Abstract
The Cry4Aa δ-endotoxin from Bacillus thuringiensis is toxic to larvae of Culex, Anopheles, and Aedes mosquitoes, which are vectors of important human tropical diseases. With the objective of designing modified toxins with improved potency that could be used as biopesticides, we determined the structure of this toxin in its functional form at a resolution of 2.8 Å. Like other Cry δ-endotoxins, the activated Cry4Aa toxin consists of three globular domains, a seven-α-helix bundle responsible for pore formation (domain I) and the following two other domains having structural similarities with carbohydrate binding proteins: a β-prism (domain II) and a plant lectin-like β-sandwich (domain III). We also studied the effect on toxicity of amino acid substitutions and deletions in three loops located at the surface of the putative receptor binding domain II of Cry4Aa. Our results indicate that one loop is an important determinant of toxicity, presumably through attachment of Cry4Aa to the surface of mosquito cells. The availability of the Cry4Aa structure should guide further investigations aimed at the molecular basis of the target specificity and membrane insertion of Cry endotoxins.
The gram-positive soil bacterium Bacillus thuringiensis subsp. israelensis produces four major insecticidal Cry proteins (Cry4Aa, Cry4Ba, Cry11Aa, and Cyt1Aa) that are toxic to larvae of insects belonging to the orders Diptera, Lepidoptera, Coleoptera, and Hymenoptera (18, 54). These proteins are of great interest for development of new bioinsecticides that cause minimal ecological damage and therefore are attractive alternatives to chemical insecticides for the control of insect pests. Engineered Cry toxins have potential applications in agriculture and also in the control of mosquitoes, which are vectors of some important tropical diseases, like malaria and viral hemorrhagic fevers (54). Cry δ-endotoxins occur as protein crystals in sporulating B. thuringiensis subsp. israelensis. These crystals are produced as inactive protoxin inclusions, which dissolve upon ingestion by insect larvae under the alkaline conditions present in the midgut lumen (54). The soluble Cry protoxin is activated by larval midgut proteases, which remove the C-terminal half and approximately 30 residues from the N terminus. This yields an active and proteolytically resistant fragment which specifically binds to receptors on the brush border membrane of the insect midgut epithelium. Conformational changes subsequently allow the activated toxin to insert into the host cell membrane and form pores. This in turn leads to ion leakage, cell lysis, and insect death (54).
The crystal structures of the following Cry toxins have been reported previously: Cry3Aa (42), Cry3Bb (25), Cry1Aa (29), Cry1Ac (41), Cry2Aa (46), and Cry4Ba (7). These toxins exhibit markedly different insect specificities in spite of their similar organizations into three structurally conserved domains. N-terminal domain I, an amphipathic α-helical bundle, is responsible for membrane insertion and pore formation (26, 50a, 63, 65). Domain II is composed of antiparallel β-strands connected by loops that vary significantly in length and amino acid sequence in different toxins. These loops are therefore thought to participate in receptor binding and hence in determining the specificity of the toxin for insect larvae (5, 35, 51, 57). C-terminal domain III, a sandwich of two antiparallel β-sheets, is believed to protect the structural integrity of active toxin molecules against further proteolytic cleavage and to promote target membrane permeabilization (45), and it could also participate in receptor recognition (11, 39). Nevertheless, the precise sequence of events that follow receptor binding by the active toxin, leading to membrane insertion and cell lysis, remains poorly defined. Extensive conformational changes of the toxin architecture are likely to be involved, possibly leading to assembly of an oligomeric toxin structure.
Among the Cry toxins, the Cry4Aa and Cry4Ba toxins are the most closely related molecules, exhibiting 35% amino acid sequence identity (13, 66) (Fig. 1). Despite their similarity, however, these toxins exhibit different levels of toxicity against various mosquito species. While Cry4Ba is highly toxic to Aedes and Anopheles larvae (vectors of dengue and yellow fever viruses and of malaria, respectively), it exhibits very little activity against Culex larvae (vector of West Nile and Japanese encephalitis viruses) (49). By contrast, Cry4Aa toxin exhibits a high level of toxicity against Culex and Aedes larvae and is slightly less toxic to Anopheles larvae (49). In addition, Cry4Aa and Cry4Ba show synergism in vivo against larvae of all three mosquito genera (2, 49). Because their interacting binding partners at the insect membrane surface are not known, the molecular basis that determines the specificity of these toxins has remained elusive. Recently, the X-ray crystal structure of the B. thuringiensis subsp. israelensis Cry4Ba toxin was published, but the 50 N-terminal residues (corresponding to helices α1 and α2) were not visible in the electron density map, presumably because of proteolysis that occurred during crystallization (7). Here we report the crystal structure of the active Cry4Aa single mutant R235Q at a resolution of 2.8 Å. Below we discuss possible mechanisms of membrane pore formation and also the structural determinants for the specificity of B. thuringiensis subsp. israelensis Cry toxin in light of site-directed mutagenesis data. Our results provide the first complete view of the three-dimensional structure of a dipteran-specific toxin and underline the importance of loop 2 of domain II for Cry4Aa toxicity.
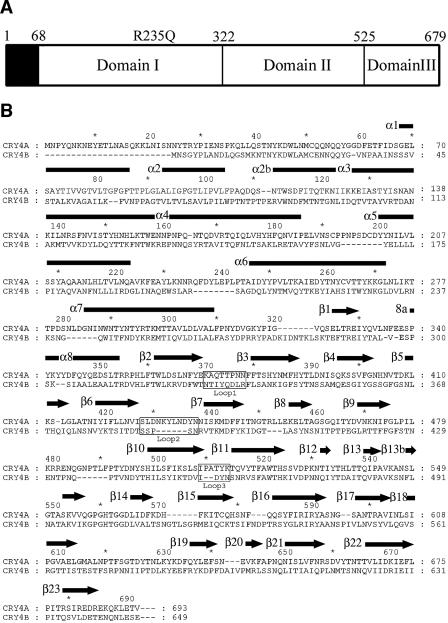
FIG. 1.
(A) Schematic representation of the three domains present in the mature Cry4Aa toxin. Amino acids 68 to 679 are visible in the final electron density map. The R235Q mutation, which makes the toxin resistant to further proteolysis, is also indicated. (B) Alignment of the amino acid sequences of the homologous dipteran-specific toxins Cry4Aa and Cry4Ba. Secondary structure elements of Cry4Aa, as determined in this work (using the Pymol program [16]), are indicated above the sequence; α helices are indicated by rectangles, and β strands are indicated by arrows. 8a is a short 310 helical segment. Residue numbers of both toxins are indicated.
MATERIALS AND METHODS
Protein purification and crystallization.
The active single-mutant protein Cry4Aa-R235Q was purified and crystallized as described previously (6). In brief, crystals were obtained at 23°C by vapor diffusion in hanging drops containing the purified protein at a concentration of 3 to 5 mg ml−1, 0.1 M Tris-acetate (pH 7.0), and 0.2 to 0.3 M KH2PO4. The crystals belong to space group C2221 with unit cell parameters a = 91.2 Å, b = 202.1 Å, and c = 98.7 Å and contain one endotoxin molecule per asymmetric unit.
Data collection, model building, refinement, and analysis.
