Abstract
The recently discovered RraA protein acts as an inhibitor of the essential endoribonuclease RNase E, and we demonstrated that ectopic expression of RraA affects the abundance of more than 700 transcripts in Escherichia coli (K. Lee, X. Zhan, J. Gao, J. Qiu, Y. Feng, R. Meganathan, S. N. Cohen, and G. Georgiou, Cell 114:623-634, 2003). We show that rraA is expressed from its own promoter, PrraA, located in the menA-rraA intergenic region. Primer extension and lacZ fusion analysis revealed that transcription from PrraA is elevated upon entry into stationary phase in a σs-dependent manner. In addition, the stability of the rraA transcript is dependent on RNase E activity, suggesting the involvement of a feedback circuit in the regulation of the RraA level in E. coli.
RNase E is an essential protein that plays a crucial role in global mRNA metabolism as well as in the maturation of functional RNAs such as rRNAs, tRNAs, tmRNA, and small regulatory RNAs (3, 9, 18, 19, 21, 28). To date, RNase E homologs have been found in more than 50 eubacteria, archaebacteria, and plants (16). The cellular level and activity of RNase E are subjected to multiple environmental controls. At one level, RNase E synthesis is autoregulated by modulating the half-life of its own mRNA (12, 26). In addition, recent studies have revealed that 5′-monophosphorylated RNA serves as an allosteric activator of the endonuclease activity (13). Furthermore, the degradation of target RNAs by RNase E is found to be affected globally by endoribonuclease-binding proteins that control the decay and abundance of individual bacterial mRNAs in trans (8, 17).
RraA (regulator of ribonuclease activity A), is an evolutionarily conserved 17.4-kDa protein with close homologs (>40% amino acid identity) in bacteria, archaea, proteobacteria, and plants. RraA binds to RNase E with an equilibrium dissociation constant (KD) in the low-micromolar range and serves as a trans-acting modulator of the endonuclease activity of the enzyme (17). High-affinity binding requires the C-terminal half region of RNase E, which acts as a scaffold for the assembly of a large multiprotein complex called the degradosome (17, 36). RraA appears to interact only with the enzyme and not with RNA substrates (17). Gene chip analysis revealed that the action of RraA results in a dramatic change in the global abundance of mRNAs in Escherichia coli, affecting over 15% of all cellular transcripts. Importantly, the gene expression profile that is obtained upon overexpression of RraA is distinct from that obtained upon depletion of RNase E or through the action of RraB, a second trans-acting RNase E inhibitor of E. coli (8).
The rraA gene is located downstream of menA, which encodes a 1,4-dihydroxy-2-naphthoic acid octaprenyltransferase that catalyzes the prenylation of the redox mediator menaquinone (32). Transcription of menA appears to occur from a σ70-dependent promoter. Earlier, Meganathan proposed that rraA (formerly designated menG) is transcribed from the menA promoter in a dicistronic mRNA (22). In this study we demonstrate that rraA is transcribed predominantly from its own promoter (PrraA) located in the intergenic region between the menA and rraA genes. Transcription from PrraA is σs dependent and is induced upon entry into stationary phase. Furthermore, we show that the synthesis of RraA is regulated at the posttranscriptional level by RNase E, suggesting the existence of a feedback regulatory circuit whereby induction of rraA transcription occurs in a σs-dependent manner and results in inhibition of RNase E activity, in turn decreasing the degradation rate of the rraA transcript.
MATERIALS AND METHODS
Bacterial strains, plasmids, and phage vectors.
The strains, plasmid, and phage vectors used in this study are listed in Table 1. The rpoS::kan mutation from strain ZK1000 was introduced into various strains by P1 transduction as described by Miller (24). The transduction of the rpoS disruption was confirmed by streaking single kanamycin-resistant transductants onto Luria-Bertani broth (LB)-kanamycin agar plates and testing the ability of catalase to hydrolyze hydrogen peroxide.
TABLE 1.
Strains, plasmids, and phage vectors
| Strain, plasmid, or phage vector | Description | Reference or source |
|---|---|---|
| Strains | ||
| JCB570 | MC1000; phoR zih12::Tn10 | 1 |
| ZK126 | W3110; ΔlacU16 tna-2 | 4 |
| ZK1000 | ZK126; rpoS::kan | 4 |
| EC-O | thi-1 relA1 Δ(pro-lac)X113[del = DE5] supE44/F42-114(FTs) lac | Lab collection |
| DY330 | W3110; ΔlacU169 gal 490 λcI857 Δ(cro-bioA) | 39 |
| MZ001 | EC-O; λ p0-lacZ | This study |
| MZ002 | EC-O; λ (nt −1076 to −1)-lacZ | This study |
| MZ003 | EC-O; λ (nt −92 to −1)-lacZ | This study |
| MZ004 | EC-O; λ (nt −1076 to −93)-lacZ | This study |
| MZ013 | MZ003; rpoS::kan | This study |
| MZ014 | MZ004; rpoS::kan | This study |
| MZ100 | JCB570; rpoS::kan | This study |
| CH1827 | MC1061; zce-726::Tn10 | 27 |
| CH1828 | CH1827; rne-1 | 25 |
| Plasmids | ||
| pSP417 | pBR ori Ampr | 29 |
| pMZ002 | pSP417 (nt −1076 to −1)-lacZ | This study |
| pMZ003 | pSP417 (nt −92 to −1)-lacZ | This study |
| pMZ004 | pSP417 (nt −1076 to −93)-lacZ | This study |
| Phage vectors | ||
| λRS45 | bla′-lacZscimm21 ind+ | 33 |
| λRS74 | placUV5-lacZ+imm21 ind+ | 33 |
| λMZ1 | Same as λRS74 but containing the p0-lacZ fusion | This study |
| λMZ2 | Same as λRS45 but containing the (nt −1076 to −1)-lacZ fusion | This study |
| λMZ3 | Same as λRS45 but containing the (nt −92 to −1)-lacZ fusion | This study |
| λMZ4 | Same as λRS45 but containing the (nt −1076 to −93)-lacZ fusion | This study |
Growth conditions.
