Abstract
Salmonella spp. are environmentally persistent pathogens that have served as one of the important models for understanding how bacteria adapt to stressful conditions. However, it remains poorly understood how they survive extreme conditions encountered outside their hosts. Here we show that the rdar morphotype, a multicellular phenotype characterized by fimbria- and cellulose-mediated colony pattern formation, enhances the resistance of Salmonella to desiccation. When colonies were stored on plastic for several months in the absence of exogenous nutrients, survival of wild-type cells was increased compared to mutants deficient in fimbriae and/or cellulose production. Differences between strains were further highlighted upon exposure to sodium hypochlorite, as cellulose-deficient strains were 1,000-fold more susceptible. Measurements of gene expression using luciferase reporters indicated that production of thin aggregative fimbriae (Tafi) may initiate formation of colony surface patterns characteristic of the rdar morphotype. We hypothesize that Tafi play a role in the organization of different components of the extracellular matrix. Conservation of the rdar morphotype among pathogenic S. enterica isolates and the survival advantages that it provides collectively suggest that this phenotype could play a role in the transmission of Salmonella between hosts.
The genus Salmonella comprises an ancient and diverse group of organisms that causes disease in, and can be commonly isolated from, a large number of animal hosts. In humans, Salmonella is the etiological agent of gastroenteritis and typhoid fever. The worldwide estimates of annual infection are 1.3 billion cases of gastroenteritis and/or diarrhea due to non-typhoidal salmonellosis, with 3 million deaths, and 16.6 million cases of typhoid fever, with nearly 600,000 deaths (24). One common feature among all Salmonella spp. is that they display enhanced survival in non-host environments, including soil and water (42). This fits into the cyclic lifestyle that has been proposed for Salmonella spp., consisting of passage through a host into the environment and back into a new host (42).
Most Salmonella isolates possess the ability to form patterned, aggregative colonies when grown on surfaces (2, 26). This multicellular phenomenon has been termed the rdar morphotype (colonies are “red, dry, and rough” when grown on media containing Congo red [9, 29]). The rdar morphotype is primarily controlled by expression of the divergent agfDEFG and agfBAC (csgDEFG/BAC) operons encoding thin aggregative fimbriae (Tafi or curli) (7, 16). AgfD is a positive regulator of Tafi production by activating transcription of agfB (16, 47). AgfD also stimulates cellulose production by transcriptional activation of adrA (47). AdrA contains the GGDEF domain involved in the synthesis of cyclic di-GMP, a signaling molecule required for activation of cellulose synthesis (14, 27, 34). Tafi and cellulose, along with additional exopolysaccharides (11, 41), form a recalcitrant extracellular matrix that serves to physically link cells together within a colony. These extracellular factors are produced by Salmonella under nutrient-limiting conditions at low osmolarity and growth temperatures below 30°C (9, 29).
We and others have hypothesized that the rdar morphotype could be important for environmental survival due to the following qualities: the recalcitrant nature of extracellular matrix components (9, 35, 41, 47), characteristic growth conditions (29), and increased resistance to antimicrobial stresses (2, 32, 35). Here, we show that the rdar morphotype enhances Salmonella survival during desiccation in the absence of nutrients. After storage on plastic for 9 months, wild-type cells of S. enterica serovar Typhimurium ATCC 14028 had 10 to 30 times increased survival compared to mutants deficient in Tafi and cellulose production. Increased resistance to desiccation would undoubtedly aid in long-term survival of Salmonella in the environment and promote passage to future hosts.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
S. enterica serovar Typhimurium strain ATCC 14028 was used as the reference strain in this study. Strains from Salmonella reference collection B (SARB) have been described previously (5). Strains were routinely grown for 20 h at 37°C with agitation in 1% tryptone, pH 7.2 (T), or Miller's Luria-Bertani broth (1.0% salt), supplemented with 50 μg/ml kanamycin (Kan), if necessary, before performing additional experiments. For growth of cells on agar, cultures were diluted to an A600 of 1.0, and 1 μl was spotted onto solid T media containing 1.5% Difco agar. For strain-mixing experiments, equal volumes of agfA, agfD, or bcsA culture were mixed before spotting on agar. Cells were grown at room temperature (RT; ∼22°C) or 28°C as indicated. To visualize the production of cellulose, cells were grown on T agar containing 200 μg/ml calcofluor white (fluorescent brightener 28; Sigma-Aldrich Canada). Colony pictures were taken with a Kodak Image Station 2,000MM camera system. For bioluminescence assays, overnight cultures were diluted 1:600 in T broth supplemented with 50 μg/ml Kan to a final volume of 150 μl in 96-well clear-bottom black plates (9520 Costar; Corning Inc.). The culture in each well was overlaid with 50 μl mineral oil prior to starting the assays. Cultures were assayed for luminescence (0.1 s) and absorbance (620 nm, 0.1 s) every 30 min during growth at 28°C with agitation in a Wallac Victor2 (Perkin-Elmer Life Sciences, Boston, Mass.).
Recombinant DNA techniques and construction of lux reporters.
Genomic DNA was prepared following the method of Walsh et al. (38). Promoter-containing regions for agfD, agfB, and adrA were PCR amplified from ATCC 14028 using primers agfD1 (GTGCTCGAGGGACTTCATTAAACATGATG [a XhoI site is underlined]) and agfD2 (GCCGGATCCTGTTTTTCATGCTGTCAC [a BamHI site is underlined]), adrA1 (GACCTCGAGAAGGCGATGACCGTAGCAAG [a XhoI site is underlined]) and adrA2 (GACGGATCCGCCTGTTCAACCGCTTTT [a BamHI site is underlined]). PCR products were purified (QIAGEN Inc.), sequentially digested with XhoI and BamHI (Invitrogen Canada Inc.), and ligated using T4 DNA ligase (Invitrogen Canada Inc.) into pCS26-Pac (XhoI-BamHI) or pU220 (BamHI-XhoI) reporter vectors containing the luxCDABE operon from Photorhabdus luminescens (4). The majority of pCS26 and pU220 cloning was performed in Escherichia coli DH10B (Invitrogen Canada Inc.). All plasmids were transformed into Salmonella strains by electroporation (Bio-Rad Laboratories Inc.). Promoter::lux plasmids were purified from Salmonella (QIAGEN Inc.), and DNA sequencing was performed by Macrogen (Seoul, South Korea) using primers pZE05 and pZE06 (4).
Construction of a sig38H4 reporter designed to measure RpoS activity.