Synchrotron data collection and molecular replacement calculations using the Cry4Ba structure as a search model (PDB code 1W99) have been reported previously (6). Model building was done using the O program (34) interspersed with cycles of electron density map improvement as described previously (6). Several cycles of molecular dynamics with a slow cooling protocol using CNS (10) were carried out. The DALI program (30) was used to search for similar three-dimensional structures in the PDB and to perform a structural comparison with Cry4Ba toxin. An alignment of the amino acid sequences of Cry4A and Cry4B was produced with the MAPS program (http://bioinfo1.mbfys.lu.se/TOP/webmaps.html). Figures 2 to 8 were produced using Pymol (16).
FIG. 2.
Ribbon representations of the Cry4Aa fold. (A) Overall view of the Cry4Aa toxin, which is composed of three domains. Pore-forming domain I is red, putative receptor binding domain II is yellow, and C-terminal domain III is blue. The secondary structure elements of each of the three domains are labeled in panels B to D. Each individual β-sheet is a different color. See Fig. 1B.
FIG. 8.
Comparison of the structures adopted by the three surface-exposed loops of Cry4Aa and Cry4Ba at the extremity of domain II. Domain II of Cry4Aa is cyan, and domain II of Cry4Ba is yellow. The locations of loops 1, 2, and 3 on domain II are indicated, and close-ups highlight the structural differences in these putative receptor binding regions. With the possible exception of loop 1 of Cry4Ba, which makes several crystalline intermolecular contacts (7), the conformations of the other loops are likely to be determined by intramolecular contacts, not by crystal packing interactions.
Mutagenesis of Cry4Aa.
Domain II of Cry4Aa and domain II of Cry4Ba were superimposed using the LSQKAB program from the CCP4 suite (14) and were displayed using the O program. Loops 1, 2, and 3 of domain II of Cry4Aa were altered by site-directed mutagenesis to mimic the corresponding loops of Cry4Ba. Site-directed mutagenesis was performed using QuickChange (Stratagene) according to the manufacturer's instructions. The mutations obtained and primers used are listed in Table 1. DNA templates were purified (QIAGEN) and replicated by PCR with the Expand long-template polymerase (Roche). The PCR products were digested with Pfu polymerase (Stratagene) at 72°C for 10 min to eliminate the single A added by the Taq polymerase and were digested with DpnI (Stratagene) at 37°C for 4 h. The ligation products (T4 ligase; New England Biolab) were transformed into Escherichia coli DH5α competent cells. All mutations were confirmed by DNA sequencing.
TABLE 1.
Sequences of primers used in site-directed mutagenesis
| Primer | Directiona | Sequence (5′→3′) | Mutant |
|---|---|---|---|
| L1TTPNN→YQDLR | F | 5′ GAAAAAGCGCAATACCAAGATCTAAGGTTTTTCACCAGCC 3′ | 4AL1YQDLR |
| R | 5′ CTGGTGAAAAACCTTAGATCTTGGTATTGCGCTTTTTCATA 3′ | ||
| L1TTPN→YQDL | F | 5′ GAAAAAGCGCAATACCAAGATCTAAATTTTTTCACCAGCC 3′ | 4AL1-YQDL |
| R | 5′ CTGGTGAAAAAATTTAGATCTTGGTATTGCGCTTTTTCAT 3′ | ||
| L1TPNN→QDLR | F | 5′ GAAAAAGCGCAAACTCAAGATCTAAGGTTTTTCACCAGCC 3′ | 4AL1-QDLR |
| R | 5′ CTGGTGAAAAACCTTAGATCTTGAGTTTGCGCTTTTTCAT 3′ | ||
| L1TPN→QDL | F | 5′ GAAAAAGCGCAAACTCAAGATCTAAATTTTTTCACCAGCC 3′ | 4AL1-QDL |
| R | 5′ CTGGTGAAAAAATTTAGATCTTGAGTTTGCGCTTTTTCAT 3′ | ||
| L1PN→DL | F | 5′ GAAAAAGCGCAAACTACTGATCTAAATTTTTTCACCAGCC 3′ | 4AL1-DL |
| R | 5′ CTGGTGAAAAAATTTAGATCAGTAGTTTGCGCTTTTTCAT 3′ | ||
| L1TP→QD | F | 5′ GAAAAAGCGCAAACTCAAGATAATAATTTTTTCACCAGCC 3′ | 4AL1-QD |
| R | 5′ CTGGTGAAAAAATTATTATCTTGAGTTTGCGCTTTTTCAT 3′ | ||
| L2deleYLN | F | 5′ GTCATAAGCTTAGATAATAAAGATTATAATAATATTAG 3′ | 4AL2-ΔYLN |
| R | 5′ CTAATATTATTATAATCTTTATTATCTAAGCTTATGAC 3′ | ||
| L2deleLN | F | 5′ CATAAGCTTAGATAATAAATATGATTATAATAATATTAGTAAAA 3′ | 4AL2-ΔLN |
| R | 5′ CATTTTACTAATATTATTATAATCATATTTATTATCTAAGCTTA 3′ | ||
| L2deleKY | F | 5′ GTCATAAGCTTAGATAATCTAAATGATTATAATAATATTAG 3′ | 4AL2-ΔKY |
| R | 5′ CTAATATTATTATAATCATTTAGATTATCTAAGCTTATGAC 3′ | ||
| L2deleKYLN | F | 5′ GATTATAATAATATTAGTAAAATGGAT 3′ | 4AL2-ΔKYLN |
| R | 5′ ATTATCTAAGCTTATGACATTTAATAA 3′ | ||
| L2-435N→Y | F | 5′ CTTAGATAATAAATATCTATACGATTATAATAATATTAGTAAAA 3′ | 4AL2-N435Y |
| R | 5′ CATTTTACTAATATTATTATAATCGTATAGATATTTATTATCTA 3′ | ||
| L3-514K→N | F | 5′ GTATCCCTGCAACATATAACACTCAAGTGTATACGTTTGC 3′ | 4AL3-K514N |
| R | 5′ GCAAACGTATACACTTGAGTGTTATATGTTGCAGGGATAC 3′ |
F, forward; R, reverse.
Toxicity assays.
Three independent mosquito larvicidal assays were carried out in triplicate with 2-day-old Aedes aegypti larvae and Culex quinquefasciatus larvae (supplied by the mosquito rearing facility of the Institute of Molecular Biology and Genetics, Mahidol University, Nakornpathom, Thailand), as well as Anopheles dirus larvae (supplied by the Armed Forces Research Institute of Medical Sciences, Thailand). The mosquito larvicidal activities of the mutant toxins were tested by diluting the toxin inclusions in water to obtain twofold serial dilutions with concentrations ranging from 12 to 0.125 μg/ml. One milliliter of diluted inclusions was added to the same volume of water with either five larvae of A. dirus or 10 larvae of A. aegypti or C. quinquefasciatus in each well of tissue culture plates (24-well plate; diameter of each well, 1.7 cm). Proteins extracted from E. coli JM109 containing the pMEx8 vector were used as a negative control. Larval mortality was recorded after incubation at 30°C for 24 h, and the concentrations of recombinant proteins that resulted in 50% mortality were calculated by a Probit method (24).
RESULTS AND DISCUSSION
Structure refinement and quality of the model.
