Abstract
Under iron-limiting conditions, Pseudomonas aeruginosa PAO1 secretes a fluorescent siderophore called pyoverdine (Pvd). After chelating iron, this ferric siderophore is transported back into the cells via the outer membrane receptor FpvA. The Pvd-dependent iron uptake pathway requires several essential genes involved in both the synthesis of Pvd and the uptake of ferric Pvd inside the cell. A previous study describing the global phenotype of a tat-deficient P. aeruginosa strain showed that the defect in Pvd-mediated iron uptake was due to the Tat-dependent export of proteins involved in Pvd biogenesis and ferric Pvd uptake (U. Ochsner, A. Snyder, A. I. Vasil, and M. L. Vasil, Proc. Natl. Acad. Sci. USA 99:8312-8317, 2002). Using biochemical and biophysical tools, we showed that despite its predicted Tat signal sequence, FpvA is correctly located in the outer membrane of a tat mutant and is fully functional for all steps of the iron uptake process (ferric Pvd uptake and recycling of Pvd on FpvA after iron release). However, in the tat mutant, no Pvd was produced. This suggested that a key element in the Pvd biogenesis pathway must be exported to the periplasm by the Tat pathway. We located PvdN, a still unknown but essential component in Pvd biogenesis, at the periplasmic side of the cytoplasmic membrane and showed that its export is Tat dependent. Our results further support the idea that a critical step of the Pvd biogenesis pathway involving PvdN occurs at the periplasmic side of the cytoplasmic membrane.
Iron is an essential element for almost all bacteria. However, under aerobic conditions at neutral pH, iron forms insoluble Fe(III) oxide hydrates and is not readily available. Many bacteria produce iron chelators, called siderophores, which make iron available to the cell. Siderophores solubilize ferric ions and transport these ions into the cells via specific outer membrane transporters. The gram-negative bacteria Pseudomonas aeruginosa produces two major siderophores. One is pyochelin (Pch), which is a derivative of salicylic acid (14), and the other is pyoverdine (Pvd), which is composed of a fluorescent chromophore and a peptide moiety (3). P. aeruginosa strains produce several Pvd proteins, which can be classified into three types (PvdI to PvdIII) and can be distinguished by their peptide amino acid sequences (38). For all Pvd proteins, the peptide and the chromophore are thought to be derived from amino acid precursors that are assembled by nonribosomal peptide synthetases (NRPSs), with other enzymes catalyzing additional reactions to complete the maturation of Pvd proteins (1, 6, 15, 25, 35, 36, 40, 53). The precise biological roles of all these enzymes in the Pvd biosynthetic pathway have not been elucidated. However, the steps of the synthesis of Pvd, especially cyclization of the chromophore, are thought to take place in the periplasm. The synthesis of the chromophore, which is a condensation product of d-tyrosine and l-2,4-diaminobutyrate (20), involves the PvdL NRPS in P. aeruginosa (41). This is the only NRPS in Pseudomonas, which is highly conserved in all of the genomes analyzed so far. PvdH is another conserved enzyme among the fluorescent Pseudomonas organisms. This enzyme, which is also required for the chromophore synthesis, is an aminotransferase that catalyzes the formation of l-2,4-diaminobutyrate from aspartate β-semialdehyde (52). The enzyme that cyclizes l-2,4-diaminobutyrate into the pyrimidine ring of the Pvd chromophore remains unknown.
The cell requires specific outer membrane transporters that actively internalize the ferric siderophore complexes (9). Transport across the outer membrane is driven by the proton motive force of the cytoplasmic membrane through a cytoplasmic membrane complex comprising TonB, ExbB, and ExbD (28, 45, 56). FptA is the Pch-specific outer membrane transporter in P. aeruginosa strains (4). The three structurally different Pvd proteins produced by P. aeruginosa strains are recognized by specific transporters in the outer membrane: FpvAI and FpvB for PvdI, FpvAII for PvdII, and FpvAIII for PvdIII (17). In the present work, PvdI and FpvAI are called Pvd and FpvA, respectively. FpvA has been well characterized using physiological, immunological, and molecular approaches (see references 11, 16, 50, and 51, among others). The structures of P. aeruginosa FptA and FpvA have been solved (12, 13). Like FhuA (21, 33), FepA (10), and FecA (22, 57), these transporters are composed of two domains: a transmembrane 22-stranded β-barrel domain and an N-terminal plug domain that fills the barrel interior.
In bacteria, protein translocation across the cytoplasmic membrane occurs via two major routes. The Sec pathway is the main route for protein export. It allows a fast translocation of nonfolded substrates (18). A second general transport pathway, called Tat, for twin-arginine translocation, has been recently described (for a review, see reference 44). The Tat machinery exports folded proteins across the cytoplasmic membrane. Most of the different Tat-secreted proteins studied are periplasmic enzymes that take part in multiprotein oxido-reduction systems involved in respiration or anaerobic growth (7, 47). Before they are secreted, Tat-dependent proteins are folded in the cytoplasm, and many bind redox cofactors (47). Both Sec and Tat-dependent proteins are synthesized in precursor form with cleavable N-terminal signal peptides that carry specific signatures for one or the other export machinery. Both signal sequences have a tripartite structure, with a basic N-terminal region preceding a longer hydrophobic part, followed by a C-terminal region containing the recognition sequence for the signal peptidase. Signal peptides that target proteins to the Tat machinery have specific additional features. The most obvious is a consensus motif containing two highly conserved arginines (R). This motif has the form S-R-R-X-Φ-Φ, where S is serine and Φ is a hydrophobic residue (leucine, phenylalanine, valine, or methionine). Tat signal peptides are usually longer and less hydrophobic than Sec signal peptides and frequently contain a basic residue in the C terminus called the “Sec avoidance motif.”
It has been shown that a P. aeruginosa tat mutant affects both Pvd biogenesis and uptake (42). The Pvd outer membrane receptor FpvA is predicted to have a putative Tat signal peptide. Consequently, it has been proposed that FpvA is transported by the Tat pathway (42). Here, we investigated the relationship between Tat export and Pvd-mediated iron uptake. We found that, in a P. aeruginosa tat mutant, ferric Pvd uptake is not altered, and FpvA is fully functional and correctly located in the outer membrane. However, we showed that at least one essential component of the Pvd biogenesis pathway, PvdN, is exported by the Tat pathway, explaining why no Pvd is synthesized in this context.
MATERIALS AND METHODS
Chemicals and siderophores.