A synthetic, 58-nucleotide, RpoS (σ38)-responsive promoter (CTCGAGATAATTCCATGCGGTTTCGCTAAAATCATGTATACTTATTATCAATTGGATCC; −35 and −10 promoter regions are underlined, XhoI and BamHI restriction sites are in boldface) was generated using overlapping oligonucleotides. Primer clone (GCGCGCTCGAGAATAATTCCATGCGGTTTCGCTAAAATCA) and sig38-reverse (GCGCGGGATCCAATTGATAATAANTATANATGATTTTAGCGAAA) were mixed, denatured at 94°C for 10 min, and allowed to anneal on ice before ligation into XhoI- and BamHI-digested pCS26. Primer sequences were designed based on consensus promoter sequences from E. coli genes that are RpoS dependent (3). The −35 promoter region was designed to be divergent from the RpoD (σ70) consensus sequence and therefore reduce binding of RpoD and subsequent transcription. Luminescence of the sig38H4 reporter in an ΔrpoS mutant strain of S. enterica serovar Typhimurium ATCC 14028 remained near background levels (<8,000 cps) during growth in 1% tryptone (data not shown).
Generation of serovar Typhimurium ATCC 14028 mutant strains.
Deletion constructs for agfA and bcsA generated from serovar Enteritidis 27655-3b DNA and cloned into pHSG415 have been described previously (39, 41). An in-frame deletion removing 612 bp in agfD (encoding amino acids 6 to 210 in AgfD) was generated using PCR primers agfD-A (GACGAATCCGTGTGTTATGCCGCCATGGG [an EcoRI site is underlined]), agfD-B (GGACTGCAGTAAACATGATG [a PstI site is underlined]), agfD-C (GCCCTGCAGCAAACGATAATCTCAGGCGG [a PstI site is underlined]), and agfD-D (GCCAAGCTTTGTCCGTGACGTTGAGCTGG [a HindIII site is underlined]). The two PCR fragments generated were directionally cloned into EcoRI- and HindIII-digested pTZ18R (Amersham Biosciences) and subcloned into pHSG415 (17). Each mutation was introduced into the chromosome of ATCC 14028 by following established procedures (40) with modifications (A. P. White, E. Ambrose, B. W. Jones, R. DeVinney, W. W. Kay, and M. G. Surette, unpublished data). Screening for strains containing the truncated agfD gene was accomplished using primers agfDko1 (CACTTGCTTTAAGATTTGTAATGGC) and agfDko2 (ATTCGCTTTCCCATTTGTCG).
Long-term survival experiments and treatment with sodium hypochlorite.
Cells from 1 μl of ATCC 14028, ΔagfD, ΔagfA, and ΔbcsA overnight cultures were inoculated onto T agar and grown for 6 days at RT. Colonies were removed from the agar surface and tested immediately (initial numbers) or stored in 24-well tissue culture plates (one colony per well) at RT for 3 or 9 months. After rehydration of colonies in 500 μl phosphate-buffered saline (PBS), pH 7.4, for 1 h at RT, 100 μl of sodium hypochlorite solution (to a final concentration of 30 to 300 ppm) or PBS (control) was added and mixed continuously using a rotating microtube mixer for 20 min at RT. Cells and colony materials were centrifuged (7,000 × g, 2 min), and supernatant was removed. Fresh PBS (500 μl) was added, and colony slurries were mixed in a tissue homogenizer until uniform turbidity was reached (∼20 s). For planktonic-grown cells, 20-h cultures in 1% tryptone were normalized to an optical density of 1 at 600 nm, and 1-ml aliquots of cells were pelleted (7,000 × g, 2 min) prior to the treatment. The homogenization step was omitted. To determine the number of viable cells remaining in each sample, cell mixtures were serially diluted in triplicate and plated in duplicate in 5-μl drops.
RESULTS
Prevalence of the rdar morphotype in Salmonella enterica subgroup I strains.
S. enterica subgroup I contains 99% of Salmonella serovars and all major human pathogenic types. To determine the prevalence of the rdar morphotype within S. enterica subgroup I, we analyzed isolates from Salmonella reference collection B (SARB) (5). Although the rdar morphotype was initially described during growth on media containing Congo red, this phenotype can be visualized during growth on standard agar by the formation of colony surface patterns (2). In addition, production of cellulose can be visualized by growing Salmonella on agar containing calcofluor (35). Three distinct phenotypic classes were observed within SARB: 80.5% of isolates formed complete patterns (rdar morphotype), 7% formed incomplete patterns, and 12.5% had no surface patterns (Table 1). For all complete pattern strains, the entire colony could be lifted off the agar surface, indicating that a strong Tafi and cellulose network had formed. Members of this group included host-restricted serovars such as Typhi and Pullorum (Table 1). Colonies from isolates in the incomplete pattern class could not be removed from the agar surface in one piece. Each of these strains produced cellulose, suggesting that production of Tafi or other extracellular matrix components was impaired. Strains without colony surface patterns were not clustered into any particular groups, except for serovar Choleraesuis (four out of four strains) (Table 1). Overall, the majority of S. enterica subgroup I isolates were able to form the rdar morphotype.
TABLE 1.
Prevalence of multicellular pattern formation (rdar morphotype) in Salmonella enterica subgroup I strains (SARB) (5)
| Serovara | No. of strains tested | No. of isolates with indicated phenotypeb
|
|||
|---|---|---|---|---|---|
| Pattern (rdar) | Incomplete pattern | No pattern | Calcofluor bindingc | ||
| Agona | 1 | 1 | 1 | ||
| Anatum | 1 | 1 | 1 | ||
| Brandenburg | 1 | 1 | 1 | ||
| Choleraesuis | 4 | 0 | 4 | 0 | |
| Decatur | 1 | 1 | 1 | ||
| Derby | 3 | 2 | 1 | 3 | |
| Dublin | 3 | 3 | 3 | ||
| Duisburg | 1 | 1 | 1 | ||
| Enteritidis | 4 | 3 | 1 | 4 | |
| Emek | 1 | 1 | 1 | ||
| Gallinarum | 1 | 1 | 1 | ||
| Haifa | 1 | 1 | 1 | ||
| Heidelberg | 2 | 2 | 2 | ||
| Indiana | 1 | 1 | 1 | ||
| Infantis | 2 | 2 | 2 | ||
| Miami | 2 | 2 | 2 | ||
| Montevideo | 2 | 2 | 2 | ||
| Muenchen | 4 | 3 | 1 | 3 | |
| Newport | 3 | 2 | 1 | 2 | |
| Panama | 3 | 3 | 3 | ||
| Paratyphi A | 1 | 1 | 1 | ||
| Paratyphi B | 5 | 1 | 3 | 1 | 4 |
| Paratyphi C | 3 | 2 | 1 | 2 | |
| Pullorum | 2 | 2 | 2 | ||
| Reading | 1 | 1 | 1 | ||
| Rubislaw | 1 | 1 | 1 | ||
| Saintpaul | 3 | 3 | 3 | ||
| Schwarzengrund | 1 | 1 | 1 | ||
| Sendai | 1 | 1 | 1 | ||
| Senftenberg | 1 | 1 | 1 | ||
| Stanley | 1 | 0 | 1 | 0 | |
| Stanleyville | 1 | 1 | 1 | ||
| Typhi | 2 | 2 | 2 | ||
| Typhimurium | 4 | 4 | 4 | ||
| Typhisuis | 2 | 2 | 2 | ||
| Wien | 2 | 2 | 2 | ||
| Total | 72 | 58 | 5 | 9 | 63 |
Strains are described in Boyd et al. (5).
