Abstract
The gene hyuP from Microbacterium liquefaciens AJ 3912 with an added His6 tag was cloned into the expression plasmid pTTQ18 in an Escherichia coli host strain. The transformed E. coli showed transport of radioisotope-labeled 5-substituted hydantoins with apparent Km values in the micromolar range. This activity exhibited a pH optimum of 6.6 and was inhibited by dinitrophenol, indicating the requirement of energy for the transport system. 5-Indolyl methyl hydantoin and 5-benzyl hydantoin were the preferred substrates, with selectivity for a hydrophobic substituent in position 5 of hydantoin and for the l isomer over the d isomer. Hydantoins with less hydrophobic substituents, cytosine, thiamine, uracil, allantoin, adenine, and guanine, were not effective ligands. The His-tagged hydantoin transport protein was located in the inner membrane fraction, from which it was solubilized and purified and its identity was authenticated.
The stereoselective hydrolyses of dl-5-monosubstituted hydantoin compounds have been widely studied for the production of optically pure d- or l-amino acids (1, 13), which are important intermediates for the production of various drugs and pharmaceuticals (2, 26). Within certain species of bacteria, the hydantoin substrate compounds are first hydrolyzed by hydantoinase enzymes to form N-carbamoyl-α-amino acids and then converted to corresponding free d- or l-amino acids catalyzed by N-carbamoyl-α-amino acid amido hydrolase (N-carbamoylase) enzymes (Fig. 1). In this process, the stereo-selective hydantoinase and/or N-carbamoylase play essential roles in determining the optical purity of the amino acid products. In addition to these hydrolyzing enzymes, there exist hydantoin racemase (HRase) enzymes that are responsible for the racemization of the substituent in position 5 of the hydantoin compounds (8, 9, 10, 23, 31, 34 and Fig. 1). HRase reacts with only one of the hydantoin substrate enantiomers resulting from the stereoselective hydrolase enzymes, so the theoretical yield of the optically pure amino acid from the original 5-substituted dl-hydantoin compounds can be aimed at 100% by judiciously combining catalysis by hydantoinase, N-carbamoylase, and HRase, a commercially important objective.
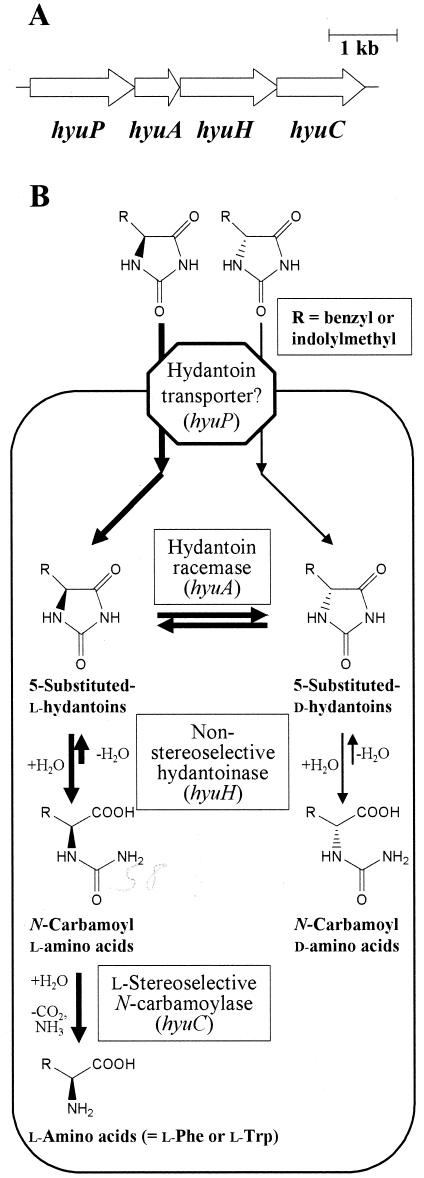
FIG. 1.
Organization of the hyu genes (A) and predicted salvage and dissimilation pathway of 5-substituted benzyl and indolylmethyl hydantoins (B) in M. liquefaciens AJ 3912. (A) The genes and their transcriptional orientation are symbolized by arrows. Encoded proteins are the hydantoin transporter (hyuP), hydantoin racemase (hyuA), hydantoinase (hyuH), and N-carbamoylase (hyuC). (B) The predicted functions of proteins encoded by hyu genes and their roles in the dissimilation pathway of 5-substituted hydantoins in M. liquefaciens AJ 3912 are illustrated. The metabolic pathways may accept a wide range of 5-substituents, but the specificities of the transporter for benzyl and indolyl groups are illustrated, leading to the synthesis of l-phenylalanine and l-tryptophan.
In spite of the intensive studies for commercialization, the physiological roles of these hydantoin-related enzymes still remain obscure. The hyu genes encoding the enzymes often form gene clusters, indicating the existence of coherent roles for the combination of hydantoinase, N-carbamoylase, and HRase (6, 24, 29, 30, 33). Among these genes for the three hydantoin metabolic enzymes, we found genes encoding putative transporters, for example in the hyu gene clusters from Pseudomonas sp. NS671 (29, 30), Arthrobacter aurescens DSM 3747 (33), and Microbacterium liquefaciens AJ 3912 (24) (Fig. 1). Another gene encoding a putative transporter was reported to be located in the region upstream of a gene encoding N-carbamoylase in Pseudomonas putida RU-KM3, although its sequence has not been published yet (11).
The putative transporters from Pseudomonas sp., A. aurescens, and M. liquefaciens were similar in their deduced amino acid sequences. Furthermore, they were also homologous to an allantoin transporter from Saccharomyces cerevisiae (22, 37). Allantoin is an intermediate of the purine metabolic pathway and has the structure of 5-ureido-hydantoin. So, the putative transporters encoded by genes located in hyu gene clusters were expected to be responsible for the uptake of hydantoin compounds, though none has been characterized so far.
