Abstract
Transcription of the Escherichia coli melAB operon is regulated by the MelR protein, an AraC family member whose activity is modulated by the binding of melibiose. In the absence of melibiose, MelR is unable to activate the melAB promoter but autoregulates its own expression by repressing the melR promoter. Melibiose triggers MelR-dependent activation of the melAB promoter and relieves MelR-dependent repression of the melR promoter. Twenty-nine single amino acid substitutions in MelR that result in partial melibiose-independent activation of the melAB promoter have been identified. Combinations of different substitutions result in almost complete melibiose-independent activation of the melAB promoter. MelR carrying each of the single substitutions is less able to repress the melR promoter, while MelR carrying some combinations of substitutions is completely unable to repress the melR promoter. These results argue that different conformational states of MelR are responsible for activation of the melAB promoter and repression of the melR promoter. Supporting evidence for this is provided by the isolation of substitutions in MelR that block melibiose-dependent activation of the melAB promoter while not changing melibiose-independent repression of the melR promoter. Additional experiments with a bacterial two-hybrid system suggest that interactions between MelR subunits differ according to the two conformational states.
The AraC family of bacterial transcription factors contains a large number of activators that regulate transcription initiation at promoters controlling genes important for virulence, stress, and metabolism (reviewed in references 7, 8, 16, and 29). Members of the AraC family are defined by an ∼110-amino-acid domain, containing two helix-turn-helix motifs, that recognizes ∼20-bp operator sequences at target promoters. Many members of the AraC family also contain an ∼170-amino-acid ligand-binding domain which regulates their activity. The Escherichia coli MelR protein appears to be a typical member of this family (33). Its function is to activate expression of the E. coli melibiose operon, melAB, in response to the availability of melibiose. MelR consists of an ∼170-amino-acid melibiose-binding N-terminal regulatory domain joined to an ∼110-amino-acid DNA-binding domain via an ∼20-amino-acid linker (17). The aim of this work was to exploit mutational analysis to understand how melibiose binding modulates the activity of MelR.
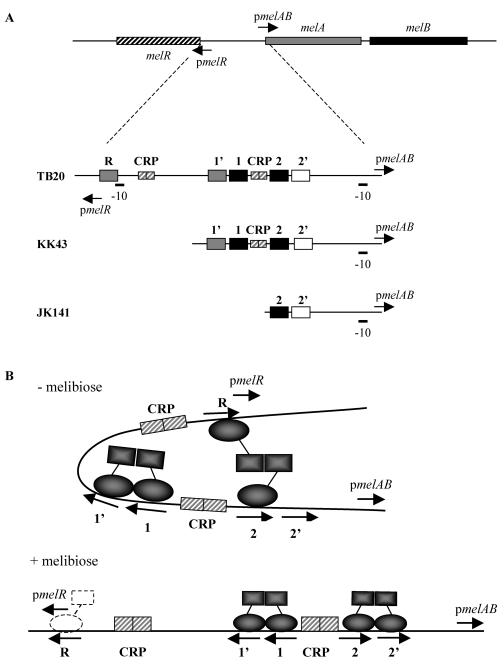
The E. coli melAB and melR genes are expressed from divergent promoters, pmelAB and pmelR, whose transcription start sites are separated by 237 bp (32). The regulatory region between the two promoters is complex and contains five 18-bp DNA sites for MelR (known as sites 1, 1′, 2, 2′, and R) and two 22-bp DNA sites for the cyclic AMP receptor protein (CRP) (2, 30, 31) (Fig. 1). Transcription initiation at pmelAB is totally dependent on MelR and melibiose. This requires the binding of MelR to operator site 2′, centered at position −42.5 upstream of the melAB transcript start site. Site 2′ overlaps the −35 element of pmelAB, and MelR bound at site 2′ activates transcription by making a direct contact with the RNA polymerase σ subunit (10). MelR binds to site 2′ only in the presence of melibiose (2). In the absence of melibiose, MelR occupies the other four sites, and this results in repression of pmelR (30). Repression of pmelR requires MelR binding to site R, which overlaps the melR promoter, but also MelR binding to site 2, located 176 bp upstream. It has been proposed (30) that repression requires the formation of a DNA loop that is stabilized by MelR binding at site R and site 2 and that the presence of melibiose breaks this loop, resulting in derepression of pmelR, occupation of site 2′, and induction of pmelAB (Fig. 1). Thus, melibiose toggles MelR between a state where it represses pmelR and is unable to activate pmelAB to a state where pmelR is derepressed and pmelAB is activated. Our aim was to understand this transition. MelR, like many AraC family members, is insoluble at higher concentrations, and structural studies have proven impossible. Hence, here we have tackled the problem using genetic approaches.
FIG. 1.
Organization of the E. coli melibiose operon regulatory region. (A) A not-to-scale illustration of the organization of the melR, melA, and melB genes, with the locations and orientation of pmelR and pmelAB. In the lower part of the figure, expanded views of the TB20, KK43, and JK141 fragments are shown, with the locations of the pmelAB and pmelR −10 elements and the different DNA sites for CRP (small hatched boxes) and MelR (larger boxes shaded according to binding hierarchy in the absence of melibiose). In this work, the TB20 fragment was cloned with EcoRI and HindIII linkers upstream and downstream of pmelR, respectively, into pRW50 to give a pmelR::lac fusion. The KK43 and JK141 fragments were cloned with EcoRI and HindIII linkers upstream and downstream of pmelAB, respectively, into pRW50 to give a pmelAB::lac fusion. (B) Interactions of MelR with the different sites in the absence and presence of melibiose as proposed by Wade et al. (30). In the absence of melibiose, MelR is unable to occupy site 2′, and an interaction between MelR bound at site 2 and site R causes strong repression of pmelR. In the presence of melibiose, MelR occupies site 2′, the interaction between site 2 and site R is broken, and the strong repression of pmelR is relieved. Weaker repression is due to residual binding of MelR to site R (dotted outline).
MATERIALS AND METHODS
Bacterial strains, plasmids, and oligonucleotide primers.
Bacterial strains and plasmids used in this work are described in Table 1, and oligonucleotide primers are listed in Table 2.
TABLE 1.
