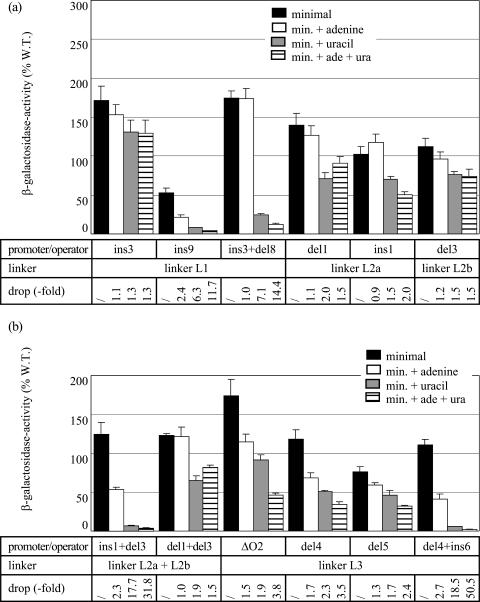
The half-turn insertion ins3 nearly abolished both the pyrimidine- and purine-mediated repression (Fig. 3). The introduction of the one-turn insertion, ins9, that increases the distance even further but restores the proper phasing, resulted in a twofold reduction of promoter activity but interfered less, though still significantly, with the regulatory processes. The pyrimidine-dependent repression was reduced about threefold with respect to the wild-type P1 construct, but in contrast, the purine-mediated regulation was intact (Fig. 3). Both the residual repression by uracil and the repression exerted by excess adenine proved to be PepA dependent, and the purine-dependent repression relies on PurR as well (data not shown). The expression levels measured with the ins3 del8 double mutant (restoration of original phasing and separation) indicate that the introduction of the 5-bp deletion del8 (deletion of TATAA from −57 to −53 [Fig. 1]) in the ins3 control region partially restored the repression by excess uracil (about sevenfold repression) (Fig. 3). This restored down regulation is similar to the repressibility of the ins9 mutant and was PepA and IHF dependent (data not shown). Notice that the ins9 insertion affects the local sequence and introduces one supplementary helical turn, whereas the ins3 del8 combination restores the original distribution of all regulatory sites with respect to P1 but results in two single-base-pair substitutions: T-A to C-G at position −60 and T-A to A-T at position −57 (Fig. 1). The net twofold negative effect of excess adenine measured in the presence of uracil (Fig. 3) suggests that PurR-mediated repression is still operational in this double mutant, at least in the presence of uracil. The lack of a net purine-dependent repression in the absence of uracil might be explained by the partial deficiency of pyrimidine-specific repression in this mutant and the tight coupling between purine- and pyrimidine-specific repression.
FIG. 3.
Effects of the cis-acting mutations and the combination of compensatory insertion and deletion mutations in the L1, L2a, L2b, and L3 linkers on P1 promoter activity and repressibility. β-Galactosidase activity is expressed as a percentage with respect to the activity of the wild-type (W. T.) construct/strain grown on minimal medium. For further details, see the legend to Fig. 2.
Combined, these results indicate that the alignment on the DNA helix of the promoter and at least some of the regulatory sites in the upstream control region is a prerequisite for the establishment of both pyrimidine- and purine-specific repression. In contrast, the exact number of helical turns and the sequence of L1 are not crucial for purine-specific repression. Moreover, the effects of the double-base-pair substitution (ins3 del8) indicate that linker L1 may comprise a previously undetected regulatory element primarily involved in pyrimidine-specific repression.
One-half and full-turn insertions and deletions in linkers L2a and L2b.