LB and M9 minimal medium (23) containing thiamine (50 μg/ml), 0.4% (wt/vol) glucose, and 0.2% (wt/vol) casein were supplemented with antibiotics, as required (50 μg/ml ampicillin, 25 μg/ml kanamycin, or 15 μg/ml tetracycline). Unless otherwise stated, cells were grown in LB medium under aeration at 37°C, and the growth was monitored by measuring the absorbance at 600 nm.
RNA methods.
For reverse transcriptase-PCR (RT-PCR) analysis, total RNA was isolated with the RNeasy kit (QIAGEN, Valencia, CA) and treated with RNase-free DNase (Ambion, Austin, TX). Fifty ng of total RNA was subjected to RT-PCR analysis using the One Step RNA PCR kit (TaKaRa, New York, NY).
Northern blot assays were performed using total RNA isolated from E. coli JCB570 grown as described above. Samples were collected at 1-hour intervals throughout the exponential and stationary phases. Five μg of total RNA per lane was loaded onto a denaturing gel containing formaldehyde and then transferred to a positively charged nylon membrane (Hybond N+; Amersham, United Kingdom). The AlkPhos direct nucleic acid labeling and detection system (Amersham, United Kingdom) was used for probe synthesis. Hybridization, washing of the membranes, and detection of signals were carried out according the manufacturer's protocol.
For primer extension, a 5′-32P-labeled oligonucleotide (5′-GCGGTTCCACGACGTTAACATCTTCTTGA-3′ or 5′-CCGTCCGCCAAAGTTGGAGAACAGC-3′) was used as a primer for the RT reaction with 5 μg of total RNA (SuperScript III RNase H− reverse transcriptase; Invitrogen, Carlsbad, CA). The primer extension products were separated on 6% polyacrylamide-7 M urea gels. The dideoxy-DNA sequence ladder from the same primer was prepared using the fmol DNA Cycle Sequencing System (Promega, Madison, WI).
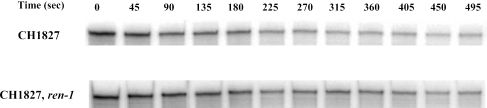
For RNase protection assays, E. coli CH1827 (MC1061; zce-726::Tn10) or CH1828 (CH1827; rne-1) was grown in LB at 30°C to an A600 of 0.4 and then half of the culture was transferred to 43.5°C and incubated for an additional 20 min. After the addition of rifampin (500 μg/ml), aliquots were withdrawn every 45 s, immediately chilled, and stored in liquid nitrogen. Total RNA was prepared from the frozen samples by using the RNeasy kit (QIAGEN, Valencia, CA), and RNase protection assays were performed using the RPA III kit (Ambion, Austin, TX) with the in vitro transcript from the complementary strand of the rraA gene (nucleotides [nt] +13 to +318 of rraA) as the probe. The band intensity was quantified using ImageQuant software.
Construction of β-galactosidase fusions.
We used PCR amplification to generate DNA fragments containing different regions upstream of rraA, i.e., nt −1076 to −1 of the rraA upstream region, which includes the PmenA, menA coding sequence, and menA-rraA intergenic region; nt −1076 to −93, which includes the PmenA and menA coding sequence; and nt −92 to −1 of the menA-rraA intergenic region, and cloned each fragment upstream of the lacZ gene in a multicopy transcriptional fusion vector, pSP417 (29). So we generated plasmids pMZ002 ([nt −1076 to −1]-lacZ), pMZ003 ([nt −92 to −1]-lacZ), and pMZ004 ([nt −1076 to −93]-lacZ); pMZ001 was the negative-control vector. The lacZ fusions in pMZ002, pMZ003, and pMZ004 were transferred onto the chromosome using the transducing lambda phage system (33). The fusions were transferred into λRS45, whereas the negative-control fusion in pMZ001 was transferred into λRS74 via a double recombination event. Plaques containing the recombinant lambda phages were isolated based on their blue plaque phenotype. The recombinant lambda phages were used to lysogenize EC-O [Δ(pro-lacZ)] and generated the following strains: MZ001 (EC-O; λ p0-lacZ), with a chromosomal promoterless lacZ gene; MZ002 (EC-O; λ [nt −1076 to −1]-lacZ), harboring nt −1076 to −1 of the rraA upstream region fused to lacZ; MZ003 (EC-O; λ [nt −92 to −1]-lacZ), harboring nt −92 to −1 of the menA-rraA intergenic region fused to lacZ; and MZ004 (EC-O; λ [nt −1076 to −93]-lacZ), harboring nt −1076 to −93 of the rraA upstream region fused to lacZ. All lysogens were tested for monolysogenization by PCR (30).