The refinement statistics are shown in Table 2. The final model comprises 598 amino acid residues spanning amino acids 68 to 679 of the Cry4Aa toxin mature sequence (Fig. 1), 252 water molecules, and one methyl-2,4-pentanediol molecule from the cryoprotecting buffer (6). Overall, the electron density map is unambiguous, and the path of the main chain is clearly defined except for residues 235 to 244 and 485 to 488, which were omitted from the final model. These segments correspond to two exposed loops connecting α-helices α5 and α6 and β-strands β9 and β10 in domains 1 and 2, respectively. The former loop is highly susceptible to proteolysis by trypsin-like enzymes (3). Elimination of the latter cleavage site by replacement of Arg-235 with a glutamine residue yielded a single mutant of Cry4Aa (Cry4Aa-R235Q; referred to below as Cry4Aa) that retained toxicity for A. aegypti larvae (8). After trypsin digestion, a 65-kDa fragment was obtained, which was crystallized (6) (Fig. 1). Thus, the absence of electron density in loop α5-α6 was probably due to conformational flexibility, not due to a nick introduced into the polypeptide chain by proteolysis (see below).
TABLE 2.
Refinement statistics
| Parameter | Data |
|---|---|
| Resolution range (Å) | 20.0-2.8 |
| Intensity cutoff [F/σ(F)] | None |
| No. of reflections: completeness (%) | 96.1 |
| Used for refinement | 20,191 |
| Used for Rfree calculation | 1,112 |
| No. of nonhydrogen atoms | |
| Protein | 4,811 |
| Water molecules | 252 |
| Rfactor (%)a | 20.25 |
| Rfree (%) | 25.91 |
| Root mean square deviations from ideality | |
| Bond lengths (Å) | 0.0064 |
| Bond angles (°) | 1.27 |
| Ramachandran plotb | |
| Residues in most favored regions (%) | 83.7 |
| Residues in additional allowed regions (%) | 15.6 |
| Residues in generously allowed regions (%) | 0.2 |
| Overall G factorc | 0.21 |
| PDB accession code | 2C9K |
Overall architecture.
The Cry4Aa toxin is a rather compact molecule composed of three distinct domains and has approximate overall dimensions of 120 by 100 by 80 Å. Domain I is a seven-helix bundle. Domain II consists of three antiparallel β-sheets with two additional short helical segments that form a β-prism structure. Domain III is an antiparallel β-sandwich with the jelly roll topology (Fig. 2).
Pore-forming domain I.
Cry4Aa N-terminal domain I (residues 68 to 321) is composed of seven amphipathic helices that are all clearly defined in our final electron density map. The most hydrophobic helix, α5, is located centrally and is surrounded by the six remaining helices. Helix α2 is interrupted by a short loop section and thus can be divided into α2 and α2b, as in Cry3Aa and Cry1Aa (29, 42) (Fig. 2). Overall, the organization of domain I is reminiscent of the organization of other pore-forming proteins composed of α-helices, such as colicin A (48) or hemolysin E from E. coli (64), with which Cry4Aa exhibits distant structural homology. A number of 128 residues of Cry4Aa and hemolysin E can be superimposed with a residual root mean square deviation of 4.2 Å, giving a Z score of 5.2. Although the architecture of the pore-forming state of a Cry toxin has not been determined yet, an “umbrella model” has been proposed to account for the toxicity. In this model, an intermediate state in the reaction pathway consists of insertion of helices α4 and α5 into the membrane as a helical hairpin structure, with the remaining helices lying at the membrane surface (26). This proposed mechanism is supported by the results of mutagenesis studies that demonstrated the crucial role of these two helices for the toxicity of Cry4Ba (61). It is also consistent with the recent finding that a crystallized Cry4Ba toxin that lacks helices 1 and 2 remains toxic (7). Insertion of a hairpin structure into the membrane, leading to pore formation, presumably follows large conformational changes in the toxin, possibly after receptor binding, proteolysis, and/or multimerization, as shown previously for Cry1Ab and Cry1Ac (4, 52, 59). The refolding of a hydrophobic hairpin motif, primed by a decrease in the pH or contact with the cytoplasmic membrane, could also play a key role in pore formation by other bacterial toxins, including colicins, as well as cholera, pertussis, and anthrax toxins (12, 21, 22). In Cry4Aa domain I, a larger number of electrostatic charges are found at the accessible surface of α-helices α1, α6, and α7 than at the surface contributed by helices α3 and α4 (Fig. 3). In the present water-soluble structure of Cry4Aa toxin, these electrostatic charges exposed at the surface of α-helices α1, α6, and α7 are largely neutralized by opposite charges located at the surface of the interacting domain II (Fig. 4). Conformational changes of the toxin (e.g., upon receptor binding) that would disrupt the interface between domains I and II are likely to expose these electrostatic charges to the solvent. Thus, their asymmetrical distribution should influence the orientation of the toxin molecule as it approaches the target membrane for α4-α5 hairpin insertion. It is possible that long-range electrostatic interactions result in an orientation of domain I with helices α4 and α5 approximately perpendicular to the lipid bilayer, as was proposed previously by Parker et al. for colicin A (48). Proteolytic cleavage between helices α5 and α6 of Cry4Aa or Cry4Ba was proposed to trigger the conformational changes required to facilitate membrane insertion (3). However, prevention of α5-α6 interhelical proteolysis by replacement of Arg-203 with Ala increases Cry4Ba larvicidal activity in vivo (1, 3). Likewise, deletion of a trypsin cleavage site at position 235 of the Cry4Aa sequence by replacement of Arg-235 with a Gln residue does not have an adverse effect on Cry4Aa in vivo toxicity for A. aegypti larvae (6). In the Cry4Aa crystal structure, we found no electron density accounting for the segment connecting helices α5 and α6, which is thus presumably flexible. However, we cannot completely rule out the possibility that in vivo, cleavage at this site by gut proteases primes the toxin for membrane penetration.
FIG. 3.
Surface representation of the electrostatic potential of pore-forming domain I of Cry4Aa (calculated using Pymol [16]). Positive electrostatic potentials are blue, and negative electrostatic potentials are red. The locations of solvent-exposed α-helices are indicated. The solvent-accessible surfaces of α-helices α1, α6, and α7 (A) show relatively higher potential and clear charge separations (including a “basic strip”) than α-helices α3 and α4 (B). Panel B is a view with domain I rotated 180° along a vertical axis; this has implications for the way that domain I approaches the target cell membrane (see text).
FIG. 4.
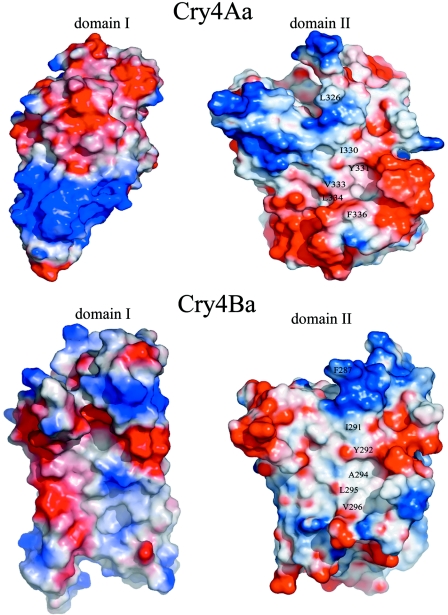
“Open book” representation of the interfaces between domains I and II of Cry4Aa and Cry4Ba. The views were obtained by rotating each of the interacting domains approximately 90° around a vertical axis. A hydrophobic patch is visible at the interface between domains I and II. Amino acids L326, I330, Y331, V333, L334, and F336 in Cry4Aa and amino acids F287, I291, Y292, A294, L295, and V296 in Cry4Ba, which are present in this hydrophobic patch, are labeled.
