Abstract
Previous work suggested that the FlgE (flagellar hook subunit) protein in Salmonella enterica serovar Typhimurium was posttranscriptionally regulated in response to the stage of flagellar assembly. Specifically, the FlgE protein could be detected in flagellar mutants defective at the stages of assembly before or after rod assembly but not in rod assembly mutants, yet flgE mRNA levels were unaffected. To elucidate posttranscriptional mechanisms involved in the coupling of flgE gene expression to hook assembly, the RNA sequences at the 5′ and 3′ ends of the flgE-containing mRNA processed from the large flgBCDEFGHIJKL operon were determined by rapid amplification of cDNA ends, and secretion of the FlgE protein in different flagellar assembly mutant strains was analyzed. The sequences 5′ and 3′ of the flgE gene where RNA processing occurred was within 15 bases upstream of the flgD stop codon and at bases 145 to 147 downstream of the flgF start codon, respectively. The ribosome binding site of the flgD gene was found to be inhibitory to flgE translation in strains deleted for the upstream flgD gene, unless the region 15 bases upstream of the flgD stop codon was present. Secretion of FlgE into the periplasm was monitored using β-lactamase (Bla) fusions as a periplasm-specific reporter, which conferred resistance to ampicillin when FlgE-Bla was secreted into the periplasm. Using this assay, we found that the effect of rod assembly mutants on FlgE levels was due to FlgE turnover in the periplasm and that the FliE rod component protein was required for efficient FlgE-Bla secretion.
Many bacteria can swim through liquid environments or crawl across surfaces by the controlled rotation of extracellular organelles called flagella. Bacteria can access favorable environments or escape from unfavorable environments using flagellum-dependent motility. Salmonella enterica serovar Typhimurium possesses 6 to 10 peritrichous flagella per cell (23). Each flagellum is composed of three distinct structures: basal body, hook, and filament (1). More than 60 genes are required for the assembly and function of the flagellum in S. enterica serovar Typhimurium (8). Flagellar gene expression is regulated temporally, and flagellar biosynthesis follows an ordered assembly process (16). The flagellar genes are expressed in a transcriptional hierarchy of three temporal promoter classes (8). At the top of the flagellar transcriptional hierarchy in S. enterica serovar Typhimurium is the flhDC master operon, which is expressed from a class 1 promoter and is sensitive to environmental and cellular signals (20, 33). Early gene products, FlhD and FlhC, form a transcriptional activator complex that directs the σ70-dependent transcription of class 2 promoters (22). The class 2 promoters mediate the transcription of genes whose products are required for the structure and assembly of the hook-basal body (HBB). In addition, two regulatory proteins, flagellum-specific sigma factor σ28 (FliA) and its cognate anti-sigma factor, FlgM, are expressed along with hook-basal body genes. The class 3 promoters transcribe genes whose products are needed late in the assembly process, including the structural components of the filament, the motor force generators, and the chemosensory genes. The σ28 protein is required for the transcription of class 3 promoters (31) but prior to HBB completion is tightly associated with anti-σ28 factor FlgM (7, 30). Thus, the transcription of class 3 promoters is inhibited until completion of the intermediate hook-basal body structure (16). Upon HBB completion, FlgM is secreted from the cell to free σ28 and to allow transcription of class 3 promoters (13, 21).
In addition to sophisticated transcriptional regulation mechanisms that control the expression of flagellar genes, a layer of posttranscriptional regulation mechanisms has recently been uncovered. A posttranscriptional control mechanism is involved in flagellar phase variation in S. enterica serovar Typhimurium (5). The FljA protein, previously thought to be a transcriptional repressor of the fliC flagellin gene, has been shown to be an RNA-binding translational regulator of fliC mRNA translation (P. Aldridge et al., unpublished results). The hook structural gene (flgE) expression also appeared to be coupled to flagellum assembly by a posttranscriptional regulatory mechanism (6). The FlgE protein was absent in strains defective in rod assembly, yet the flgE mRNA level was unaffected. Recently, similar data were reported for fimA regulation of Porphyromonas gingivalis (32) and flaA regulation of Borrelia burgdorferi (29). The mRNA levels of fimA and flaA in PG2131 and flaB mutants, respectively, did not change, but their protein levels were decreased compared with those of their wild types, consistent with a posttranscriptional control mechanism.
Little is known about S. enterica serovar Typhimurium flgE mRNA translation in coordination with hook assembly. The flgB-L operon has 11 genes, and the stoichiometry of individual proteins per flagellum is highly variable (24). The stoichiometry of the FlgE protein is 5 to 20 times higher than that of other flgB-L operon proteins. Northern blot analysis of Rhodobacter sphaeroides flgE suggested a role for mRNA processing in the regulation of flgB-J operon expression and identified two different flgE-containing transcripts (4).