In order to establish, for the first time, the function of these putative transporters encoded in hyu gene clusters, we have cloned and expressed one of them, “Mhp” from M. liquefaciens AJ3912, in Escherichia coli. This protein is found to transport primarily 5-indolyl-methyl-hydantoin and 5-benzyl-hydantoin. The His6-tagged protein MhpH6 was purified to a homogeneity sufficient for crystallization trials.
MATERIALS AND METHODS
Chemicals.
5-Substituted hydantoins were synthesized from corresponding amino acids (25). For the synthesis of 3H-5-benzyl-l-hydantoin (3H-l-BH) and 3H-5-indolymethyl-l-hydantoin (3H-l-IMH), the corresponding 3H-labeled phenylalanine (ring-2,4-3H, 1.4 TBq/mmol; ICN Biomedicals, Irvine, CA) or tryptophan (ring-5-3H, 1.0 TBq/mmol; Amersham Biosciences, Piscataway, NJ) was used. The specific radioactivities of the synthesized compounds were 236 MBq/mmol (3H-l-BH) and 241 MBq/mmol (3H-l-IMH), respectively. The concentrations of l-BH and l-IMH were determined by using the following absorbance coefficients: for l-BH, ɛ257 was 184 M−1, and for l-IMH, ε280 was 5,440−1cm−1.
Bacterial strains and plasmids.
M. liquefaciens AJ 3912 was used as gene source. An expression vector for the production of MhpH6 protein was constructed by using the vector pTTQ18 (5, 16, 21). The gene hyuP encoding the Mhp protein was amplified from the chromosomal DNA of the strain by PCR using the following primers: hyuP upstream primer with EcoRI site, 5′-CGTCAATGAATTCGACACCCATCGAAGAGGCT-3′, and hyuP downstream primer with PstI site, 5′-TCCTTCTCCTGCAGGGTACTGCTTCTCGGTGGG-3′.
The amplified fragment was then digested with EcoRI and PstI and inserted between the corresponding sites of plasmid JLC1, an expression vector originally including a glucose transporter gene gluP fused in frame with an RGSH6-tag on the C terminus (3), which had been excised by EcoRI-PstI digestion. Then the E. coli BLR host strain (Novagen) was transformed with the vector thus constructed (pSHP11). This recombinant strain (E. coli/pSHP11) was used as the MhpH6 protein producer.
Culture media and conditions.
CM2G medium, containing 10 g/liter d-glucose, 5 g/liter yeast extracts, 5 g/liter peptone, and 10 g/liter NaCl (pH 7.0), was used for the cultivation of M. liquefaciens to obtain a source of chromosomal DNA. The strain was cultured in this medium at 30°C for 18 h. For the cultivation of E. coli/pSHP11, liquid mineral (LM) medium containing 20 mM glycerol, 10 g/liter peptone, 5 g/liter yeast extract, and 5 g/liter NaCl or M9 medium containing 20 mM glycerol, 0.2% (wt/vol) Casamino Acids, 6 g/liter Na2HPO4, 3 g/liter KH2HPO4, 1 g/liter NH4Cl, 0.5 g/liter NaCl, 2 mM MgSO4, and 0.2 mM CaCl2 was used. Carbenicillin was added into the media at a concentration of 0.2 mg/ml to maintain selection for the β-lactamase-encoding plasmids when required. Refreshed E. coli/pSHP11 was inoculated into 2 ml of LM medium and cultured at 37°C for 16 h. A 1-ml portion of the culture broth was inoculated into 50 ml of fresh M9-Casamino Acids-carbenicillin medium and cultured at 37°C. When the absorbance at 600 nm reached approximately 0.4 cm−1, a final concentration of 0.2 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added into the culture medium. Then the culture was continued at 27°C for an extra 12 h or at 30°C for an extra 4 to 6 h.
Uptake assays.
The uptakes of radioisotope-labeled hydantoins into E. coli cells were assayed by a method modified from those of West (32) and Henderson and Macpherson (4) as follows. The harvested cells of induced or uninduced E. coli/pSHP11 were washed three times with assay buffer containing 150 mM KCl and 5 mM MES [2-(N-morpholino)ethanesulfonic acid (pH 6.6)] and then adjusted to a cell density of A660 of approximately 2.0. Samples of the cell suspensions containing 20 mM glycerol were aerated at 25°C for 3 min, and the reaction was initiated by the addition of 3H-l-BH (236 MBq/mmol) or 3H-l-IMH (241 MBq/mmol) at a final concentration of 25 μM unless indicated otherwise. An aliquot of the reaction mixture was taken and applied at precisely timed intervals onto a 0.45-μm-pore-size filter, which was then immediately washed with excess assay buffer. The radioactivity incorporated in the cells was measured using a scintillation counter and converted into the amount (nanomolar) of substrates taken into 1 mg of dry cells using a formula of A660 = 1.47 mg−1ml of dry cells. All these assays are averages of triplicate measurements used to calculate standard deviations. The amounts of substrates taken into cells were measured using either induced or uninduced cells, and the differences between the uptakes by each type of cell were regarded as the effects caused by MhpH6. This was done to correct for any unspecific binding of the substrates to cell walls.
To determine the sensitivity of uptake of E. coli/pSHP11 to the substrate concentrations, the concentrations of 3H-l-IMH or 3H-l-BH differed between 0.1 and 250 μM. The amount of uptake at 3 min after the initiation of each reaction was measured.