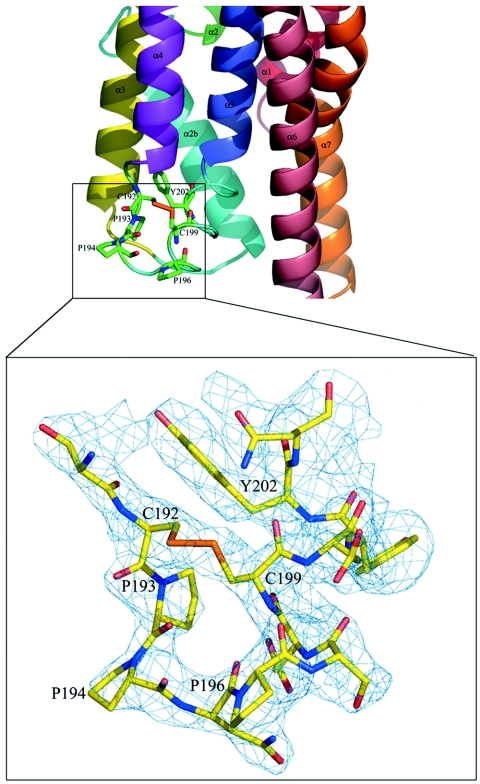
For efficient insertion of the toxin into the target membrane, an α4-loop-α5 hairpin structure is required (27). Interestingly, the 15-residue α4-α5 loop of Cry4Aa has a unique structure compared to other Cry toxins, with several proline residues located at positions 193, 194, and 196 (Fig. 5). Moreover, cysteine residues present at positions 192 and 199 form a disulfide bridge, thus restricting the flexibility of this potentially mobile segment (Fig. 1 and 5). Both the unique disulfide bridge (Cys192-Cys199) and the proline-rich motif (Pro193-Pro-Asn-Pro196) play essential roles in Cry4Aa toxin activity, conceivably by maintaining the α4-α5 loop structural integrity, which may be required for efficient membrane insertion of the α4-α5 transmembrane hairpin (58). Further studies of the role of the α4-α5 hairpin of Cry4Aa by using mutagenesis demonstrated that the A. aegypti larvicidal activity was completely abolished when the strictly conserved aromatic residue Tyr-202 was replaced by an aliphatic amino acid, while replacement by an aromatic side chain, phenylalanine, did not affect the toxicity (50). This is consistent with the crucial role played by the corresponding residue Tyr-170 for the larvicidal activity of the Cry4Ba toxin (36). Indeed, aromatic Trp and Tyr residues tend to specifically interact with the lipid membrane outer leaflets, as was previously shown structurally for the fusion loops of class II viral envelope glycoproteins (9). A large number of hydrophobic residues exposed to solvent are also found in other pore-forming toxins, including hemolysin E from E. coli (64) and aerolysin (47). These residues were proposed to interact with hydrophobic lipid tails. A number of conserved aromatic amino acids, including Tyr-202, could interact with the phospholipid head groups, thus anchoring the toxin next to the membrane-water interface. A functional analysis of the channel activities of various Cry toxins has been described previously. Membrane perturbation revealed that the activated Cry4Aa toxin could form ion channels with a conductance of 127 pS, a value which is comparable to the values for Cry4Ba (∼68 to 127 pS) (50a) and Cry1Ca (∼120 pS) (56) but lower than the values for Cry1Aa (∼450 pS) (29) and Cry1Ac (∼457 pS) (56). Channels formed by Cry4Aa are cation selective and voltage independent, like the channels induced by other Cry toxins (50a, 55). Unlike the homologous toxin Cry4Ba, the 65-kDa activated Cry4Aa toxin was unable to induce the release of entrapped calcein from pure liposomes (W. Pornwaroon, personal communication). The different characteristics of membrane perturbation induced by Cry4Aa and Cry4Ba may be related to the striking differences in the length and conformation of their α4-α5 membrane insertion loops.
FIG. 5.
View of the α4-α5 loop in domain I of Cry4Aa. This segment is thought to play an important role in membrane insertion (see text). At the bottom is a close-up of the region containing the α4-α5 loop indicated by the rectangle. The final electron density map is displayed at a contour of 1σ. There is a disulfide bridge between Cys-192 and Cys-199.
Domain II: receptor binding domain.
Domain II consists of three antiparallel β-sheets packed through formation of a central hydrophobic core. Both sheet 1 comprising strands β5, β2, β3, and β4 and sheet 2 (strands β8, β7, β6, and β9) adopt a “Greek key” topology. Three β strands, β1, β11, and β10, form sheet 3, which is augmented by α-helix α8 and a short 310 helix 8a and adopts a fold similar to that of other known Cry protein structures (Fig. 2). The interface between domains I and II is composed of a mixture of salt bridges, hydrogen bonds, and van der Waals interactions. Since insertion into the lipid bilayer presumably involves structural rearrangements in order to engage the hairpin in the target membrane, disruption of these interdomain interactions must occur prior to pore formation (55). In this respect, an interesting feature is the presence of a conserved hydrophobic patch on the molecular surfaces corresponding to the domain I-domain II interface of Cry4Aa and Cry4Ba (Fig. 4). Hydrophobic patches on protein surfaces are generally determinants of protein-protein or protein-ligand interactions (43). The biological relevance of these hydrophobic patches of the Cry4Aa and Cry4Ba toxins requires further investigation.
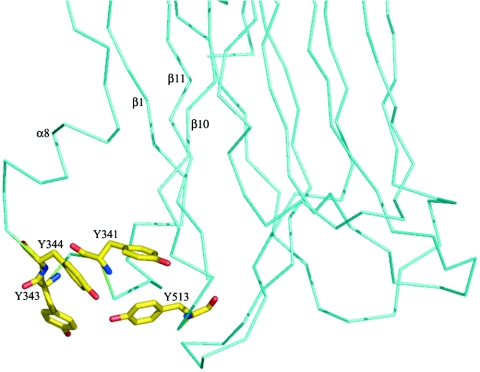
A search for homologous three-dimensional structures in the PDB revealed that domain II of Cry4Aa is structurally similar to the plant lectin jacalin (53) and to Maclura pomifera agglutinin (40). The architecture shared with lectins suggests that domain II probably binds to the carbohydrate moiety of a glycoprotein receptor of the target insect membrane. This notion is further reinforced by the finding that Caenorhabditis elegans resistance to Cry5B toxicity is linked to the loss of a gene encoding a galactosyltransferase (28). Indeed, loss of this carbohydrate-modifying enzyme could directly affect the binding of the toxin to its receptor(s) (28). Two proteins, aminopeptidase N and cadherin-like protein, have been characterized as candidate receptors for lepidopteran-specific Cry1 toxins (37, 44, 52, 62). By contrast, no receptor protein for dipteran-specific toxins has been identified so far. Comparison with the M. pomifera agglutinin structure (PDB code 1JOT) (40), whose interaction with a disaccharide is mediated by three aromatic amino acids, revealed that Cry4Aa also possesses a cluster of aromatic amino acids, Tyr-341, Tyr-343, Tyr-344, and Tyr-348 in the β1-α8 loop and Tyr-513 in loop 3 (Fig. 1 and 6). These five aromatic amino acids form a potential binding site with dimensions that could accommodate a short oligosaccharide. The functional importance of this region of domain II was further established by mutating two amino acids in the β1-α8 loop of Cry11A (23) and Tyr-455 in loop 3 of Cry4Ba (60). A significant loss of toxicity for A. aegypti was observed.