β-Galactosidase assays.
Cultures grown with aeration at 37°C, in rich or M9 medium overnight, were subcultured into the fresh medium. Samples were collected at exponential and stationary phases. The samples were collected at 4°C and resuspended in an appropriate volume of ice-cold Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM M KCl, 1 mM MgSO4; pH 7.0) (23) to give an A600 in the range from 0.6 to 0.9. β-Galactosidase activities were determined from at least three independent experiments, as previously described (24).
Western immunoblotting.
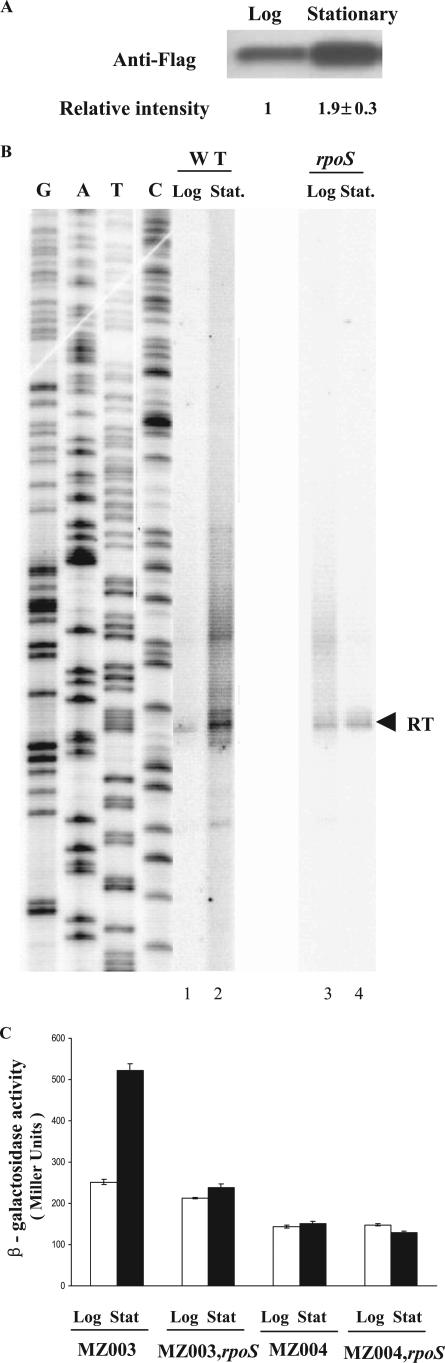
The cells were harvested by centrifugation, resuspended in phosphate-buffered saline, and lysed by passage through a French press (2,000 lb/in2). The lysate was centrifuged for 15 min at 4°C to remove cell debris, and the protein concentration in the supernatant was determined by the Bradford assay (Bio-Rad Laboratories, Hercules, CA). One μg of total protein was separated by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis. Western blot assays were performed by standard methods using monoclonal anti-FLAG-M2-peroxidase (horseradish peroxidase) antibody conjugate (Sigma, St. Louis, MO). Signal intensities were measured, quantified by molecular analysis software (Quantity One; Bio-Rad), and displayed below the blot image.
RESULTS AND DISCUSSION
Identification of the PrraA promoter.
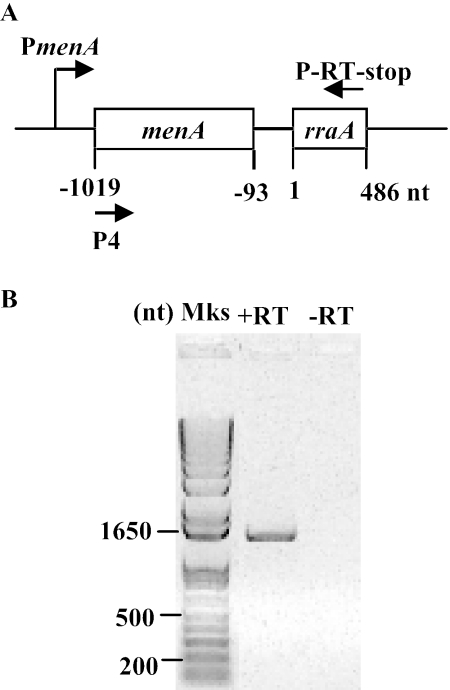
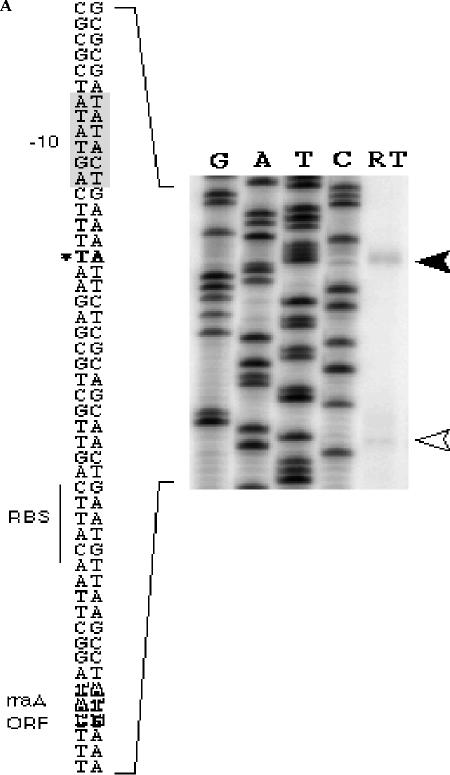
Meganathan had proposed that rraA is transcribed as a dicistronic mRNA from the PmenA promoter (22). The transcription initiation site for this putative dicistronic transcript was identified at 57 bp upstream of the menA translation start site (34). Consistent with this hypothesis, a menA-rraA transcript of the expected size was detected by RT-PCR analysis (Fig. 1), suggesting that rraA is transcribed together with the preceding menA as a dicistronic mRNA. However, Northern blot analysis with two different rraA-specific probes revealed that the predominant RNA in samples in either exponential or stationary phase corresponds to a shorter species, ca. 550 bases in length (data not shown). Primer extension of RNA isolated from E. coli JCB570 using the 5′-32P-labeled oligonucleotide probe 32P-GCGGTTCCACGACGTTAACATCTTCTTGA revealed an rraA transcript that is initiated at an A residue 28 nt upstream from the ATG codon (Fig. 2A). In addition to this major transcript, the data in Fig. 2A also showed a faint band at the position of the initiation codon of rraA (indicated by an open arrowhead). However, this band was not detected in a primer extension assay using an alternative primer (32P-CCGTCCGCCAAAGTTGGAGAACAGC) that anneals to a different location within the rraA RNA (located 23 bp upstream from the first primer), suggesting that the minor band likely corresponds to a premature stop product of the reverse transcriptase.
FIG. 1.