55FeCl3 was from Perkin Elmer Life and Analytical Sciences (Boston, Mass.). Pyoverdins (Pvd, Pvd-Fe, and Pvd-55Fe) were prepared as described previously (2, 19, 49). Anti-FpvA and anti-FptA polyclonal antisera were prepared from purified FpvA and FptA. New Zealand rabbits were immunized with 150 μg of receptor in 1 ml of phosphate-buffered saline and Freund's complete adjuvant V.
Bacterial strains and growth media.
The strains used in this study are the wild-type strains P. aeruginosa PAK and PAO1 and their respective isogenic mutants that lack functional Tat machinery: PAKΔtatC (54), PAOΔtatABC, PAO1ΔfpvA (50), and PAO1ΔpvdN (30). The PAOΔtatABC mutant was constructed as previously described (5). Briefly, 500-bp sections upstream and downstream of the target genes were PCR amplified. The oligonucleotides were designed for amplifying fragments with overlapping 3′ and 5′ ends. Both fragments were ligated by using an overlapping PCR. This was done by using the most-upstream and -downstream primers in a second PCR with a mix of the two fragments as the matrix. The resulting PCR product was cloned into the PCR2.1 plasmid (TA cloning kit; Invitrogen). A 1,000-bp BamHI-EcoRV DNA fragment was then subcloned into the suicide pKNG101 vector (27). The resulting construct was transferred to P. aeruginosa by mobilization. The strains in which the chromosomal integration event occurred were selected on Pseudomonas isolation agar plates containing 2,000 μg of streptomycin per ml. Excision of the plasmid, resulting in the deletion of the chromosomal target gene, was performed after selection on Luria-Bertani (LB) plates containing 5% sucrose. Clones that became sucrose resistant and streptomycin sensitive were confirmed to contain the gene deletion by PCR analysis. The recombinant plasmid pMMB-PvdNV5H6 was introduced into P. aeruginosa using the conjugative properties of pRK2013 and clones selected on Pseudomonas isolation agar (Difco Laboratories) plates containing 300 μg/ml carbenicillin. Strains were grown at 37°C with aeration in different media: succinate (or iron limiting) (19), LB, and phosphate limiting (proteose peptone broth).
Construction of pMMB-PvdNV5H6.
pMMB-PvdNV5H6 encoding PvdNV5H6 was constructed using the Gateway PAO1 collection (29). Basically, all PAO1 open reading frames (ORF) were cloned by PCR and inserted into an entry vector constituting the PAO1 Gateway bank. Subsequently, any ORF can be rapidly moved into the desired destination vector by using phage attR and attL recombination sites flanking the cloned gene in the entry vector and in the destination vectors to allow phage recombinase-mediated recloning. We moved ORF 2394 encoding PvdN into the destination vector pET-DEST42 (Invitrogen) to produce a C-terminal V5-hexahistidine (V5H6)-tagged PvdN, called PvdNV5H6. The pvdNV5H6 gene fusion was then reinserted into the broad-host-range vector pMMB67EH (24) using XbaI and SmaI restriction sites. This fusion gene was placed under the control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible tac promoter to give pMMB-PvdNV5H6.
Cell fractionation.
Cellular fractions used for the FpvA localization shown in Fig. 1 were prepared as follows: bacteria were collected by centrifugation after overnight growth in iron-limited medium, frozen, and stored overnight at −80°C in 10 mM Tris-HCl (pH 8) containing a Complete EDTA-free (Roche) protease inhibitor cocktail. The cell samples were subjected to ultrasonic disintegration, and unlysed cells were removed by centrifugation (15 min at 12,000 × g). The cell envelope fractions were collected by ultracentrifugation (30 min at 125,000 × g) and dissolved in 10 mM Tris-HCl (pH 8). Supernatants correspond to soluble fractions (cytoplasmic and periplasmic). Cytoplasmic and outer membrane proteins were separated by differential sodium lauryl sarcosyl (SLS) solubilization. Cell envelope fractions were incubated with 2% SLS for 25 min at room temperature with gentle shaking. SLS-insoluble outer membrane proteins were separated from soluble cytoplasmic membrane proteins by ultracentrifugation (30 min at 125,000 × g).
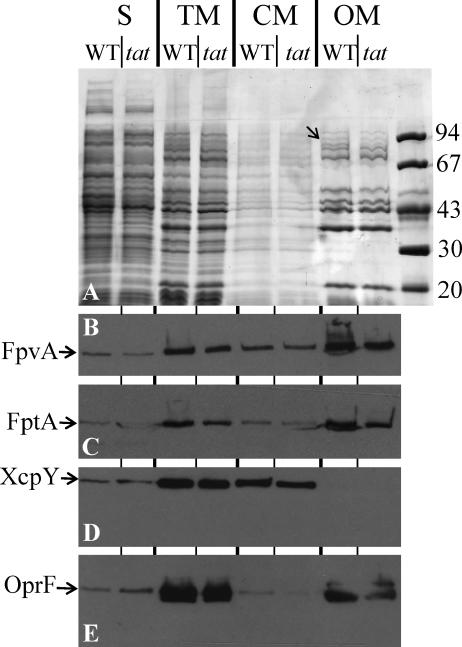
FIG. 1.
FpvA is transported into the outer membrane in a Tat-independent manner. Bacterial cell culture of wild-type PAO1 (WT) and its isogenic tatABC mutant (tat) grown in iron-limited medium were subjected to subcellular fractionation. S, soluble fraction containing cytoplasmic and periplasmic protein; TM, whole cell envelope; CM, cytoplasmic membrane; OM, outer membrane. The equivalent of 0.1 OD600 unit of the cultures was loaded onto a 9% SDS-PAGE gel and then stained with Coomassie blue (A). Alternatively, proteins were blotted onto nitrocellulose and revealed using anti-FpvA antibody (B), anti-FptA antibody (C), anti-XcpY antibody (D), or anti-OprF antibody (E). Molecular mass markers (kDa) are shown on the right. The position of FpvA, identified by mass spectrometry, is shown by an arrow in panel A.
Outer membrane preparations used for fluorescence resonance energy transfer (FRET) experiments presented in Fig. 2 were prepared as previously described (50).
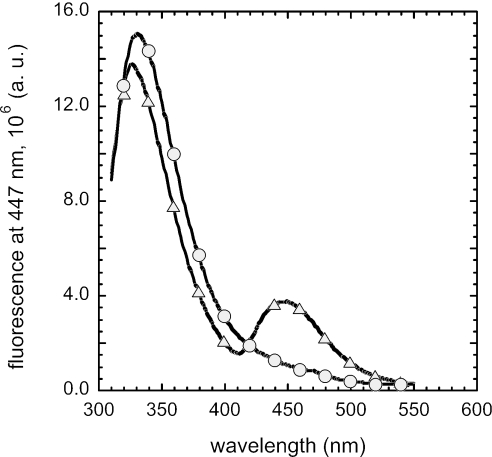
FIG. 2.