Previously, we have shown that the 5-bp deletion del3 (deleting AATCT from −148 to −144) in linker L2b, between the PEPA2 and PUR boxes, abolishes the pyrimidine- and purine-specific repression (12). Shortening or increasing the distance between PEPA1 and PUR (linker L2a) by one-half turn, as in the del1 (deleting TTAGA from −113 to −109) and ins1 (insertion of GCTGC between −111 and −112) mutants, respectively, proved to have similar effects (Fig. 1 and 3). In contrast, the ins1 del3 double mutant, harboring the combination of an insertion and deletion of equal length, in L2a and L2b, respectively, was fully repressible by pyrimidines as well as purines. Further analyses in the pepA::Tn10 and ihfA::Tn10 genetic backgrounds indicate that repression in the ins1 del3 double mutant is PepA and IHF dependent (data not shown). In this double mutant, the positions of the PepA and IHF binding sites are unchanged relative to each other and to the promoter. In contrast, the PUR box is displaced by one-half turn with respect to the promoter and to both PEPA sites. Nevertheless, as shown previously (12), the purine-specific repression was normal. Because repression in this double mutant was fully restored compared to the single mutants, we may conclude that the sites of the ins1, del1, and del3 mutations do not constitute a sequence-specific protein-DNA recognition site. Unlike the ins1 del3 mutations, the double mutations del1 del3, combining two 5-bp deletions within the linkers L2a and L2b, respectively, resulted in a severely hampered pyrimidine-specific repression (10-fold drop) and completely abolished purine-specific repression (Fig. 3). This combination of half-turn deletions also restores the proper phasing of all sites except PUR but differs from the ins1 del3 combination in the total length of L2, which is shorter by one helical turn. As a consequence, PEPA2 and the IHF box are brought one turn closer to PEPA1 and to the promoter. Thus, the total length of L2 is critical for both pyrimidine- and purine-specific repression, but the position of the PurR box within linker L2 is tolerant of change.
To better evaluate the sensitivity of the pyrimidine- and purine-specific components of P1 repression to perturbations of the phasing and separation of regulatory sites, we have generated a homogenous series of insertion mutations of systematically increasing length (1 to 11 bp and 15 bp). As the extra sequences were introduced at the site of ins1 (see above), they should not disrupt any protein binding site. The full set of data is provided (see Fig. S1 in the supplemental material).
The introduction of even a single base pair had significant effects on P1 activity and repressibility. The 1-bp insertion resulted in a twofold increase in promoter activity on minimal medium to a level comparable to that of wild-type P1 activity in a pepA strain, and an approximately twofold reduction of the pyrimidine-specific control.
The pyrimidine-dependent repression was further reduced in the 2-bp insertion mutant (eightfold reduction) and nearly vanished in the insertion mutants of 3 bp and more. As the insertions reached the length of a nearly full helical turn (10- and 11-bp insertions), the repressibility increased slightly but remained extremely low (about two to threefold). Thus, pyrimidine-dependent repression of P1 activity is highly sensitive to alterations in the distance separating the PEPA sites.
Excess adenine resulted in different effects in the various insertion mutants. For the 1-, 8-, and 15-bp insertion mutants, we could still observe an approximately 1.5-fold down regulation by excess adenine in the presence of uracil. This effect was PurR dependent, suggesting that PurR still binds and partially represses these mutant promoters. In all other mutants, the purine-dependent repression was abolished.
Deletions in linker L3.
The removal of the upstream part of the carAB control region (positions −419 to −271), including the IHF box and part of linker L3, as in the P1-ΔO2 construct (Fig. 1), severely affected the pyrimidine-dependent repression (10-fold reduction) compared to wild-type P1 repression but hardly affected the purine-mediated repression (Fig. 3). Shortening the L3 linker by 5 bp (deleting AAATT from −257 to −253) in the del4 mutant (Fig. 1) proved to have little effect on the purine-mediated repression but severely hampered the pyrimidine-specific repression (Fig. 3). Similarly, the 10-bp deletion del5 (deleting TTTTTAAATT from −262 to −253 [Fig. 1]) reduced the pyrimidine-mediated repression to less than a factor of two but also reduced the effect of excess adenine more so than the del4 mutation did (Fig. 3). The full-range, PepA- and IHF-dependent repression observed for the deletion/insertion del4 ins6 double mutant (Fig. 1 and 3) indicates that the 5-bp deletion does not destroy a sequence-specific recognition element for protein binding. As expected, disruption of the pepA gene further reduced the effect of excess uracil and abolished the purine-specific repression of all linker L3 mutants (data not shown).
Combinations of 5-bp insertions and deletions in different linkers.