For the determination of the effect of NaCl, the assay buffer was changed to include 140 mM KCl and 10 mM NaCl instead of 150 mM KCl. To determine the effect of dinitrophenol (DNP), 2 mM DNP at final concentration was added to the reaction mixture 3 min prior to the initiation of the reaction. To determine the optimal pH for the uptake by MhpH6, 10 mM potassium acetate (pH 4.0), 5 mM MES (pH 4.9, 6.1, 6.6, 7.1, and 7.9), 10 mM Tris-HCl (pH 8.0), and 10 mM glycine-NaOH (pH 10.0) were used instead of 5 mM MES (pH 6.6) in each assay buffer. In these assays, 3H-l-IMH was used as the substrate. The initial rate of uptake at each pH was calculated by the amount of uptake in the first 15 s of the reaction.
Substrate and stereoselectivity of MhpH6.
The substrate and stereoselectivity of MhpH6 were determined by competitive inhibition of the uptake of 3H-l-BH (25 μM) caused by the addition of unlabeled potential alternative substrates (250 μM) using the standard conditions above. The amount of uptake of 3H-l-BH at 3 min after the initiation of each reaction was measured in triplicate and represented as a percentage of the uptake without any other compounds added except 3H-l-BH. When using d- and l-BH and l-IMH, additional dose-response measurements were also carried out with concentrations of cold substrate of between 0.25 to 250 μM, corresponding to a ratio of 0.01 to 10 in relation to the concentration of 3H-l-BH.
Preparation of membranes.
Membranes were prepared by the water lysis method or by explosive decompression using a French press (28). In each case, the cells harvested by centrifugation were washed first with 0.2 M Tris-HCl buffer (pH 8.0) and then used for the preparations. Inner and outer membrane fractions were further prepared from French press membranes by sucrose density gradient centrifugation (28).
Solubilization and purification of MhpH6.
The inner membrane fraction of E. coli/pSHP11 adjusted to approximately 5 mg protein per milliliter was incubated in 20 mM Tris-HCl buffer (pH 8.0), including 20 mM imidazole, 20% (vol/vol) glycerol, 0.3 M NaCl, and 1% (wt/vol) n-dodecyl-β-d-maltoside (DDM) on ice, for 1 h. The supernatant obtained after centrifugation at 160,000 × g for 30 min was used as the soluble fraction; the precipitate was suspended in the same volume of H2O as was the supernatant and used as the insoluble fraction. The supernatant obtained above was incubated with Ni-nitrilotriacetic acid (NTA) agarose (QIAGEN, Hilden, Germany) preequilibrated with buffer W, containing 20 mM Tris-HCl (pH 8.0), 20 mM imidazole, 10% (vol/vol) glycerol, and 0.05% (wt/vol) DDM, at 4°C for 3 h. The resin was separated from unbound protein solution by centrifugation and then washed with 10 times its volume of buffer W. The washed resin was then packed in a small column and the absorbed proteins were eluted by the addition of 200 mM imidazole (pH 8.0), 20% (vol/vol) glycerol, and 0.05% (wt/vol) DDM. The eluted fractions were examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the pure fractions of MhpH6 were used for further experiments.
Analytical methods.
Prediction of transmembrane helices in Mhp protein was carried out using the program TMHMM version 2.0 (Technical University of Denmark). Protein concentration was measured by the method of Schaffner and Weissman (19) using bovine serum albumin as a standard. SDS-PAGE was carried out using 13% (wt/vol) acrylamide by the Laemmli method (7) with molecular weight markers (SDS-7; Sigma Aldrich, St. Louis, MO). Proteins were visualized by staining with Coomassie brilliant blue R. Proteins with His6-tags were visualized also by Western blotting with anti-RGS His6 antibody (QIAGEN) as described in the manufacturer's manual using a His6 protein ladder (QIAGEN) for molecular weight markers. For analysis of the N-terminal amino acid sequence of MHPH6, the purified protein was blotted from the SDS-PAGE gel onto a polyvinylidene difluoride membrane and sequenced by the Edman degradation method.
RESULTS
A structural model of the Mhp protein.
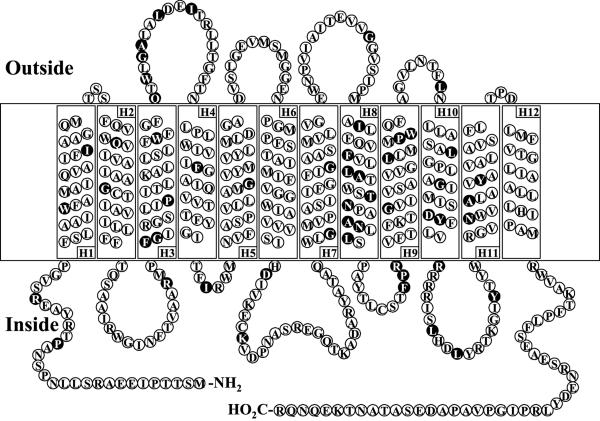
Twelve transmembrane helices were strongly predicted from the hydropathic profile of the amino acid sequence of Mhp using the program TMHMM as illustrated in Fig. 2. The model shown is consistent with the positive-inside rule of von Heijne and Gavel (27), which is particularly evident in the loop between helix H10 and helix H11. The residues that are identical among putative hydantoin transport proteins from Arthrobacter aurescens DSM 3747 and Pseudomonas sp. NS761 and the allantoin transporter from Saccharomyces cerevisiae are indicated in the figure. Particular concentrations of these conserved residues are observed in helix H8 and the loop between the helices H3 and H4.
FIG. 2.
The predicted structure of MHP. The structure model of MHP is illustrated on the basis of the prediction of transmembrane helices by the TMHMM program. The predicted 12 helices are numbered from the N terminus to the C terminus and shown as H1 to H12, respectively. The identical residues among MHP, putative hydantoin permease proteins from Arthrobacter aurescens DSM 3747 (GenBank accession no. AAG02128), from Pseudomonas sp. NS761 (GenBank accession number E42594), and allantoin permease from Saccharomyces cerevisiae (GenBank accession no. CAA78826) are indicated in reverse type.
