Abstract
Bacterial cells sense the extracellular environment and adapt to that environment by activating gene regulation circuits, often by means of signaling molecules. The Serratia marcescens hemophore is a signaling molecule that acts as an extracellular heme-scavenging protein. The heme-loaded hemophore interacts with its cognate receptor (HasR), triggering transmembrane signaling and turning on transcription of hemophore-dependent heme uptake genes. We investigated the features of the holo-hemophore, the only HasR ligand known to act as an inducer. We used a hemophore mutant that does not deliver its heme and a HasR mutant that does not bind heme, and we showed that heme transfer from the hemophore to the receptor is necessary for induction. Using a hemophore mutant that does not bind heme and that blocks heme transport, we demonstrated that two molecules that do not interact (heme and the mutant hemophore) may nonetheless induce this system. These findings suggest that hemophore-mediated induction and heme transport involve different mechanisms. The hemophore region important for induction was precisely localized to amino acids 50 to 55, which lie in one of the two HasR-binding hemophore regions. This bipartite stimulus probably corresponds to a physiological process because heme is transferred to the receptor before apo-hemophore release. This bipartite regulation mechanism may allow the bacterium to adjust its heme transport mechanism to the perceived environmental heme concentration.
Most organisms, including bacteria, require iron. Bacteria encounter various types of environmental iron sources, including ferric and ferrous iron, iron-containing siderophores, heme, heme carrier proteins, and hemophores (10). In most cases, bacterial iron needs are covered by tightly regulated iron/heme uptake systems encoded by genes clustered in operons expressed only under iron deficiency conditions (2). In the presence of high intracellular iron concentrations, these operons are repressed by a common regulator, the iron-loaded Fur protein (3, 17).
However, not all iron acquisition systems are simultaneously induced in response to iron limitation. Many systems are also positively regulated by the availability of a specific iron source (9, 33). In gram-negative bacteria, iron/heme ligands are recognized by specific outer membrane proteins that play a key role in active transport and ligand-mediated positive regulation (13). These receptors have similar overall structures, with a membrane-inserted β barrel closed by an N-terminal plug region (15). The activity of these receptors is dependent on the TonB-ExbB-ExbD inner membrane protein complex, with which they interact via a short conserved region, named the TonB box, near the receptor's N terminus (6).
Ligand receptor binding, even in the absence of transport, is sufficient to induce transcription of the corresponding ligand uptake genes (12). This response to the extracellular environment is mediated by extracytoplasmic function (ECF) sigma factors and membrane-bound anti-sigma factors (4, 5, 21). Binding of the iron ligand to its cognate outer membrane receptor releases the anti-sigma factor-mediated inhibition of the ECF sigma factor, inducing transcription of the target operons (18). The TonB-ExbB-ExbD complex is also required for transmembrane signaling. All the outer membrane receptors known to be regulated in this manner have a 50- to 100-amino-acid N-terminal regulatory extension beyond the TonB box; this extension is essential for sigma activation (32).
The Serratia marcescens hemophore-dependent heme-acquisition system (Has) is subject to this type of regulation (31). The Has system consists of a Fur-regulated operon encoding HasR, the hemophore-specific outer membrane receptor, HasA, the hemophore, and HasD and HasE, the specific inner membrane hemophore secretion proteins (16). The final gene of the has operon, hasB, encodes a TonB homolog involved in heme uptake via the Has system (29). Two genes immediately upstream of the has operon encode HasI, an ECF sigma factor, and HasS, an anti-sigma factor (31).
The heme-loaded hemophore is a globular 19-kDa protein with H32 and Y75 double iron coordination. The H83 residue is an alternative ligand of heme iron (24). The seven β strands form an antiparallel β-sheet on one face of the protein, and the four α helices are positioned on the other face (1).
The hemophore receptor, HasR, resembles other outer membrane receptors that are dependent on TonB for energy. It is predicted to have a similar three-dimensional structure, with a plug and a β barrel (26). HasR also functions as a heme receptor, taking up free and hemoglobin-bound heme, albeit less efficiently than it takes up hemophore-bound heme (16). HasR contains two histidine residues (H189 and H603) that have been shown to be conserved in several heme receptors (8) and to be required for free and hemophore-bound heme uptake (F. Huché, N. Izadi-Pruneyre, A. Lecroisey, R. Gilli, C. Wandersman, and P. Delepelaire, unpublished data). HasR binds free heme with a Kd around 1 μM. It binds apo- and holo-hemophores at the same or overlapping sites with similar apparent Kd values (5 nM) (24). This interaction is mediated by two independent hemophore-binding regions, β strands 51-60 and 95-105 (22). The binding of heme or hemophore and the transfer of heme from the hemophore to the receptor are energy independent. In contrast, heme transport and the release of heme-free hemophore require energy, with hemophore release demanding the most energy (23).
The has operon is induced by a heme-loaded hemophore (holo-HasA). This signaling process is dependent both on HasR and on HasI and HasS (31) and has been fully reconstituted in Escherichia coli (7). Neither heme nor apo-HasA can induce hasR expression, raising questions about the nature of the induction stimulus (7).