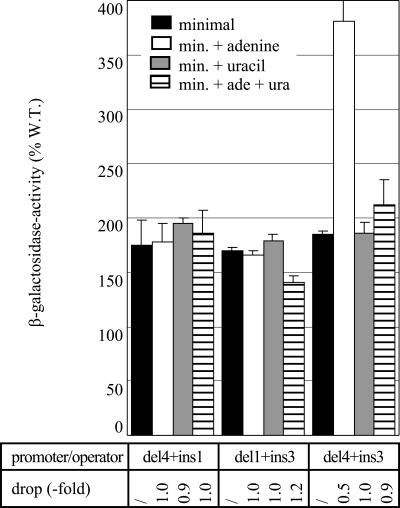
We have shown above that constructs bearing the combination of an insertion and deletion of equal length within a single linker are nearly fully (L2 and L3) or at least significantly (L1) repressible by both excess uracil and adenine. This is clearly not the case for combinations of insertions and deletions situated in different linkers. All three combinations analyzed, del4 plus ins1 (L3 and L2a), del1 plus ins3 (L2a and L1), and del4 plus ins3 (L3 and L1), exhibited constitutive expression at a high level (Fig. 4). Remarkably, the del4 ins3 combination showed an increase in promoter activity in the presence of excess adenine, similar to the observation for certain insertions in linker L2a (3-, 7- and 11-bp insertions, see Fig. S1 in the supplemental material).
FIG. 4.
Effects of combinations of 5-bp insertions and deletions located in different linkers of the carP1 control region: L3 plus L2a (del4+ins1), L2a plus L1 (del1+ins3), and L3 plus L1 (del4+ins3). All constructs were analyzed in the genetic context of strain FW102 (wild type). β-Galactosidase activity is expressed as a percentage with respect to the activity of the wild-type (W. T.) P1 construct grown on minimal medium. For further details, see the legend to Fig. 2.
PepA and IHF binding to mutant and truncated operator fragments.
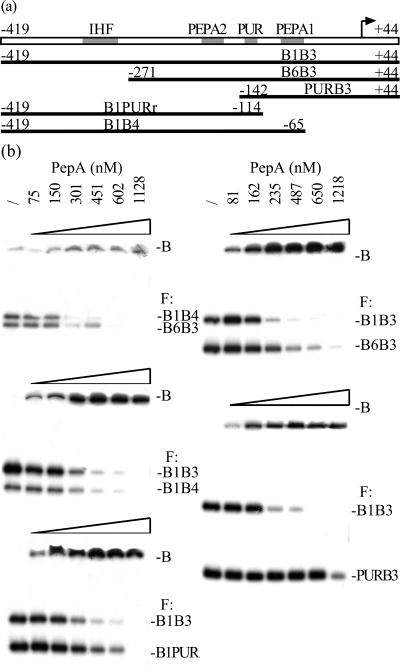
From all trans-acting DNA-binding proteins involved in P1 regulation, PepA makes the most extended contacts with the operator DNA and has the largest structural impact on the DNA configuration. Moreover, the elimination of PepA from the cell results in the most pronounced drop in repressibility by excess pyrimidines and purines (Fig. 2). But what are the requirements for PepA binding? To delimit the global area of the P1 operator that influences PepA binding, we have performed mobility shift experiments with six overlapping fragments that cover different parts of the control region (Fig. 5a). Binding assays were performed with different pairs of purified fragments. Figure 5b shows a representative set of mobility shift assays. Binding to the B6B3 fragment (−271 to +44) was reduced approximately twofold with respect to the intact operator (B1B3 fragment). Therefore, the removal of the upstream part of the control region (corresponding to the ΔO2 deletion) negatively affects complex formation, indicating that the region upstream of PEPA2 participates in complex formation. In contrast, binding to the B1B3 and B1B4 fragments occurred with the same apparent affinity, indicating that the removal of the L1 linker has no significant effect on complex formation. Similarly, the reduced binding to the B1PUR and PURB3 fragments (2-fold and 4.5-fold increases in KD, respectively) indicates that the PEPA1-PEPA2 zone is important for PepA binding.
FIG. 5.
(a) Schematic diagram of the different fragments of the carAB operator region used for PepA binding. (b) Representative set of autoradiographs of mobility shift assay with PepA binding to pairs of fragments, as indicated. The protein concentration (/, none) and the positions of free DNA (F) and of PepA-DNA complexes (B) are indicated. PepA-DNA complexes hardly penetrate the gel (6); therefore, complexes formed with fragments of different length are not resolved. Binding was calculated on the basis of the decrease in the intensity of the free DNA bands.