Colony morphology and associated phenotypes were recorded after growth on T agar at 28°C for up to 10 days: pattern (rdar), colonies had complete surface patterns and could be lifted intact from the agar surface; incomplete pattern, colonies could not be lifted from the agar surface; no pattern, colonies were smooth and mucoid.
Fluorescence of colonies grown on T agar supplemented with 200 μg/ml calcofluor was observed under a 366-nm UV light source and compared to ATCC 14028 positive control and ΔbcsA negative control strains.
Real-time pattern formation during formation of the rdar morphotype.
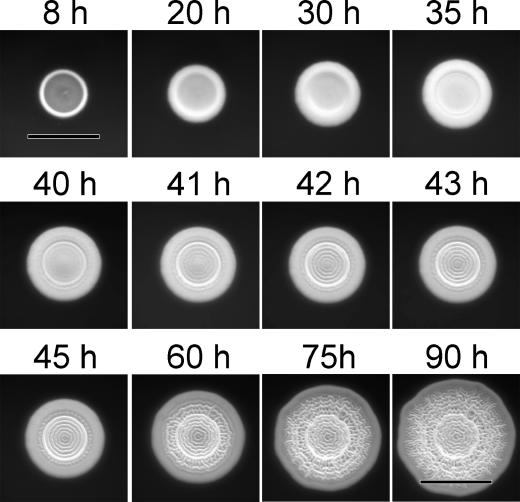
During analysis of SARB colony morphologies, we observed unusual concentric ring morphologies in colonies from several isolates. To determine if this was an intermediate stage in the formation of a complete surface pattern, we examined the growth of S. enterica serovar Montevideo (SARB30) by time-lapse photography (Fig. 1 and supplementary movie 1 at www.med.ucalgary.ca/webs/bprg/Surette/Surette.html). The surface of the colony was smooth for the initial 35 h of growth (Fig. 1, top panels). Between 40 and 41 h of growth, there was a rapid transition in morphology resulting from the formation of regular, concentric rings in the center of the colony (Fig. 1, middle panels). For the remainder of the growth period, new rings were formed between the colony center and colony edge until the entire surface was covered with a symmetrical pattern (Fig. 1, bottom panels). Similar growth patterns were observed for most SARB isolates and serovar Typhimurium ATCC 14028, although there was some variability in the observed pattern morphology (data not shown). The rapidity and uniformity of initial pattern formation suggested that individual cells within the colony were organized. This demonstrated that pattern formation in the rdar morphotype was a coordinated multicellular behavior, consistent with a developmental shift in the growing colony (33).
FIG. 1.
Multicellular pattern formation during growth of Salmonella in colonies of the rdar morphotype. S. enterica serovar Montevideo (SARB 30) cells were inoculated on T agar and incubated at room temperature for 90 h. Colony images were recorded every 30 min. Bars represent 5 mm. The entire sequence of 170 pictures can be viewed as a movie (www.med.ucalgary.ca/webs/bprg/Surette/Surette.html).
Ordering the expression of genes required for Salmonella multicellular aggregation.
Expression of key gene promoters involved in Salmonella multicellular aggregation was monitored during growth in 96-well plates (Fig. 2). This growth state is different than Salmonella growth on surfaces. However, when liquid cultures are overlaid with mineral oil, a microaerophilic environment is created (C. M. Southward and M. G. Surette, unpublished data), and this was shown to be one of the main requirements for agfD expression in liquid culture (15). Transcription of agfB and adrA, leading to the production of Tafi and cellulose, respectively, is known to be dependent on AgfD. Expression of agfD is dependent on activity of the stationary-phase sigma factor, RpoS (25). To examine the timing of expression of operons involved in this pathway, agfD, agfB, and adrA promoters were cloned in front of a promoterless lux reporter (4, 45). In order to monitor RpoS activity during growth, we constructed a synthetic RpoS (σ38)-responsive promoter::lux fusion.
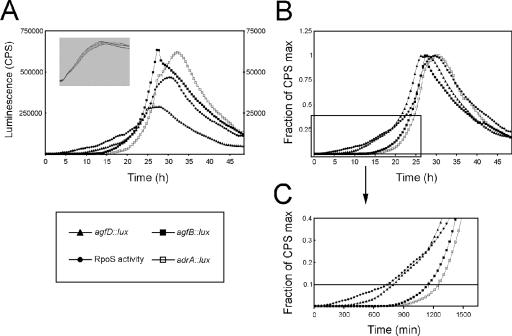
FIG. 2.
Temporal expression of genes required for formation of the rdar morphotype in serovar Typhimurium. Expression of agfD, agfB, adrA, or RpoS (σ38)-responsive promoter::lux fusions during ATCC 14028 growth in 1% tryptone at 28°C with agitation. A. Light production as a function of time. Luminescence (counts per second [cps]) measurements for agfD and agfB::lux fusions are on the left axis; measurements for adrA and RpoS-responsive lux fusions are on the right axis. Representative expression curves from one experiment are shown. The inset shows cell density measurements (A620) for each reporter strain as a function of time. B. Light production of lux reporter strains as a function of time, normalized by the maximum cps of each strain. To ensure accurate timing of expression during growth, each assay was repeated at least six times and individual growth curves were matched. The expression curves represent the average from these experiments. C. Expression curves from panel B were enlarged to highlight the temporal resolution of gene expression. The line is representative of the 10% gene activation level.