We used a collection of HasR and HasA mutants and showed that induction may result from the concomitant addition of two molecules that do not interact directly, heme and a mutant hemophore that does not bind heme. The apo-hemophore region required for induction is a five-amino-acid region located between positions 50 and 55 in one of the two HasR-binding regions of the hemophore.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotype or relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| C600 | F−thr leu fhuA lacY thi supE | Laboratory collection |
| XL10 Gold ultracompetent cells | Tetr Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac Hte [F′ proAB lacIqZΔM15 Tn10(Tetr) Amy Camr] | Laboratory collection |
| POP3 | araD139 ΔlacU169 rpsl relA thi | Laboratory collection |
| POP3BF1 | POP3 fur::Cm attλ::hasR-lacZ (Ampr Cmr) | 7 |
| POP3 hasR-lacZ hemA(pAMhasISR) | POP3 attλ::hasR-lacZ ΔhemA::Km pAMhasISR (Ampr Kmr Spcr) | This study |
| POP3 hasR-lacZ hemA fur(pAMhasISR) | POP3 attλ::hasR-lacZ ΔhemA::Km fur::Cm pAMhasISR (Ampr Kmr Cmr Spcr) | This study |
| Plasmids | ||
| pAMrbshasISR | pAM238 carrying hasI, hasS, and hasR, Spcr | 7 |
| pAMrbshasISR Δ45-125 | pAM238 carrying hasI, hasS, and hasR Δ45-125, Spcr | 7 |
| pACYC-hasDE | pACYC184 E. coli cloning vector carrying hasDE, Cmr | 24 |
| X134pAM | pAM238 carrying hasA, Spcr | 25 |
| H32A pAM | pAM238 carrying hasAH32A, Spcr | 24 |
| Y75A pAM | pAM238 carrying hasAY75A, Spcr | 24 |
| 76T pAM | pAM238 carrying hasA76T, Spcr | 22 |
| S51P pAM | pAM238 carrying hasAS51P, Spcr | 22 |
| S51A pAM | pAM238 carrying hasAS51A, Spcr | 22 |
| G95V pAM | pAM238 carrying hasAG95V, Spcr | 22 |
| T20 pAM | pAM238 carrying hasAT20, Spcr | 22 |
| T46 pAM | pAM238 carrying hasAT46, Spcr | 22 |
| T53 pAM | pAM238 carrying hasAT53, Spcr | 22 |
| T55 pAM | pAM238 carrying hasAT55, Spcr | 22 |
| T58 pAM | pAM238 carrying hasAT58, Spcr | 22 |
| T59 pAM | pAM238 carrying hasAT59, Spcr | 22 |
| T68 pAM | pAM238 carrying hasAT68, Spcr | 22 |
| T99 pAM | pAM238 carrying hasAT99, Spcr | 22 |
| T107 pAM | pAM238 carrying hasAT107, Spcr | 22 |
| T181 pAM | pAM238 carrying hasAT181, Spcr | 22 |
| H32AY75AH83A pAM | pAM238 carrying hasAH32AY75AH83A, Spcr | 24 |
| pBAD24-hasR | pBAD24 cloning vector carrying hasR, Ampr | 22 |
| pBAD24-hasRH1H2 | pBAD24 cloning vector carrying hasR containing His 189-to-Ala and His603-to-Ala mutations, Ampr | Laboratory collection |
| pAMrbshasISRH1H2 | pAM238 carrying hasI, hasS, and hasR containing His 189-to-Ala and His603-to-Ala mutations, Spcr | This study |
| H32AY75AH83AG95V pAM | pAM238 carrying hasAH32AY75AH83AG95V, Spcr | This study |
| H32AY75AH83AS51P pAM | pAM238 carrying hasAH32AY75AH83AS51P, Spcr | This study |
| T49 pAM | pAM238 carrying hasAT49, Spcr | This study |
| T50 pAM | pAM238 carrying hasAT50, Spcr | This study |
| T56 pAM | pAM238 carrying hasAT56, Spcr | This study |
| G95P pAM | pAM238 carrying hasAG95P, Spcr | This study |
Media and growth conditions.
Bovine hemoglobin, δ-aminolevulinic acid, and 2,2′dipyridyl were obtained from Sigma. The hemoglobin concentration was calculated based on the levels of heme monomer. Hemoglobin and δ-aminolevulinic acid solutions were filter sterilized by using filters with 0.45-μm pores. Bacteria were grown aerobically at 37°C in LB rich medium or in M63B1 or M9 minimal medium (28) supplemented with 0.4% glucose or 0.4% glycerol as a source of carbon. When required, ammonium iron(III) citrate was added to a final concentration of 5 μM. The iron chelator 2,2′dipyridyl was used at a final concentration of 0.2 mM, and δ-aminolevulinic acid was used at a final concentration of 20 μg · ml−1. Antibiotics were added to the following final concentrations: ampicillin, 50 μg · ml−1; spectinomycin, 100 μg · ml−1; chloramphenicol, 20 μg · ml−1; and kanamycin, 25 μg · ml−1. Solid media contained 1.5% Difco agar.
Genetic techniques.
Cells were transformed by the calcium chloride method (27) or by electroporation (14).
DNA manipulation and molecular cloning.
Large-scale plasmid DNA preparations were obtained with a QIAfilter plasmid midi kit (QIAGEN Inc.), used according to the manufacturer's instructions. Small-scale plasmid DNA preparations were obtained with a QIAprep spin miniprep kit (QIAGEN Inc.), used according to the manufacturer's instructions. Restriction, modification, and ligation were carried out according to the recommendations of the kit manufacturers. Nucleotide sequencing was performed by Genome Express.
Site-directed mutagenesis.
Site-directed mutagenesis was carried out with a Stratagene QuickChange XL site-directed mutagenesis kit (catalog no. 200516) and was verified by sequencing. Mutants T49, T50, T56, G95P, H32AY75AH83AS51P, and H32AY75AH83AG95V were generated in pAM238-hasA by using the primers described in Table 2. The pentapeptide insertions generated in all the HasA mutants used are described in Table 3.
TABLE 2.