FIG. 6.
View of the exposed cluster of aromatic amino acids (indicated by sticks) at the surface of domain II of Cry4Aa (only a partial view of the α-carbon chain of domain II is shown). This cluster is proposed to function as a carbohydrate binding site (see text). Secondary structure elements are indicated.
Domain III.
C-terminal domain III (residues 525 to 679) contains two antiparallel β-sheets that adopt a β-sandwich fold with the jelly roll topology. The outer sheet is composed of six strands, β12, β13b, β16, β22, β18, and β19, which are exposed to the solvent. The inner sheet, containing seven strands (β20, β17, β23, β21, β13, β14, and β15), faces the other two domains (Fig. 2). Domains II and III are associated via the intersheet connection through hydrogen bonds and hydrophobic interactions. Superposition of domain III of Cry4Aa and domain III of Cry4Ba revealed close structural similarity except for some loops exposed to the solvent. In Cry4Aa, the loops connecting β15 to β16 and β19 to β20 are shorter than the loops in Cry4Ba, and the loop connecting β17 to β18 is longer.
Mutations in domain III of Cry1Aa toxin had an effect on both ion channel activity and membrane permeability (56, 67). Domain III could play a role in protecting the toxin against further cleavage by gut proteases (42). Domain swapping experiments suggested that domain III is also involved in determining insect specificity (17, 18, 19, 20).
A three-dimensional structural similarity search revealed that domain III closely resembles the N-terminal cellulose binding domain of a protein from the bacterium Cellulomonas fimi (33) and a xylanase from Clostridium thermocellum (15). This suggests that like domain II, domain III could also bind carbohydrate residues. Although the level of amino acid sequence identity between domain III of Cry4Aa and the xylanase is only 11.5%, the structural similarity is quite extensive since the two polypeptide segments can be superimposed with a residual root mean square deviation of 2.2 Å, giving a Z score of 12.2 (Fig. 7). Thus, one possibility is that insect specificity is determined by protein-protein or protein-carbohydrate interactions mediated by both domains II and III of the toxin. So far, the importance of carbohydrates in dipteran-specific toxins has not been demonstrated. However, lectin-like domain III of the lepidopteran-specific Cry1Ac toxin was shown previously to bind N-acetylgalactosamine (11, 31, 32).
FIG. 7.
Superposition of C-terminal domain III of Cry4Aa with xylanase from C. thermocellum, highlighting the common fold. Only the α-carbon traces are shown (magenta for Cry4Aa and green for xylanase).
Site-directed mutagenesis of residues in domain II.
Mutagenesis and loop swapping experiments with Cry toxins have identified regions of domain II as major determinants of insect specificity (1, 51, 68, 69).
Like the complementarity-determining regions of immunoglobulins, three loops that have various lengths and amino acid sequences connect a conserved framework of β-strands. These loops, which are clustered at the extremity of domain II opposite its N and C termini (Fig. 2 and 8), connect strands β2 and β3 (residues 372 to 379; loop 1), strands β6 and β7 (residues 431 to 438; loop 2), and strands β10 and β11 (residues 511 to 514; loop 3) of domain II (42). Superposition of the corresponding segments of Cry4Aa and Cry4Ba revealed significant structural divergence (Fig. 8). In particular, loop 2 in Cry4Aa is six residues longer than the corresponding loop in Cry4Ba (Fig. 8). Before the experimental structures were reported, Abdullah and coworkers introduced residues from Cry4Aa into the three loops of Cry4Ba domain II. Exchanging loop 3 significantly enhanced the toxicity of Cry4Ba for Culex, while the activity against Anopheles and Aedes larvae was maintained (1). This suggested that an important determinant of the specificity of the Cry4Aa toxin for Culex was located in the loop 3 region and that important determinants of the toxicity of Cry4Ba for Anopheles and Aedes were in loops 1 and 2.
Here, we used site-directed mutagenesis based on the crystal structure to identify residues crucial for the toxicity of Cry4Aa. We selected exposed residues in the three loops and replaced them with the structurally corresponding residues of Cry4Ba toxin or performed a deletion if the corresponding segment was shorter in Cry4Ba (e.g., loop 2). The results for the larvicidal activities of the Cry4Aa toxin mutants are summarized in Table 3.
TABLE 3.
Toxicity assays for variant Cry4Aa toxins with three species of mosquitoes
| Toxin | 50% Lethal concn (ng/ml)
|
||
|---|---|---|---|
| A. aegypti | A. dirus | C. quinquefasciatus | |
| Cry4A | 2,390 (1,103-5,136)a | 159 (115-199) | 345 (285-410) |
| 4AL1YQDL | 1,696 (698-3,958) | 352 (299-409) | 874 (709-1,125) |
| 4AL1QDLR | 6,640 (3,400-12,900) | 2,270 (270-4,770) | 1,180 (0-2,380) |
| 4AL1QDL | 2,320 (1,520-3,410) | 588 (77-3,795) | 350 (170-580) |
| 4AL1DL | 3,160 (2,190-4,390) | 455 (60-2,110) | 450 (6-1,170) |
| 4AL2ΔKY | 20,160 (10,358-96,661) | >50,000b | 9,655 (2,309-20,200) |
| 4AL2N435Y | 1,520 (1,086-1,964) | 1,187 (910-1,494) | 126 (89-161) |
| 4AL3K514N | 3,500 (1,760-6,130) | 508 (424-609) | 295 (224-3,430) |
The values in parentheses are 95% confidence limits.
The 95% confidence limits could not be determined.
Five Cry4Aa mutants, including three deletion mutants with mutations in loop 2 (4AL1YQDLR, 4AL1QD, 4AL2ΔKYLN, 4AL2ΔYLN, and 4AL2ΔLN), were expressed very poorly and could not be studied further.
The seven other mutants of Cry4Aa could be expressed in E. coli at levels comparable to the level of the wild-type toxin. Replacement of residues of Cry4Aa in loop 1 with the corresponding residues of Cry4Ba did not dramatically alter the toxicity for larvae of members of the three genera of insects. As many as four residues (with the amino acid sequence TTPN) of loop 1 could be replaced (mutant 4AL1YQDL), leading to only a marginal decrease (approximately twofold) in the toxicity of the corresponding mutant for A. dirus and C. quinquefasciatus, while full toxicity for A. aegypti larvae was retained (Table 3).
This suggests that there are no strong structural constraints imposed on loop 1 for the binding of Cry4Aa to its receptors or that this loop does not participate in binding. By contrast, introduction of three residues of loop 1 of Cry4Aa into Cry4Ba (mutant 4BL1QTT) led to a severely impaired Cry4Ba toxin (1).
In Cry4Aa mutant 4AL2ΔKY two residues of the long loop2 were deleted to mimic the shorter loop present in Cry4Ba. Large amounts of this protein could be expressed in E. coli, and digestion of it by trypsin yielded a pattern similar to the pattern for the wild-type toxin (data not shown). However, the toxicity for the three insect genera was dramatically reduced (Table 3); for example, the toxicity for A. dirus was reduced about 300-fold. This suggests that there is direct participation of this loop in receptor binding, although we cannot formally rule out the possibility that there are some conformational change effects, which would alter the integrity of the protein structure. Interestingly, a single substitution within loop 2, replacement of Asn-435 by a tyrosine residue, preserved most of the toxicity, indicating that the length of the loop rather than its precise sequence is an important determinant of specificity. Likewise, replacement of Lys-514 in loop 3 by an Asn residue, as found in Cry4Ba, resulted in an active toxin.