Bacterial strains and plasmids used in this work
| Bacterial strain or plasmid | Characteristics or descriptiona | Origin |
|---|---|---|
| E. coli strains | ||
| WAM131 | E. coli K-12 ara thi pro Δlac | Belyaeva et al. (2) |
| WAM132 | ΔmelR derivative of WAM131 | Belyaeva et al. (2) |
| WAM1321 | ΔmelA derivative of WAM132 | This work |
| BTH101 | E. coli K-12 cya-99 | Ladant (14) |
| Plasmids | ||
| pKD3 | Carries Cmr flanked by flp sites | Datsenko and Wanner (4) |
| pKD46 | Encodes λ red functions | Datsenko and Wanner (4) |
| pCP20 | Encodes FLP recombinase | Datsenko and Wanner (4) |
| pJW15 | Carries melR and Ampr | Wade et al. (30) |
| pJW15 derivatives | Carrying different mutant melR alleles | This work |
| pJW15ΔmelR | pJW15 derivative with deletion of melR | Wade et al. (30) |
| pRW50 | Lac expression vector, carries Tetr | Lodge et al. (15) |
| KK43-pRW50 | pRW50 carrying KK43 pmelAB fragment | Belyaeva et al. (2) |
| JK141-pRW50 | pRW50 carrying JK141 pmelAB fragment | This work |
| TB20-pRW50 | pRW50 carrying TB20 pmelR fragment | Wade et al. (30) |
| pK-T25 | Carries T25 adenylate cyclase fragment; Kanr | Karimova et al. (13) |
| pK-T25-zip | Carries T25:leucine zipper fusion | Karimova et al. (13) |
| pK-T25-MelR | Carries T25::melR fusion | This work |
| pK-T25-MelR derivatives | Carries T25::melR derivative fusions | This work |
| pU-T18C | Carries T18 adenylate cyclase fragment; Ampr | Karimova et al. (14) |
| pU-T18C-zip | Carries T18::leucine zipper fusion | Karimova et al. (14) |
| pU-T18C-MelR | Carries T18::melR fusion | This work |
| pU-T18C-MelR derivatives | Carries T18::melR derivative fusions | This work |
Cm, chloramphenicol; Amp, ampicillin; Tet, tetracycline; Kan, kanamycin.
TABLE 2.
Oligonucleotide primers
| Name | Sequence |
|---|---|
| D42648 | 5′-CTGCCATGATGAAGTTATTCAAGCAAGCCAGGAGA TCTGCGTGTAGGCTGGAGCTGCTTC-3′ |
| D42649 | 5′-CCGGAATAAATCCCCCGGCGCAGTAGCGTTTAGTC GCGTTCATATGAATATCCTCCTTAG-3′ |
| D49091 | 5′-GCAGAATTCGATCTGAGTTTAT-3′ |
| D10527 | 5′-GCAGGTCGTTGAACTGAGCCTGAAATTCAGG-3′ |
| D37452 | 5′-GCTAGTCTAGAGATGAATACAGATACGTTTATG-3′ |
| D37453 | 5′-GCGGGGTACCCGTTAGCCGGGAAACGTCTGGCG-3′ |
| D5431 | 5′-ACCTGACGTCTAAGAAACC-3′ |
| D4600 | 5′-GTAGTCGGTGTGTTCAC-3′ |
| D38456 | 5′-GATATGCGGTGCGCGAAACTCAAT-3′ |
| D38392 | 5′-GCGAATTTGCTCGTTCGGACTGTTT-3′ |
| D37665 | 5′-CGCCATGCGCAATCTTATGTTAGCC-3′ |
WAM1321 is a derivative of WAM132 carrying an in-frame deletion of the melA gene, constructed by the Datsenko and Wanner (4) method. This construction was made by using PCR primers D42648 and D42649 to amplify a DNA fragment carrying the cat gene from pKD3 flanked by upstream and downstream melA sequences. The fragment was electroporated into WAM132 carrying pKD46 encoding phage λ red functions, and crossovers were selected as chloramphenicol-resistant colonies. After checking the insertion of the cat gene using PCR, pKD46 was cured. The FLP recombinase, carried by pCP20, was then used to remove the cat insert to generate an in-phase deletion in the melA gene.
JK141-pRW50 was derived from KK43-pRW50 using PCR with primers D49091 and D10527 to construct a shorter pmelAB fragment (Fig. 1). The resulting PCR product was restricted with EcoRI and HindIII and cloned into pRW50.
Plasmids pK-T25-MelR and pU-T18C-MelR and derivatives were constructed by cloning an XbaI-KpnI fragment encoding full-length or mutant MelR into pK-T25 or pU-T18C plasmid that had been digested with XbaI and KpnI. The fragments were generated by PCR, using primers D37452 and D37453 with pJW15 encoding wild-type or mutant MelR as template, followed by digestion of the product with XbaI and KpnI.
Generation of random mutations in melR.
Error-prone PCR was used to amplify an EcoRI-HindIII fragment encoding melR using pJW15 as a template and primers D5431 and D4600. In these experiments, we used Taq DNA polymerase and buffer conditions as described by Barne et al. (1). Fragments from different reactions were digested with EcoRI and HindIII, purified, and recloned into pJW15 to generate independent libraries of mutations. DNA from the libraries was electroporated into tester strains carrying pmelAB::lac fusions as described below in Results. Transformants were screened either on minimal medium plates containing 20 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) or MacConkey lactose plates, with 80 μg/ml ampicillin and 35 μg/ml tetracycline. For minimal medium, we used M9 containing 0.3% fructose and 0.1% Casamino Acids, as previously described (32). After selection of each pJW15 derivative, encoding mutant MelR, the entire EcoRI-HindIII fragment encoding melR was sequenced in the University of Birmingham Functional Genomics laboratory (http://www.genomics.bham.ac.uk/) using primer D5431.
Combination of different melR mutations.
Most derivatives of pJW15 encoding MelR with two or more substitutions were made by exploiting the unique NsiI site corresponding to codon 100. The YD25 FY53, KE123 DG256, and NI183 FS191 double mutants were made by megaprimer PCR using the D38456, D38392, and D37665 primers, respectively, together with the flanking D5431 or D4600 primers. The QR238 TA277 derivative was isolated as a spontaneous pJW15 mutant during screening for MelR mutants competent for melibiose-independent activation of pmelAB.
Assays for activation and repression by mutant MelR derivatives.