Primers used for site-directed mutagenesis
| Mutation | Forward primer | Reverse primer |
|---|---|---|
| H32AY75AH83AG95V | 5′-ATTCCCTGTCCTTCGTCGACGGTTGAGCGGT-3′ | 5′-ACCGCTCAAACCGTCGACGAAGGACAGGGAATC-3′ |
| H32AY75AH83AS5P | 5′-TTCTATGGCGGCAGCCTGCCCGGTAGTCAGTA TGCC-3′ | 5′-ATGGCATACTGACTACCGGGCAGGCTGCCGCC ATAG-3′ |
| T50 | 5′-TTCTATGGCGGCAGCCTGGGGTACCCCAGTATGTCCGGTAGTCAGTATGCC-3′ | 5′-ATACTGACTACCGGACATACTGGGGTACCCCAGGCTGCCGCCATAGAAGCC-3′ |
| T56 | 5′-TCCGGTAGTCAGTATGCCGGGTACCCCACTATGATCAGTAGCACGGCAAACC-3′ | 5′-TTTGCCGTGCTACTGATCATACTGGGGTACCCGGCATACTGACTACCGG-3′ |
| T49 | 5′-TTCTATGGCGGCAGCGGGTACCCCAGTATGCTGTCCGGTAGTCAGTATGCC-3′ | 5′-ATACTGACTACCGGACAGCATACTGGGGTACCCGCTGCCGCCATAGAAGCC-3′ |
| G95P | 5′-ATTCCCTGTCCTTCCCCGACGGTTTGAGCGGTG-3′ | 5′-ACCGCTCAAACCGTCGGGGAAGGACAGGGAATC-3′ |
TABLE 3.
Pentapeptides inserted in the HasA mutant formsa
| Name of insertion | Pentapeptide inserted |
|---|---|
| T20 (in vivo) | L20-GYPYL-G21 |
| T46 (in vivo) | Y46-GGYPY-G47 |
| T49 (in vitro) | S49-GYPSM-L50 |
| T50 (in vitro) | L50-GYPSM-S51 |
| T53 (in vivo) | S53-RGTPS-Q54 |
| T55 (in vivo) | Y55-GVPQY-A56 |
| T56 (in vitro) | A56-GYPSM-I57 |
| T58 (in vivo) | S58-RGTPS-S59 |
| T59 (in vivo) | S59-RGTPS-T60 |
| T68 (in vitro) | V68-GYPSM-A69 |
| T99 (in vitro) | S99-GYPSM-G100 |
| T107 (in vitro) | S107-GYPSM-I108 |
| T181 (in vivo) | D181-RGTPD-S182 |
The numbers indicate the position of the last amino acid preceding the pentapeptide insertion. Mutants obtained with the Tn4430 transposon are indicated by “in vivo” (22); “in vitro” indicates the mutants obtained by site-directed mutagenesis leading to a pentapeptide insertion.
Plasmid construction.
We constructed pAMrbshasISRH1H2 by replacing the mutant copy of the hasR gene (D1, a large part of the hasR sequence deleted) in pAMrbshasISRD1 with the hasR gene of interest (H1H2). The donor plasmid pBAD24hasRH1H2 was cut with FseI and KpnI, which cleaved the ends of the hasR gene. The FseI-KpnI fragment containing hasRH1H2 was then purified and ligated with the recipient plasmid, pAMrbshasISRD1, which was previously digested with FseI and KpnI. The resulting construct was used to transform cells, and clones containing the pAM recombinant plasmid (pAMhasISRH1H2) were selected on plates containing spectinomycin and screened in digestion experiments.
Electrophoresis and immunological techniques.
Proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by Coomassie blue staining. Dot blot HasA binding assays were performed with cultures of C600(pBAD24) and C600(pFR2hasR), as previously described (24). Briefly, aliquots (50 μl; approximately 107 CFU) of washed cell pellets were applied to nitrocellulose filters. Dot blotting with serial dilutions of wild-type or mutant apo-HasA proteins or no HasA was performed as described by Letoffe et al. (25).
Preparation of apo-HasA proteins and loading with heme.
Apo-HasA proteins were obtained from the culture supernatant of strain POP3(pACYC184-hasDE, pAM 238-hasA) grown at 30°C in M9 glycerol medium supplemented with 5 μM ammonium iron(III) citrate. The supernatant was collected and concentrated by precipitation with 65% ammonium sulfate. A 5- to 10-ml aliquot of the sample was then extensively dialyzed in a cassette (Slide-A-Lyser dialysis cassette [extra strength] from Pierce) against 1 liter of TN buffer (50 mM Tris-HCl [pH 7.5], 100 mM NaCl) containing protease inhibitors (Complete EDTA-free protease inhibitor cocktail tablets from Roche). Aliquots of the dialyzed preparation were frozen and stored at −20°C. The heme content of the protein, as determined from absorbance at the Soret band wavelength, was less than 0.5% in all cases. The purity of protein preparations was more than 99%, as shown by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Apo-HasA was loaded with heme by mixing 1 ml of 5 × 10−6 M apo-HasA in TN buffer with 1 ml of 4.5 × 10−6 M heme in TN buffer and incubating the preparation for 15 min at room temperature. Holo-HasA heme loading was determined based on Soret band intensity.
β-Galactosidase assay.
β-Galactosidase was assayed as described by Pardee et al. (30).
RESULTS
Heme transfer from HasA to HasR is required for induction of the has signaling cascade. (i) Study of a HasA mutant affected in heme release.