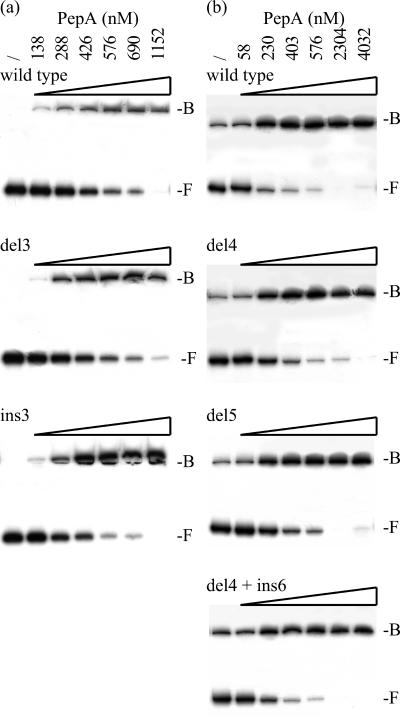
PepA binding to the wild-type B1B3 fragment and derivatives carrying a half-turn deletion or insertion in the L1 (ins3) and L2 (ins1, del1, and del3) linkers was similar (Fig. 6a). The 5- and 10-bp deletions del4 and del5 in linker L3 resulted in a slight increase in the apparent KD (approximately 1.3-fold). Binding to the del4 ins6 double mutant was as efficient as to the wild type (Fig. 6b). Similarly, we demonstrated that the del4 and del5 deletions in linker L3 had no significant effect on the binding of IHF (not shown).
FIG. 6.
(a) Autoradiographs of PepA binding to the wild-type B1B3 fragment and derivatives carrying the del3 or ins3 mutation. (b) Autoradiographs of a comparative PepA binding assay to the wild-type fragment and derivatives carrying the del4, del5, or del4 ins6 mutations. The PepA concentrations used (/, none) and the positions of free DNA (F) and of PepA-DNA complexes (B) are indicated.
Since all these mutations severely hampered the pyrimidine- and/or purine-specific repression in vivo, we may conclude that the regulatory deficiencies of the insertion and deletion mutants analyzed in this work (with the exception of the large deletions ΔO1 and ΔO2, which are not restricted to linker regions but eliminate one or several identified binding sites) do not result from an important reduction of PepA or IHF binding but likely result from a deficiency in the further assembly of the regulatory complex or its communication with the initiating RNA polymerase-promoter complex.
DISCUSSION
The analyses of half- and full-turn insertion and deletion mutants in different zones of the operator indicate that the pyrimidine-dependent repression of P1 activity is extremely sensitive to perturbations in the distribution of regulatory targets on the DNA helix. The detrimental effect of all half-turn modifications indicates that a proper phasing of all sites is required to establish significant pyrimidine-specific repression. Moreover, the systematic analysis of the L2 linker length indicates that the separation of the two PEPA boxes is strictly constrained for both pyrimidine- and purine-mediated repression. In contrast, the length (number of full helical turns) of linkers L1 and L3 is not constrained for purine-dependent regulation, although L3 is clearly constrained in length for the establishment of the pyrimidine-specific response.
Our results also demonstrate that PepA is the key element that assures the functional and structural coupling between pyrimidine- and purine-mediated repression. Previously, we have shown that the ihfA::Tn10 and pyrH41 mutations interfere less with purine-specific repression than the disruption of the pepA or purR gene does (12) (Fig. 2). Here we demonstrate that the introduction of a half-turn deletion (del4) in linker L3 or the elimination of the IHF binding site (ΔO2) severely hampers the pyrimidine-dependent regulation but hardly affects the purine-mediated repression. Quite the opposite, all the half-turn insertions or deletions in linkers L2a (ins1 and del1), L2b (del3), and L1 (ins3), which inevitably result in an improper phasing of one or both PEPA boxes with respect to the promoter, destroy both the pyrimidine- and purine-specific regulation. As indicated by the in vitro binding assays and the normal repressibility observed in the compensatory insertion/deletion ins1 del3 mutant, the lack of repression of the half-turn mutants cannot be ascribed to a failure in PepA binding. In the ins1 del3 double mutant, the PUR box is still displaced by one half-turn with respect to the promoter and to each one of the PEPA boxes, but PEPA1 and PEPA2 are correctly aligned with respect to each other and the promoter. Therefore, we may conclude that the position of the PEPA boxes with respect to the promoter is the most important, not only for pyrimidine-specific repression but also for PurR-mediated purine-dependent regulation. The position of the PUR box seems to be less constrained. This conclusion is underlined by the behavior of the ins9 mutation where a full helical turn is inserted in linker L1 without directly affecting the PurR action, even though the pyrimidine-specific regulation is reduced by approximately threefold.