Light production from each of the reporter strains was measured during growth of ATCC 14028 (Fig. 2A). The strength of the promoters varied over 10-fold; agfD and agfB were the strongest with maximal light production of 285,000 cps and 650,000 cps, respectively. adrA had weaker expression with a maximal light production of 60,000 cps. Despite magnitude differences, the peak activity of these three reporters appeared to be synchronized (Fig. 2A). To better resolve temporal differences, expression curves were normalized by the maximum light production from each reporter (Fig. 2B). Peak expression occurred after 25 h, indicating that all promoters were transcriptionally active during the stationary phase of growth. Furthermore, activation of each gene corresponded with increased RpoS activity in the cell as measured by our synthetic promoter (Fig. 2B).
We compared normalized promoter expression profiles at the 10% activation level to determine the relative order of transcription (Fig. 2C) (20, 45). An increase in RpoS activity from basal levels was measured between 8 and 12 h of growth. This increase was followed immediately by activation of agfD transcription. There was a 5- to 6-h lag period before activation of agfB and adrA reporters, both of which had steep activation profiles. agfB was activated 90 to 120 min before adrA, on average (Fig. 2C). This suggests that fimbrial biosynthesis may be an initiating event in formation of the Salmonella extracellular matrix. The sharp expression peaks observed for agfB and adrA correspond to the sharp transition in pattern formation during colony growth (Fig. 1, 40 to 42 h).
Extracellular complementation of colony surface patterns.
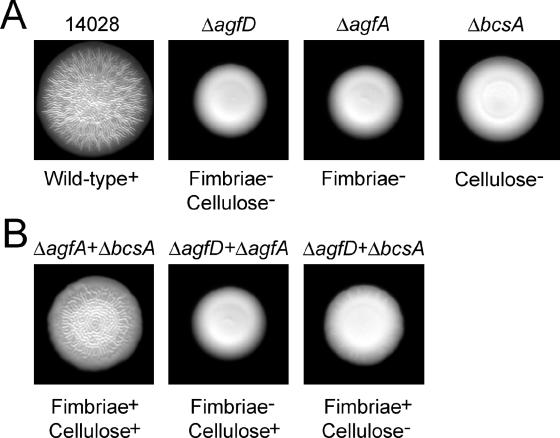
ATCC 14028 forms large, patterned colonies (Fig. 3A) that can be peeled off the agar surface intact (29). An isogenic ΔagfD strain lacking both Tafi and cellulose production formed smaller colonies that remained smooth and mucoid (Fig. 3A) (28, 47). Strains lacking either Tafi production (ΔagfA) (8, 25) or cellulose production (ΔbcsA) (47) also formed smooth colonies (Fig. 3A), but these colonies were not entirely mucoid; Tafi provide short-range cell-cell interactions that yield an adhesive texture, and cellulose provides long-range cell-cell interactions that yield a sticky texture (28). To highlight the extracellular interactions leading to pattern formation, mutant strains lacking Tafi, cellulose, or both components (ΔagfA, ΔbcsA, or ΔagfD) were mixed prior to spotting on T agar. When ΔagfA and ΔbcsA cells were combined, the Tafi and cellulose polymers produced from different cells were able to restore pattern formation on the colony surface (Fig. 3B). These colonies had an adhesive texture, indicating that a partial multicellular network had formed. In contrast, mixtures of cells lacking either Tafi or cellulose formed colonies that remained smooth and partially mucoid (Fig. 3B). Thus, extracellular interactions between individual cells are responsible for the formation of colony surface patterns in the rdar morphotype of Salmonella.
FIG. 3.
Tafi and cellulose interact extracellulary to form surface patterns in Salmonella rdar morphotype colonies. A. Colony morphologies of ATCC 14028 and agfD, agfA, and bcsA deletion strains grown on 1% tryptone (T) agar at 28°C for 48 h. B. Colony morphologies of mixtures of equal numbers of ΔagfD, ΔagfA, or ΔbcsA cells after growth on T agar at 28°C for 48 h.
Long-term survival of Salmonella in colonies of the rdar morphotype.
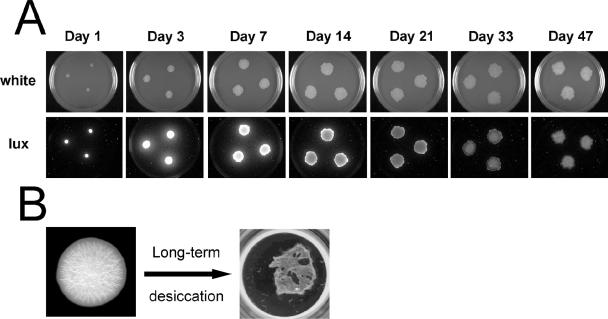
Preliminary experiments with rdar morphotype colonies of ATCC 14028 indicated that cells were viable after several weeks to months of growth at RT. Since RpoS is known to control the rdar morphotype (25) along with the general stress response (18), we monitored RpoS activity during rdar colony growth. Peak RpoS activity was observed between 3 and 7 days (Fig. 4A). After this time, luciferase production was gradually reduced but remained above detectable levels even after 47 days (Fig. 4A). This was in contrast to a synthetic RpoD (σ70)-responsive promoter that had peak expression at day 1 and dropped below detectable limits after 7 days (data not shown). These data indicated that cells were alive and respiring after nearly 2 months. This led us to investigate the fate of cells in colonies that were peeled off from the agar surface and dried out in the plastic wells of a multiwell plate (Fig. 4B). Surprisingly, a large number of cells remained viable even after several months of storage. We hypothesized from these initial experiments that the rdar morphotype may contribute to the long-term survival of Salmonella.
FIG. 4.
Long-term survival of serovar Typhimurium in colonies of the rdar morphotype. A. RpoS (σ38) activity in growing cells was monitored using the sig38H4 promoter::lux fusion. Colonies were inoculated with 105 cells from an overnight culture and were grown at RT (∼22°C) for 50 days. Pictures were taken at the times indicated to record colony morphology (white) or luminescence production (lux). B. Individual rdar colonies peeled off the surface of T agar were stored for 3 to 9 months at RT in plastic wells of a 24-well plate in the absence of nutrients prior to testing survival and resistance to bleach treatment.
Survival of Salmonella during long-term desiccation.