Interestingly, loops 2 and 3 are clustered in the vicinity of the β1-α8 loop, close to the interface between domain II and domain I, whereas loop 1 is some 25 Å away (Fig. 2). This region appears to be crucial for receptor binding by Cry4Aa, an event which could trigger conformational changes that lead to disruption of the interface between domains I and II and prime domain I for insertion into the host membrane.
Thus, taken together, our data suggest that different regions of Cry4Aa and Cry4Ba are important for toxicity, possibly by using distinct binding sites for interaction with the host receptors. While these results are consistent with the synergistic effects of the two toxins that have been observed (49), they do not support approaches in which fragments of one toxin could be easily grafted onto the other toxin to produce an engineered Cry toxin having a broadened spectrum.
However, we hope that the availability of an experimental structure for a complete active fragment of Cry4Aa will accelerate the development of engineered Cry toxins that have increased potency and prolonged stability. This structure should be useful if insect resistance to the existing Cry toxins emerges, which seems likely. It should also stimulate further mechanistic studies aimed at dissecting the molecular steps leading to pore formation.
Acknowledgments
We are grateful for the provision of excellent beam time and expert advice at E.S.R.F. beam line ID29 (Grenoble, France). We thank Somsri Sakdee for expert technical assistance and J. Torres for reading the manuscript.
The laboratory of J.L. is supported by grants from N.T.U. (grant RG 29/05), the Singapore Biomedical Research Council (grants 03/1/21/20/291 and 02/1/22/17/043), and the Singapore National Medical Research Council (grant NMRC/SRG/001/2003). P.B. acknowledges support from the Thailand Research Fund and the National Science and Technology Development Agency, Thailand.
REFERENCES
- 1.Abdullah, M. A., O. Alzate, M. Mohammad, R. J. McNall, M. J. Adang, and D. H. Dean. 2003. Introduction of Culex toxicity into Bacillus thuringiensis Cry4Ba by protein engineering. Appl. Environ. Microbiol. 69:5343-5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angsuthanasombat, C., N. Crickmore, and D. J. Ellar. 1992. Comparison of Bacillus thuringiensis subsp. israelensis CryIVA and CryIVB cloned toxins reveals synergism in vivo. FEMS Microbiol. Lett. 73:63-68. [DOI] [PubMed] [Google Scholar]
- 3.Angsuthanasombat, C., N. Crickmore, and D. J. Ellar. 1993. Effects on toxicity of eliminating a cleavage site in a predicted interhelical loop in Bacillus thuringiensis CryIVB delta-endotoxin. FEMS Microbiol. Lett. 111:255-261. [DOI] [PubMed] [Google Scholar]
- 4.Aronson, A. I., C. Geng, and L. Wu. 1999. Aggregation of Bacillus thuringiensis Cry1A toxins upon binding to target insect larval midgut vesicles. Appl. Environ. Microbiol. 65:2503-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ballester, V. V., F. Granero, R. A. de Maagd, D. Bosch, J. L. Mensua, and J. Ferre. 1999. Role of Bacillus thuringiensis toxin domains in toxicity and receptor binding in the diamondback moth. Appl. Environ. Microbiol. 65:1900-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boonserm, P., C. Angsuthanasombat, and J. Lescar. 2004. Crystallization and preliminary crystallographic study of the functional form of the Bacillus thuringiensis mosquito-larvicidal Cry4Aa mutant toxin. Acta Crystallogr. Sect. D. 60:1315-1318. [DOI] [PubMed] [Google Scholar]
- 7.Boonserm, P., P. Davis, D. J. Ellar, and J. Li. 2005. Crystal structure of the mosquito-larvicidal toxin Cry4Ba and its biological implications. J. Mol. Biol. 348:363-382. [DOI] [PubMed] [Google Scholar]
- 8.Boonserm, P., W. Pornwiroon, G. Katzenmeier, S. Panyim, and C. Angsuthanasombat. 2004. Optimised expression in Escherichia coli and purification of the functional form of the Bacillus thuringiensis Cry4Aa delta-endotoxin. Protein Expr. Purif. 35:397-403. [DOI] [PubMed] [Google Scholar]
- 9.Bressanelli, S., K. Stiasny, S. L. Allison, E. A. Stura, S. Duquerroy, J. Lescar, F. X. Heinz, and F. A. Rey. 2004. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 23:728-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 11.Burton, S. L., D. J. Ellar, J. Li, and D. J. Derbyshire. 1999. N-acetylgalactosamine on the putative insect receptor aminopeptidase N is recognised by a site on the domain III lectin-like fold of a Bacillus thuringiensis insecticidal toxin. J. Mol. Biol. 287:1011-1022. [DOI] [PubMed] [Google Scholar]
- 12.Cabiaux, V., C. Wolff, and J. M. Ruysschaert. 1997. Interaction with a lipid membrane: a key step in bacterial toxins virulence. Int. J. Biol. Macromol. 21:285-298. [DOI] [PubMed] [Google Scholar]
- 13.Chungjatupornchai, W., H. Hofte, J. Seurinck, C. Angsuthanasombat, and M. Vaeck. 1988. Common features of Bacillus thuringiensis toxins specific for Diptera and Lepidoptera. Eur. J. Biochem. 173:9-16. [DOI] [PubMed] [Google Scholar]
- 14.Collaborative Computational Project Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 15.Czjzek, M., D. N. Bolam, A. Mosbah, J. Allouch, C. M. Fontes, L. M. Ferreira, O. Bornet, V. Zamboni, H. Darbon, N. L. Smith, G. W. Black, B. Henrissat, and H. J. Gilbert. 2001. The location of the ligand-binding site of carbohydrate-binding modules that have evolved from a common sequence is not conserved. J. Biol. Chem. 276:48580-48587. [DOI] [PubMed] [Google Scholar]
- 16.DeLano, W. L. 2002. The PyMOL user's manual. DeLano Scientific, San Carlos, CA.
- 17.de Maagd, R. A., P. L. Bakker, L. Masson, M. J. Adang, S. Sangadala, W. Stiekema, and D. Bosch. 1999. Domain III of the Bacillus thuringiensis delta-endotoxin Cry1Ac is involved in binding to Manduca sexta brush border membranes and to its purified aminopeptidase N. Mol. Microbiol. 31:463-471. [DOI] [PubMed] [Google Scholar]
- 18.de Maagd, R. A., A. Bravo, and N. Crickmore. 2001. How Bacillus thuringiensis has evolved specific toxins to colonize the insect world. Trends Genet 17:193-199. [DOI] [PubMed] [Google Scholar]
- 19.de Maagd, R. A., M. S. Kwa, H. van der Klei, T. Yamamoto, B. Schipper, J. M. Vlak, W. J. Stiekema, and D. Bosch. 1996. Domain III substitution in Bacillus thuringiensis delta-endotoxin CryIA(b) results in superior toxicity for Spodoptera exigua and altered membrane protein recognition. Appl. Environ. Microbiol. 62:1537-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Maagd, R. A., M. Weemen-Hendriks, W. Stiekema, and D. Bosch. 2000. Bacillus thuringiensis delta-endotoxin Cry1C domain III can function as a specificity determinant for Spodoptera exigua in different, but not all, Cry1-Cry1C hybrids. Appl. Environ. Microbiol. 66:1559-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evans, L. J., M. L. Goble, K. A. Hales, and J. H. Lakey. 1996. Different sensitivities to acid denaturation within a family of proteins: implications for acid unfolding and membrane translocation. Biochemistry 35:13180-13185. [DOI] [PubMed] [Google Scholar]
- 22.Falnes, P. O., and K. Sandvig. 2000. Penetration of protein toxins into cells. Curr. Opin. Cell Biol. 12:407-413. [DOI] [PubMed] [Google Scholar]
- 23.Fernandez, L. E., C. Perez, L. Segovia, M. H. Rodriguez, S. S. Gill, A. Bravo, and M. Soberon. 2005. Cry11Aa toxin from Bacillus thuringiensis binds its receptor in Aedes aegypti mosquito larvae through loop alpha-8 of domain II. FEBS Lett. 579:3508-3514. [DOI] [PubMed] [Google Scholar]
- 24.Finney, D. 1971. Probit analysis, 3rd ed. Cambridge University Press, London, United Kingdom.