Cloning and heterologous expression of transport activity associated with the hyuP gene.
The hyuP gene from M. liquefaciens was ligated into the EcoRI/PstI-cut vector pJLC1. The resulting plasmid, designated pSHP11 and containing hyuP downstream of the IPTG-inducible tac promoter, was transformed into the E. coli BLR host strain, which was grown in the presence or absence of IPTG. The washed cells of the recombinant strain (E. coli/pSHP11) were examined for the uptake of radiolabeled l-BH or l-IMH (Fig. 3).
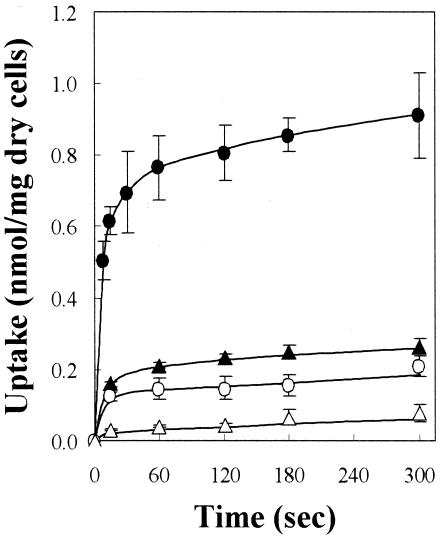
FIG. 3.
Time course of uptakes of 5-substituted hydantoins by E. coli/pSHP11. 3H-l-IMH (circles) and 3H-l-BH (triangles) were used as the substrates. The washed cells of E. coli/pSHP11 obtained by growth on M9 Casamino Acids medium with (closed symbols) or without (open symbols) induction by IPTG were used for the uptake assay carried out in standard conditions (5 mM MES buffer [pH 6.6], 150 mM KCl, and 25 μM substrate at 25°C). Error bars indicate standard deviations.
When using either l-BH or l-IMH as the substrate, higher uptake was observed in the cells of the IPTG-induced culture than was observed in those of the uninduced culture (Fig. 3 and Table 1). Using induced cells of E. coli BLR/pTTQ18 without any inserts, the uptake of substrates was less than that observed in uninduced cells of E. coliBLR/pSHP11 (data not shown), demonstrating that the host strain did not significantly accumulate these hydantoins. The extents of uptake kept increasing even after 300 s, but the rates of uptake for both of the substrates progressively decreased as the reactions proceeded. Judging from both the initial uptake rates and the extents of uptake in 300 s, l-IMH was a better substrate for MhpH6 than was l-BH.
TABLE 1.
Effects of NaCl, DNP, and pH on the uptake of l-IMH
| pH | Additive | Initial rate of uptake (nmol/min/mg dry cells)a
|
||
|---|---|---|---|---|
| Inducedb | Uninducedb | Differenceb | ||
| 6.6 | None | 2.48 | 0.50 | 1.98 |
| 6.6 | 10 mM NaClc | 2.40 | 0.35 | 2.05 |
| 6.6 | 2 mM DNPd | 1.15 | 0.34 | 0.81 |
| 4.0 | None | 0.34 | 0.23 | 0.11 |
| 4.9 | None | 1.57 | 0.17 | 1.40 |
| 6.1 | None | 2.29 | 0.43 | 1.87 |
| 7.1 | None | 2.19 | 0.37 | 1.82 |
| 7.9 | None | 2.02 | 0.53 | 1.49 |
| 9.0 | None | 0.93 | 0.06 | 0.86 |
| 10.0 | None | 0.23 | 0.03 | 0.20 |
The uptake rate of cells was measured at standard conditions.
Cells cultivated with IPTG (induced) and without IPTG (uninduced) were used and the difference (difference) in uptake rates of both cells was calculated by the subtraction of the rate of uninduced from that of induced.
The assay buffer including 140 mM KCl and 10 mM NaCl was used instead of 150 mM KCl.
A total of 2 mM DNP of final concentration was added in reaction mixture 3 min prior to the initiation of the reaction.
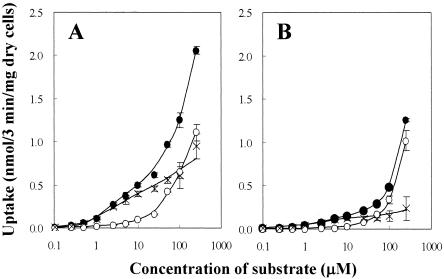
Next, the sensitivity of uptake activity to the concentration of substrate was tested (Fig. 4). Hydantoins are relatively hydrophobic molecules, and it is to be expected that they can partition into cell membranes and enter cells to an extent without the benefit of a transport protein, at least at higher concentrations. Accordingly, in these assays over a wide range of substrate concentrations (0.1 to 250 μM), cells obtained from both induced and uninduced cultures were examined. There were substantial differences in the amount of uptake between induced and uninduced cells. The uptake of l-IMH by MhpH6 was significant at concentrations below 1 μM, uptake by uninduced cells was observed at only concentrations above 10 μM l-IMH. To extract the uptake caused by the production of MhpH6, the differences between uptakes of both of the cells were also plotted in Fig. 4A. The concentration of l-IMH for a 50% extent of accumulation in induced cells was about 3 μM, whereas that for uninduced cells was >1 mM. Similar results were also observed using l-BH as the substrate (Fig. 4B).
FIG. 4.
The sensitivity of uptake to the concentration of substrates. The uptakes at various substrate concentrations were observed by using the induced (•) and uninduced cells (○) of E. coli/pSHP11. The differences of uptake between both the cultures were also plotted (×). The substrate used was either l-IMH (A) or l-BH (B). The assays were carried out at standard conditions with various concentrations of substrates, and the amounts of uptake for 3 min were measured. Error bars indicate standard deviations.