To determine if Tafi, cellulose, and other matrix components were contributing to long-term survival, colonies of ATCC 14028 and isogenic agfD-, agfA-, and bcsA-deleted strains were compared directly after 3 and 9 months of storage on plastic. Despite the difference in colony sizes (Fig. 3A), each colony type had similar numbers of viable cells before storage on plastic (Table 2). After 3 months of storage, the ATCC 14028 reference strain, capable of producing Tafi and cellulose, had the highest rate of survival at 68% (Table 2). In contrast, cell numbers for ΔagfD, ΔagfA, and ΔbcsA colonies were significantly reduced to 6.2%, 11%, and 21% of initial numbers, respectively (Table 1). After 9 months of storage, survival of ATCC 14028 cells was nearly 10%, whereas cell numbers for ΔagfD and ΔagfA colonies were further reduced to 0.27% and 1.2%, respectively (Table 1). Survival of ΔbcsA cells was also reduced compared to ATCC 14028, but the difference was not statistically significant (Table 1). These data demonstrated that production of Tafi and cellulose enhanced long-term survival of Salmonella under these conditions. The ΔbcsA and ΔagfA mutants had different survival characteristics, demonstrating that these two extracellular polymers collectively enhance survival, with cellulose being slightly less important.
TABLE 2.
Survival of Salmonella after months of desiccation in the absence of exogenous nutrients
| Strain (phenotype) | CFU/colonya | % Survivalb |
|---|---|---|
| ATCC 14028 (wild type+) | ||
| Initial colony | 2.3 × 109 ± 0.14 × 109 | |
| 3 mo desiccation | 1.6 × 109 ± 0.15 × 109 | 68 |
| 9 mo desiccation | 2.2 × 108 ± 0.19 × 108 | 9.7 |
| ΔagfD (fimbriae−, cellulose−) | ||
| Initial colony | 3.3 × 109 ± 0.14 × 109 | |
| 3 mo desiccation | 2.0 × 108 ± 0.28 × 108 | 6.2 |
| 9 mo desiccation | 8.7 × 106 ± 1.4 × 106 | 0.27 |
| ΔagfA (fimbriae−) | ||
| Initial colony | 3.2 × 109 ± 0.64 × 108 | |
| 3 mo desiccation | 3.4 × 108 ± 0.12 × 108 | 11 |
| 9 mo desiccation | 3.8 × 107 ± 0.34 × 107 | 1.2 |
| ΔbcsA (cellulose−) | ||
| Initial colony | 2.3 × 109 ± 0.18 × 109 | |
| 3 mo desiccation | 4.8 × 108 ± 0.37 × 108 | 21 |
| 9 mo desiccation | 1.6 × 108 ± 0.17 × 107 | 6.8 |
Average number of viable cells (CFU) per dried colony plus or minus the standard error. At least four individual colonies were tested.
Percent survival of cells in desiccated colonies determined relative to values from initial colonies.
Resistance of cells in rdar morphotype colonies to treatment with sodium hypochlorite.
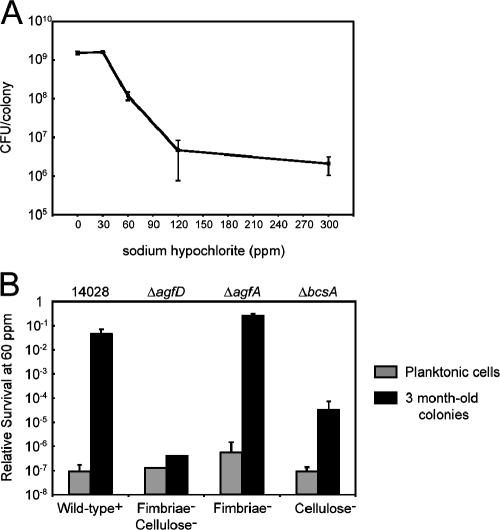
Survival in a desiccated environment without exogenous nutrients would aid in the persistence of Salmonella in non-host environments. However, this is just one aspect of survival. To examine how cells respond to additional stresses, ATCC 14028 rdar colonies that had been stored for 3 months were rehydrated and exposed to sodium hypochlorite for 20 min (Fig. 5A). No reductions in CFU were measured using a concentration of 30 ppm (Fig. 5A). At 300 ppm, a concentration in excess of normal sanitization levels (30), only a 3-log reduction in survival was observed (0.1% survival). At 1,000 ppm or 0.1% sodium hypochlorite, all cells were killed (data not shown).
FIG. 5.
Survival of serovar Typhimurium cells in colonies of the rdar morphotype following a 20-min exposure to sodium hypochlorite. A. ATCC 14028 rdar colonies stored for 3 months on plastic were rehydrated and treated with different concentrations of sodium hypochlorite. Each point represents the average number of surviving cells (CFU) per colony and standard errors corresponding to at least two individual colonies. B. Survival of cells from 3-month-old colonies of ATCC 14028, ΔagfD, ΔagfA, and ΔbcsA was compared to that of planktonic cells grown at 37°C for 20 h (stationary phase) after treatment with 60 ppm sodium hypochlorite. Bars represent the average relative survival (log CFU after treatment/log CFU before treatment) and standard errors from at least four individual colonies or aliquots of planktonic cells. CFUs for colonies before treatment are displayed in Table 1; for planktonic cells, the initial values ranged from 4.2 × 108 to 5.5 × 108 CFU.
To determine if Tafi, cellulose, and other extracellular matrix components conferred protection to sodium hypochlorite, ATCC 14028, ΔagfD, ΔagfA, and ΔbcsA cells were compared after growth in conditions when Tafi and cellulose are not produced (i.e., stationary-phase planktonic cells at 37°C) versus rdar colonies that had been stored for 3 months (Fig. 5B). Planktonic cells from all strains were susceptible to 60 ppm sodium hypochlorite and showed greater than 6-log reductions in CFU after treatment (Fig. 5B, gray bars). In contrast, cells in dried colonies of the ΔagfA strain were resistant with less than 1-log reduction in CFU after treatment. This level of resistance was similar to that of rdar colonies from ATCC 14028 (Fig. 5A). Cells in dried colonies from the cellulose-deficient ΔbcsA strain were susceptible and showed greater than a 4-log reduction in CFU (<0.01% survival; Fig. 5B). Mucoid colonies of the ΔagfD strain were most susceptible; survival was reduced to levels similar to that of planktonic-grown cells (Fig. 5B). These results demonstrated that cellulose and additional components regulated by AgfD confer protection to sodium hypochlorite.