One mutant from our pentapeptide insertion mutant collection, HasA 76T, had the following properties: pentapeptide insertion between residues 76 and 77 (Fig. 1), heme binding with a Kd of 10−9 M (the Kd for wild-type HasA was 10−11 M), and binding to the receptor with an affinity similar to that of the wild-type protein (22). These properties and the results of a circular dichroism analysis suggested that the mutant and wild-type proteins had similar conformations (data not shown). However, the mutant was unable to supply heme to cells expressing HasR. Also, it displayed no heme transfer from holo-HasA 76T to apo-HasR “in vitro” (Huché, unpublished data). The mutation therefore seemed to involve heme release. All our studies of hasR-lacZ induction were carried out in E. coli strain POP3BF1 (POP3 fur::Cm attλ::hasR-lacZ) and its derivatives. Addition of holo-HasA 76T to strain POP3BF1(pAMrbshasISR) in M63 glucose medium did not induce expression of the hasR-lacZ fusion, whereas a holo-HasA wild-type hemophore induced expression of the fusion by a factor of about 15, as previously reported (7) (Fig. 2). The heme-loading status of holo-HasA 76T was monitored during the incubation; unlike wild-type holo-HasA, the mutant protein continued to display 100% heme loading throughout the experiment (data not shown). HasA 76T had a lower affinity for heme than the wild-type protein, and the insertion affected the heme pocket. We therefore evaluated the induction activities of heme pocket mutants with affinities for heme of 10−11 M (wild-type HasA and HasAH32A), 5 × 10−9 M (HasAY75A), and 10−7 M (HasAH32AY75A) (24). These mutants supply heme to HasR-expressing cells (24). Mutant proteins loaded with heme were added to strain POP3BF1(pAMrbshasISR). All the heme pocket mutants tested induced the hasR-lacZ fusion about 30% less efficiently than the wild type, implying that the heme pocket per se is not involved in hasR induction (Fig. 2).
FIG. 1.
Location of single amino acid change (yellow) and insertion site of the pentapeptide (purple) in the three-dimensional structure of HasA.
FIG. 2.
Effect of HasA mutants, affected in heme binding or release, on hasR-lacZ expression. Strain POP3BF1(pAMrbshasISR) was grown in M63B1 glucose medium in the presence and absence of the heme-loaded form of wild-type or mutant hemophores at a concentration of 10−7 M. β-Galactosidase activity was assayed as described in Materials and Methods. The β-galactosidase activities are means of at least three experiments; the error bars indicate one standard deviation. WT, wild type.
(ii) HasR mutant study.
We used a HasR mutant unable to use free or hemophore-bound heme. This mutant carries the mutations H189A and H603A (H1H2) and has no affinity for heme (Huché et al., unpublished). E. coli strain POP3BF1 was transformed with pAMrbshasISR (a plasmid containing wild-type hasR) or pAMrbshasISRH1H2 (encoding the HasR mutant protein). Production of the mutant protein was assessed by immunodetection with anti-HasR antibodies (data not shown). Similar amounts of the mutant and wild-type proteins were produced. The presence of the mutant at the cell surface and its reactivity with HasA were confirmed by dot blot assays with whole cells (data not shown). Strains POP3BF1(pAMrbshasISR) and POP3BF1(pAMrbshasISRH1H2) were grown in M63 glucose medium. They displayed β-galactosidase activity at subinduction levels. Holo-HasA increased hasR-lacZ expression by a factor of about 15 with the wild-type HasR receptor (Fig. 3), as previously reported (7). In contrast, the hasR-lacZ fusion was not induced following the addition of holo-HasA to cells producing HasRH1H2. Heme release from the hemophore was measured by determining the decrease in the cell culture supernatant holo-HasA Soret band intensity during the course of the induction experiment. Holo-hemophore incubated with the strain expressing the wild-type receptor was entirely unloaded, but holo-hemophore incubated with the strain expressing the mutant receptor displayed 100% loading throughout the experiment (data not shown).
FIG. 3.
Effect of heme transfer from the hemophore to the receptor on hasR-lacZ expression. Strain POP3BF1(pAMrbshasISR) and a derivative of this strain containing a mutant copy of hasR(H1-H2) were grown in M63B1 glucose medium in the presence and absence of wild-type heme-loaded HasA (holo-HasA) at a concentration of 10−7 M. β-Galactosidase activity was assayed as described in Materials and Methods. The β-galactosidase activities are means of at least three experiments; the error bars indicate one standard deviation. WT, wild type.
This set of experiments with a HasA mutant unable to deliver heme and a HasR mutant unable to accept heme showed that holo-hemophore binding to HasR is not sufficient for induction; heme must reach the receptor for initiation of the has signaling cascade. However, heme alone is not sufficient for induction, suggesting that all or part of the HasA protein is also required for induction. We therefore tried to identify the hemophore regions required for hasR induction.
Identification of hemophore regions required for hasR induction.
The interaction between HasA and HasR involves two HasA regions, a region between positions 51 and 60 and a region between positions 95 and 105. Single mutants S51P and G95V both bound HasR with high affinity, whereas the double mutant had no measurable affinity. All these mutants bound heme with high affinity and had the same overall structure as the wild type (22).
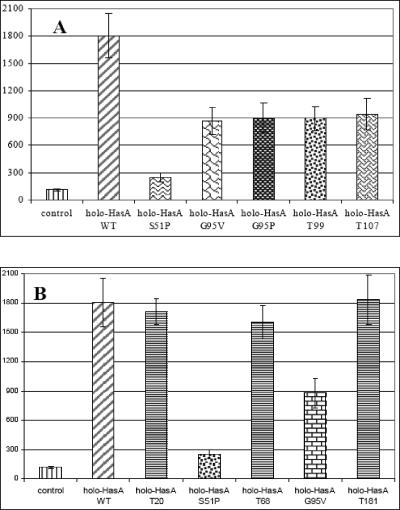
We assessed the ability of the two previously described single mutants, S51P and G95V (22), to induce hasR expression. Holo-HasA S51P and holo-HasA G95V were added to the test strain POP3BF1(pAMrbshasISR). Addition of holo-HasA G95V induced expression of the hasR-lacZ fusion about sevenfold. Holo-HasA S51P induced hasR-lacZ expression by a factor of two (Fig. 4A). Other proteins with single changes in the two HasA binding regions were also tested, and holo-HasA S51A (not affected in HasA binding to HasR) fully induced the hasR-lacZ fusion (data not shown). Holo-HasA G95P induced the hasR-lacZ fusion to an extent similar to the extent observed with holo-HasA G95V (Fig. 4A). Thus, the binding region lying between residues 95 and 105 can be deformed by a proline without significantly changing the HasA induction properties.