RT-PCR analysis. A. The annealing positions of the reverse primer (P-RT-stop) and forward primer (P4) used in the RT-PCR analysis of the menA-rraA region. B. RT-PCR analysis of rraA transcript using the primers P4 and P-RT-stop. −RT, the negative control containing the same amounts of RNA, primers, and Taq polymerase but no reverse transcriptase. Lane Mks, molecular size markers.
FIG. 2.
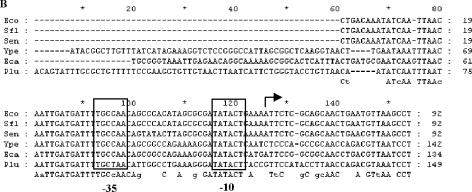
Identification of the PrraA promoter. A. Primer extension. Five μg total RNA isolated from E. coli JCB570 cells grown to log phase (A600 = 0.7) was reverse transcribed using the primer (5′-GCGGTTCCACGACGTTAACATCTTCTTGA-3′) complementary to nucleotides +36 to +64 of rraA. Filled arrowhead, the reverse transcription product; open arrowhead, the premature stop product of the reverse transcriptase. RBS, ribosomal binding site; ORF, open reading frame. B. Multiple sequence alignment of the menA-rraA intergenic sequence: Abbreviations: Eco, Escherichia coli K-12; Sfl, Shigella flexner I; Sen, Salmonella enterica; Ype, Yersinia pestis; Eca, Erwinia carotovora; Plu, Photorhabdus luminescens. The transcript start site is indicated by a solid arrow. −35 and −10 sites are indicated by open boxes.
DNA sequence analysis of the menA-rraA intergenic region suggested the existence of a putative promoter (PrraA) (Fig. 2B). rraA and menA are separated by a 92-bp intergenic region that shows a significant degree of conservation among closely related bacteria such as Shigella flexneri (92/92 bp, 100% identity in the intergenic region) and Salmonella enterica (86/92 bp, 93% identity in the intergenic region). A lower degree of sequence identity was observed for phylogenetically more distant bacteria, Yersinia pestis, Erwinia carotovora, and Photorhabdus luminescens. Analysis using GENETYX-MAC 11.2.5 identified regions that match the σ70 consensus −35 and −10 sequences, centered at 32 and 8 nucleotides, respectively, upstream of the transcription initiation site (Fig. 2B). Multiple sequence alignment, using CLUSTAL X 1.8, among the bacterial species which have close homologs (>80%) of E. coli K-12 rraA revealed that the sequence of the PrraA promoter is conserved among gammaproteobacteria. As shown in Fig. 2B, the −35 and −10 regions are identical, except for Photorhabdus luminescens, whose −35 region differs from that of E. coli K-12 by 1 bp.
Genetic analysis of expression of PrraA using lacZ transcriptional fusions.
We used PCR amplification to generate DNA fragments containing different regions upstream of rraA extending up to, and including, the menA promoter, as shown in Fig. 3A, and cloned each fragment upstream of the lacZ gene in a multicopy transcriptional fusion vector, pSP417 (29). In this way, we generated plasmids pMZ002 ([nt −1076 to −1]-lacZ), pMZ003 ([nt −92 to −1]-lacZ), and pMZ004 ([nt −1076 to −93]-lacZ); pMZ001 was the negative-control vector.