When equal numbers of ΔbcsA and ΔagfA cells were combined prior to growth, ΔbcsA cells in the hybrid rdar colonies (Fig. 3B) showed increased resistance to sodium hypochlorite (data not shown). The degree of protection correlated with the number of cellulose-positive (ΔagfA) cells present, indicating that survival was dependent on the overall amount of cellulose present in the colony. This reinforced the correlation between the extracellular interactions leading to the rdar morphotype and the inherent resistance of this multicellular state.
DISCUSSION
Salmonella has evolved specialized mechanisms for exploiting many host environments, yet these pathogens also have a remarkable ability to persist in extreme environments outside the host (42). Here we have demonstrated that coordinated multicellular behavior leading to the rdar morphotype can be linked to the long-term survival of Salmonella. The uniformity of pattern formation in colonies of the rdar morphotype indicates a high level of cell-cell organization. We hypothesize that the rdar morphotype is an example of a microbial developmental process (33). Presumably, colony surface patterns arise because Salmonella senses its environment and triggers production of extracellular matrix components, such as Tafi, cellulose, and additional exopolysaccharides (41). Under laboratory conditions, the environmental triggers for this phenotype appear to mimic life outside a host (i.e., limiting nutrients, low temperatures, and low osmolarity).
The rdar morphotype was formed by 58 of 72 (80.5%) natural isolates in the SARB collection. Previous studies to determine the prevalence of the rdar morphotype within S. enterica subgroup I have primarily focused on human-associated serovars Enteritidis and Typhimurium (26, 35). More recent work has focused on a variety of serovars isolated from clinical and food sources (36). In each of these studies, rdar morphotype formation was detected in greater than 70% of isolates. Given that the SARB represents a comprehensive strain collection, we conclude that the ability to form the rdar morphotype is prevalent throughout S. enterica subgroup I. Serovars Choleraesuis and Typhi appear to be exceptions, since all 36 isolates tested by Romling et al. (26) did not express the rdar morphotype. However, both serovar Typhi isolates tested in this study were capable of forming rdar colonies. The reason for this discrepancy is unclear but is presumed to represent diversity within serovar Typhi.
Our experiments demonstrate an important role for the rdar morphotype in the enhanced long-term survival and persistence of Salmonella. The enhanced survival under conditions of desiccation would be advantageous in many natural environments and may also contribute to the contamination of dried food products (19). The finding that almost 10% of ATCC 14028 cells were alive after 9 months of storage illustrates the remarkable survival properties of Salmonella. The increased resistance of 3-month-old rdar colonies to sodium hypochlorite has obvious implications for the sanitization of contaminated surfaces (i.e., in the agricultural and food-processing industries). Our observations were comparable to those of a recent study performed on cells forming pellicles at the air-liquid interface in standing liquid culture (32). This was not surprising, since the same ATCC 14028 strain was analyzed and Tafi and cellulose are required for pellicle formation (14). These and other studies (2, 21, 31) stress the differences between studying planktonic cells grown in typical laboratory conditions and cells growing in more resistant physiological states.
Individual extracellular matrix components had different contributions to long-term survival and persistence. The Tafi-deficient strain (ΔagfA) had a surprising reduction in survival of desiccation compared to the cellulose-deficient strain (ΔbcsA). Our gene expression analyses indicated that Tafi production may be an initiating event during the formation of the rdar morphotype. The temporal difference between agfB (Tafi production) and adrA (cellulose production) activation was approximately 90 to 120 min. Overexpression of adrA from a multicopy plasmid resulted in a more rapid appearance of the rdar phenotype, although the colony morphology was visibly altered (data not shown). Therefore, the significance of the temporal difference between agfB and adrA is unknown. Tafi production is required for the increased surface spreading observed in rdar colonies; ΔbcsA (Cellulose−, Tafi+) colonies are consistently larger than ΔagfA (Cellulose+, Tafi−) colonies (Fig. 3A). From these different observations, we hypothesize that Tafi plays a critical role in organizing the extracellular matrix, perhaps leading to an optimal spatial arrangement of cells (47) and increased survival. Cellulose, on the other hand, was not as important for survival but conferred protection against sodium hypochlorite. Cellulose-positive strains (wild type and ΔagfA) had greater than 4 orders of magnitude increased survival compared to that of cellulose-deficient strains (ΔbcsA and ΔagfD). This confirmed previous sodium hypochlorite tests with broth-grown cells of Salmonella (35). The protective nature of cellulose could apply to other reactive compounds, such as hydrogen peroxide (2). Cellulose polymers may directly remove or neutralize reactive species, as suggested by Scher et al. (32). The presence of cellulose may also trap additional Salmonella polysaccharides on the cell surface (41) to enhance protection.
Long-term survival during desiccation and resistance to sodium hypochlorite was most reduced in the agfD mutant strain lacking Tafi and cellulose production (47). The absence of these two polymers did not entirely account for the reduction measured, however. Survival of ΔagfD was four times less than that of the Tafi-deficient (ΔagfA) strain after 9 months of storage and 2 logs less than that of the cellulose-deficient strain (ΔbcsA) after treatment with sodium hypochlorite. AgfD is a global transcriptional regulator known to regulate the expression of several genes related to biofilm formation (6) as well as newly identified components of the extracellular matrix, such as capsular polysaccharides (D. L. Gibson, A. P. White, S. D. Snyder, C. Heiss, P. Azadi, M. G. Surette, and W. W. Kay, unpublished) and BapA, a large cell surface protein containing repeated sequence motifs (23). It is hypothesized that ΔagfD cells have increased susceptibility to desiccation stress and sodium hypochlorite due to lack of these additional extracellular components.
The relationship between the Salmonella rdar morphotype and survival described here may be just one example of a generalized survival strategy used by many other microorganisms, both pathogens and nonpathogens. The genes for Tafi (agf or csg) and cellulose (bcs or yhj) are conserved throughout the salmonellae (http://globin.cse.psu.edu/enterix/enteric/enteric.html) (12), and these polymers are expressed by the vast majority of isolates. Curli and cellulose can also be produced by environmental E. coli isolates (10) and organisms commonly isolated from the human digestive tract (46). In addition, cellulose is produced by numerous soilborne organisms and plant pathogens, such as Agrobacterium tumefaciens (1), Pseudomonas fluorescens (37), and Erwinia chrysanthemi (43), and contributes to the fitness of these organisms in their natural environments. P. aeruginosa (13), Candida spp. (22), and Vibrio cholerae (44) all display coordinated multicellular behavior when grown on solid media. Unlike Salmonella spp., none of these microorganisms are known to produce Tafi or cellulose. However, in each case, protein and sugar polymers are involved in the formation of an extracellular matrix that encases individual cells and leads to aggregation. This is likely the principle function of Tafi and cellulose in Salmonella, to enable individual cells to stick together on surfaces and withstand stresses in the environment. This would allow for the smooth transition of Salmonella between hosts.