L1 is the only linker in which the combination of a 5-bp insertion and deletion does not fully restore the pyrimidine-specific repression. The ins3 del8 double mutant operator differs from the wild type at only two positions: G-C instead of T-A at position −60, and a switch from A-T to T-A at position −57. It is noteworthy that these substitutions are located within a 17-bp stretch exclusively composed of A and T residues. The fact that P1 repressibility in the ins3 del8 double mutant, where all previously identified regulatory sites and the promoter are correctly positioned, is not fully restored suggests that this promoter-proximal region participates in a sequence-specific manner in the establishment of pyrimidine-dependent regulation. This region could be contacted by a pyrimidine-specific component of the regulatory complex, possibly PyrH that is believed to be the sensor of the system (18). Protein-protein contacts might play an important role in the recruitment of PyrH to the operator and/or in the further stabilization of the higher-order nucleoprotein complex. The establishment of the potential PyrH-DNA contact, as suggested here, might also require the remodelling of the operator by PepA and possibly by IHF. The recent determinations of the structures of E. coli and P. furiosus UMP kinase (3, 19) have shown that the organization and complementary surface charge potentials of the hexameric UMP kinase and PepA enzymes might sustain their interaction with the threefold axes aligned (19). Most interestingly, the previously isolated pyrH41 mutant deficient for pyrimidine-specific repression of P1 but not affected in the catalytic activity of the UMP kinase bears a single-amino-acid substitution (A94E) that is localized in this contact area (18, 19). In this hypothesis, both PyrH and PepA would establish protein-DNA and protein-protein contacts. The L1 region was also shown to be covered by the P1-bound RNA polymerase in the closed complex (10). Therefore, it is plausible that the substitutions at −57 and −60 interfere with the pyrimidine-dependent repression by altering a specific protein-DNA contact or by influencing the cross talk of the regulatory complex with the RNA polymerase through a DNA conformation-mediated mechanism. Scanning mutagenesis and further in vitro experiments are required to determine the limits of this novel promoter-proximal regulatory target and to unravel the molecular details of its mode of action in the establishment of pyrimidine-specific repression.
The analysis of a carP1 derivative (P1-ΔO1) disconnected from its downstream tandem partner (carP2) and devoid of its upstream control region has allowed us (i) to demonstrate that the A+T-rich region upstream of the −35 element does not function as a UP element, (ii) to distinguish the major protein-dependent pyrimidine-specific repression from other direct and indirect regulatory effects, and (iii) to reveal the antagonistic effects—PurR-dependent repression and PurR-independent stimulation—of excess adenine on P1 activity. The latter, weak (about twofold) but physiologically relevant effect had not yet been observed, as it is overruled by the major pyrimidine- and purine-specific repression mechanisms in the wild-type construct and in most mutants analyzed so far. The existence of this purine-dependent stimulatory effect automatically implies that the PurR-dependent repression of P1 activity is about twice as important as previously estimated. Our findings therefore lead to a reappraisal of purine-specific regulation of P1 activity.
In a pepA mutant and with the P1-ΔO1 construct, where all protein-dependent mechanisms are eliminated, a weak residual pyrimidine-dependent regulation of promoter activity was detected. Could this be ascribed to UTP-sensitive reiterative transcription (stuttering)? UTP-sensitive reiterative transcription relies on the presence of a triplet of T residues immediately downstream of the P1 initiation site (16). It is therefore expected to be intact in P1-ΔO1 and in pepA mutants. However, this transcriptional control mechanism is mainly operational at UTP concentrations typically found in pyrimidine auxotrophs grown under conditions of pyrimidine limitation, which are below the ones present in autotrophs grown on minimal medium or in the presence of excess uracil (16). Moreover, Han and Turnbough (16) have reported a similar 1.4-fold down regulation of a mutant P1 promoter disconnected from the main protein-mediated control and affected in the UTP-sensitive reiterative transcription control. Therefore, the observed down regulation is most likely due to an as-yet unidentified direct or indirect effect triggered by excess uracil.