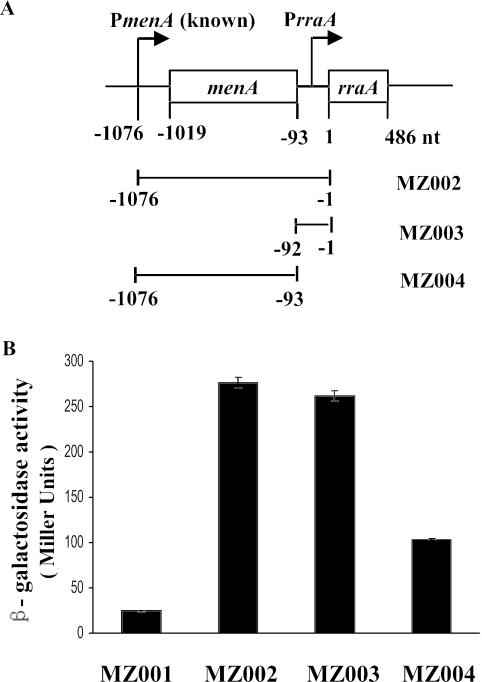
FIG. 3.
lacZ transcriptional fusions. A. Schematic of the transcriptional rraA-lacZ fusions used in this study. B. β-Galactosidase activities in MZ001, MZ002, MZ003, and MZ004 cells. MZ001 (EC-O; λ p0-lacZ) has a chromosomal promoterless lacZ gene; MZ002 (EC-O; λ [nt −1076 to −1]-lacZ) has nt −1076 to −1 of the rraA upstream region fused to lacZ; MZ003 (EC-O; λ [nt −92 to −1]-lacZ) has nt −92 to −1 of the menA-rraA intergenic region fused to lacZ; MZ004 (EC-O; λ [nt −1076 to −93]-lacZ) has nt −1076 to −93 of the rraA upstream region fused to lacZ. Cells were grown in LB under aeration at 37°C and harvested in log phase (A600 = 0.5 to 0.8). Samples were normalized by optical density, and enzymatic activities were measured in Miller units. The data presented are averages of at least three independent determinations, and error bars correspond to the standard deviations.
To rule out the possibility that differences in the β-galactosidase activity expressed from the above transcriptional fusions might be partially due to plasmid copy number effects, we then made single chromosomal copy isolates of each construct using the transducing lambda phage system (33). The lacZ fusions in pMZ001, pMZ002, pMZ003, and pMZ004 were transferred into either λRS74 or λRS45 via a double recombination event to generate λMZ1, λMZ2, λMZ3, and λMZ4, respectively. Subsequently, E. coli EC-O was lysogenized with the recombinant phages to generate strains carrying a single copy of each promoter-lacZ fusion. As shown in Fig. 3B, strain MZ003 (EC-O; λ [nt −92 to −1]-lacZ), with only the menA-rraA intergenic region fused to lacZ, showed levels of β-galactosidase activity identical to those observed in MZ002 (EC-O; λ [nt −1076 to −1]-lacZ), which contains a fusion to the rraA upstream region, including the PmenA and menA coding sequence. In contrast, MZ004 (EC-O; λ [nt −1076 to −93]-lacZ), containing a fusion to nt −1076 to −93 of the rraA upstream region that lacked the menA-rraA intergenic region, showed 67% lower activity.
rraA expression in stationary phase.
Earlier microarray studies had shown that the rraA mRNA level is increased upon entrance to stationary phase (https://asap.ahabs.wisc.edu/annotation/php/ASAP1.htm). The abundance of the RraA protein in exponential- and stationary-phase cells was examined by Western blotting using strain DY330-rraASPA (5, 39), in which sequential peptide affinity (SPA) tags are fused to the C terminus of the rraA open reading frame within the chromosome. We found that the level of RraA protein increased by about twofold in stationary phase (Fig. 4A). Many genes that are upregulated in stationary phase are part of the rpoS regulon. The E. coli rpoS regulon comprises more than 100 genes that are transcribed by the sigma factor σs (or σ38, RpoS) and are upregulated in stationary phase or under certain stress conditions such as hyperosmolarity, low pH, and oxidative and temperature stresses and function largely in stress response, carbon metabolism, and cell envelope biogenesis (10). In order to determine if rraA transcription is dependent on the growth phase, we examined the rraA promoter activity in exponential- or stationary-phase cells grown in LB medium at 37°C. Primer extension analysis of RNA from stationary-phase cells revealed a marked increase in the rraA transcript in stationary-phase cells (Fig. 4B). In addition to the rraA transcript, several minor bands were also evident in samples from stationary-phase cells. These correspond to premature termination or misannealing products since neither could they be detected consistently nor were they detected in a primer extension assay using a different primer. No increase in the level of rraA transcript in stationary-phase cultures was observed in the rpoS mutant strain MZ100 (JCB570; rpoS::kan). Collectively, these results indicate that transcription from the PrraA promoter increases in stationary phase and this effect is dependent on the transcription factor σs.