Fluorescent emission spectra of FpvA in P. aeruginosa wild-type and tat mutant. Outer membrane fractions of P. aeruginosa PAK (▵) and PAKΔtatC (○) were dissolved in 50 mM Tris-HCl at pH 8.0. Excitation was at 290 nm, and fluorescence was recorded at 447 nm. a.u., arbitrary units.
Spheroplasts used in the trypsin digestion experiment (see Fig. 6) were prepared as follow: 1 × 1010 bacteria (optical density at 600 nm [OD600] of 10) were collected by centrifugation and resuspended in 150 μl of buffer A (200 mM Tris-HCl, pH 5.5, 0.5 mM EDTA, 500 mM sucrose) and 2.2 ml of buffer B (30 mM Tris-HCl, pH 7.6, 10 mM EDTA, 750 mM sucrose). Lysozym (3,300 U) (Euromedex) resuspended in 100 μl of buffer B was slowly added to the cells, and the preparation was slowly shaken at room temperature for 40 min. The spheroplasts were then ready for trypsin digestion.
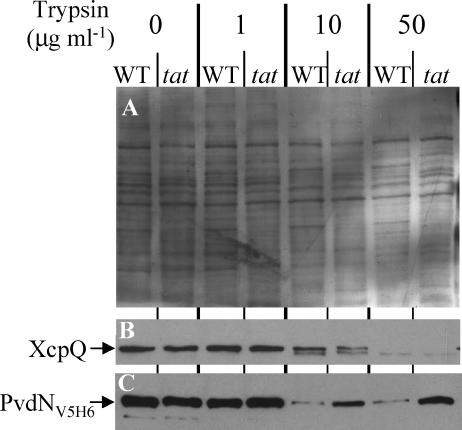
FIG. 6.
Protease accessibility of PvdN in spheroplasts. Wild-type PAO1 and PAOΔtatABC mutant strains expressing pvdNV5H6 were grown in phosphate-limited medium. pvdNV5H6 expression was induced in the exponential phase by incubation with 0.1 mM IPTG for 2 h. Cells were then collected and treated for spheroplast preparation. Wild-type (WT) and tatABC (tat) spheroplast samples equivalent to 0.1 OD600 unit of the original cultures were further treated with increasing concentrations of trypsin and analyzed by 11% SDS-PAGE, followed by Western blotting using anti-XcpQ (B) and anti-V5 (C) antibodies. Panel A is a red Ponceau-stained nitrocellulose membrane used for Western blotting. This shows that for each trypsin concentration, the same amount of protein is loaded for the wild-type strain and the tat mutant, indicating that the smaller amount of PvdN observed in the wild-type strain compared to the tat mutant is not due to overloading. This experiment was repeated twice.
Trypsin digestion.
We tested the trypsin sensitivity of PvdNV5H6 by incubating 250 μl (1 × 109 bacteria) of the spheroplast preparation at various trypsin concentrations for 10 min at room temperature. Protease activity was inhibited by adding 1 mM phenylmethylsulfonyl fluoride.
SDS-PAGE and immunoblotting.
For protein analysis, protein samples were resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (34) with 2% SDS and heated for 10 min at 95°C. The equivalent of 1 × 108 bacteria (OD600 of 0.1) was loaded onto the gels. Electrophoresis was carried out in the SDS-polyacrylamide gels at 25 mA per gel at room temperature. For Western blotting, proteins were transferred from the gel onto nitrocellulose membranes. The membranes were blocked by incubation overnight in phosphate-buffered saline (pH 7.6)-0.1% dried milk-0.01% Tween 20 and then incubated in blocking buffer with primary antibodies against XcpQ (8), β-lactamase (QIAGEN), V5 epitope (Bethyl), FpvA, FptA, XcpY (39), or OprF (23), followed by a second incubation in blocking buffer with horseradish peroxidase-conjugated goat anti-rabbit or anti-mouse immunoglobulin G antibodies. Blots were developed using an enhanced chemiluminescence protocol (Amersham).
Iron uptake.
Iron uptake assays were carried out as reported previously for the FpvA/Pvd system (48). After overnight growth in iron-limited medium, bacteria were prepared at an OD600 of 0.5 in 50 mM Tris-HCl (pH 8.0) and incubated at 37°C. Transport assays were initiated by adding 100 nM Pvd-55Fe. Aliquots (100 μl) of the suspension were removed at different times and filtered, and the retained radioactivity was counted. The experiment was repeated in the presence of 200 μM carbonyl cyanide m-chlorophenylhydrazone (CCCP; Sigma).
Fluorescence spectroscopy.
Fluorescence experiments were carried out using a Photon Technology International TimeMaster (Bioritech) spectrofluorometer. To show that Pvd is recycled on FpvA, the experiments were carried out as previously described (48). After overnight growth in iron-limited medium, the cells were resuspended at an OD600 of 0.5 in 50 mM Tris-HCl (pH 8.0). The bacterial suspension (995 μl) was stirred at 29°C in a 1-ml cuvette. Pvd-Fe (5 μl) was added to 995 μl of the bacterial suspension to give a final concentration of 100 nM Pvd-Fe. The fluorescence at 447 nm (excitation wavelength set to 290 nm) was measured every second for 30 min. Cell stability at 290 nm was checked by repeating the same experiments in the absence of the siderophore. For the FRET spectra on outer membranes, the membranes were resuspended in 50 mM Tris-HCl (pH 8.0) at a concentration of 160 μg/ml total protein. The excitation wavelength was set to 290 nm.
RESULTS
The Tat system is not required for FpvA transport to the outer membrane.