WAM132 or WAM1321 cells carrying pRW50 with either pmelAB::lac or pmelR::lac fusions and pJW15 encoding melR were grown aerobically overnight at 30°C in M9 medium containing 0.3% fructose, 0.1% Casamino Acids, 80 μg/ml ampicillin, and 35 μg/ml tetracycline as previously described (32). The next day, 100-μl aliquots were inoculated into 5 ml of fresh culture either without or with added melibiose (10 mM). These cultures were grown aerobically at 30°C for several hours until the A600 reached 0.3 to 0.4. At this point, cultures were lysed with toluene, and β-galactosidase activities were measured as described by Miller (18). Activities were used to measure MelR-dependent activation of pmelAB and MelR-dependent repression of pmelR. Note that these assays were performed at 30°C to avoid complications due to the thermosensitivity of the MelB melibiose permease (28).
Bacterial two-hybrid assays.
The bacterial adenylate cyclase two-hybrid (BACTH) assay, as described by Karimova et al. (13), was used to monitor interactions between melR fused to the T18 and T25 segments of the Bordetella pertussis adenylate cyclase. The E. coli cya strain BTH101 was transformed by derivatives of pU-T18C and pK-T25, and transformants were plated on MacConkey lactose or MacConkey maltose plates containing 100 μg/ml ampicillin and 50 μg/ml kanamycin. For β-galactosidase assays, transformants were grown aerobically at 30°C in LB medium containing 100 μg/ml ampicillin and 50 μg/ml kanamycin to an A600 of 0.3 to 0.4. Cultures were lysed with toluene, and activities were measured as described by Miller (18).
RESULTS
Melibiose-independent activation of pmelAB by MelR.
We previously showed that MelR-dependent activation of pmelAB could be readily monitored using the low-copy-number, broad-host-range lac fusion plasmid pRW50, carrying the KK43 pmelAB promoter fragment (KK43-pRW50) (2). The KK43 pmelAB fragment carries two pairs of DNA sites for MelR (sites 1 and 1′ and sites 2 and 2′) separated by a DNA site for CRP (Fig. 1). In the absence of melibiose, MelR and CRP bind to form a complex, but site 2′ is not occupied. The occupation of site 2′, and concomitant transcription activation, is triggered by melibiose (2). To screen for MelR mutants with altered regulatory properties, we used the E. coli ΔmelR Δlac strain, WAM132, transformed with KK43-pRW50. Plasmid pJW15 was used to supply MelR to activate expression of the pmelAB::lac fusion carried by KK43-pRW50. Mutant melR libraries were made by using error-prone PCR to amplify a DNA fragment encoding the melR gene and recloning this DNA into pJW15.
WAM132 cells carrying KK43-pRW50 and pJW15 encoding wild-type melR score as Lac+ on X-Gal indicator plates containing melibiose and score as Lac− in the absence of melibiose. Table 3 lists β-galactosidase activity measurements, which showed that pmelAB activity is very low in the absence of melibiose and that melibiose triggers a >50-fold increase in pmelAB activity that is MelR dependent. DNA from eight independent preparations of pJW15 carrying randomly mutated melR was electroporated into WAM132 cells containing KK43-pRW50. Transformants were plated onto X-Gal indicator plates in the absence of melibiose. We reasoned that colonies exhibiting an enhanced Lac+ phenotype must carry pJW15 plasmids encoding MelR mutants (designated MelR*) able to activate pmelAB in the absence of melibiose. After screening and purification of such colonies, extraction of pJW15 DNA, and back-transformation into the test strain, we isolated 107 mutant pJW15 derivative candidates. DNA sequencing revealed that, among these, 29 carry single base changes that give rise to single amino acid substitutions in MelR. These changes are spread throughout the entire length of MelR and are listed in Table 3.
TABLE 3.
β-Galactosidase activity in WAM132 ΔmelR Δlac cellsa
| pJW15 derivative encoding melR | β-Galactosidase activity pmelAB::lac (KK43)
|
Melibiose concn (μM) required for 50% induction | |
|---|---|---|---|
| No melibiose | 10 mM melibiose | ||
| No melR | 11 | 11 | |
| Wild-type melR | 17 | 980 | 4 ± 2 |
| Single mutants | |||
| QE14 | 51 | 997 | |
| EV24 | 40 | 1,225 | |
| YD25 | 232 | 1,070 | 0.2 ± 0.1 |
| YH25 | 38 | 1,048 | |
| YN25 | 42 | 1,078 | |
| RP27 | 190 | 598 | |
| IT37 | 33 | 960 | |
| SN41 | 32 | 1,254 | |
| FY53 | 124 | 1,080 | 0.4 ± 0.1 |
| IV68 | 34 | 1,000 | |
| GD71 | 241 | 1,098 | |
| IT73 | 33 | 1,080 | |
| GS119 | 85 | 923 | |
| KE123 | 77 | 941 | 0.5 ± 0.1 |
| QR128 | 131 | 980 | |
| QR140 | 31 | 1,080 | |
| EG156 | 133 | 1,421 | |
| ST167 | 105 | 1,077 | |
| SF167 | 133 | 970 | 0.5 ± 0.1 |
| KR182 | 68 | 1,068 | 0.5 ± 0.1 |
| NI183 | 140 | 990 | 0.5 ± 0.1 |
| NS183 | 70 | 1,400 | |
| QR190 | 163 | 774 | |
| FL191 | 32 | 902 | |
| FS191 | 34 | 931 | |
| SG194 | 35 | 1,049 | |
| QR238 | 30 | 995 | |
| DG256 | 190 | 1,215 | |
| TA277 | 30 | 1,030 | |
| Double mutants | |||
| YD25 FY53 | 310 | 1,209 | |
| FY53 SF167 | 210 | 1,113 | |
| FY53 KR182 | 440 | 1,150 | |
| FY53 DG256 | 594 | 1,261 | |
| KE123 DG256 | 742 | 1,279 | |
| NI183 FS191 | 170 | 1,090 | |
| QR238 TA277 | 318 | 1,035 | |
| Quadruple mutants | |||
| YD25 FY53 | 870 | 1,023 | |
| QR238 TA277 | |||
| YD25 FY53 | 920 | 1,034 | |
| NI183 FS191 | |||
The first column of the table lists the substitutions in different MelR derivatives selected as competent for melibiose-independent activation of pmelAB. Activities are expressed in the units described by Miller (18) and were determined using the Miller protocol. The data in columns 2 and 3 are averages of at least four independent determinations that differed by no more than 10%. Cells were grown aerobically in defined medium with fructose as a carbon source either with or without 10 mM melibiose and were harvested in exponential phase at an optical density at 600 nm of ∼0.3. Column 4 lists the melibiose concentration required for 50% of the melibiose-dependent activation of pmelAB. These estimates are derived from assays of pmelAB::lac expression in the WAM1321 ΔmelA strain grown in media with different concentrations of melibiose (0.1, 1.0, and 10 μM and 0.1, 1.0, and 10 mM).