FIG. 4.
rraA expression in stationary phase. A. Western immunoblot analysis of RraA level in exponential and stationary phases. Strain DY330-rraASPA, bearing a sequential peptide affinity (SPA) tag at the C terminus of the rraA open reading frame, was cultured in LB medium at 32°C and harvested during exponential growth (A600 = 0.4) and stationary phase (A600 = 3.5). RraA is expressed at an endogenous level from its chromosomal promoter and detected by the highly specific anti-FLAG antibody. Blots were replicated from three independent protein preparations. Replicate measurements were made on the same membrane to determine the reproducibility of the analysis. The intensity of the signal was observed to be linear by using a dilution series of total protein. The data are the means and errors of three separate blots. B. Primer extension analysis of the transcription from PrraA in growth phases. Total RNA was prepared from isogenic strains JCB570 (WT, wild type) and MZ100 (JCB570; rpoS::kan) grown to log phase (A600 = 0.6) and stationary phase (A600 = 2.5). Equivalent amounts of RNA (5 μg) were reverse transcribed using the primer (5′-GCGGTTCCACGACGTTAACATCTTCTTGA-3′) complementary to nucleotides +36 to +64 of rraA. The position of the reverse transcription product is shown by the arrowhead. C. Effect of σs level on the transcriptional activity of the various PrraA-lacZ fusions in growth phases. Strains MZ003 (EC-O; λ [nt −92 to −1]-lacZ), MZ004 (EC-O; λ [nt −1076 to −93]-lacZ), MZ013 (MZ003; rpoS::kan), and MZ014 (MZ004; rpoS::kan) were grown in log (A600 = 0.6) and stationary (A600 = 2.5) phases in M9 medium supplemented with 0.2% casein with aeration, at 37°C. Samples were normalized by optical density, and enzymatic activities were measured in Miller units. The data presented are averages of at least three independent determinations, and error bars correspond to the standard deviations.
The effect of rpoS on the transcriptional activity of the PrraA promoter was also evaluated using lacZ fusions transcribed from the PrraA promoter. Specifically, β-galactosidase activity was determined in strains MZ003, MZ004, MZ013 (MZ003; rpoS::kan), and MZ014 (MZ004; rpoS::kan) grown to either exponential or stationary phases. As shown in Fig. 4C, in strain MZ003, the β-galactosidase activity increased more than twofold in stationary-phase cultures (A600 ≈ 2.5) compared with log-phase cells (A600 ≈ 0.6). In the isogenic rpoS mutant MZ013, the β-galactosidase activity was comparable to the level observed in parental strain MZ003 grown in exponential phase, but the induction of lacZ expression in stationary phase was abolished. As a control, the β-galactosidase activity of MZ004 and MZ014 cells where lacZ is transcribed from the upstream menA promoter was neither growth phase dependent nor affected by rpoS. These results clearly suggest that σs is responsible for the increased transcription from PrraA upon entrance into stationary phase. Recently, using microrarray analysis Hengge-Aronis's group identified over 400 genes which are positively controlled by σs in E. coli. While rraA was not recognized as a σs-dependent gene in this study, we note that 33 out of the 87 genes which had been experimentally shown to be σs controlled in earlier studies also failed to be detected in the microarray experiments (37).
Many σs promoters contain an extended −10 region, KCTAYRCTTAA (nucleotides −14 to −4; K stands for T or G, Y stands for T or C, and R stands for A or G) (37). Particularly, C at −13 has been shown to interact directly with lysine 173 in the 2.5 region of σs (2, 7, 31, 37). Nevertheless, despite its importance, C at −13 is not absolutely essential for more efficient promoter utilization by σs than by σ70, as shown with mutant variants of the aidB promoter (11, 15). The extended −10 motif is found only on the genes which are σs dependent under all three growth and stress conditions (i.e., transition into stationary-phase growth, hyperosmotic shift, and low pH). They account for less than 30% of the total genes which are positively controlled by rpoS and were named the “core set” of σs-dependent genes (37). Genes that display σs dependence only under certain conditions do not contain this extended −10 sequence (37). In PrraA, the extended −10 region (GATATACT) contains an A rather than a C at −13, suggesting that additional factors may be involved in sigma factor selectivity. For example, the well-characterized σs-controlled gene csiD, which is mainly induced by carbon starvation, contains an A at −13 (−10 sequence, GATATTTT) (20, 38) and requires cyclic AMP (cAMP)-cAMP receptor protein for sigma factor selectivity.
We could not identify sequences resembling either the distal or the proximal UP element, which has been proposed to enhance σs selectivity (35). The absence of an apparent UP element is not surprising, as such sequences are absent in more than 50% of the known σs-dependent promoters (35). It should be noted that extensive earlier studies have revealed that σs promoter selectivity is affected by a number of factors including DNA supercoiling, topology, and the action of additional protein regulators (6, 14, 31, 37).
The decay of the rraA transcript is dependent on RNase E.
Since the function of RraA is to modulate the endonuclease activity of RNase E, it was of interest to examine whether RNase E plays a role in the decay of the rraA transcript. Because RNase E is an essential protein, the stability of the rraA mRNA was examined in E. coli CH1828, which contains the temperature-sensitive rne-1 allele. Cells were grown at 30°C in LB medium under aerobic conditions to mid-log phase, followed by a temperature upshift to 43.5°C for 20 min. Rifampin was added to inhibit transcription, samples were withdrawn at different times, and the level of rraA was determined by RNase protection assays (Fig. 5). In the parental strain CH1827, the half-life of rraA, determined by fitting the linear regression model, was virtually identical at 30°C and at 43.5°C (t1/2 = 2.4 ± 0.3 and 2.9 ± 0.2 min, respectively). However, incubation of the rne-1 mutant strain at the nonpermissive temperature resulted in a significant (twofold) stabilization of the rraA transcript, whose t1/2 increased to 6.1 ± 1.1 min. Since RraA acts as an inhibitor of RNase E activity, the finding that the processing of the rraA transcript is RNase E dependent suggests the existence of a feedback mechanism: induction of rraA synthesis, possibly by environmental triggers that stimulate σs-dependent transcription, would be expected to result in higher accumulation of RraA protein, in turn inhibiting RNase E activity and thus leading to stabilization of the rraA transcript. The net result would be a further increase in the level of RraA protein and greater inhibition of RNase E activity.