In the P. aeruginosa PAO1 strain, the Pvd receptor FpvA contains two consecutive arginines within a long N-terminal region in its signal peptide. These may be part of a Tat signal peptide. Therefore, we studied the cellular location of FpvA in a PAOΔtatABC mutant to check whether FpvA transport is Tat dependent. Strain PAOΔtatABC and the parental strain PAO1 were grown under iron-limited conditions. We prepared cell fractions (cytoplasmic membrane, outer membrane, and soluble fraction containing both cytoplasmic and periplasmic proteins) as described in Materials and Methods and separated the proteins in polyacrylamide gel containing SDS. The proteins were stained by Coomassie blue (Fig. 1A) or were transferred onto nitrocellulose membranes and identified using specific antibodies (Fig. 1B to E). We first observed that a 90-kDa protein, identified by mass spectrometry as FpvA, was more abundant in the outer membrane fractions of the wild-type strain than the tat mutant (tat) (Fig. 1A). As tat mutants do not produce Pvd, this lower level of FpvA is consistent with previous data (6) showing that a Pvd-deficient strain produces less FpvA than the wild-type strain. However, immunoblotting with specific antibodies against FpvA indicated that the receptor was located in the outer membrane of a tat mutant (Fig. 1B) or a wild-type strain. We obtained exactly the same results when we analyzed the Pch transporter FptA (Fig. 1C). However, unlike FpvA, this siderophore transporter has no typical Tat signal peptide. A small fraction of the outer membrane protein OprF was recovered in the cytoplasmic membrane fraction (Fig. 1E). This suggests some contamination of the cytoplasmic membrane by proteins of the outer membrane, thus explaining the small amount of FpvA and FptA recovered in the cytoplasmic membrane fraction. More importantly, XcpY, a cytoplasmic membrane protein, was only recovered from the cytoplasmic membrane fraction (Fig. 1D), indicating no contamination of the outer membrane fraction by cytoplasmic membrane proteins. This experiment clearly shows that FpvA is properly located in the outer membrane of the tat mutant.
The Tat system is not required for FpvA function.
We further investigated the functionality of FpvA in the outer membrane of a tat mutant. Previous studies have shown that FpvA receptors of cells producing Pvd were all loaded with iron-free Pvd (apo-Pvd) and that this FpvA-Pvd complex is the normal transporter state under iron-depleted conditions (50). The loaded status of FpvA can be determined by the fluorescent properties of Pvd (50). Apo-Pvd can undergo FRET with the tryptophan present in the protein. Upon excitation at 290 nm, the FpvA-Pvd complex fluoresces at 447 nm, as shown in Fig. 2, when the outer membrane fraction of the P. aeruginosa strain PAK (50) is used. Outer membrane fractions prepared from tat mutant (PAKΔtatC) strains showed no fluorescence at 447 nm upon excitation at 290 nm, indicating that no binding of Pvd to FpvA had occurred (Fig. 2). This is not surprising because tat mutants are Pvd-deficient cells.
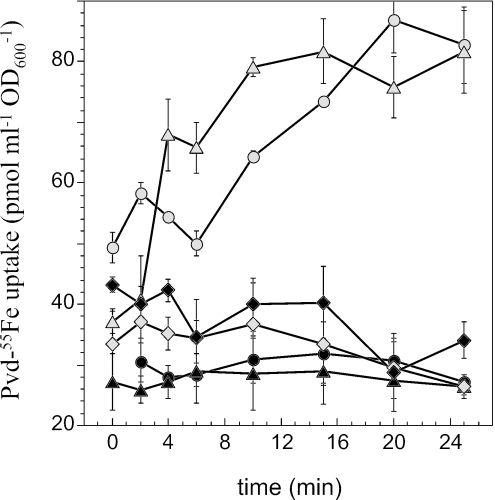
The ability of FpvA to transport iron in PAOΔtatABC cells was studied by incubating cells grown under iron-deficient conditions with Pvd-55Fe and monitoring the uptake of iron. FpvA expressed in a tat mutant was functional and could transport iron as well as FpvA expressed in wild-type cells (Fig. 3). This iron uptake involves specifically the FpvA receptor, since no iron was transported in an fpvA mutant. When the PAOΔtatABC cells were treated with the protonophore CCCP, transport was inhibited as in wild-type cells (Fig. 3). This supports the TonB-dependent mechanism characteristic of siderophore receptors. The same results were obtained with the PAKΔtatC strain (data not shown).
FIG. 3.
Iron uptake in P. aeruginosa wild-type and fpvA and tatABC mutants. P. aeruginosa PAO1 (▵), PAOΔtatABC (○), and PAOΔfpvA (⋄) cells at an OD600 of 0.5 were incubated for 15 min in 50 mM Tris-HCl (pH 8.0) before transport assays were started by adding 100 nM Pvd-55Fe. Aliquots (100 μl) of the suspension were removed at different times and filtered, and the retained radioactivity was counted. The experiment was repeated in the presence of 200 μM CCCP (▴, P. aeruginosa PAO1; •, PAOΔtatABC; and ⧫, PAOΔfpvA). The data presented in this figure are the results of three experiments (error bars are indicated). Similar results were obtained with P. aeruginosa PAK and PAKΔtatC.
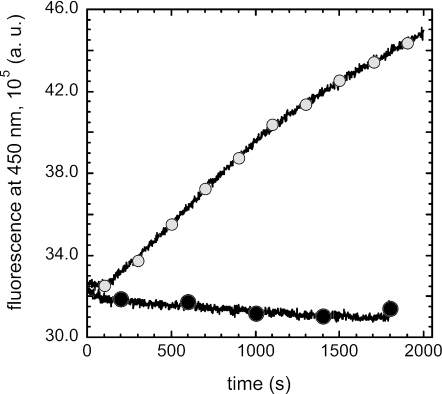
We have previously shown that once iron is released inside the cells, Pvd is recycled on FpvA and in the extracellular medium (48). Therefore, we tested whether FpvA in a tat mutant is fully functional for all steps of the iron acquisition process, including the recycling of Pvd. Cells were incubated in the presence of Pvd-Fe, and the fluorescence at 447 nm was monitored (FRET with excitation at 290 nm). We observed an increase in fluorescence at 447 nm when PAKΔtatC cells were incubated in the presence of Pvd-Fe (Fig. 4), indicating binding of apo-Pvd to FpvA after iron release in the cells. There is no FRET signal when Pvd-Fe binds to free binding sites on FpvA. As previously described (48), when higher concentrations of Pvd-Fe were used, we observed recycling of Pvd on FpvA and in the extracellular medium (data not shown).
FIG. 4.
Pvd recycling in P. aeruginosa wild-type and tat mutant strains. PAKΔtatC cells at an OD600 of 0.6 were incubated in 50 mM Tris-HCl (pH 8.0) at 29°C. The assay was started by adding 100 nM Pvd-Fe to the cells (○). The increase in fluorescence at 447 nm was monitored by measuring the fluorescence at 450 nm (excitation wavelength, 290 nm) every second for 30 min. The same fluorescence measurements were repeated in the absence of the siderophore (•). a.u., arbitrary units.
The Tat system is required for PvdN biogenesis.
In the P. aeruginosa tatC mutant, we observed no specific Pvd fluorescence (data not shown and reference 42), suggesting that at least one key element in the Pvd biogenesis pathway is exported in the periplasm by the Tat pathway. Among the different proteins known to be directly or indirectly involved in Pvd biogenesis, three of them, PA2389, PvdP (PA2392), and PvdN (PA2394), contain two consecutives arginines in their signal peptides. However, the new TatP1.0 prediction program (http://www.cbs.dtu.dk/services/TatP/), together with a recent study (32), suggests that only PvdN, a putative class V aminotransferase, contains a valid Tat signal peptide.