FIG. 4.
Role of the HasR-binding regions of HasA in hasR-lacZ induction. Strain POP3BF1(pAMrbshasISR) was grown in M63B1 glucose medium in the presence and absence of heme-loaded wild-type or variant hemophores having mutations in the binding regions of HasA (A) or all along HasA (B) at a concentration of 10−7 M. β-Galactosidase activity was assayed as described in Materials and Methods. The β-galactosidase activities are means of at least three experiments; the error bars indicate one standard deviation. WT, wild type.
Two other mutants from the laboratory collection with pentapeptide insertions near residues 95 and 105 (Table 3), HasAT99 and HasAT107, (22), were tested for hasR induction. Both of these mutants induced the hasR-lacZ fusion to an extent similar to the extent observed with holo-HasA G95V (Fig. 4A). The binding region of HasA between residues 51 and 60 is therefore clearly more important for induction than the binding region between residues 95 and 105 is.
Other HasA mutants carrying pentapeptide insertions at various positions in the HasA protein outside the two binding regions (positions 51 to 60 and 95 to 107) were also loaded with heme and tested. All of these mutants induced hasR to an extent similar to the extent observed with wild-type holo-HasA (Fig. 4B), indicating the absence of significant involvement of any other HasA region in the initiation of signal transduction.
Scanning the 51-60 binding region for induction properties with pentapeptide insertion mutants.
The failure of the HasA S51P mutant to induce hasR-lacZ suggested that the region between residues 51 and 60 in HasA is important for the initiation of has signaling.
We studied a collection of mutants (22) obtained by insertion of a pentapeptide into this region (see Materials and Methods) for precise identification of the residues important for holo-HasA-dependent hasR-lacZ induction. These mutant proteins were loaded with heme and tested for hasR-lacZ induction in the test strain. Little or no induction of the hasR-lacZ fusion was observed with holo-HasA T50, holo-HasA T53, and holo-HasA T55. In contrast, holo-HasA T46, holo-HasA T49, holo-HasA T56, holo-HasA T58, and holo-HasA T59 all strongly induced expression of the hasR-lacZ fusion (Fig. 5). Holo-HasA T46 and holo-HasA T49 induced hasR-lacZ expression to an extent similar to the extent observed with the wild type, indicating that the region upstream of residue 50 is not required for hasR induction. Pentapeptide scanning analysis therefore implicated residues 50 to 55 of the 51-60 β-strand in the initiation of has signaling.
FIG. 5.
Effect of pentapeptide insertions in HasA binding region 51-60 on hasR-lacZ expression. Strain POP3BF1(pAMrbshasISR) was grown in M63B1 glucose medium in the presence and absence of heme-loaded wild-type or mutant HasA hemophore at a concentration of 10−7 M. β-Galactosidase activity was assayed as described in Materials and Methods. The β-galactosidase activities are means of at least three experiments; the error bars indicate one standard deviation. WT, wild type.
Noninteracting heme and HasA are able to initiate the has signaling cascade.
Both heme delivery and residues 50 to 55 of the 51-60 binding region of HasA were required for hasR-lacZ induction. We investigated whether these two induction factors could act separately by determining whether supplying free heme with HasA led to hasR induction. To prevent heme binding to HasA, a HasA H32AY75AH83A triple mutant unable to bind heme was used.
Addition of HasA H32AY75AH83A or heme alone had no effect on hasR-lacZ expression in our test strain (Fig. 6). Full hasR-lacZ expression, as observed with wild-type holo-HasA, was induced in the presence of both heme and HasA H32AY75AH83A. The has signaling cascade may therefore be initiated in two ways: by holo-HasA and heme transfer to HasR or by concomitant addition of heme and the mutant hemophore unable to interact with heme. Heme binds to HasR on cells loaded with HasA H32AY75AH83A, inducing hasR-lacZ.
FIG. 6.
Induction of hasR-lacZ by two noninteracting molecules, either heme and triple mutant HasA H32AY75AH83A, heme and quadruple mutant HasA H32AY75AH83AS51P, or heme and quadruple mutant HasA H32AY75AH83AG95V. Strain POP3BF1(pAMrbshasISR) was grown in M63B1 glucose medium in the presence and absence of heme-loaded HasA (holo-HasA) at a concentration of 10−7 M. β-Galactosidase activity was assayed as described in Materials and Methods. The β-galactosidase activities are means of at least three experiments; the error bars indicate one standard deviation. Bar A, wild-type holo-HasA at a concentration 10−7 M; bar B, heme at a concentration of 10−5 M; bar C, apo-HasA H32AY75AH83A at a concentration of 10−7M; bar D, apo-HasA H32AY75AH83A at a concentration of 10−7 M plus heme at a concentration of 10−7 M; bar E, apo-HasA H32AY75AH83AG95V at a concentration of 10−7 M; bar F, apo-HasA H32AY75AH83AG95V at a concentration of 10−7 M plus heme at a concentration of 10−7 M; bar G, apo-HasA H32AY75AH83AS51P at a concentration of 10−7 M; bar H, apo-HasA H32AY75AH83AS51P at a concentration of 10−7 M plus heme at a concentration of 10−7 M.