FIG. 5.
Degradation profile of the rraA mRNA in wild-type and rne-1 strains. RNase protection assays and total RNA isolation were done as described in Materials and Methods on strains CH1827 (MC1061; zce-726::Tn10) and CH1828 (CH1827; rne-1). Time points in seconds were sampled after rifampin addition. Equivalent amounts of RNA (5 μg) were used in RNA protection assays and loaded into each lane of a 6% polyacrylamide-7 M urea gel. The band intensity was quantified using ImageQuant software. Degradation rates were determined by fitting the linear regression model and represent the averages of two independent determinations.
The microarray data of Lee et al. (17) revealed that either overexpression or deletion of rraA affect the steady-state abundance of a number of transcripts from σs-dependent genes. It is noteworthy that the rpoS transcript and its regulators rseABC have been shown to be stabilized by either RraA overexpression or RNase E depletion (17). Accordingly, a possible biological role for the σs-dependent PrraA activity in early stationary phase may be to provide a means for the protection of σs-dependent transcripts from the decay catalyzed by RNase E.
Acknowledgments
We are grateful to C. Jain and I. J. Molineux for many useful suggestions. We thank R. W. Simons, R. Kolter, and A. Emili for their gifts of strains, plasmids, and phage vectors. We are especially grateful to C. Jain for reading the manuscript.
This work was supported by NIH grant GM55090-05.
REFERENCES
- 1.Bardwell, J. C., K. McGovern, and J. Beckwith. 1991. Identification of a protein required for disulfide bond formation in vivo. Cell 67:581-589. [DOI] [PubMed] [Google Scholar]
- 2.Becker, G., and R. Hengge-Aronis. 2001. What makes an Escherichia coli promoter σs dependent? Role of the −13/−14 nucleotide promoter positions and region 2.5 of σs. Mol. Microbiol. 39:1153-1165. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein, J. A., A. B. Khodursky, P. H. Lin, S. Lin-Chao, and S. N. Cohen. 2002. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc. Natl. Acad. Sci. USA 99:9697-9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bohannon, D. E., N. Connell, J. Keener, A. Tormo, M. Espinosa-Urgel, M. M. Zambrano, and K. Roberto. 1991. Stationary-phase-inducible “gearbox” promoters: differential effects of katF mutations and role of σ70. J. Bacteriol. 173:4482-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butland, G., J. M. Peregrin-Alvarez, J. Li, W. Yang, X. Yang, V. Canadien, A. Starostine, D. Richards, B. Beattie, N. Krogan, M. Davey, J. Parkinson, J. Greenblatt, and A. Emili. 2005. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 433:531-537. [DOI] [PubMed] [Google Scholar]
- 6.Colland, F., M. Barth, R. Hengge-Aronis, and A. Kolb. 2000. Sigma factor selectivity of Escherichia coli RNA polymerase: role for CRP, IHF and Lrp transcription factors. EMBO J. 19:3028-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Espinosa-Urgel, M., C. Chamizo, and A. Tormo. 1996. A consensus structure for σs-dependent promoters. Mol. Microbiol. 21:657-659. [DOI] [PubMed] [Google Scholar]
- 8.Gao, J., K. Lee, M. Zhao, J. Qiu, X. Zhan, A. Saxena, C. J. Moore, S. N. Cohen, and G. Georgiou. Submitted for publication.
- 9.Ghora, B. K., and D. Apirion. 1978. Structural analysis and in vitro processing to p5 rRNA of a 9S RNA molecule isolated from an rne mutant of E. coli. Cell 15:1055-1066. [DOI] [PubMed] [Google Scholar]
- 10.Hengge-Aronis, R. 2002. Signal transduction and regulatory mechanisms involved in control of the σs (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66:373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hengge-Aronis, R. 2002. Stationary phase gene regulation: what makes an Escherichia coli promoter σs-selective? Curr. Opin. Microbiol. 5:591-595. [DOI] [PubMed] [Google Scholar]
- 12.Jain, C., and J. G. Belasco. 1995. RNase E autoregulates its synthesis by controlling the degradation rate of its own mRNA in Escherichia coli: unusual sensitivity of the rne transcript to RNase E activity. Genes Dev. 9:84-96. [DOI] [PubMed] [Google Scholar]
- 13.Jiang, X., and J. G. Belasco. 2004. Catalytic activation of multimeric RNase E and RNase G by 5′-monophosphorylated RNA. Proc. Natl. Acad. Sci. USA 101:9211-9216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kusano, S., Q. Ding, N. Fujita, and A. Ishihama. 1996. Promoter selectivity of Escherichia coli RNA polymerase Eσ70 and Eσ38 holoenzymes. Effect of DNA supercoiling. J. Biol. Chem. 271:1998-2004. [DOI] [PubMed] [Google Scholar]
- 15.Lacour, S., A. Kolb, A. B. Zehnder, and P. Landini. 2002. Mechanism of specific recognition of the aidB promoter by σs-RNA polymerase. Biochem. Biophys. Res. Commun. 292:922-930. [DOI] [PubMed] [Google Scholar]