Energization of hydantoin uptake.
To investigate energization of the observed uptakes, their sensitivities to NaCl, the uncoupling agent DNP, and pH were examined. The uptake of l-IMH was not accelerated by the addition of NaCl (Table 1), indicating that Mhp is not a solute:Na+ symporter. The uptake was inhibited by the addition of DNP (Table 1), indicating the requirement for an electrochemical proton gradient for this system. This experiment does not exclude the participation of ATP in energization, but this participation is unlikely since the protein does not contain nucleotide binding sequence motifs, nor is there an ATP binding cassette protein encoded by the operon (20). The inhibition of uptake by DNP was also observed for the uptake of allantoin (5-ureido-hydantoin) by a protein homologous to Mhp in S. cerevisiae (22).
Consequently, the sensitivity of the uptake to reaction pH was determined. The uptake activity was profoundly affected by reaction pH and showed an optimum between pH 6 and 7 (Table 1). This is typical of a proton-linked energization mechanism (4, 12, 14).
Specificity.
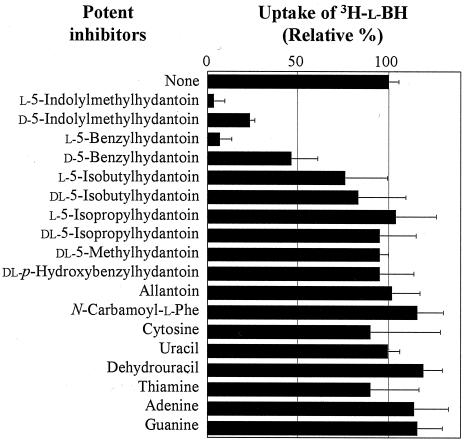
Substrate and/or inhibitor specificity of MhpH6 was examined by investigating competition to the uptake of 3H-l-BH caused by unlabeled potential substrates (Fig. 5). The uptake of 3H-l-BH was clearly inhibited by the addition of unlabeled d- or l-BH and d- or l-IMH, indicating that these 5-substituted hydantoin compounds are all good substrates/inhibitors for MhpH6. In addition, a slight inhibition was observed by the addition of l- or dl-5-isobutyl-hydantoin, indicating that this compound also might be a ligand of MhpH6. Other compounds tested (cytosine, guanine, adenine, dihydrouracil, uracil, thiamine, allantoin, N-carbamoyl-l-phenylalanine, and various hydantoins) (Fig. 5) seemed not to be substrates or inhibitors of MhpH6.
FIG. 5.
Substrate specificity of MhpH6. The substrate specificity of MHPH6 was estimated by the competitive inhibitions to the uptake of 3H-l-BH caused by the potential substrates. The assays were carried out under standard conditions with 25 μM 3H-l-BH and 250 μM potent cold substrates. The amount of uptake of 3H-l-BH at 3 min after the initiation of each reaction was measured and represented as a percentage of the uptake without any substrates added other than 3H-l-BH. Error bars indicate standard deviations.
In addition to the specificity described above, differences in the extent of inhibition were observed related to chirality, since the l forms of both BH and IMH were more effective than the d forms (Fig. 5). In order to quantitate these stereoselectivities, full dose-response analyses were performed between 0.25 to 250 μM, corresponding to d-/l- ratios of 0.01 to 10.0 (data not shown). This analysis indicated an approximately 5-times higher preference for the l isomer to the d isomer for both BH and IMH.
Amplified expression and identification of the MhpH6 protein.
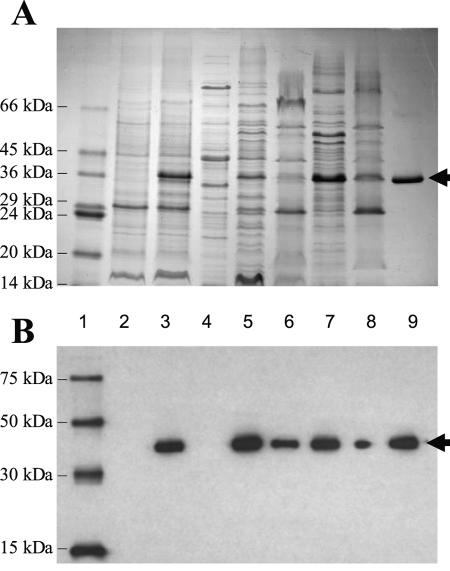
MhpH6, the Mhp protein containing a His6 tag at its C terminus, was heterologously expressed by the construct pSHP11 in an E. coli host (see above). The level of expression was substantial, so the produced protein was putatively identified as an extra protein by staining with Coomassie blue in membranes from induced, but not uninduced, cells (Fig. 6A, lanes 2 and 3). The protein was not observed in the soluble fraction (Fig. 6A, lane 4). After further separation of inner and outer membrane fractions from the total membrane fraction, the localization of MhpH6 was mainly observed in the inner membrane fraction (Fig. 6A, lanes 5 and 6). The apparent molecular mass of the produced protein was estimated to be approximately 36 kDa by SDS-PAGE, although its predicted molecular mass from the sequence is 54,580 Da. It is common for membrane transport proteins to migrate at anomalous molecular mass values lower than the values predicted from the amino acid sequences (17, 28). The identification of the overexpressed protein as MhpH6 was authenticated by the Western blot to the RGSH6 epitope (Fig. 6B), in which the appearance of a positive reaction correlated with the molecular mass and appearance/disappearance of the putative MhpH6 in the Coomassie blue-stained gel (Fig. 6A).
FIG. 6.