To quantify the effects of the different substitutions, the β-galactosidase activities in cultures of WAM132 cells containing KK43-pRW50 and each mutant pJW15 derivative, grown in the absence of melibiose, were measured. The data in Table 3 show that the changes increase pmelAB activity by factors ranging from 1.5-fold to 14-fold. None of the substitutions resulted in full melibiose-independent activation, and many caused only marginal increases in expression. The biggest effect, seen with the GD71 substitution, corresponds to only ∼25% of the full activation. Results in Table 3 show that, with each mutant MelR, the addition of melibiose increases the activity of pmelAB. In most cases, the observed expression is similar to that observed with wild-type MelR in the presence of melibiose.
For some of the substitutions, we measured the concentration of melibiose required to increase expression of the pmelAB::lac fusion. To do this, the assays were made in strain WAM1321, a derivative of WAM132 carrying an in-phase deletion of the melA gene. Recall that melA encodes the α-galactosidase that hydrolyzes melibiose to glucose and galactose. Data in Table 3 show that, with pJW15 encoding wild-type MelR, 4 μM melibiose is required for 50% of the melibiose-dependent induction of pmelAB activity. With pJW15 encoding the YD25, FY53, KE123, SF167, KR182, or NI183 MelR* substitutions, only 0.2 to 0.5 μM melibiose is required.
In the next series of experiments, we investigated whether combining different substitutions located in different parts of MelR would result in a mutant protein better able to activate pmelAB in the absence of melibiose. Thus, we constructed pJW15 derivatives that encode the following double substitutions: YD25 FY53, FY53 SF167, FY53 KR182, FY53 DG256, KE123 DG256, NI183 FS191, and QR238 TA277. Results in Table 3 show that, in combination, the effects of the different substitutions were additive, and in some cases synergy was found. Hence, with pJW15 carrying the double substitutions, pmelAB activity in the absence of melibiose ranges from 17 to 75% of the activity with wild-type MelR in the presence of melibiose. Finally, pJW15 derivatives were constructed encoding MelR YD25 FY53 together with the NI183 FS191 or QR238 TA277 changes. Further data listed in Table 3 show that, with pJW15 carrying these quadruple substitutions, pmelAB activity in the absence of melibiose rises to >90% of the activity seen with wild-type MelR in the presence of melibiose.
Repression of pmelR by MelR* mutants.
The simplest explanation of our data is that melibiose switches MelR from a conformation that is unable to activate pmelAB to a conformation that is able to activate pmelAB and that the different MelR* substitutions favor adoption of the latter conformation to different extents. In our previous work (30), we showed that, in the absence of melibiose, the expression of a pmelR::lac fusion could be repressed >10-fold by MelR and that this repression required MelR binding to site R, overlapping pmelR, and to site 2, located 176 bp upstream. Since repression is greatly reduced by melibiose, we investigated whether it is reduced by the different MelR* substitutions. To do this we used the ΔmelR Δlac strain, WAM132, transformed with pRW50 carrying the TB20 pmelR promoter fragment (TB20-pRW50). The TB20 pmelR fragment carries the melR promoter and upstream sequences, including MelR site 2 (Fig. 1). Plasmid pJW15 was used to supply wild-type or mutant MelR to repress expression of the pmelR::lac fusion carried by TB20-pRW50.
WAM132 cells containing TB20-pRW50 and pJW15 encoding wild-type MelR score as Lac− on MacConkey or X-Gal indicator plates. A Lac+ phenotype is observed if pJW15 is replaced by empty vector plasmid. Table 4 lists β-galactosidase activity measurements, which confirmed that, in the absence of melibiose, pmelR activity is repressed >10-fold by wild-type MelR. With one exception, each of the different single MelR* substitutions results in a small reduction in the MelR-dependent repression of pmelR. The exception is the GS119 MelR* substitution, which results in a much greater reduction in repression. Table 4 also shows measurements of the repression of pmelR by MelR carrying different combinations of substitutions. Double-substituted MelR represses pmelR less efficiently, and repression by MelR carrying YD25 FY53 together with the NI183 FS191 or QR238 TA277 changes is minimal.
TABLE 4.
β-Galactosidase activity in WAM132 ΔmelR Δlac cells grown in the absence of melibiosea
| pJW15 derivative encoding melR | β-Galactosidase activity
|
|
|---|---|---|
| pmelR::lac (TB20) | pmelAB::lac (KK43) | |
| No melR | 274 | 11 |
| Wild-type melR | 25 | 17 |
| Single mutants | ||
| QE14 | 37 | 51 |
| EV24 | 35 | 40 |
| YD25 | 65 | 232 |
| YH25 | 45 | 38 |
| YN25 | 45 | 42 |
| RP27 | 50 | 190 |
| IT37 | 27 | 33 |
| SN41 | 38 | 32 |
| FY53 | 60 | 124 |
| IV68 | 38 | 34 |
| GD71 | 60 | 241 |
| IT73 | 37 | 33 |
| GS119 | 154 | 85 |
| KE123 | 50 | 77 |
| QR128 | 42 | 131 |
| QR140 | 36 | 31 |
| EG156 | 49 | 133 |
| ST167 | 39 | 105 |
| SF167 | 40 | 133 |
| KR182 | 37 | 68 |
| NI183 | 51 | 140 |
| NS183 | 40 | 70 |
| QR190 | 60 | 163 |
| FL191 | 29 | 32 |
| FS191 | 31 | 34 |
| SG194 | 25 | 35 |
| QR238 | 65 | 30 |
| DG256 | 52 | 190 |
| TA277 | 37 | 30 |
| Double mutants | ||
| YD25 FY53 | 205 | 310 |
| FY53 SF167 | 140 | 210 |
| FY53 KR182 | 135 | 440 |
| FY53 DG256 | 231 | 594 |
| KE123 DG256 | 143 | 742 |
| NI183 FS191 | 55 | 170 |
| QR238 TA277 | 103 | 318 |
| Quadruple mutants | ||
| YD25 FY53 | 263 | 870 |
| QR238 TA277 | ||
| YD25 FY53 | 269 | 920 |
| NI183 FS191 | ||
The first column of the table lists the substitutions in different MelR derivatives selected as competent for melibiose-independent activation of pmelAB. The second column lists the measured β-galactosidase activities in WAM132ΔmelR Δlac cells carrying a pmelR::lac fusion and the different MelR derivatives. The third column lists the measured β-galactosidase activities in cells carrying a pmelAB::lac fusion and different MelR derivatives (as in Table 3). Activities, expressed in the units described by Miller (18), are the averages of at least four independent determinations that differed by no more than 10%. Cells were grown aerobically in defined medium without melibiose, with fructose as a carbon source, and were harvested in exponential phase at an optical density at 600 nm of ∼0.3.