The similar promoter activities and residual regulatory effects measured with the P1-ΔO1 construct and with the wild-type promoter in the pepA::Tn10 background underline that the inactivation of PepA completely abolishes the major protein-dependent pyrimidine-specific regulation and highlight the preponderant role of PepA in the control of P1 activity. The PepA-induced remodelling of the operator region is also the key element in the structural coupling of the liganded PurR-dependent repression to the pyrimidine-specific control. Therefore, although PepA is a structural component of the regulatory complex rather than a specific sensor, its binding to the car operator is an absolute prerequisite for the establishment of both pyrimidine- and purine-specific repression.
Supplementary Material
Acknowledgments
This research was supported by a Research Programme of the Research Foundation-Flanders (FWO-Vlaanderen), the Research Council of the Vrije Universiteit Brussel (OZR-VUB), and the Vlaamse Gemeenschapscommissie.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alèn, C., D. Sherratt, and S. Commoms. 1997. Direct interaction of aminopeptidase A with recombination site DNA in Xer site-specific recombination. EMBO J. 16:5188-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bouvier, J., J. C. Patte, and P. Stragier. 1984. Multiple regulatory signals in the control region of the Escherichia coli carAB operon. Proc. Natl. Acad. Sci. USA 81:4139-4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briozzo, P., C. Evrin, P. Meyer, L. Assairi, N. Joly, O. Bârzu, and A. M. Gilles. 2005. Structure of Escherichia coli UMP kinase differs from that of other NMP kinases and sheds new light on enzyme regulation. J. Biol. Chem. 280:25533-25540. [DOI] [PubMed] [Google Scholar]
- 4.Charlier, D., D. Gigot, N. Huysveld, M. Roovers, A. Piérard, and N. Glansdorff. 1995. Pyrimidine regulation of the Escherichia coli and Salmonella typhimurium carAB operons: CarP and integration host factor (IHF) modulate the methylation status of a GATC site present in the control region. J. Mol. Biol. 250:383-391. [DOI] [PubMed] [Google Scholar]
- 5.Charlier, D., and N. Glansdorff. September. 2004, posting date. Chapter 3.6.1.10, Biosynthesis of arginine and polyamines. In R. Curtiss III et al., EcoSal--Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, D.C. [Online.] http://www.ecosal.org. [DOI] [PubMed]
- 6.Charlier, D., G. Hassanzadeh, A. Kholti, D. Gigot, A. Piérard, and N. Glansdorff. 1995. carP, involved in pyrimidine regulation of the Escherichia coli carbamoylphosphate synthetase operon encodes a sequence-specific DNA-binding protein identical to XerB and PepA, also required for resolution of ColE1 multimers. J. Mol. Biol. 250:392-406. [DOI] [PubMed] [Google Scholar]
- 7.Charlier, D., A. Kholti, N. Huysveld, D. Gigot, D. Maes, T.-L. Thia-Toong, and N. Glansdorff. 2000. Mutational analysis of Escherichia coli PepA, a multifunctional DNA-binding aminopeptidase. J. Mol. Biol. 302:411-426. [Online.] doi: 10.1006/jmbi.2000.4067. [DOI] [PubMed] [Google Scholar]
- 8.Charlier, D., M. Roovers, D. Gigot, N. Huysveld, A. Piérard, and N. Glansdorff. 1993. Integration host factor (IHF) modulates the expression of the pyrimidine-specific promoter of the carAB operons of Escherichia coli K12 and Salmonella typhimurium LT2. Mol. Gen. Genet. 237:273-286. [DOI] [PubMed] [Google Scholar]
- 9.Charlier, D., M. Roovers, F. Van Vliet, A. Boyen, R. Cunin, Y. Nakamura, N. Glansdorff, and A. Piérard. 1992. Arginine regulon of Escherichia coli K-12. A study of repressor-operator interactions and of in vitro binding affinities versus in vivo repression. J. Mol. Biol. 226:367-386. [DOI] [PubMed] [Google Scholar]
- 10.Charlier, D., G. Weyens, M. Roovers, J. Piette, C. Boquet, A. Piérard, and N. Glansdorff. 1988. Molecular interactions in the control region of the carAB operon encoding Escherichia coli carbamoylphosphate synthetase. J. Mol. Biol. 204:867-877. [DOI] [PubMed] [Google Scholar]