Localization, solubilization, and purification of MhpH6 heterologously expressed in E. coli. The expression of MhpH6 in E. coli was analyzed by SDS-PAGE (A, stained with Coomassie brilliant blue R) and Western blotting (B, detected with anti-RGS His antibody). Lane 1, marker proteins indicated the positions by thin lines (with molecular masses). The protein fractions were prepared from E. coli/pSHP11 cells obtained from induced (lane 3 to 6) or uninduced culture (lane 2) using LM medium. Lanes 2 and 3, total membrane fraction; lane 4, soluble fraction; lane 5, inner membrane fraction; lane 6, outer membrane fraction. The proteins in the inner membrane fraction of induced culture were solubilized by DDM, and then soluble (lane 7) and insoluble (lane 8) fractions were obtained. MhpH6 was then purified to electrophoretical homogeneity by using Ni-NTA resin (lane 9). Thick arrows indicate the position of MhpH6.
Solubilization and purification of MhpH6.
The solubilization of MhpH6 was efficiently promoted by treatment with 1% (wt/vol) DDM (Fig. 6A and B, lanes 7 and 8). Finally, the purification of MhpH6 to electrophoretic homogeneity was achieved (see Materials and Methods) by using Ni-NTA resin (Fig. 6A, lane 9). Through all the purification steps, the molecular mass of MhpH6 was observed at 36 kDa (Fig. 6A and B, lanes 5, 7, 8, and 9), and no other proteins were observed in the Western blot analysis (Fig. 6B).
The N-terminal amino acid sequence of the purified MhpH6 was determined for 10 residues as MNSTPIEEAR, corresponding exactly to what was expected from the nucleotide sequence in the constructed plasmid. Coupled with the integrity of the RGSH6 C-terminal end of the protein shown by the Western blot this verifies that the purified polypeptide chain of MhpH6 is intact.
DISCUSSION
In this paper, the Mhp protein encoded by the gene hyuP in the hyu gene cluster of M. liquefaciens AJ3912 (Fig. 1) was expressed in E. coli and shown to be a transporter that is functionally responsible for the uptake of 5-substituted hydantoin compounds. This outcome illustrates the power of comparative genomics to infer functions of genes, in this case, from a combination of both the prediction that an encoded protein is a transporter by its clustering by long-range homology in the NCS-1 family and also the occurrence of its gene in an operon encoding putatively related metabolic enzymes. The preferred substrates were found to be l-5-indolylmethyl-hydantoin and l-5-benzyl-hydantoin, consistent with the chiral specificity for the l form of both the associated hydantoinase and N-carbamoylase in M. liquefaciens AJ3912 (24, 36). Thus, the transport protein is deduced to play a role in the metabolism of tryptophan, phenylalanine, and probably tyrosine. Consistently, nucleobases thiamine and allantoin were found not to be substrates or inhibitors, though they are substrates for other members of the NCS-1 subfamily 2.A.39 (15, 22) of major facilitator superfamily transport proteins dispersed among gram-positive and gram-negative bacteria and yeasts (15, 18), to which Mhp is deduced to belong from the similarities of their amino acid sequences. Transport of their substrates is thought to be energized by the transmembrane electrochemical gradient of protons (14, 17); their sensitivity to the uncoupling agent and pH dependence (Table 1) of hydantoin transport are also consistent with those for the Mhp protein.
The function of the putative transporter from P. putida has not been characterized but was predicted not to be involved in the transport of hydantoin since disruption of the gene did not affect hydantoinase activity in resting cells (11). The substrate used in that report was hydantoin without any 5-sustitututions, so the protein from P. putida may still be a hydantoin transporter with specificity for 5-aromatic-substituted hydantoins like Mhp (Fig. 5). Alternatively, the disruption of the transporter gene in P. putida might have been compensated by an additional hydantoin transport system in the cells, as observed in E. coli cells (Fig. 4).
The MhpH6 protein was produced heterologously in E. coli in a substantial amount (Fig. 6), extending the success of using plasmid pTTQ18 for the expression of various kinds of membrane transporters (16, 28). The produced MhpH6 was targeted to the inner membrane fraction, and the slight appearance in the outer membrane fraction might be caused by the contamination of outer membranes with inner membrane fractions. The protein was effectively solubilized from the membrane by treatment with DDM and then purified to apparent homogeneity by using Ni-NTA resin. The retention of the predicted N-terminal amino acid sequence and positive response to the anti-His-tag antibody shows the protein must be intact and comprise the 501 amino acid residues deduced from the corresponding nucleotide sequence, despite the molecular mass of about 36,000 Da observed by SDS-PAGE, which was smaller than that calculated from the sequence. This may be caused by retention of the tertiary structure of the protein even with SDS, as reported for other membrane transporters (17), resulting from the strong hydrophobicity of these proteins.
The inhibition by the uncoupling agent and the level of accumulation suggested that uptakes of 5-substituted-hydantoin compounds by Mhp were energized to achieve higher concentrations inside the cell, probably by the proton motive force (Fig. 3 and Table 1). The uptake rate dropped quickly, probably because the solubility of both the substrates used is low, roughly estimated as 5 mM (l-BH) and 2 mM (l-IMH) at 25°C, so that internal substrates might reach saturation inside the cell. The authentic producers of hydantoin transporters may avoid such a problem by the conversion of the substrate taken into the cells to more soluble compounds, i.e., N-carbamoyl α-amino acids and/or α-amino acids. Consistently, the affinity for external hydantoins was high, with an apparent Km value of Mhp for l-IMH estimated in the micromolar range (Fig. 4), comparable to 15 μM for the Km value of the allantoin transporter from S. cerevisiae (22). Another uptake system with very high Km value (in the order of millimolars) was observed in the E. coli host cells (Fig. 4), but its affinity was insufficient for the efficient uptake of IMH. Importantly, it was reported that the uptake of hydantoin with low solubility became rate limiting in the production of amino acids by a whole-cell biocatalyst of E. coli producing hydantoin racemase, hydantoinase, and N-carbamoylase (35), so the expression of Mhp in such an E. coli host is expected to accelerate amino acid production for commercial exploitation.