Acknowledgments
The sig38H4 luciferase reporter vector was designed and constructed by Kanti Pabbaraju and M.G.S. We thank Ken Sanderson for providing access to SARB and SARC strains at the Salmonella Genetic Stock Centre (University of Calgary) and K. Sanderson, M. Elowitz, B. Bassler, and E. Crump for critical reading of the manuscript.
This work was supported by grants from the National Sciences and Engineering Research Council (NSERC) to W.W.K. and Canadian Institutes of Health Research to M.G.S. and through Genome Prairie, Genome BC, and Inimex Pharmaceuticals through the “Functional Pathogenomics of Mucosal Immunity” project. M.G.S. is supported as an Alberta Heritage Foundation for Medical Research (AHFMR) Senior Scholar and Canada Research Chair in Microbial Gene Expression. A.P.W. is supported by a postdoctoral fellowship from AHFMR.
REFERENCES
- 1.Amikam, D., and M. Benziman. 1989. Cyclic diguanylic acid and cellulose synthesis in Agrobacterium tumefaciens. J. Bacteriol. 171:6649-6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anriany, Y. A., R. M. Weiner, J. A. Johnson, C. E. De Rezende, and S. W. Joseph. 2001. Salmonella enterica serovar Typhimurium DT104 displays a rugose phenotype. Appl. Environ. Microbiol. 67:4048-4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker, G., and R. Hengge-Aronis. 2001. What makes an Escherichia coli promoter sigma(S) dependent? Role of the −13/−14 nucleotide promoter positions and region 2.5 of sigma(S). Mol. Microbiol. 39:1153-1165. [DOI] [PubMed] [Google Scholar]
- 4.Bjarnason, J., C. M. Southward, and M. G. Surette. 2003. Genomic profiling of iron-responsive genes in Salmonella enterica serovar Typhimurium by high-throughput screening of a random promoter library. J. Bacteriol. 185:4973-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd, E. F., F. S. Wang, P. Beltran, S. A. Plock, K. Nelson, and R. K. Selander. 1993. Salmonella reference collection B (SARB): strains of 37 serovars of subspecies I. J. Gen. Microbiol. 139:1125-1132. [DOI] [PubMed] [Google Scholar]
- 6.Brombacher, E., C. Dorel, A. J. Zehnder, and P. Landini. 2003. The curli biosynthesis regulator CsgD co-ordinates the expression of both positive and negative determinants for biofilm formation in Escherichia coli. Microbiology 149:2847-2857. [DOI] [PubMed] [Google Scholar]
- 7.Collinson, S. K., S. C. Clouthier, J. L. Doran, P. A. Banser, and W. W. Kay. 1996. Salmonella enteritidis agfBAC operon encoding thin, aggregative fimbriae. J. Bacteriol. 178:662-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collinson, S. K., P. C. Doig, J. L. Doran, S. Clouthier, T. J. Trust, and W. W. Kay. 1993. Thin, aggregative fimbriae mediate binding of Salmonella enteritidis to fibronectin. J. Bacteriol. 175:12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collinson, S. K., L. Emody, K. H. Muller, T. J. Trust, and W. W. Kay. 1991. Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. J. Bacteriol. 173:4773-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collinson, S. K., L. Emody, T. J. Trust, and W. W. Kay. 1992. Thin aggregative fimbriae from diarrheagenic Escherichia coli. J. Bacteriol. 174:4490-4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Rezende, C. E., Y. Anriany, L. E. Carr, S. W. Joseph, and R. M. Weiner. 2005. Capsular polysaccharide surrounds smooth and rugose types of Salmonella enterica serovar Typhimurium DT104. Appl. Environ. Microbiol. 71:7345-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Florea, L., M. McClelland, C. Riemer, S. Schwartz, and W. Miller. 2003. EnteriX 2003: visualization tools for genome alignments of Enterobacteriaceae. Nucleic Acids Res. 31:3527-3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedman, L., and R. Kolter. 2004. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51:675-690. [DOI] [PubMed] [Google Scholar]
- 14.Garcia, B., C. Latasa, C. Solano, F. Garcia-del Portillo, C. Gamazo, and I. Lasa. 2004. Role of the GGDEF protein family in Salmonella cellulose biosynthesis and biofilm formation. Mol. Microbiol. 54:264-277. [DOI] [PubMed] [Google Scholar]
- 15.Gerstel, U., and U. Romling. 2001. Oxygen tension and nutrient starvation are major signals that regulate agfD promoter activity and expression of the multicellular morphotype in Salmonella typhimurium. Environ. Microbiol. 3:638-648. [DOI] [PubMed] [Google Scholar]
- 16.Hammar, M., A. Arnqvist, Z. Bian, A. Olsen, and S. Normark. 1995. Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Mol. Microbiol. 18:661-670. [DOI] [PubMed] [Google Scholar]
- 17.Hashimoto-Gotoh, T., F. C. Franklin, A. Nordheim, and K. N. Timmis. 1981. Specific-purpose plasmid cloning vectors. I. Low copy number, temperature-sensitive, mobilization-defective pSC101-derived containment vectors. Gene 16:227-235. [DOI] [PubMed] [Google Scholar]
- 18.Hengge-Aronis, R. 2002. Recent insights into the general stress response regulatory network in Escherichia coli. J. Mol. Microbiol. Biotechnol. 4:341-346. [PubMed] [Google Scholar]
- 19.Hiramatsu, R., M. Matsumoto, K. Sakae, and Y. Miyazaki. 2005. Ability of Shiga toxin-producing Escherichia coli and Salmonella spp. to survive in a desiccation model system and in dry foods. Appl. Environ. Microbiol. 71:6657-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalir, S., J. McClure, K. Pabbaraju, C. Southward, M. Ronen, S. Leibler, M. G. Surette, and U. Alon. 2001. Ordering genes in a flagella pathway by analysis of expression kinetics from living bacteria. Science 292:2080-2083. [DOI] [PubMed] [Google Scholar]