MelR mutants that are less able to activate pmelAB.
The substitutions in the MelR* mutants described above appear to bias MelR towards a conformation that can activate pmelAB but is unable to repress pmelR. To find complementary mutants, stabilized in the conformation that represses pmelR but unable to activate pmelAB, we screened the eight libraries of mutated pJW15 using two steps. In the first step, pJW15 DNA was electroporated into the WAM131 Δlac strain containing plasmid KK43-pRW50 carrying a pmelAB::lac fusion. Since strain WAM131 is melR+, colonies score as Lac+ on MacConkey lactose indicator plates either with or without pJW15 encoding MelR. However, some melR alleles unable to activate pmelAB result in Lac− colonies, and 50 such colonies were selected. In the second step, pJW15 DNA was purified from each colony and retransformed into WAM132 cells containing TB20-pRW50. Most of the 50 pJW15 mutant DNAs gave rise to Lac+ colonies. These DNAs, which encode mutant MelR that is unable to repress pmelR, presumably due to a defect in DNA binding, were discarded. However, this second screening step identified 11 pJW15 derivatives encoding MelR that were still able to repress pmelR, despite being defective in the activation of pmelAB. Each of these mutant MelR derivatives carried a single substitution. The different changes, listed in Table 5, are distributed throughout MelR.
TABLE 5.
β-Galactosidase activity in WAM132 ΔmelR Δlac cells carrying MelR mutants defective in activation of pmelABa
| pJW15 derivative encoding melR | β-Galactosidase activity
|
||
|---|---|---|---|
| pmelAB::lac (KK43)
|
pmelR::lac (TB20), no melibiose | ||
| No melibiose | 10 mM melibiose | ||
| No melR | 11 | 12 | 274 |
| Wild-type melR | 16 | 1,021 | 25 |
| Single mutants | |||
| NS50 | 12 | 10 | 25 |
| FL53 | 16 | 220 | 52 |
| PS81 | 17 | 263 | 64 |
| IT95 | 11 | 11 | 22 |
| TA117 | 10 | 187 | 21 |
| TI117 | 6 | 12 | 62 |
| ND183 | 7 | 137 | 44 |
| AT201 | 6 | 6 | 30 |
| MT243 | 5 | 4 | 27 |
| LS251 | 6 | 13 | 61 |
| SR271 | 5 | 9 | 26 |
The first column of the table lists the substitutions in different MelR derivatives selected as defective for melibiose-dependent activation of pmelAB. The second and third columns list the measured β-galactosidase activities in WAM132 ΔmelR Δlac cells carrying a pmelAB::lac fusion and the different MelR derivatives. Cells were grown aerobically in defined medium without (column 2) or with (column 3) 10 mM melibiose, with fructose as a carbon source, and were harvested in exponential phase at an optical density at 600 nm of ∼0.3. The fourth column lists the measured β-galactosidase activities in WAM132 ΔmelR Δlac cells carrying a pmelR::lac fusion and the different MelR derivatives. Activities, expressed in the units described by Miller (18), are the averages of at least four independent determinations that differed by no more than 10%.
To quantify the effects of the different changes, the β-galactosidase activities in cultures of WAM132 cells containing KK43-pRW50 and each mutant pJW15 derivative were measured without and with melibiose. Data in Table 5 show that, for seven of the mutants (NS50, IT95, TI117, AT201, MT243, LS251, and SR271), 10 mM melibiose is unable to induce pmelAB activity. In contrast, with four of the mutants (FL53, PS81, TA117, and ND183), melibiose induces pmelAB activity, but to a lesser level than with wild-type MelR. Titrations with melibiose revealed that the concentration of melibiose required for 50% induction is increased by five- to eightfold compared to wild-type MelR (C. L. Webster, unpublished data). Interestingly, other substitutions of F53 or N183 create MelR* derivatives that are triggered at lower melibiose concentrations (Table 3).
To quantify the effects of the 11 changes on repression of pmelR, the β-galactosidase activities in cultures of WAM132 cells containing TB20-pRW50 and each mutant pJW15 derivative were measured in the absence of melibiose. These assays showed that each of the different mutant MelR derivatives was able to repress pmelR (Table 5). Maximum repression was found with the NS50, IT95, TA117, MT243, and SR271 mutants. Since the NS50, IT95, MT243, and SR271 substitutions result in noninducible (NI) MelR, we conclude that these changes lock MelR in the minus-melibiose conformation.
Interactions between MelR subunits measured by BACTH.
We reasoned that the difference between the form of MelR unable to activate pmelAB but competent for repression of pmelR and the alternative form that can activate pmelAB but is unable to repress pmelR might be, in part, due to subunit-subunit interactions. Since, to date, purified MelR has been refractory to biophysical investigation, we sought to study these interactions using the well-characterized BACTH assay (13). This relies on the observation that Bordetella pertussis adenylate cyclase consists of two independently folding domains and that this adenylate cyclase can become active when the two domains are brought together in the cell. Thus, when the T18 and T25 fragments are expressed as separate entities, host cells score as negative for adenylate cyclase activity, but if T18 and T25 are fused to interacting partners, hosts can score as a positive. Recall that adenylate cyclase catalyzes the synthesis of cyclic AMP, whose levels in E. coli can be monitored by plate assays of maltose phenotypes or enzyme assays of β-galactosidase activity. Thus, E. coli strain BTH101, which is defective for adenylate cyclase (cya), was transformed with plasmids pU-T18C and pK-T25 that express, respectively, the Bordetella pertussis adenylate cyclase T18 and T25 fragments. Transformants score as Mal− and contain low levels of β-galactosidase. However, results summarized in Table 6 show that cells transformed with pU-T18C and pK-T25 derivatives encoding fusions of the T18 and T25 fragments to wild-type MelR score as Mal+ and contain significantly increased levels of β-galactosidase. The explanation for this is that MelR self-associates and brings together the T18 and T25 fragments to generate adenylate cyclase activity. This association is unchanged by the MT243 substitution, which appears to freeze MelR in its melibiose-free conformation. In contrast, when MelR carrying the YD25, FY53, NI183, and FS191 substitutions is fused to the T18 and T25 fragments, cells score as Mal− and contain low levels of β-galactosidase. Since the combination of the YD25, FY53, NI183, and FS191 substitutions converts MelR to its melibiose-triggered state, we conclude that the association between MelR subunits must differ according to their conformation.