- 25.Galitsky, N., V. Cody, A. Wojtczak, D. Ghosh, J. R. Luft, W. Pangborn, and L. English. 2001. Structure of the insecticidal bacterial delta-endotoxin Cry3Bb1 of Bacillus thuringiensis. Acta Crystallogr. Sect. D 57:1101-1109. [DOI] [PubMed] [Google Scholar]
- 26.Gazit, E., P. La Rocca, M. S. Sansom, and Y. Shai. 1998. The structure and organization within the membrane of the helices composing the pore-forming domain of Bacillus thuringiensis delta-endotoxin are consistent with an “umbrella-like” structure of the pore. Proc. Natl. Acad. Sci. USA 95:12289-12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerber, D., and Y. Shai. 2000. Insertion and organization within membranes of the delta-endotoxin pore-forming domain, helix 4-loop-helix 5, and inhibition of its activity by a mutant helix 4 peptide. J. Biol. Chem. 275:23602-23607. [DOI] [PubMed] [Google Scholar]
- 28.Griffitts, J. S., J. L. Whitacre, D. E. Stevens, and R. V. Aroian. 2001. Bt toxin resistance from loss of a putative carbohydrate-modifying enzyme. Science 293:860-864. [DOI] [PubMed] [Google Scholar]
- 29.Grochulski, P., L. Masson, S. Borisova, M. Pusztai-Carey, J. L. Schwartz, R. Brousseau, and M. Cygler. 1995. Bacillus thuringiensis CryIA(a) insecticidal toxin: crystal structure and channel formation. J. Mol. Biol. 254:447-464. [DOI] [PubMed] [Google Scholar]
- 30.Holm, L., and C. Sander. 1993. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233:123-138. [DOI] [PubMed] [Google Scholar]
- 31.Jenkins, J. L., M. K. Lee, S. Sangadala, M. J. Adang, and D. H. Dean. 1999. Binding of Bacillus thuringiensis Cry1Ac toxin to Manduca sexta aminopeptidase-N receptor is not directly related to toxicity. FEBS Lett. 462:373-376. [DOI] [PubMed] [Google Scholar]
- 32.Jenkins, J. L., M. K. Lee, A. P. Valaitis, A. Curtiss, and D. H. Dean. 2000. Bivalent sequential binding model of a Bacillus thuringiensis toxin to gypsy moth aminopeptidase N receptor. J. Biol. Chem. 275:14423-14431. [DOI] [PubMed] [Google Scholar]
- 33.Johnson, P. E., M. D. Joshi, P. Tomme, D. G. Kilburn, and L. P. McIntosh. 1996. Structure of the N-terminal cellulose-binding domain of Cellulomonas fimi CenC determined by nuclear magnetic resonance spectroscopy. Biochemistry 35:14381-14394. [DOI] [PubMed] [Google Scholar]
- 34.Jones, T. A., J. Y. Zou, S. W. Cowan, and Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. Sect. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 35.Jurat-Fuentes, J. L., and M. J. Adang. 2001. Importance of Cry1 delta-endotoxin domain II loops for binding specificity in Heliothis virescens (L.). Appl. Environ. Microbiol. 67:323-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanintronkul, Y., I. Sramala, G. Katzenmeier, S. Panyim, and C. Angsuthanasombat. 2003. Specific mutations within the alpha4-alpha5 loop of the Bacillus thuringiensis Cry4B toxin reveal a crucial role for Asn-166 and Tyr-170. Mol. Biotechnol. 24:11-20. [DOI] [PubMed] [Google Scholar]
- 37.Knight, P. J., B. H. Knowles, and D. J. Ellar. 1995. Molecular cloning of an insect aminopeptidase N that serves as a receptor for Bacillus thuringiensis CryIA(c) toxin. J. Biol. Chem. 270:17765-17770. [DOI] [PubMed] [Google Scholar]
- 38.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 39.Lee, M. K., B. A. Young, and D. H. Dean. 1995. Domain III exchanges of Bacillus thuringiensis CryIA toxins affect binding to different gypsy moth midgut receptors. Biochem. Biophys. Res. Commun. 216:306-312. [DOI] [PubMed] [Google Scholar]
- 40.Lee, X., A. Thompson, Z. Zhang, H. Ton-that, J. Biesterfeldt, C. Ogata, L. Xu, R. A. Johnston, and N. M. Young. 1998. Structure of the complex of Maclura pomifera agglutinin and the T-antigen disaccharide, Galbeta1,3GalNAc. J. Biol. Chem. 273:6312-6318. [DOI] [PubMed] [Google Scholar]
- 41.Li, J., D. J. Derbyshire, B. Promdonkoy, and D. J. Ellar. 2001. Structural implications for the transformation of the Bacillus thuringiensis delta-endotoxins from water-soluble to membrane-inserted forms. Biochem. Soc. Trans. 29:571-577. [DOI] [PubMed] [Google Scholar]
- 42.Li, J. D., J. Carroll, and D. J. Ellar. 1991. Crystal structure of insecticidal delta-endotoxin from Bacillus thuringiensis at 2.5 A resolution. Nature 353:815-821. [DOI] [PubMed] [Google Scholar]
- 43.Lijnzaad, P., H. J. Berendsen, and P. Argos. 1996. Hydrophobic patches on the surfaces of protein structures. Proteins 25:389-397. [DOI] [PubMed] [Google Scholar]
- 44.Masson, L., Y. J. Lu, A. Mazza, R. Brousseau, and M. J. Adang. 1995. The CryIA(c) receptor purified from Manduca sexta displays multiple specificities. J. Biol. Chem. 270:20309-20315. [DOI] [PubMed] [Google Scholar]
- 45.Masson, L., B. E. Tabashnik, A. Mazza, G. Prefontaine, L. Potvin, R. Brousseau, and J. L. Schwartz. 2002. Mutagenic analysis of a conserved region of domain III in the Cry1Ac toxin of Bacillus thuringiensis. Appl. Environ. Microbiol. 68:194-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morse, R. J., T. Yamamoto, and R. M. Stroud. 2001. Structure of Cry2Aa suggests an unexpected receptor binding epitope. Structure 9:409-417. [DOI] [PubMed] [Google Scholar]
- 47.Parker, M. W., J. T. Buckley, J. P. Postma, A. D. Tucker, K. Leonard, F. Pattus, and D. Tsernoglou. 1994. Structure of the Aeromonas toxin proaerolysin in its water-soluble and membrane-channel states. Nature 367:292-295. [DOI] [PubMed] [Google Scholar]
- 48.Parker, M. W., F. Pattus, A. D. Tucker, and D. Tsernoglou. 1989. Structure of the membrane-pore-forming fragment of colicin A. Nature 337:93-96. [DOI] [PubMed] [Google Scholar]
- 49.Poncet, S., A. Delecluse, A. Klier, and G. Rapoport. 1995. Evaluation of synergistic interactions among the CryIVA, CryIVB, and CryIVD toxic components of B. thuringiensis subsp. israelensis crystals. J. Invertebr. Pathol. 66:131-135. [Google Scholar]