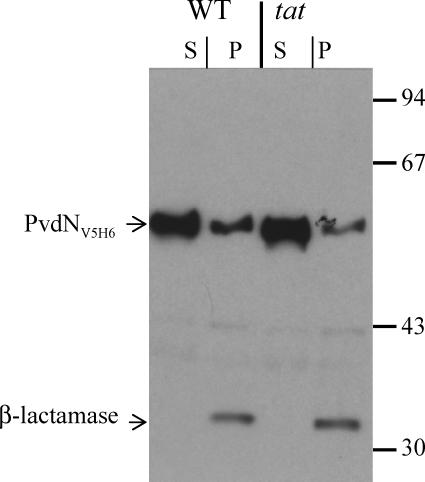
The cellular location of PvdN in a P. aeruginosa tat mutant was investigated to demonstrate directly its export by the Tat pathway. A C-terminally tagged PvdN (PvdNV5H6) was engineered (see Materials and Methods), and the chimera was produced in the PAO1 and PAOΔtatABC strains. The analysis of the subcellular location of PvdNV5H6 by separating spheroplasts and periplasm revealed that most PvdN is present in the spheroplast fraction in both wild-type and Tat-deficient strains, whereas the periplasmic marker β-lactamase was fully recovered in the periplasmic fraction (Fig. 5). Therefore, in order to determine the PvdN translocation status in the spheroplast fraction, protease accessibility experiments were carried out (Fig. 6). Spheroplasts were prepared for either wild-type or tatABC mutant strains expressing pvdNV5H6, and increasing concentrations of trypsin were added. Proteins were separated in SDS gel containing 11% polyacrylamide, transferred onto nitrocellulose membrane, and stained by Ponceau red (Fig. 6A). Specific proteins were subsequently immunoblotted using specific antibodies (Fig. 6B and C). Unlike XcpQ, an outer membrane protein involved in type II secretion (8) that is exported independently of the Tat system and presents similar protease sensitivity in wild-type and Tat-deficient strains, the trypsin degradation profile of PvdNV5H6 was dependent on a functional Tat system. PvdNV5H6 appeared significantly more resistant to trypsin in the Tat-deficient strain than in the wild-type strain (Fig. 6C). PvdNV5H6 protease protection in PAOΔtatABC spheroplasts is consistent with its presence in the cytoplasm and, thus, with Tat-dependent export of this protein. The insolubility and higher sensitivity of the protein in the wild-type spheroplasts suggest that PvdN is located on the periplasmic side of the cytoplasmic membrane.
FIG. 5.
PvdNV5H6 is recovered in the spheroplast fraction in both wild-type and tat-deficient strains. Wild-type (WT) and tatABC (tat) P. aeruginosa mutant strains expressing pvdNV5H6 were grown in LB medium. pvdNV5H6 expression was induced in the exponential phase by 0.1 mM IPTG for 1 h. Cells were further collected and treated for spheroplast preparation. Wild-type and tatABC spheroplasts were further centrifuged for 10 min at 8,000 RPM. The pellet corresponds to the spheroplast (S), whereas the supernatant corresponds to periplasm (P). Samples equivalent to 0.1 OD600 unit of original cultures were further analyzed by 11% SDS-PAGE, followed by Western blotting using anti-V5 and anti-β-lactamase antibodies. Molecular mass markers (kDa) are shown on the right.
DISCUSSION
Previous studies have shown that the Tat system is involved in the export of many proteins. These include extracellular proteins involved in virulence, such as hemolytic and nonhemolytic phospholipases (54), or periplasmic enzymes that take part in multiprotein oxido-reduction systems and are involved in respiration or anaerobic growth (7, 47). P. aeruginosa tat mutants have been described as being unable to produce the siderophore Pvd and as having an affected ferric Pvd uptake process (42). In this study, we have shown that the outer membrane location (Fig. 1) and function (Fig. 2 to 4) of this siderophore receptor are not altered in a tat mutant despite the presence of a possible Tat signal peptide at the N terminus of FpvA. In a tat mutant, the FpvA receptor is fully functional for all the different steps of Pvd-Fe uptake (Pvd-Fe uptake and Pvd recycling on FpvA after iron release). These findings demonstrated that FpvA does not require the Tat pathway for its export across the cytoplasmic membrane. This is not in agreement with a previous study of the Tat-dependent transport of FpvA (42). This discrepancy can be explained by the different experimental approaches used. Ochsner et al. found that Tat-deficient cells were not rescued by cross-feeding Pvd and concluded that Tat deficiency affected the uptake of Pvd-Fe (42). They also used a mutant strain producing an FpvA protein having an altered Tat recognition motif (FpvA-R18K). Pvd production was reduced in this mutant, and it failed to grow in the presence of the iron chelator ethylenediamine di(o-hydroxyphenyl)acetic acid (42). The authors finally concluded that FpvA transport was Tat dependent. However, they did not show any direct evidence for the presence or the absence of FpvA in the outer membranes of these two mutants (ΔtatC and fpvA-R18K), and, therefore, no direct proof of Tat-dependent export of FpvA was given. In the present study, we used two direct and complementary approaches that demonstrated the correct function and cellular location of FpvA in a tat mutant.
FpvA is not transported by the Tat machinery, despite the presence of two consecutives arginines in its signal peptide. Tat-independent transport of predicted Tat substrates has already been reported previously (26). This suggests that although the known specific features of the Tat signal peptide are necessary, these features cannot confidently predict Tat substrates. Consequently, prediction of Tat signal peptides appears to be difficult and must always be experimentally confirmed. There are several other arguments against a Tat-dependent transport of FpvA. FpvA is not known to bind any cofactor requiring folding into the cytoplasm and, therefore, does not require Tat translocation. Like porins, FpvA (12) is a member of the β-barrel outer membrane protein family for which no cytoplasmic folding is required. During outer membrane protein biogenesis, the folding steps leading to the β-barrel formation start in the periplasm and finish in the outer membrane (55). Finally, FpvA is the only Pvd transporter to have two arginines in its signal peptide. FpvAII and FpvAIII do not have this motif and therefore reach the outer membrane via a Sec-dependent route. This is also the case for FptA, the Pch siderophore receptor of P. aeruginosa, which possesses a typical Sec signal peptide and is transported, like FpvA, in a Tat-independent manner (Fig. 1). In the P. aeruginosa genome, a recent, improved prediction study has revealed possible Tat substrates including another TonB-dependent receptor, FepD (32), for which Tat dependency still remains to be demonstrated.