TABLE 6.
β-Galactosidase activity in BTH101 cya cells containing pK-T25 and pU-T18C derivativesa
| pK-T25 derivative | pU-T18C derivative | β-Galactosidase activity | Phenotype on MacConkey maltose plates |
|---|---|---|---|
| pK-T25 | pU-T18C | 50 | White (Mal−) |
| pK-T25-zip | pU-T18C-zip | 690 | Red (Mal+) |
| pK-T25-MelR | pU-T18C-MelR | 271 | Red |
| pK-T25-MelR YD25 | pU-T18C-MelR | 73 | White |
| FY53 NI183 FS191 | YD25 FY53 NI183 FS191 | ||
| pK-T25-MelR MT243 | pU-T18C-MelR MT243 | 248 | Red |
Activities are expressed in the units described by Miller and were determined using the Miller protocol (18). The data shown are the averages of at least four independent measurements that differed by no more than 10%. Cells were grown aerobically at 30°C in LB medium containing 50 μg/ml kanamycin and 100 μg/ml ampicillin and were harvested in exponential phase at an optical density at 600 nm of ∼0.4. Column 4 lists the observed phenotype of the starting colony on MacConkey maltose plates after overnight growth at 30°C.
Mutant MelR that is active in the absence of melibiose has less need for CRP.
When triggered by melibiose, wild-type MelR requires the assistance of CRP to activate transcription at pmelAB (31). The likely explanation for this is that a nucleoprotein complex of DNA-MelR and CRP is needed for melibiose-activated MelR to occupy site 2′. To examine whether MelR that is frozen in the activating conformation has the same requirement for CRP, we constructed the JK141 promoter, which lacks MelR-binding sites 1′ and 1 and the pmelAB DNA site for CRP (Fig. 1), and cloned the resulting fragment in pRW50 to give plasmid JK141-pRW50. Results in Table 7 show that wild-type MelR is unable to activate expression from the JK141 promoter. This was expected, since the CRP requirement for MelR-dependent activation of pmelAB is well established (31). However, MelR carrying the YD25, FY53, NI183, and FS191 substitutions, as well as being able to activate pmelAB carried by the KK43 fragment, is also able to activate the JK141 promoter. This shows that MelR carrying these substitutions has a reduced requirement for CRP to activate pmelAB.
TABLE 7.
β-Galactosidase activity in WAM132 ΔmelR Δlac cells carrying KK43 or JK141 pmelAB::lac fusions and pJW15a
| MelR encoded by pJW15 derivative | β-Galactosidase activity
|
|||
|---|---|---|---|---|
| KK43-pRW50
|
JK141-pRW50
|
|||
| No melibiose | With melibiose | No melibiose | With melibiose | |
| Wild type | 23 | 1043 | 3 | 28 |
| YD25 FY53 NI183 FS191 | 875 | 929 | 234 | 703 |
Activities are expressed in the units described by Miller and were determined using the Miller protocol (18). The data shown are the averages of at least four independent values that differed by no more than 10%. Cells were grown aerobically in defined medium with fructose as a carbon source either with or without 10 mM melibiose and were harvested in exponential phase at an optical density at 600 nm of ∼0.3.
DISCUSSION
Melibiose is needed for wild-type MelR to activate transcription initiation at pmelAB. This is because activation requires MelR binding to site 2′ that overlaps the pmelAB −35 hexamer element, and melibiose is required for wild-type MelR to occupy this site (2). However, in the absence of melibiose, MelR still binds at site 2 and at site R, and this results in strong repression of pmelR (30). This strong repression is relieved by melibiose and, thus, melibiose toggles MelR between two alternative states, one that activates pmelAB and one that represses pmelR (Fig. 1B). Our goal was to use genetic analysis to investigate these two states.
The best-understood AraC family member is the Escherichia coli AraC protein itself, which is also toggled between two states by its ligand, arabinose (reviewed in references 23 and 24). AraC-dependent transcription regulation has been most studied at the araBAD and araC genes, which are expressed from divergent promoters, paraBAD and paraC, whose transcription start sites are separated by 166 bp. Activation of paraBAD requires AraC to bind at adjacent 20-bp operator sites, I1 and I2, centered at positions −63.5 and −43.5 upstream of the transcription start site. The I2 site overlaps the −35 element of paraBAD, and AraC normally occupies this site only in the presence of arabinose. In the absence of arabinose, AraC binds to site I1 and an upstream site O2, and this results in repression of paraC. Thus, arabinose converts AraC from a form that binds to distal targets (O2 and I1) to a form that binds to adjacent targets (I1 and I2). To explain this, Schleif and his colleagues proposed the light switch model (23, 24), which was derived from X-ray structural analyses of the AraC N-terminal arabinose-binding domain. These studies (26, 27) showed that the AraC N-terminal domain contains a cupin fold that carries the binding site for arabinose and that the extreme N-terminal arm (AraC residues 1 to 20) folds over bound arabinose. Schleif and coworkers have found that, in the absence of arabinose, this N-terminal arm switches to interacting with the AraC DNA-binding domain (9, 25). Hence, in the X-ray structure of the AraC N-terminal domain without arabinose, this arm is unstructured and cannot be seen. The light switch model proposes that, in the absence of arabinose, the interaction of the AraC N-terminal arm with the C-terminal DNA-binding domain constrains the subunits of the AraC dimer in an orientation that makes it energetically favorable to bind to distal (O2 and I1) rather than adjacent (I1 and I2) targets (11, 25). This model is supported by genetic analyses, notably, mutations that alter amino acids in the AraC N-terminal arm that result in AraC-dependent activation of paraBAD in the absence of arabinose (19, 21, 22, 34, 35).