HasA H32AY75AH83A has been used to study heme uptake via HasR (23). HasA H32AY75AH83A does not bind heme and blocks free heme uptake (23). We compared the abilities of the wild-type and H32AY75AH83A hemophores to block heme uptake in the POP3 hasR-lacZ hemA(pAMrbshasISR) strain and its fur derivative. The H32AY75AH83A hemophore blocked heme uptake in both POP3 hasR-lacZ hemA(pAMrbshasISR) and its fur derivative. The H32AY75AH83A hemophore can therefore induce hasR expression while blocking heme uptake; heme transport is not required for hasR induction. Like HasA H32AY75AH83A, HasA 76T blocks heme transport. However, unlike HasA H32AY75AH83A, HasA 76T does not induce the hasR-lacZ fusion in the presence of heme.
HasA regions involved in induction in the presence of separately provided heme and hemophore.
We introduced S51P and G95V mutations separately into HasA H32AY75AH83A to investigate the involvement of HasA binding regions 51-60 and 95-105 in the induction of has signaling in the presence of noninteracting heme and hemophore. The HasA quadruple mutants H32AY75AH83AS51P and H32AY75AH83AG95V were tested for hasR-lacZ induction (Fig. 6). Neither 10−5 M heme nor 10−7 M apo quadruple mutant supplied separately induced hasR. Concomitant addition of HasA H32AY75AH83AG95V and heme induced hasR by a factor of five, whereas concomitant addition of HasA H32AY75AH83AS51P and heme did not induce hasR expression. In both cases, the amounts of the mutant proteins were similar to the amounts of the wild-type hemophore in controls at the end of the incubation period, as shown by immunodetection (data not shown). The quadruple mutant HasA H32AY75AH83AS51P interacted normally with HasR, as shown by dot blotting (data not shown). Thus, residue 51 is also required for hasR induction in the absence of an interaction between heme and the hemophore. The same region (region 50-55) of HasA is probably required for induction by holo-HasA and for induction by noninteracting, separately supplied heme and hemophore.
DISCUSSION
Expression of the has operon of S. marcescens, which allows heme acquisition, is positively regulated by a transmembrane signaling cascade. This cascade is initiated by the binding of heme-loaded hemophore to HasR, its cognate receptor, which leads to specific sigma factor activation. HasR has a number of ligands, heme, apo-hemophore, and holo-hemophore, but only holo-hemophore initiates the signaling cascade. This raises questions concerning the nature of the inducing stimulus. This stimulus may be a particular holo-HasA conformation. Indeed, the three-dimensional structures of holo-HasA and apo-HasA indicate that heme binding to HasA substantially modifies the conformation of the heme pocket, without changing the α-β fold of the protein (1) (PDB accession number PDB ID 1YBJ). Alternatively, induction may result from the extraction of heme from HasA, heme landing on HasR, or empty hemophore recycling.
We used two types of mutants blocked in heme transfer from HasA to HasR: HasA mutants unable to deliver heme and a HasR mutant unable to receive heme. Both types of mutation abolish holo-HasA-mediated hasR induction. Thus, no particular holo-HasA conformation acts as the stimulus for induction. Interaction between heme and the receptor is necessary for induction. However, no induction was observed with heme alone, suggesting that induction requires either heme transfer or the concomitant presence of heme and apo-HasA on the receptor. The addition of heme together with a HasA mutant unable to bind heme (HasA H32AY75AH83A) led to full hasR induction, demonstrating that a direct interaction between heme and the hemophore is not required to trigger induction. Thus, the extraction of heme from the hemophore is not the stimulus required for induction.
Our findings also indicate that HasA H32AY75AH83A binding to HasR does not prevent heme from reaching HasR. However, HasA H32AY75AH83A binding to the receptor blocks free heme uptake (23). Similar results were obtained with a hemA isogenic mutant of POP3BF1(pAMhasISR), the strain used for the induction experiments (data not shown). This induction by heme in the presence of HasA H32AY75AH83A led us to reject our initial hypothesis that HasA H32AY75AH83A prevents heme from gaining access to HasR (23). Purified heme-free HasA H32AY75AH83A-HasR complexes bind heme in vitro, confirming that heme can reach a hemophore-loaded receptor (N. Izadi, personal communication). HasA H32AY75AH83A may remain on the receptor, blocking heme transport but not heme binding. The mechanism by which heme transport is inhibited remains unclear. It is possible that the wild-type hemophore dissociates from the receptor after its heme is transferred, whereas no such dissociation is observed with a mixture of heme and the H32AY75AH83A mutant hemophore. Hemophore recycling may require heme transfer from the hemophore to the receptor. This process is not possible for HasA molecules not loaded with heme. The induction-incompetent HasA 76T molecule, which cannot deliver its heme, also inhibits heme uptake. These findings are consistent with the hypothesis that heme transfer is necessary for hemophore recycling. Alternatively, a small amount of heme may reach HasR in the presence of HasA H32AY75AH83A, and this may be sufficient for induction but not growth.
Whatever the mechanism of inhibition, this study demonstrated that induction and transport are distinct mechanisms, as they are in several other systems (12, 20).