The Tat pathway is involved in the biogenesis of Pvd because a tat mutant does not produce Pvd (reference 42 and data not shown). A total of at least 15 genes have now been identified that are essential for Pvd synthesis in P. aeruginosa PAO1, but little is known concerning the Pvd biosynthesis pathway. Both the chromophore (41) and the peptide chain of this siderophore (31, 37) are synthesized by NRPS, with other enzymes catalyzing additional reactions to complete the maturation of Pvd proteins (1, 6, 15, 25, 35, 36, 40, 53). The precise biological function of these enzymes and in what order they play a role in the Pvd biosynthetic pathway are still not clear. Currently, the steps of the synthesis of Pvd, especially cyclization of the chromophore, are thought to take place in the periplasm.
We have shown here that PvdN is transported into the periplasm by the Tat pathway and binds in this cellular compartment to the cytoplasmic membrane. At the periplasmic side of the cytoplasmic membrane, PvdN could be part of a protein complex involved in the final steps of Pvd biogenesis. Many genes in the different Pvd clusters encode proteins that are anchored in the cytoplasmic membrane or have a predicted signal peptide, although their functions in Pvd biosynthesis are still hypothetical; examples are PvdM (PA2393), a predicted membrane-bound dipeptidase, and PvdE, which belongs to the family of the ATP-binding cassette transporters (36, 46). When these proteins are mutated, a decrease in the production of Pvd is observed (30, 42, 43).
Previous studies indicated that Pvd biogenesis is not fully abolished in a pvdN mutant, whereas there is no Pvd production in the tat mutant (30, 42, 43). This discrepancy may indicate that at least another enzyme important for the synthesis of the siderophore must be Tat dependent. Since PA2389 and PvdP have no valid Tat signal peptide (32), it is likely that there is another Tat-dependent protein whose involvement in Pvd biosynthesis has not been shown yet. Further experimental studies will be thus necessary to clearly understand the complete Tat dependence of the synthesis and production of Pvd.
In conclusion, our study has shown that despite the presence of a twin arginine in the sequence signal of FpvA, this transporter is not transported via the Tat machinery. However, PvdN, an enzyme involved in the production of Pvd, is exported in a Tat-dependent way. This enzyme is transported in the periplasmic compartment and binds to the cytoplasmic membrane, where it is probably part of a multiprotein complex involved in the synthesis of Pvd. The identification of the other components of this multiprotein complex and an explanation of why PvdN needs to be exported by the Tat pathway will be goals for future studies.
ADDENDUM IN PROOF
In a recent paper, Caldelari et al. (I. Caldelari, S. Mann, C. Crooks, and T. Palmer, Mol. Plant Microbe Interact. 19:200-212, 2006) describe the role of the Tat system in the plant pathogen Pseudomonas syringae strain DC3000. In this study, the authors showed Tat-dependent transport of PSPTO2155, a putative aminotransferase involved in the synthesis of a siderophore of this bacterium. Moreover, in the same study the authors showed that, in spite of the presence of two consecutive arginines in their signal sequences, the outer membrane localization of two putative TonB-dependent siderophore receptors found in P. syringae (PSPTO3294 and PSPTO3574) is Tat independent. All together, these results are in full agreement with our findings.
Acknowledgments
We thank G. Ball for providing strain PAOΔtatABC construction, D. Moinier for mass spectrometry analysis, and F. Hoegy for technical assistance.
This work was partially supported by the Fondation Bettencourt-Schueller and the European Union grant “Tat-machine (FP6-2003-LIFESCIHEALTH-I05257),” by the Centre National de la Recherche Scientifique (Programme Physique et Chimie du Vivant), and the Ministère de l'Enseignement Supérieure de la Recherche et de la Technologie (ACC-SDV5).
REFERENCES
- 1.Ackerley, D. F., T. T. Caradoc-Davies, and I. L. Lamont. 2003. Substrate specificity of the nonribosomal peptide synthetase PvdD from Pseudomonas aeruginosa. J. Bacteriol. 185:2848-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albrecht-Garry, A. M., S. Blanc, N. Rochel, A. Z. Ocacktan, and M. A. Abdallah. 1994. Bacterial iron transport: coordination properties of pyoverdin PaA, a peptidic siderophore of Pseudomonas aeruginosa. Inorg. Chem. 33:6391-6402. [Google Scholar]
- 3.Ankenbauer, R., S. Sriyosachati, and C. D. Cox. 1985. Effects of siderophores on the growth of Pseudomonas aeruginosa in human serum and transferrin. Infect. Immun. 49:132-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ankenbauer, R. G., and H. N. Quan. 1994. FptA, the Fe(III)-pyochelin receptor of Pseudomonas aeruginosa: a phenolate siderophore receptor homologous to hydroxamate siderophore receptors. J. Bacteriol. 176:307-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ball, G., E. Durand, A. Lazdunski, and A. Filloux. 2002. A novel type II secretion system in Pseudomonas aeruginosa. Mol. Microbiol. 43:475-485. [DOI] [PubMed] [Google Scholar]
- 6.Beare, P. A., R. J. For, L. W. Martin, and I. L. Lamont. 2003. Siderophore-mediated cell signalling in Pseudomonas aeruginosa: divergent pathways regulate virulence factor production and siderophore receptor synthesis. Mol. Microbiol. 47:195-207. [DOI] [PubMed] [Google Scholar]
- 7.Berks, B. C. 1996. A common export pathway for proteins binding complex redox cofactors. Mol. Microbiol. 22:393-404. [DOI] [PubMed] [Google Scholar]
- 8.Bitter, W., M. Koster, M. Latijnhouwers, H. de Cock, and J. Tommassen. 1998. Formation of oligomeric rings by XcpQ and PilQ, which are involved in protein transport across the outer membrane of Pseudomonas aeruginosa. Mol. Microbiol. 27:209-219. [DOI] [PubMed] [Google Scholar]