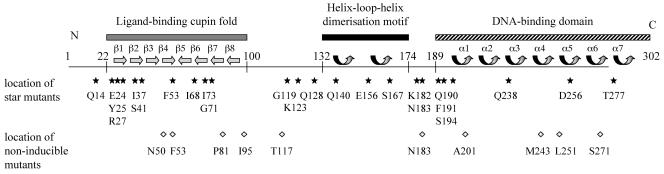
The striking parallels between AraC and MelR led us to consider whether the light switch model applies to MelR. Although we have no structural data for MelR, its domain organization appears to be similar to AraC (Fig. 2). In particular, amino acid sequence similarities argue that MelR residues 25 to 100 constitute a ligand-binding cupin fold (5, 6) and MelR residues 190 to 302 fold as an AraC family DNA-binding domain (29). In preliminary experiments, we targeted mutations to the segment of melR encoding the 20 N-terminal amino acids of MelR, but we were unable to find any changes that resulted in MelR capable of melibiose-independent activation of pmelAB (T. A. Belyaeva, unpublished data). The subsequent random mutational analysis of the entire melR gene, presented in this paper, argues that the light switch model cannot apply to MelR.
FIG. 2.
Domain organization of the E. coli MelR protein. The 302 amino acids of MelR are illustrated as a horizontal line, annotated with different structural features. The locations of a ligand-binding cupin fold, consisting of eight β-sheet elements (5, 6) and a helix-loop-helix dimerization motif, deduced from similarities with AraC and published AraC structures (26, 27), are shown. The location of the DNA-binding domain, consisting of seven α-helix elements and deduced from similarities with MarA and the published MarA structure (20), is shown. The lower part of the figure shows the locations of different substitutions that confer the ability to partially activate pmelAB in the absence of melibiose (stars) and the locations of substitutions that interfere with melibiose-dependent activation (diamonds).
Our analysis revealed that substitutions resulting in melibiose-independent activation of pmelAB fall at loci throughout MelR and do not cluster in its N-terminal or its DNA-binding domain. Individual substitutions confer only incremental degrees of melibiose independence, though full melibiose independence is found when different substitutions are combined. In all the cases that were tested, the melibiose independence due to different substitutions was additive, suggesting that the conversion to the activating state of MelR requires changes in all of the different segments of the protein (Fig. 2). Substitutions in different parts of MelR must be combined to create fully active melibiose-independent MelR. Thus, we suppose that melibiose binding to the MelR cupin fold triggers a concerted series of conformation changes that affect the N-terminal domain, the C-terminal domain, and the connecting linker. These changes appear to be mimicked by the different MelR* substitutions that we isolated.
Our results are most easily interpreted by a two-state model for MelR in which one state is fully competent for activation of pmelAB but is unable to repress pmelR, and vice versa for the other state. Thus, all the changes that confer melibiose independence for pmelAB activation relieve repression of pmelR in the absence of melibiose, and there is a clear correlation between the two functions (Table 4). In accord with this, we identified four NI mutants of MelR, NS50, IT95, MT243, and SR271, which appear to lock MelR in the minus-melibiose conformation and are fully competent for repression of pmelR (Table 5). Due to the difficulty of working with purified MelR, we resorted to using an artificial bacterial two-hybrid system to investigate the two states of MelR. Since it is likely that MelR-MelR interactions are important for both activation of pmelAB and repression of pmelR, we suppose that that differences recorded in Table 5 are due to differences in these interactions. Presumably, in one state but not the other, the MelR-MelR interaction results in a productive interaction between the T18 and T25 adenylate cyclase fragments.
From our study, we can conclude that, though the ligand-free forms of both E. coli AraC and MelR proteins strongly repress expression from their own promoters, different mechanisms are used for ligand-dependent switching to a state that can activate transcription. For AraC in the absence of ligand, the N-terminal arm constrains the DNA-binding C-terminal domain (23, 24). Arabinose removes this constraint, and the C-terminal domain is then able to activate transcription at paraBAD. Consistent with this, Bustos and Schleif (3) showed that the isolated AraC C terminal is competent for this activation. In contrast, the corresponding C-terminal domain of MelR alone is unable to activate pmelAB (12, 17). Thus, the binding of melibiose to the MelR N-terminal domain is required to transmit an activatory signal to the MelR C-terminal domain, and both the N- and C-terminal domains are required for activation of pmelAB expression. The nature of this signal is not understood, but the scattering of substitutions that affect switching suggests that all segments of MelR are involved. Interestingly, some of the substitutions on our MelR* mutants fall in the linker that joins the N-terminal ligand-binding domain and the C-terminal DNA-binding domain. This suggests that the interdomain linker may play more than a neutral role as signals are passed between the two domains.
Acknowledgments
This project was funded by the UK BBSRC with project grant number BB/C501484/1 and a Ph.D. studentship to C.K.
Thanks are due to Daniel Ladant and Lars Westblade for assistance with the bacterial two-hybrid experiments, to Jim Dunwell for information on cupin folds, and to David Grainger, Georgina Lloyd, and Robert Schleif for commenting on the manuscript.