- 11.Dagert, M., and S. D. Ehrlich. 1979. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene 6:23-28. [DOI] [PubMed] [Google Scholar]
- 12.Devroede, N., T.-L. Thia-Toong, D. Gigot, D. Maes, and D. Charlier. 2004. Purine and pyrimidine-specific repression of the Escherichia coli carAB operon are functionally and structurally coupled. J. Mol. Biol. 336:25-42. [Online.] doi: 10.1016/jmb.2003.12.024. [DOI] [PubMed] [Google Scholar]
- 13.Filutowicz, M., H. Grimek, and K. Appelt. 1994. Purification of the Escherichia coli integration host factor (IHF) in one chromatographic step. Gene 147:149-150. [DOI] [PubMed] [Google Scholar]
- 14.Glansdorff, N. 1965. Topography of cotransducible arginine mutations in Escherichia coli K-12. Genetics 51:167-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guhathakurta, A., I. Viney, and D. Summers. 1996. Accessory proteins impose site selectivity during ColE1 dimer resolution. Mol. Microbiol. 20:613-620. [DOI] [PubMed] [Google Scholar]
- 16.Han, X., and C. L. Turnbough, Jr. 1998. Regulation of carAB expression in Escherichia coli occurs in part through UTP-sensitive reiterative transcription. J. Bacteriol. 180:705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Higuchi, R., B. Krummel, and R. K. Saiki. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16:7351-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kholti, A., D. Charlier, D. Gigot, N. Huysveld, M. Roovers, and N. Glansdorff. 1998. pyrH-encoded UMP-kinase directly participates in pyrimidine-specific modulation of promoter activity in Escherichia coli. J. Mol. Biol. 280:571-582. [DOI] [PubMed] [Google Scholar]
- 19.Marco-Marín, C., F. Gil-Ortiz, and V. Rubio. 2005. The crystal structure of Pyrococcus furiosus UMP kinase provides insight into catalysis and regulation in microbial pyrimidine nucleotide biosynthesis. J. Mol. Biol. 352:436-454. [Online.] doi: 1016/jmb.2005.07.045. [DOI] [PubMed] [Google Scholar]
- 20.McCulloch, R., M. Burke, and D. Sherratt. 1994. Peptidase activity of Escherichia coli aminopeptidase A is not required for its role in Xer site-specific recombination. Mol. Microbiol. 12:241-251. [DOI] [PubMed] [Google Scholar]
- 21.Piette, J., H. Nyunoya, C. Lusty, R. Cunin, G. Weyens, M. Crabeel, D. Charlier, N. Glansdorff, and A. Piérard. 1984. DNA sequence of the carA gene and the control region of carAB: tandem promoters, controlled by arginine and the pyrimidines, regulate the synthesis of carbamoyl-phosphate synthetase in Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 81:4134-4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reijns, M., Y. Lu, S. Leach, and S. Colloms. 2005. Mutagenesis of PepA suggests a new model for the Xer/cer synaptic complex. Mol. Microbiol. 57:927-941. [DOI] [PubMed] [Google Scholar]
- 23.Stirling, S., S. Colloms, J. Collins, G. Szatmari, and D. Sherratt. 1989. xerB, an Escherichia coli gene required for plasmid ColE1 site-specific recombination, is identical to pepA, encoding aminopeptidase A, a protein with substantial similarity to bovine lens leucine aminopeptidase. EMBO J. 8:1623-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sträter, N., D. Sherratt, and S. Commoms. 1999. X-ray structure of aminopeptidase A from Escherichia coli and a model for the nucleoprotein complex in Xer site-specific recombination. EMBO J. 18:4513-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang, H., N. Glansdorff, and D. Charlier. 1998. The arginine repressor of Escherichia coli K-12 makes direct contacts to minor and major groove determinants of the operator. J. Mol. Biol. 277:805-824. [DOI] [PubMed] [Google Scholar]
- 26.Whipple, F. W. 1998. Genetic analysis of prokaryotic and eukaryotic DNA-binding proteins in Escherichia coli. Nucleic Acids Res. 26:3700-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.