- 9.Braun, V. 2003. Iron uptake by Escherichia coli. Front. Biosci. 8:s1409-s1421. [DOI] [PubMed] [Google Scholar]
- 10.Buchanan, S. K., B. S. Smith, L. Venkatramani, D. Xia, L. Esser, M. Palmetkar, R. Chakraborti, D. Van der Helm, and J. Deisenhofer. 1999. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat. Struct. Biol. 6:56-63. [DOI] [PubMed] [Google Scholar]
- 11.Clément, E., P. J. Mesini, F. Pattus, M. A. Abdallah, and I. J. Schalk. 2004. The binding mechanism of pyoverdin with the outer membrane receptor FpvA in Pseudomonas aeruginosa is dependent on its iron-loaded status. Biochemistry 43:7954-7965. [DOI] [PubMed] [Google Scholar]
- 12.Cobessi, D., H. Célia, N. Folschweiller, I. J. Schalk, M. A. Abdallah, and F. Pattus. 2005. The crystal structure of the pyoverdin outer membrane receptor FpvA from Pseudomonas aeruginosa at 3.6 angstroms resolution. J. Mol. Biol. 34:121-134. [DOI] [PubMed] [Google Scholar]
- 13.Cobessi, D., H. Celia, and F. Pattus. 2005. Crystal structure at high resolution of ferric-pyochelin and its membrane receptor FptA from Pseudomonas aeruginosa. J. Mol. Biol. 352:893-904. [DOI] [PubMed] [Google Scholar]
- 14.Cox, C. D., K. L. Rinehart, Jr., M. L. Moore, and J. C. Cook, Jr. 1981. Pyochelin: novel structure of an iron-chelating growth promoter for Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 78:4256-4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cunliffe, H. E., T. R. Merriman, and I. L. Lamont. 1995. Cloning and characterization of pvdS, a gene required for pyoverdine synthesis in Pseudomonas aeruginosa: PvdS is probably an alternative sigma factor. J. Bacteriol. 177:2744-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dean, C. R., and K. Poole. 1993. Expression of the ferric enterobactin receptor (PfeA) of Pseudomonas aeruginosa: involvement of a two-component regulatory system. Mol. Microbiol. 8:1095-1103. [DOI] [PubMed] [Google Scholar]
- 17.de Chial, M., B. Ghysels, S. A. Beatson, V. Geoffroy, J. M. Meyer, T. Pattery, C. Baysse, P. Chablain, Y. N. Parsons, C. Winstanley, S. J. Cordwell, and P. Cornelis. 2003. Identification of type II and type III pyoverdine receptors from Pseudomonas aeruginosa. Microbiology 149:821-831. [DOI] [PubMed] [Google Scholar]
- 18.de Keyzer, J., C. van der Does, and A. J. Driessen. 2003. The bacterial translocase: a dynamic protein channel complex. Cell Mol. Life Sci. 60:2034-2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Demange, P., S. Wendenbaum, C. Linget, C. Mertz, M. T. Cung, A. Dell, and M. A. Abdallah. 1990. Bacterial siderophores: structure and NMR assignment of pyoverdins PaA, siderophores of Pseudomonas aeruginosa ATCC 15692. Biol. Metals 3:155-170. [Google Scholar]
- 20.Dorrestein, P. C., K. Poole, and T. P. Begley. 2003. Formation of the chromophore of the pyoverdine siderophores by an oxidative cascade. Org. Lett. 5:2215-2217. [DOI] [PubMed] [Google Scholar]
- 21.Ferguson, A. D., J. Breed, K. Diederichs, W. Welte, and J. W. Coulton. 1998. An internal affinity-tag for purification and crystallization of the siderophore receptor FhuA, integral outer membrane protein from Escherichia coli K-12. Protein Sci. 7:1636-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferguson, A. D., R. Chakraborty, B. S. Smith, L. Esser, D. van der Helm, and J. Deisenhofer. 2002. Structural basis of gating by the outer membrane transporter FecA. Science 295:1715-1719. [DOI] [PubMed] [Google Scholar]
- 23.Finnen, R. L., N. L. Martin, R. J. Siehnel, W. A. Woodruff, M. Rosok, and R. E. Hancock. 1992. Analysis of the Pseudomonas aeruginosa major outer membrane protein OprF by use of truncated OprF derivatives and monoclonal antibodies. J. Bacteriol. 174:4977-4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furste, J. P., W. Pansegrau, R. Frank, H. Blocker, P. Scholz, M. Bagdasarian, and E. Lanka. 1986. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48:119-131. [DOI] [PubMed] [Google Scholar]
- 25.Handfield, M., D. E. Lehoux, F. Sanschagrin, M. J. Mahan, D. E. Woods, and R. C. Levesque. 2000. In vivo-induced genes in Pseudomonas aeruginosa. Infect. Immun. 68:2359-2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jongbloed, J. D., H. Antelmann, M. Hecker, R. Nijland, S. Bron, U. Airaksinen, F. Pries, W. J. Quax, J. M. van Dijl, and P. G. Braun. 2002. Selective contribution of the twin-arginine translocation pathway to protein secretion in Bacillus subtilis. J. Biol. Chem. 277:44068-44078. [DOI] [PubMed] [Google Scholar]
- 27.Kaniga, K., I. Delor, and G. R. Cornelis. 1991. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109:137-141. [DOI] [PubMed] [Google Scholar]
- 28.Koebnik, R. 2005. TonB-dependent trans-envelope signalling: the exception or the rule? Trends Microbiol. 13:343-347. [DOI] [PubMed] [Google Scholar]
- 29.Labaer, J., Q. Qiu, A. Anumanthan, W. Mar, D. Zuo, T. V. Murthy, H. Taycher, A. Halleck, E. Hainsworth, S. Lory, and L. Brizuela. 2004. The Pseudomonas aeruginosa PA01 gene collection. Genome Res. 14:2190-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamont, I. L., and L. W. Martin. 2003. Identification and characterization of novel pyoverdine synthesis genes in Pseudomonas aeruginosa. Microbiology 149:833-842. [DOI] [PubMed] [Google Scholar]
- 31.Lehoux, D. E., F. Sanschagrin, and R. C. Levesque. 2000. Genomics of the 35-kb pvd locus and analysis of novel pvdIJK genes implicated in pyoverdine biosynthesis in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 190:141-146. [DOI] [PubMed] [Google Scholar]
- 32.Lewenza, S., J. L. Gardy, F. S. Brinkman, and R. E. Hancock. 2005. Genome-wide identification of Pseudomonas aeruginosa exported proteins using a consensus computational strategy combined with a laboratory-based PhoA fusion screen. Genome Res. 15:321-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Locher, K. P., B. Rees, R. Koebnik, A. Mitschler, L. Moulinier, J. P. Rosenbusch, and D. Moras. 1998. Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes. Cell 95:771-778. [DOI] [PubMed] [Google Scholar]