The hemophore (plus heme) is required for induction, implicating HasR-binding regions of HasA (S51 to Y60 and G95 to P105) in induction. Mutations affecting the region between residues 95 and 105 of HasA halved induction. Mutations in the 50-55 sequence of HasA abolished induction. Mutation between Y55 and T60 did not decrease induction but modified the binding properties of HasA (22). Thus, only one half of the 51-60 binding region, corresponding to a bend, is required for induction. We investigated the roles of the two HasA binding regions in the presence of independently supplied heme and apo-HasA, using the quadruple mutants HasA H32AY75AH83AS51P and HasA H32AY75AH83AG95V. These mutants have mutations in one binding region and are unable to bind heme. HasA H32AY75AH83AS51P did not induce hasR in the presence of heme, demonstrating that the 51-60 binding region of HasA is important for induction both by holo-HasA and by heme plus apo-HasA. A synthetic peptide corresponding to the β strand between residues 51 and 60 competed with the corresponding HasA region for binding to the receptor (22). Nevertheless, concomitant addition of heme and this HasA peptide did not induce hasR (data not shown). This suggests that the 51-60 region adopts a particular conformation for induction that is different from the conformation required for binding and that the synthetic peptide does not adopt this conformation. The slightly-weaker-than-wild-type induction by HasA proteins carrying mutations in the 95-105 binding site is consistent with the hypothesis that HasA protein integrity is required to ensure the correct conformation of residues 50 to 55.
We broke the induction stimulus down into two elements, which may correspond to physiological steps in heme delivery. Studies with purified HasA-HasR complexes have shown that heme transfer in vitro requires neither an energy supply nor complex dissociation (Huché et al., unpublished). HasA/HasR complex dissociation does not occur in vitro, but it is the most energy-demanding step in vivo. Thus, heme is transferred from HasA to HasR before release of the apo-hemophore from the receptor, and this is the signal perceived by the bacterium. TonB is required for hasR induction, acting at and playing a role in a subsequent step. According to the FecA model, ligand binding generates a TonB-dependent conformational change in the receptor regulatory region, releasing the sigma factor from anti-sigma factor inhibition (12).
The bipartite regulation of the has system allows the bacterium to perceive the environmental heme concentration and to adjust its heme transport mechanisms accordingly. Under iron limitation conditions and in the absence of heme, S. marcescens produces basal levels of HasR and HasA, which form complexes at the cell surface. This may prevent hemophore dilution and inactivation in the surrounding medium. In the presence of low concentrations of heme, preformed HasA-HasR complexes switch the system on, facilitating heme scavenging at nanomolar concentrations. When the relative heme/HasA ratio is high, such as during growth in the bloodstream, heme transport leads to apo-hemophore recycling, with free heme binding more frequently than loaded hemophores to receptors. This binding of free heme does not induce the signaling cascade. Heme is taken up via residual HasR receptors or by other S. marcescens heme uptake systems induced in iron-chelated medium.
Like HasA, several iron ligands (ferric dicitrate, pyochelin, and pyoverdin) (19, 34) bind to their specific outer membrane receptors as iron-free molecules. Other ligands are positively regulated by transmembrane signaling mechanisms, like that of the canonical Fec system (11). For citrate, as for HasA, only the iron-loaded form triggers the signal (11, 31). Crystal structures of two siderophore receptors have shown that very different conformational changes are induced by the binding of apo- and holo-siderophores (34). However, the iron carried by the siderophore is not bound directly to the receptor, and the loaded molecule is transported as a whole. Thus, for siderophore-dependent iron transport, the metal ion and its organic chelator do not have different functions in initiation of the signaling cascade.
Acknowledgments
We thank Sylvie Letoffe, Philippe Delepelaire, and Jean-Marc Ghigo for helpful practical and conceptual discussions and for critical reading of the manuscript.
REFERENCES
- 1.Arnoux, P., R. Haser, N. Izadi, A. Lecroisey, M. Delepierre, C. Wandersman, and M. Czjzek. 1999. The crystal structure of HasA, a hemophore secreted by Serratia marcescens. Nat. Struct. Biol. 6:516-520. [DOI] [PubMed] [Google Scholar]
- 2.Bagg, A., and J. B. Neilands. 1987. Molecular mechanism of regulation of siderophore-mediated iron assimilation. Microbiol. Rev. 51:509-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baichoo, N., and J. D. Helmann. 2002. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J. Bacteriol. 184:5826-5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bashyam, M. D., and S. E. Hasnain. 2004. The extracytoplasmic function sigma factors: role in bacterial pathogenesis. Infect. Genet. Evol. 4:301-308. [DOI] [PubMed] [Google Scholar]
- 5.Beare, P. A., R. J. For, L. W. Martin, and I. L. Lamont. 2003. Siderophore-mediated cell signalling in Pseudomonas aeruginosa: divergent pathways regulate virulence factor production and siderophore receptor synthesis. Mol. Microbiol. 47:195-207. [DOI] [PubMed] [Google Scholar]
- 6.Bell, P. E., C. D. Nau, J. T. Brown, J. Konisky, and R. J. Kadner. 1990. Genetic suppression demonstrates interaction of TonB protein with outer membrane transport proteins in Escherichia coli. J. Bacteriol. 172:3826-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biville, F., H. Cwerman, S. Letoffe, M. S. Rossi, V. Drouet, J. M. Ghigo, and C. Wandersman. 2004. Haemophore-mediated signalling in Serratia marcescens: a new mode of regulation for an extra cytoplasmic function (ECF) sigma factor involved in haem acquisition. Mol. Microbiol. 53:1267-1277. [DOI] [PubMed] [Google Scholar]