Acknowledgments
We thank N. G. Rutherford, J. O'Reilly (for overall technical support and discussions), R. B. Herbert, A. Rajakarier, S. G. Patching (for technical support and discussions of the synthesis of radiolabeled compounds), and J. Keen (for analysis of N-terminal amino acid sequence). We also thank K. Yokozeki and K. Watanabe in Ajinomoto Co., Inc. for their encouragement.
We thank Ajinomoto Co., Inc. and BBSRC for financial support of this research.
REFERENCES
- 1.Altenbuchner, J., M. Siemann-Herzberg, and C. Syldatk. 2001. Hydantoinases and related enzymes as biocatalysts for the synthesis of unnatural chiral amino acids. Curr. Opin. Biotechnol. 12:559-563. [DOI] [PubMed] [Google Scholar]
- 2.Bommarius, A. S., M. Schwarm, and K. Drauz. 1998. Biocatalysis to amino acid-based chiral pharmaceuticals—examples and perspectives. J. Mol. Catal. B Enzym. 5:1-11. [Google Scholar]
- 3.Clough, J. L. 2001. Structure-function relationships of the l-fucose-H+ symport protein (FucP) of Escherichia coli, and the homologous d-glucose transport protein (GluP) of Brucella abortus. Ph.D. thesis. University of Leeds, Leeds, United Kingdom.
- 4.Henderson, P. J. F., and A. J. S. Macpherson. 1986. Assay, genetics, proteins, and reconstitution of proton-linked galactose, arabinose, and xylose transport systems of Escherichia coli. Methods Enzymol. 125:387-429. [DOI] [PubMed] [Google Scholar]
- 5.Henderson, P. J. F., C. K. Hoyle, and A. Ward. 2000. Expression, purification and properties of multidrug efflux proteins. Biochem. Soc. Trans. 28:513-517. [PubMed] [Google Scholar]
- 6.Hils, M., P. Munch, J. Altenbuchner, C. Syldatk, and R. Mattes. 2001. Cloning and characterization of genes from Agrobacterium sp. IP I-671 involved in hydantoin degradation. Appl. Microbiol. Biotechnol. 57:680-688. [DOI] [PubMed] [Google Scholar]
- 7.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 8.Las Heras-Vazquez, F. J., S. Martinez-Rodriguez, L. Mingorance-Cazorla, J. M. Clemente-Jimenez, and F. Rodriguez-Vico. 2003. Overexpression and characterization of hydantoin racemase from Agrobacterium tumefaciens C58. Biochem. Biophys. Res. Commun. 303:541-547. [DOI] [PubMed] [Google Scholar]
- 9.Martinez-Rodriguez, S., F. J. Las Heras-Vazquez, J. M. Clemente-Jimenez, and F. Rodriguez-Vico. 2004. Biochemical characterization of a novel hydantoin racemase from Agrobacterium tumefaciens C58. Biochimie 86:77-81. [DOI] [PubMed] [Google Scholar]
- 10.Martinez-Rodriguez, S., F. J. Las Heras-Vazquez, L. Mingorance-Cazorla, J. M. Clemente-Jimenez, and F. Rodriguez-Vico. 2004. Molecular cloning, purification, and biochemical characterization of hydantoin racemase from the legume symbiont Sinorhizobium meliloti CECT 4114. Appl. Environ. Microbiol. 70:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matcher, G. F., S. G. Burton, and R. A. Dorrington. 2004. Mutational analysis of the hydantoin hydrolysis pathway in Pseudomonas putida RU-KM3S. Appl. Microbiol. Biotechnol. 65:391-400. [DOI] [PubMed] [Google Scholar]
- 12.Muiry, J. A. R., T. C. Gunn, T. P. McDonald, S. A. Bradley, C. G. Tate, and P. J. F. Henderson. 1993. Proton-linked l-rhamnose transport, and its comparison with l-fucose transport in Enterobacteriaceae. Biochem. J. 290:833-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogawa, J., and S. Shimizu. 1997. Diversity and versatility of microbial hydantoin-transforming enzymes. J. Mol. Catal. B Enzym. 2:163-176. [Google Scholar]
- 14.Ramos, S., and R. Kaback. 1977. The relationship between the electrochemical proton gradient and active transport in Escherichia coli membrane vesicles. Biochemistry 16:854-859. [DOI] [PubMed] [Google Scholar]
- 15.Ren, Q., K. H. Kang, and I. T. Paulsen. 2004. Transport DB: a relational database of cellular membrane transport systems. Nucleic Acids Res. 32:D284-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saidijam, M., G. Psakis, J. L. Clough, J. Meuller, S. Suzuki, C. J. Hoyle, S. L. Palmer, S. M. Morrison, M. K. Pos, R. C. Essenberg, M. C. J. Maiden, A. Abu-bakr, S. G. Baumberg, A. A. Neyfakh, J. K. Griffith, M. J. Stark, A. Ward, J. O'Reilly, N. G. Rutherford, M. K. Phillips-Jones, and P. J. F. Henderson. 2003. Collection and characterisation of bacterial membrane proteins. FEBS Lett. 555:170-175. [DOI] [PubMed] [Google Scholar]
- 17.Saidijam, M., K. E. Bettaney, G. Szakonyi, G. Psakis, K. Shibayama, S. Suzuki, J. L. Clough, V. Blessie, A. Abu-Bakr, S. Baumberg, J. Muller, C. K. Hoyle, S. L. Palmer, P. Butaye, K. Walravens, S. G. Patching, J. O'Reilly, N. G. Rutherford, R. M. Bill, D. I. Roper, M. K. Phillips-Jones, and P. J. Henderson. 2005. Active membrane transport and receptor proteins from bacteria. Biochem. Soc. Trans. 33:867-872. [DOI] [PubMed] [Google Scholar]