- 50.Pornwiroon, W., G. Katzenmeier, S. Panyim, and C. Angsuthanasombat. 2004. Aromaticity of Tyr-202 in the alpha4-alpha5 loop is essential for toxicity of the Bacillus thuringiensis Cry4A toxin. J. Biochem. Mol. Biol. 37:292-297. [DOI] [PubMed] [Google Scholar]
- 50a.Puntheeranurak, T., P. Uawithya, L. Potvin, C. Angsuthanasombat, and J. L. Schwartz. 2004. Ion channels formed in planar lipid bilayers by the dipteran-specific Cry4B Bacillus thuringiensis toxin and its alpha1-alpha5 fragment. Mol. Membr. Biol. 21:67-74. [DOI] [PubMed] [Google Scholar]
- 51.Rajamohan, F., O. Alzate, J. A. Cotrill, A. Curtiss, and D. H. Dean. 1996. Protein engineering of Bacillus thuringiensis delta-endotoxin: mutations at domain II of CryIAb enhance receptor affinity and toxicity toward gypsy moth larvae. Proc. Natl. Acad. Sci. USA 93:14338-14343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sangadala, S., F. S. Walters, L. H. English, and M. J. Adang. 1994. A mixture of Manduca sexta aminopeptidase and phosphatase enhances Bacillus thuringiensis insecticidal CryIA(c) toxin binding and 86Rb(+)-K+ efflux in vitro. J. Biol. Chem. 269:10088-10092. [PubMed] [Google Scholar]
- 53.Sankaranarayanan, R., K. Sekar, R. Banerjee, V. Sharma, A. Surolia, and M. Vijayan. 1996. A novel mode of carbohydrate recognition in jacalin, a Moraceae plant lectin with a beta-prism fold. Nat. Struct. Biol. 3:596-603. [DOI] [PubMed] [Google Scholar]
- 54.Schnepf, E., N. Crickmore, J. Van Rie, D. Lereclus, J. Baum, J. Feitelson, D. R. Zeigler, and D. H. Dean. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:775-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schwartz, J. L., M. Juteau, P. Grochulski, M. Cygler, G. Prefontaine, R. Brousseau, and L. Masson. 1997. Restriction of intramolecular movements within the Cry1Aa toxin molecule of Bacillus thuringiensis through disulfide bond engineering. FEBS Lett. 410:397-402. [DOI] [PubMed] [Google Scholar]
- 56.Schwartz, J. L., L. Potvin, X. J. Chen, R. Brousseau, R. Laprade, and D. H. Dean. 1997. Single-site mutations in the conserved alternating-arginine region affect ionic channels formed by CryIAa, a Bacillus thuringiensis toxin. Appl. Environ. Microbiol. 63:3978-3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smedley, D. P., and D. J. Ellar. 1996. Mutagenesis of three surface-exposed loops of a Bacillus thuringiensis insecticidal toxin reveals residues important for toxicity, receptor recognition and possibly membrane insertion. Microbiology 142:1617-1624. [DOI] [PubMed] [Google Scholar]
- 58.Tapaneeyakorn, S., W. Pornwiroon, G. Katzenmeier, and C. Angsuthanasombat. 2005. Structural requirements of the unique disulphide bond and the proline-rich motif within the alpha4-alpha5 loop for larvicidal activity of the Bacillus thuringiensis Cry4Aa delta-endotoxin. Biochem. Biophys. Res. Commun. 330:519-525. [DOI] [PubMed] [Google Scholar]
- 59.Tigue, N. J., J. Jacoby, and D. J. Ellar. 2001. The alpha-helix 4 residue, Asn135, is involved in the oligomerization of Cry1Ac1 and Cry1Ab5 Bacillus thuringiensis toxins. Appl. Environ. Microbiol. 67:5715-5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tuntitippawan, T., P. Boonserm, G. Katzenmeier, and C. Angsuthanasombat. 2005. Targeted mutagenesis of loop residues in the receptor-binding domain of the Bacillus thuringiensis Cry4Ba toxin affects larvicidal activity. FEMS Microbiol. Lett. 242:325-332. [DOI] [PubMed] [Google Scholar]
- 61.Uawithya, P., T. Tuntitippawan, G. Katzenmeier, S. Panyim, and C. Angsuthanasombat. 1998. Effects on larvicidal activity of single proline substitutions in alpha3 or alpha4 of the Bacillus thuringiensis Cry4B toxin. Biochem. Mol. Biol. Int. 44:825-832. [DOI] [PubMed] [Google Scholar]
- 62.Vadlamudi, R. K., E. Weber, I. Ji, T. H. Ji, and L. A. Bulla, Jr. 1995. Cloning and expression of a receptor for an insecticidal toxin of Bacillus thuringiensis. J. Biol. Chem. 270:5490-5494. [DOI] [PubMed] [Google Scholar]
- 63.Von Tersch, M. A., S. L. Slatin, C. A. Kulesza, and L. H. English. 1994. Membrane-permeabilizing activities of Bacillus thuringiensis coleopteran-active toxin CryIIIB2 and CryIIIB2 domain I peptide. Appl. Environ. Microbiol. 60:3711-3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wallace, A. J., T. J. Stillman, A. Atkins, S. J. Jamieson, P. A. Bullough, J. Green, and P. J. Artymiuk. 2000. E. coli hemolysin E (HlyE, ClyA, SheA): X-ray crystal structure of the toxin and observation of membrane pores by electron microscopy. Cell 100:265-276. [DOI] [PubMed] [Google Scholar]
- 65.Walters, F. S., S. L. Slatin, C. A. Kulesza, and L. H. English. 1993. Ion channel activity of N-terminal fragments from CryIA(c) delta-endotoxin. Biochem. Biophys. Res. Commun. 196:921-926. [DOI] [PubMed] [Google Scholar]
- 66.Ward, E. S., and D. J. Ellar. 1988. Cloning and expression of two homologous genes of Bacillus thuringiensis subsp. israelensis which encode 130-kilodalton mosquitocidal proteins. J. Bacteriol. 170:727-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wolfersberger, M. G., X. J. Chen, and D. H. Dean. 1996. Site-directed mutations in the third domain of Bacillus thuringiensis delta-endotoxin CryIAa affect its ability to increase the permeability of Bombyx mori midgut brush border membrane vesicles. Appl. Environ. Microbiol. 62:279-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu, S. J., C. N. Koller, D. L. Miller, L. S. Bauer, and D. H. Dean. 2000. Enhanced toxicity of Bacillus thuringiensis Cry3A delta-endotoxin in coleopterans by mutagenesis in a receptor binding loop. FEBS Lett. 473:227-232. [DOI] [PubMed] [Google Scholar]
- 69.Wu, S. J., and D. H. Dean. 1996. Functional significance of loops in the receptor binding domain of Bacillus thuringiensis CryIIIA d-endotoxin. J. Mol. Biol. 255:628-640. [DOI] [PubMed] [Google Scholar]