- 21.Kim, W., T. Killam, V. Sood, and M. G. Surette. 2003. Swarm-cell differentiation in Salmonella enterica serovar Typhimurium results in elevated resistance to multiple antibiotics. J. Bacteriol. 185:3111-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laffey, S. F., and G. Butler. 2005. Phenotype switching affects biofilm formation by Candida parapsilosis. Microbiology 151:1073-1081. [DOI] [PubMed] [Google Scholar]
- 23.Latasa, C., A. Roux, A. Toledo-Arana, J. Ghigo, C. Gamazo, J. R. Penadés, and I. Lasa. 2005. BapA, a large secreted protein required for biofilm formation and host colonization of Salmonella enterica serovar Enteritidis. Mol. Microbiol. 58:1522-1539. [DOI] [PubMed] [Google Scholar]
- 24.Pang, T., Z. A. Bhutta, B. B. Finlay, and M. Altwegg. 1995. Typhoid fever and other salmonellosis: a continuing challenge. Trends Microbiol. 3:253-255. [DOI] [PubMed] [Google Scholar]
- 25.Romling, U., Z. Bian, M. Hammar, W. D. Sierralta, and S. Normark. 1998. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J. Bacteriol. 180:722-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romling, U., W. Bokranz, W. Rabsch, X. Zogaj, M. Nimtz, and H. Tschape. 2003. Occurrence and regulation of the multicellular morphotype in Salmonella serovars important in human disease. Int. J. Med. Microbiol. 293:273-285. [DOI] [PubMed] [Google Scholar]
- 27.Romling, U., M. Gomelsky, and M. Y. Galperin. 2005. C-di-GMP: the dawning of a novel bacterial signalling system. Mol. Microbiol. 57:629-639. [DOI] [PubMed] [Google Scholar]
- 28.Romling, U., M. Rohde, A. Olsen, S. Normark, and J. Reinkoster. 2000. AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol. Microbiol. 36:10-23. [DOI] [PubMed] [Google Scholar]
- 29.Romling, U., W. D. Sierralta, K. Eriksson, and S. Normark. 1998. Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol. Microbiol. 28:249-264. [DOI] [PubMed] [Google Scholar]
- 30.Russell, S. M., and S. P. Axtell. 2005. Monochloramine versus sodium hypochlorite as antimicrobial agents for reducing populations of bacteria on broiler chicken carcasses. J. Food Prot. 68:758-763. [DOI] [PubMed] [Google Scholar]
- 31.Ryu, J. H., and L. R. Beuchat. 2005. Biofilm formation by Escherichia coli O157:H7 on stainless steel: effect of exopolysaccharide and Curli production on its resistance to chlorine. Appl. Environ. Microbiol. 71:247-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scher, K., U. Romling, and S. Yaron. 2005. Effect of heat, acidification, and chlorination on Salmonella enterica serovar Typhimurium cells in a biofilm formed at the air-liquid interface. Appl. Environ. Microbiol. 71:1163-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shapiro, J. A. 1998. Thinking about bacterial populations as multicellular organisms. Annu. Rev. Microbiol. 52:81-104. [DOI] [PubMed] [Google Scholar]
- 34.Simm, R., M. Morr, A. Kader, M. Nimtz, and U. Romling. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 53:1123-1134. [DOI] [PubMed] [Google Scholar]
- 35.Solano, C., B. Garcia, J. Valle, C. Berasain, J. M. Ghigo, C. Gamazo, and I. Lasa. 2002. Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol. Microbiol. 43:793-808. [DOI] [PubMed] [Google Scholar]
- 36.Solomon, E. B., B. A. Niemira, G. M. Sapers, and B. A. Annous. 2005. Biofilm formation, cellulose production, and curli biosynthesis by Salmonella originating from produce, animal, and clinical sources. J. Food Prot. 68:906-912. [DOI] [PubMed] [Google Scholar]
- 37.Spiers, A. J., J. Bohannon, S. M. Gehrig, and P. B. Rainey. 2003. Biofilm formation at the air-liquid interface by the Pseudomonas fluorescens SBW25 wrinkly spreader requires an acetylated form of cellulose. Mol. Microbiol. 50:15-27. [DOI] [PubMed] [Google Scholar]
- 38.Walsh, P. S., D. A. Metzger, and R. Higuchi. 1991. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. BioTechniques 10:506-513. [PubMed] [Google Scholar]
- 39.White, A. P., S. K. Collinson, P. A. Banser, D. L. Gibson, M. Paetzel, N. C. Strynadka, and W. W. Kay. 2001. Structure and characterization of AgfB from Salmonella enteritidis thin aggregative fimbriae. J. Mol. Biol. 311:735-749. [DOI] [PubMed] [Google Scholar]
- 40.White, A. P., S. K. Collinson, J. Burian, S. C. Clouthier, P. A. Banser, and W. W. Kay. 1999. High efficiency gene replacement in Salmonella enteritidis: chimeric fimbrins containing a T-cell epitope from Leishmania major. Vaccine 17:2150-2161. [DOI] [PubMed] [Google Scholar]
- 41.White, A. P., D. L. Gibson, S. K. Collinson, P. A. Banser, and W. W. Kay. 2003. Extracellular polysaccharides associated with thin aggregative fimbriae of Salmonella enterica serovar Enteritidis. J. Bacteriol. 185:5398-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winfield, M. D., and E. A. Groisman. 2003. Role of nonhost environments in the lifestyles of Salmonella and Escherichia coli. Appl. Environ. Microbiol. 69:3687-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yap, M. N., C. H. Yang, J. D. Barak, C. E. Jahn, and A. O. Charkowski. 2005. The Erwinia chrysanthemi type III secretion system is required for multicellular behavior. J. Bacteriol. 187:639-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yildiz, F. H., and G. K. Schoolnik. 1999. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. USA 96:4028-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zaslaver, A., A. E. Mayo, R. Rosenberg, P. Bashkin, H. Sberro, M. Tsalyuk, M. G. Surette, and U. Alon. 2004. Just-in-time transcription program in metabolic pathways. Nat. Genet. 36:486-491. [DOI] [PubMed] [Google Scholar]
- 46.Zogaj, X., W. Bokranz, M. Nimtz, and U. Romling. 2003. Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract. Infect. Immun. 71:4151-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zogaj, X., M. Nimtz, M. Rohde, W. Bokranz, and U. Romling. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 39:1452-1463. [DOI] [PubMed] [Google Scholar]