- 16.Lee, K., and S. N. Cohen. 2003. A Streptomyces coelicolor functional orthologue of Escherichia coli RNase E shows shuffling of catalytic and PNPase-binding domains. Mol. Microbiol. 48:349-360. [DOI] [PubMed] [Google Scholar]
- 17.Lee, K., X. Zhan, J. Gao, J. Qiu, Y. Feng, R. Meganathan, S. N. Cohen, and G. Georgiou. 2003. RraA. a protein inhibitor of RNase E activity that globally modulates RNA abundance in E. coli. Cell 114:623-634. [PubMed] [Google Scholar]
- 18.Li, Z., and M. P. Deutscher. 2002. RNase E plays an essential role in the maturation of Escherichia coli tRNA precursors. RNA 8:97-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, Z., S. Pandit, and M. P. Deutscher. 1999. RNase G (CafA protein) and RNase E are both required for the 5′ maturation of 16S ribosomal RNA. EMBO J. 18:2878-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marschall, C., V. Labrousse, M. Kreimer, D. Weichart, A. Kolb, and R. Hengge-Aronis. 1998. Molecular analysis of the regulation of csiD, a carbon starvation-inducible gene in Escherichia coli that is exclusively dependent on σS and requires activation by cAMP-CRP. J. Mol. Biol. 276:339-353. [DOI] [PubMed] [Google Scholar]
- 21.Masse, E., N. Majdalani, and S. Gottesman. 2003. Regulatory roles for small RNAs in bacteria. Curr. Opin. Microbiol. 6:120-124. [DOI] [PubMed] [Google Scholar]
- 22.Meganathan, R. 1996. Biosynthesis of the isoprenoid quinones menaquinone (vitamin K2) and ubiquinone (coenzyme Q), p. 642-656. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. American Society for Microbiology, Washington, D.C. [Google Scholar]
- 23.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Mudd, E. A., A. J. Carpousis, and H. M. Krisch. 1990. Escherichia coli RNase E has a role in the decay of bacteriophage T4 mRNA. Genes Dev. 4:873-881. [DOI] [PubMed] [Google Scholar]
- 26.Mudd, E. A., and C. F. Higgins. 1993. Escherichia coli endoribonuclease RNase E: autoregulation of expression and site-specific cleavage of mRNA. Mol. Microbiol. 9:557-568. [DOI] [PubMed] [Google Scholar]
- 27.Ono, M., and M. Kuwano. 1979. A conditional lethal mutation in an Escherichia coli strain with a longer chemical lifetime of messenger RNA. J. Mol. Biol. 129:343-357. [DOI] [PubMed] [Google Scholar]
- 28.Ow, M. C., and S. R. Kushner. 2002. Initiation of tRNA maturation by RNase E is essential for cell viability in E. coli. Genes Dev. 16:1102-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Podkovyrov, S. M., and T. J. Larson. 1995. A new vector-host system for construction of lacZ transcriptional fusions where only low-level gene expression is desirable. Gene 156:151-152. [DOI] [PubMed] [Google Scholar]
- 30.Powell, B. S., M. P. Rivas, D. L. Court, Y. Nakamura, and C. L. Turnbough. 1994. Rapid confirmation of single copy lambda prophage integration by PCR. Nucleic Acids Res. 22:5765-5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Repoila, F., and S. Gottesman. 2001. Signal transduction cascade for regulation of RpoS: temperature regulation of DsrA. J. Bacteriol. 183:4012-4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shineberg, B., and I. Young. 1976. Biosynthesis of bacterial menaquinones: the membrane-associated 1,4-dihydroxy-2-naphthoate octaprenyltransferase of Escherichia coli. Biochemistry 15:2754-2758. [DOI] [PubMed] [Google Scholar]
- 33.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 34.Suvarna, K., D. Stevenson, R. Meganathan, and M. E. Hudspeth. 1998. Menaquinone (vitamin K2) biosynthesis: localization and characterization of the menA gene from Escherichia coli. J. Bacteriol. 180:2782-2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Typas, A., and R. Hengge-Aronis. 2005. Differential ability of σSand σ70 of Escherichia coli to utilize promoters containing half or full UP-element sites. Mol. Microbiol. 55:250-260. [DOI] [PubMed] [Google Scholar]
- 36.Vanzo, N. F., Y. S. Li, B. Py, E. Blum, C. F. Higgins, L. C. Raynal, H. M. Krisch, and A. J. Carpousis. 1998. Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev. 12:2770-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weber, H., T. Polen, J. Heuveling, V. F. Wendisch, and R. Hengge-Aronis. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 187:1591-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weichart, D., R. Lange, N. Henneberg, and R. Hengge-Aronis. 1993. Identification and characterization of stationary phase-inducible genes in Escherichia coli. Mol. Microbiol. 10:407-420. [PubMed] [Google Scholar]
- 39.Zeghouf, M., J. Li, G. Butland, A. Borkowska, V. Canadien, D. Richards, B. Beattie, A. Emili, and J. F. Greenblatt. 2004. Sequential peptide affinity (SPA) system for the identification of mammalian and bacterial protein complexes. J. Proteome Res. 3:463-468. [DOI] [PubMed] [Google Scholar]