REFERENCES
- 1.Barne, K. A., J. A. Bown, S. J. Busby, and S. D. Minchin. 1997. Region 2.5 of the Escherichia coli RNA polymerase sigma70 subunit is responsible for recognition of the extended −10′ motif at promoters. EMBO J. 16:4034-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belyaeva, T. A., J. T. Wade, C. L. Webster, V. J. Howard, M. S. Thomas, E. I. Hyde, and S. J. Busby. 2000. Transcription activation at the Escherichia coli melAB promoter: the role of MelR and the cyclic AMP receptor protein. Mol. Microbiol. 36:211-222. [DOI] [PubMed] [Google Scholar]
- 3.Bustos, S. A., and R. F. Schleif. 1993. Functional domains of the AraC protein. Proc. Natl. Acad. Sci. USA 90:5638-5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunwell, J. M., A. Culham, C. E. Carter, C. R. Sosa-Aguirre, and P. W. Goodenough. 2001. Evolution of functional diversity in the cupin superfamily. Trends Biochem. Sci. 26:740-746. [DOI] [PubMed] [Google Scholar]
- 6.Dunwell, J. M., A. Purvis, and S. Khuri. 2004. Cupins: the most functionally diverse protein superfamily? Phytochemistry 65:7-17. [DOI] [PubMed] [Google Scholar]
- 7.Egan, S. M. 2002. Growing repertoire of AraC/XylS activators. J. Bacteriol. 184:5529-5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallegos, M.-T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. AraC/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghosh, M., and R. F. Schleif. 2001. Biophysical evidence of arm-domain interactions in AraC. Anal. Biochem. 295:107-112. [DOI] [PubMed] [Google Scholar]
- 10.Grainger, D. C., C. L. Webster, T. A. Belyaeva, E. I. Hyde, and S. J. Busby. 2004. Transcription activation at the Escherichia coli melAB promoter: interactions of MelR with its DNA target site and with domain 4 of the RNA polymerase sigma subunit. Mol. Microbiol. 51:1297-1309. [DOI] [PubMed] [Google Scholar]
- 11.Harmer, T., M. Wu, and R. Schleif. 2001. The role of rigidity in DNA looping-unlooping by AraC. Proc. Natl. Acad. Sci. USA 98:427-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howard, V. J., T. A. Belyaeva, S. J. Busby, and E. I. Hyde. 2002. DNA binding of the transcription activator protein MelR from Escherichia coli and its C-terminal domain. Nucleic Acids Res. 30:2692-2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karimova, G., J. Pidoux, A. Ullmann, and D. Ladant. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. USA 95:5752-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karimova, G., A. Ullmann, and D. Ladant. 2001. Protein-protein interaction between Bacillus stearothermophilus tyrosyl-tRNA synthetase subdomains revealed by a bacterial two-hybrid system. J. Mol. Microbiol. Biotechnol. 3:73-82. [PubMed] [Google Scholar]
- 15.Lodge, J., J. Fear, S. Busby, P. Gunasekaran, and N.-R. Kamini. 1992. Broad host range plasmids carrying the Escherichia coli lactose and galactose operons. FEMS Microbiol. Lett. 74:271-276. [DOI] [PubMed] [Google Scholar]
- 16.Martin, R. G., and J. L. Rosner. 2001. The AraC transcriptional activators. Curr. Opin. Microbiol. 4:132-137. [DOI] [PubMed] [Google Scholar]
- 17.Michán, C. M., S. J. Busby, and E. I. Hyde. 1995. The Escherichia coli MelR transcription activator: production of a stable fragment containing the DNA-binding domain. Nucleic Acids Res. 23:1518-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Reed, W. L., and R. F. Schleif. 1999. Hemiplegic mutations in AraC protein. J. Mol. Biol. 294:417-425. [DOI] [PubMed] [Google Scholar]
- 20.Rhee, S., R. G. Martin, J. L. Rosner, and D. R. Davies. 1998. A novel DNA-binding motif in MarA: the first structure for an AraC family transcription activator. Proc. Natl. Acad. Sci. USA 95:10413-10418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ross, J. J., U. Gryczynski, and R. Schleif. 2003. Mutational analysis of residue roles in AraC function. J. Mol. Biol. 328:85-93. [DOI] [PubMed] [Google Scholar]
- 22.Saviola, B., R. Seabold, and R. Schleif. 1998. Arm-domain interactions in AraC. J. Mol. Biol. 278:539-548. [DOI] [PubMed] [Google Scholar]
- 23.Schleif, R. 2000. Regulation of the L-arabinose operon of Escherichia coli. Trends Genet. 16:559-565. [DOI] [PubMed] [Google Scholar]
- 24.Schleif, R. 2003. AraC protein: a love-hate relationship. Bioessays 25:274-282. [DOI] [PubMed] [Google Scholar]
- 25.Seabold, R. R., and R. F. Schleif. 1998. Apo-AraC actively seeks to loop. J. Mol. Biol. 278:529-538. [DOI] [PubMed] [Google Scholar]
- 26.Soisson, S. M., B. Macdougall-Shackleton, R. Schleif, and C. Wolberger. 1997. Structural basis for ligand-regulated oligomerization of AraC. Science 276:421-425. [DOI] [PubMed] [Google Scholar]
- 27.Soisson, S. M., B. Macdougall-Shackleton, R. Schleif, and C. Wolberger. 1997. The 1.6 Å crystal structure of the AraC sugar-binding and dimerization domain complexed with D-fucose. J. Mol. Biol. 273:226-237. [DOI] [PubMed] [Google Scholar]
- 28.Tamai, E., T. Shimamoto, M. Tsuda, T. Mizushima, and T. Tsuchiya. 1998. Conversion of temperature-sensitive to -resistant gene expression due to mutations in the promoter region of the melibiose operon in Escherichia coli. J. Biol. Chem. 273:16860-16864. [DOI] [PubMed] [Google Scholar]
- 29.Tobes, R., and J. L. Ramos. 2002. AraC-XylS database: a family of positive transcriptional regulators in bacteria. Nucleic Acids Res. 30:318-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wade, J. T., T. A. Belyaeva, E. I. Hyde, and S. J. Busby. 2000. Repression of the Escherichia coli melR promoter by MelR: evidence that efficient repression requires the formation of a repression loop. Mol. Microbiol. 36:223-229. [DOI] [PubMed] [Google Scholar]
- 31.Wade, J. T., T. A. Belyaeva, E. I. Hyde, and S. J. Busby. 2001. A simple mechanism for co-dependence on two activators at an Escherichia coli promoter. EMBO J. 20:7160-7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Webster, C., K. Kempsell, I. Booth, and S. Busby. 1987. Organisation of the regulatory region of the Escherichia coli melibiose operon. Gene 59:253-263. [DOI] [PubMed] [Google Scholar]
- 33.Webster, C., L. Gardner, and S. Busby. 1989. The Escherichia coli melR gene encodes a DNA-binding protein with affinity for specific sequences located in the melibiose operon regulatory region. Gene 83:207-213. [DOI] [PubMed] [Google Scholar]
- 34.Wu, M., and R. Schleif. 2001. Mapping arm-DNA binding domain interactions in AraC. J. Mol. Biol. 307:1001-1009. [DOI] [PubMed] [Google Scholar]
- 35.Wu, M., and R. Schleif. 2001. Strengthened arm-dimerization domain interactions in AraC. J. Biol. Chem. 276:2562-2564. [DOI] [PubMed] [Google Scholar]