- 8.Bracken, C. S., M. T. Baer, A. Abdur-Rashid, W. Helms, and I. Stojiljkovic. 1999. Use of heme-protein complexes by the Yersinia enterocolitica HemR receptor: histidine residues are essential for receptor function. J. Bacteriol. 181:6063-6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braun, V. 1997. Surface signaling: novel transcription initiation mechanism starting from the cell surface. [Review.] Arch. Microbiol. 167:325-331. [DOI] [PubMed] [Google Scholar]
- 10.Braun, V., and H. Killmann. 1999. Bacterial solutions to the iron-supply problem. Trends Biochem. Sci. 24:104-109.10203757 [Google Scholar]
- 11.Braun, V., and S. Mahren. 2005. Transmembrane transcriptional control (surface signalling) of the Escherichia coli Fec type. FEMS Microbiol. Rev. 29:673-684. [DOI] [PubMed] [Google Scholar]
- 12.Braun, V., S. Mahren, and M. Ogierman. 2003. Regulation of the FecI-type ECF sigma factor by transmembrane signalling. Curr. Opin. Microbiol. 6:173-180. [DOI] [PubMed] [Google Scholar]
- 13.Crosa, J. H. 1997. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol. Mol. Biol Rev. 61:319-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dower, W. J., J. F. Miller, and C. W. Radgale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferguson, A. D., and J. Deisenhofer. 2004. Metal import through microbial membranes. Cell 116:15-24. [DOI] [PubMed] [Google Scholar]
- 16.Ghigo, J. M., S. Letoffe, and C. Wandersman. 1997. A new type of hemophore-dependent heme acquisition system of Serratia marcescens reconstituted in Escherichia coli. J. Bacteriol. 179:3572-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hantke, K. 2001. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4:172-177. [DOI] [PubMed] [Google Scholar]
- 18.Helmann, J. D. 2002. The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 46:47-110. [DOI] [PubMed] [Google Scholar]
- 19.Hoegy, F., H. Celia, G. L. Mislin, M. Vincent, J. Gallay, and I. J. Schalk. 2005. Binding of iron-free siderophore, a common feature of siderophore outer membrane transporters of Escherichia coli and Pseudomonas aeruginosa. J. Biol. Chem. 280:20222-20230. [DOI] [PubMed] [Google Scholar]
- 20.James, H. E., P. A. Beare, L. W. Martin, and I. L. Lamont. 2005. Mutational analysis of a bifunctional ferrisiderophore receptor and signal-transducing protein from Pseudomonas aeruginosa. J. Bacteriol. 187:4514-4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamont, I. L., P. A. Beare, U. Ochsner, A. I. Vasil, and M. L. Vasil. 2002. Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 99:7072-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Letoffe, S., L. Debarbieux, N. Izadi, P. Delepelaire, and C. Wandersman. 2003. Ligand delivery by haem carrier proteins: the binding of Serratia marcescens haemophore to its outer membrane receptor is mediated by two distinct peptide regions. Mol. Microbiol. 50:77-88. [DOI] [PubMed] [Google Scholar]
- 23.Letoffe, S., P. Delepelaire, and C. Wandersman. 2004. Free and hemophore-bound heme acquisitions through the outer membrane receptor HasR have different requirements for the TonB-ExbB-ExbD complex. J. Bacteriol. 186:4067-4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Letoffe, S., C. Deniau, N. Wolff, E. Dassa, P. Delepelaire, A. Lecroisey, and C. Wandersman. 2001. Haemophore-mediated bacterial haem transport: evidence for a common or overlapping site for haem-free and haem-loaded haemophore on its specific outer membrane receptor. Mol. Microbiol. 41:439-450. [DOI] [PubMed] [Google Scholar]
- 25.Letoffe, S., F. Nato, M. E. Goldberg, and C. Wandersman. 1999. Interactions of HasA, a bacterial haemophore, with haemoglobin and with its outer membrane receptor HasR. Mol. Microbiol. 33:546-555. [DOI] [PubMed] [Google Scholar]
- 26.Letoffe, S., K. Wecker, M. Delepierre, P. Delepelaire, and C. Wandersman. 2005. Activities of the Serratia marcescens heme receptor HasR and isolated plug and beta-barrel domains: the beta-barrel forms a heme-specific channel. J. Bacteriol. 187:4637-4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maniatis, T., E. F. Fritch, and J. Sambrook. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Paquelin, A., J. M. Ghigo, S. Bertin, and C. Wandersman. 2001. Characterization of HasB, a Serratia marcescens TonB-like protein specifically involved in the haemophore-dependent haem acquisition system. Mol. Microbiol. 42:995-1005. [DOI] [PubMed] [Google Scholar]
- 30.Pardee, A. B., F. Jacob, and J. Monod. 1959. The genetic control and cytoplasmic of inducibility in the synthesis of β-galactosidase of Escherichia coli. J. Mol. Biol. 1:165-178. [Google Scholar]
- 31.Rossi, M. S., A. Paquelin, J. M. Ghigo, and C. Wandersman. 2003. Haemophore-mediated signal transduction across the bacterial cell envelope in Serratia marcescens: the inducer and the transported substrate are different molecules. Mol. Microbiol. 48:1467-1480. [DOI] [PubMed] [Google Scholar]
- 32.Schalk, I. J., W. W. Yue, and S. K. Buchanan. 2004. Recognition of iron-free siderophores by TonB-dependent iron transporters. Mol. Microbiol. 54:14-22. [DOI] [PubMed] [Google Scholar]
- 33.Visca, P., L. Leoni, M. J. Wilson, and I. L. Lamont. 2002. Iron transport and regulation, cell signalling and genomics: lessons from Escherichia coli and Pseudomonas. Mol. Microbiol. 45:1177-1190. [DOI] [PubMed] [Google Scholar]
- 34.Yue, W. W., S. Grizot, and S. K. Buchanan. 2003. Structural evidence for iron-free citrate and ferric citrate binding to the TonB-dependent outer membrane transporter FecA. J. Mol. Biol. 332:353-368. [DOI] [PubMed] [Google Scholar]