- 34.Lugtenberg, B., J. Meijers, R. Peters, P. van der Hoek, and L. van Alphen. 1975. Electrophoretic resolution of the “major outer membrane protein” of Escherichia coli K12 into four bands. FEBS Lett. 58:254-258. [DOI] [PubMed] [Google Scholar]
- 35.McMorran, B. J., H. M. Kumara, K. Sullivan, and I. L. Lamont. 2001. Involvement of a transformylase enzyme in siderophore synthesis in Pseudomonas aeruginosa. Microbiology 147:1517-1524. [DOI] [PubMed] [Google Scholar]
- 36.McMorran, B. J., M. E. Merriman, I. T. Rombel, and I. L. Lamont. 1996. Characterisation of the pvdE gene which is required for pyoverdine synthesis in Pseudomonas aeruginosa. Gene 176:55-59. [DOI] [PubMed] [Google Scholar]
- 37.Merriman, T. R., M. E. Merriman, and I. L. Lamont. 1995. Nucleotide sequence of pvdD, a pyoverdine biosynthetic gene from Pseudomonas aeruginosa: PvdD has similarity to peptide synthetases. J. Bacteriol. 177:252-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyer, J. M., A. Stintzi, D. De Vos, P. Cornelis, R. Tappe, K. Taraz, and H. Budzikiewicz. 1997. Use of siderophores to type pseudomonads: the three Pseudomonas aeruginosa pyoverdine systems. Microbiology 143:35-43. [DOI] [PubMed] [Google Scholar]
- 39.Michel, G., S. Bleves, G. Ball, A. Lazdunski, and A. Filloux. 1998. Mutual stabilization of the XcpZ and XcpY components of the secretory apparatus in Pseudomonas aeruginosa. Microbiology 144:3379-3386. [DOI] [PubMed] [Google Scholar]
- 40.Miyazaki, H., H. Kato, T. Nakazawa, and M. Tsuda. 1995. A positive regulatory gene, pvdS, for expression of pyoverdin biosynthetic genes in Pseudomonas aeruginosa PAO. Mol. Gen. Genet. 248:17-24. [DOI] [PubMed] [Google Scholar]
- 41.Mossialos, D., U. Ochsner, C. Baysse, P. Chablain, J. P. Pirnay, N. Koedam, H. Budzikiewicz, D. U. Fernandez, M. Schafer, J. Ravel, and P. Cornelis. 2002. Identification of new, conserved, non-ribosomal peptide synthetases from fluorescent pseudomonads involved in the biosynthesis of the siderophore pyoverdine. Mol. Microbiol. 45:1673-1685. [DOI] [PubMed] [Google Scholar]
- 42.Ochsner, U., A. Snyder, A. I. Vasil, and M. L. Vasil. 2002. Effects of the twin-arginine translocase on secretion of virulence factors, stress response, and pathogenesis. Proc. Natl. Acad. Sci. USA 99:8312-8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ochsner, U. A., P. J. Wilderman, A. I. Vasil, and M. L. Vasil. 2002. GeneChip expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol. Microbiol. 45:1277-1287. [DOI] [PubMed] [Google Scholar]
- 44.Palmer, T., F. Sargent, and B. C. Berks. 2005. Export of complex cofactor-containing proteins by the bacterial Tat pathway. Trends Microbiol. 13:175-180. [DOI] [PubMed] [Google Scholar]
- 45.Postle, K., and R. J. Kadner. 2003. Touch and go: tying TonB to transport. Mol. Microbiol. 49:869-882. [DOI] [PubMed] [Google Scholar]
- 46.Ravel, J., and P. Cornelis. 2003. Genomics of pyoverdine-mediated iron uptake in pseudomonads. Trends Microbiol. 11:195-200. [DOI] [PubMed] [Google Scholar]
- 47.Robinson, C., and A. Bolhuis. 2001. Protein targeting by the twin-arginine translocation pathway. Nat. Rev. Mol. Cell Biol. 2:350-356. [DOI] [PubMed] [Google Scholar]
- 48.Schalk, I. J., M. A. Abdallah, and F. Pattus. 2002. Recycling of pyoverdin on the FpvA receptor after rerric pyoverdin uptake and dissociation in Pseudomonas aeruginosa. Biochemistry 41:1663-1671. [DOI] [PubMed] [Google Scholar]
- 49.Schalk, I. J., C. Hennard, C. Dugave, K. Poole, M. A. Abdallah, and F. Pattus. 2001. Iron-free pyoverdin binds to its outer membrane receptor FpvA in Pseudomonas aeruginosa: a new mechanism for membrane iron transport. Mol. Microbiol. 39:351-360. [DOI] [PubMed] [Google Scholar]
- 50.Schalk, I. J., P. Kyslik, D. Prome, A. van Dorsselaer, K. Poole, M. A. Abdallah, and F. Pattus. 1999. Copurification of the FpvA ferric pyoverdin receptor of Pseudomonas aeruginosa with its iron-free ligand: implications for siderophore-mediated iron transport. Biochemistry 38:9357-9365. [DOI] [PubMed] [Google Scholar]
- 51.Schons, V., R. A. Atkinson, C. Dugave, R. Graff, G. L. Mislin, L. Rochet, C. Hennard, B. Kieffer, M. A. Abdallah, and I. J. Schalk. 2005. The structure-activity relationship of ferric pyoverdine bound to its outer membrane transporter: implications for the mechanism of iron uptake. Biochemistry 44:14069-14079. [DOI] [PubMed] [Google Scholar]
- 52.Vandenende, C. S., M. Vlasschaert, and S. Y. Seah. 2004. Functional characterization of an aminotransferase required for pyoverdine siderophore biosynthesis in Pseudomonas aeruginosa PAO1. J. Bacteriol. 186:5596-5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Visca, P., A. Ciervo, and N. Orsi. 1994. Cloning and nucleotide sequence of the pvdA gene encoding the pyoverdin biosynthetic enzyme l-ornithine N5-oxygenase in Pseudomonas aeruginosa. J. Bacteriol. 176:1128-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Voulhoux, R., G. Ball, B. Ize, M. L. Vasil, A. Lazdunski, L. F. Wu, and A. Filloux. 2001. Involvement of the twin-arginine translocation system in protein secretion via the type II pathway. EMBO J. 20:6735-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Voulhoux, R., and J. Tommassen. 2004. Omp85, an evolutionarily conserved bacterial protein involved in outer-membrane-protein assembly. Res. Microbiol. 155:129-135. [DOI] [PubMed] [Google Scholar]
- 56.Wiener, M. C. 2005. TonB-dependent outer membrane transport: going for Baroque? Curr. Opin. Struct. Biol. 15:394-400. [DOI] [PubMed] [Google Scholar]
- 57.Yue, W. W., S. Grizot, and S. K. Buchanan. 2003. Structural evidence for iron-free citrate and ferric citrate binding to the TonB-dependent outer membrane transporter FecA. J. Mol. Biol. 332:353-368. [DOI] [PubMed] [Google Scholar]