- 18.Saier, M. H., Jr., B. H. Eng, S. Fard, J. Garg, D. A. Haggerty, W. J. Hutchinson, D. L. Jack, E. C. Lai, H. J. Liu, D. P. Nusinew, A. M. Omar, S. S. Pao, I. T. Paulsen, J. A. Quan, M. Sliwinski, T.-T. Tseng, S. Wachi, and G. B. Young. 1999. Phylogenetic characterization of novel transport protein families revealed by genome analyses. Biochim. Biophys. Acta 1422:1-56. [DOI] [PubMed] [Google Scholar]
- 19.Schaffner, W., and C. Weissman. 1973. A rapid, sensitive, and specific method for the determination of protein in dilute solution. Anal. Biochem. 56:502-514. [DOI] [PubMed] [Google Scholar]
- 20.Schmitt, L., and R. Tampe. 2002. Structure and mechanism of ABC transporters. Curr. Opin. Struct. Biol. 12:754-760. [DOI] [PubMed] [Google Scholar]
- 21.Stark, M. J. R. 1987. Multicopy expression vectors carrying the lac repressor gene for regulated high-level expression of genes in Escherichia coli. Gene 51:255-267. [DOI] [PubMed] [Google Scholar]
- 22.Sumrada, R., and T. G. Cooper. 1977. Allantoin transport in Saccharomyces cerevisiae. J. Bacteriol. 131:839-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki, S., N. Onishi, and K. Yokozeki. 2005. Purification and characterization of hydantoin racemase from Microbacterium liquefaciens AJ 3912. Biosci. Biotechnol. Biochem. 69:530-536. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki, S., Y. Takenaka, N. Onishi, and K. Yokozeki. 2005. Molecular cloning and expression of hyu genes from Microbacterium liquefaciens AJ 3912, responsible for the conversion of 5-substituted hydantoins to α-amino acids, in Escherichia coli. Biosci. Biotechnol. Biochem. 69:1473-1482. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki, T., K. Igarashi, K. Hase, and K. Tuzimura. 1973. Optical rotatory dispersion and circular dichroism of amino acid hydantoins. Agric. Biol. Chem. 37:411-416. [Google Scholar]
- 26.Syldatk, C., A. Laufer, R. Muller, and H. Hoke. 1990. Production of optically pure d- and l-α-amino acids by bioconversion of d,l-5-monosustituted hydantoin derivatives. Adv. Biochem. Eng. Biotechnol. 41:29-75. [Google Scholar]
- 27.von Heijne, G., and Y. Gavel. 1988. Topogenic signals in integral membrane proteins. Eur. J. Biochem. 174:671-678. [DOI] [PubMed] [Google Scholar]
- 28.Ward, A., N. M. Sanderson, J. O'Reilly, N. G. Rutherford, B. Poolman, and P. J. F. Henderson. 2000. The amplified expression, identification, purification, assay and properties of histidine-tagged bacterial membrane transport proteins, p. 141-166. In S. A. Baldwin (ed.), Membrane transport—a practical approach. Blackwell's Press, Oxford, United Kingdom.
- 29.Watabe, K., T. Ishikawa, Y. Mukohara, and H. Nakamura. 1992. Cloning and sequencing of the genes involved in the conversion of 5-substituted hydantoins to the corresponding l-amino acids from the native plasmid of Pseudomonas sp. strain NS671. J. Bacteriol. 174:962-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watabe, K., T. Ishikawa, Y. Mukohara, and H. Nakamura. 1992. Identification and sequencing of a gene encoding a hydantoin racemase from the native plasmid of Pseudomonas sp. strain NS671. J. Bacteriol. 174:3461-3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watabe, K., T. Ishikawa, Y. Mukohara, and H. Nakamura. 1992. Purification and characterization of the hydantoin racemase of Pseudomonas sp. strain NS671 expressed in Escherichia coli. J. Bacteriol. 174:7989-7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.West, I. C. 1970. Lactose transport coupled to proton movements in Escherichia coli. Biochem. Biophys. Res. Commun. 41:655-661. [DOI] [PubMed] [Google Scholar]
- 33.Wiese, A., C. Syldatk, R. Mattes, and J. Altenbuchner. 2001. Organization of genes responsible for the stereospecific conversion of hydantoins to α-amino acids in Arthrobacter aurescens DSM 3747. Arch. Microbiol. 176:187-196. [DOI] [PubMed] [Google Scholar]
- 34.Wiese, A., M. Pietzsch, C. Syldatk, R. Mattes, and J. Altenbuchner. 2000. Hydantoin racemase from Arthrobacter aurescens DSM 3747: heterologous expression, purification and characterization. J. Biotechnol. 80:217-230. [DOI] [PubMed] [Google Scholar]
- 35.Wilms, B., A. Wiese, C. Syldatk, R. Mattes, and J. Altenbuchner. 2001. Development of an Escherichia coli whole cell biocatalyst for the production of l-amino acids. J. Biotechnol. 86:19-30. [DOI] [PubMed] [Google Scholar]
- 36.Yokozeki, K., Y. Hirose, and K. Kubota. 1987. Mechanism of asymmetric production of l-aromatic amino acids from the corresponding hydantoins by Flavobacterium sp. Agric. Biol. Chem. 51:737-746. [Google Scholar]
- 37.Yoo, H. S., T. S. Cunningham, and T. G. Cooper. 1992. The allantoin and uracil permease gene sequences of Saccharomyces cerevisiae are nearly identical. Yeast 8:997-1006. [DOI] [PubMed] [Google Scholar]