In the present study, we investigated the posttranscriptional mechanism of flgE in S. enterica serovar Typhimurium. We used rapid amplification of cDNA ends (RACE) (3) to map the mRNA processing sites of flgB-L mRNA in S. enterica serovar Typhimurium. In addition, an FlgE-β-lactamase (FlgE-Bla) fusion protein was constructed in order to assay whether the FlgE protein was secreted into the periplasm in rod mutant strains. We found that the FlgE-Bla protein was secreted into the periplasm and that the activity of FlgE-Bla was fully dependent on the flagellar basal structure. We also present evidence that the proximal rod FliE protein is required for efficient FlgE-Bla secretion but is not absolutely required, as was previously suggested (27).
MATERIALS AND METHODS
Strains and plasmids.
All bacterial strains and plasmids used in this work are listed in Table 1.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Genotype | Source or referencea |
|---|---|---|
| Strains | ||
| S. enterica serovar Typhimurium | ||
| LT2 | Wild type | J. Roth |
| TH4702 | pKD46/LT2 | 5 |
| TH6733 | flgE5315::MudA | |
| TH8547 | ΔflgD6219(Δaa2-232) fljBe,n,xvh2 | |
| TH9924 | ΔflgD6534(Δaa57-232) | |
| TH9925 | ΔflgD6558(Δaa57-232)::tetRA ΔpyrC::FRT-Km-FRT | |
| TH9926 | ΔflgD6535(Δaa34-232) | |
| TH9927 | ΔflgD6536(Δaa14-232) | |
| TH9928 | pflgD/ΔflgD6558(Δaa57-232)::tetRA ΔpyrC::FRT-Km-FRT | |
| TH9929 | ΔflgD6537(Δaa2-56) | |
| TH9930 | ΔflgD6538(Δaa2-177) | |
| TH9931 | ΔflgD6539(Δaa2-200) | |
| TH9932 | ΔflgD6540(Δaa2-220) | |
| TH9933 | ΔflgD6541(Δaa2-224) | |
| TH9934 | ΔflgD6542(Δaa2-228) | |
| TH9935 | ΔflgD6543(Δaa2-231) | |
| TH9936 | pflgD/ΔflgD6219(Δaa2-232) flgC6544(G828T Mot+) fljBe,n,xvh2 | |
| TH9937 | pflgD/ΔflgD6219(Δaa2-232) flg-6545(Mot+) fljBe,n,xvh2 | |
| TH9938 | pflgD/ΔflgD6219(Δaa2-232) flgC6546(G827A Mot+) (Mot+) fljBe,n,xvh2 | |
| TH9939 | pflgD/ΔflgD6219(Δaa2-232) flgC6547(A829T Mot+) (Mot+) fljBe,n,xvh2 | |
| TH9940 | pflgD/ΔflgD6219(Δaa2-232) flg-6548(Mot+) fljBe,n,xvh2 | |
| TH9941 | pflgD/ΔflgD6219(Δaa2-232) flg-6549(Mot+) fljBe,n,xvh2 | |
| TH9942 | pKD46/ΔflgD6219(Δaa2-232) fljBe,n,xvh2 | |
| TH9943 | ΔflgD6550(Δaa2-227) | |
| TH9944 | ΔflgD6551(Δaa2-232)::165 bp of tetA 3′ region | |
| TH9945 | ΔflgD6219(Δaa2-232) flgC6552(G828T) | |
| TH9946 | ΔflgD6219(Δaa2-232) flgC6553(G827A) | |
| TH9947 | pflgD/ΔflgD6219(Δaa2-232) fljBe,n,xvh2 | |
| TH9948 | flgE6554::bla | |
| TH9949 | flgE6554::bla ΔflgBC6557 | |
| TH9950 | flgE6554::bla ΔflgBC6557 fliF5036::Tn10dCm | |
| TH9951 | flgE6554::bla ΔflgBC6557 flhD5704::Tn10dTc | |
| TH9952 | flgE6554::bla ΔflgBC6557 ΔfliE6555::tetRA | |
| Plasmids | ||
| pTrc99A | Ptrc expression vector | Pharmacia |
| pKD46 | pBAD γ β exo | 9 |
| pflgD | flgD+ in pTrc99A | |
| pflgDE | flgD+E+ in pTrc99A |
Unless otherwise indicated, all strains and plasmids were constructed for this work.
Growth conditions and bacterial genetic and molecular methods.
Growth conditions, media, P22 transductional methods, λ-Red swaps, and motility assay were performed as described previously (9, 10).
Construction of the flgE-bla fusions.
The flgE-bla fusions were constructed by the replacement of chromosomally targeted tetRA elements with specific DNA fragments as described previously (15). The tetRA element from transposon Tn10 includes the tetracycline resistance gene, tetA, and the adjacent, divergently transcribed tetA repressor gene, tetR (17). The tetRA element was inserted just before the stop codon of flgE in strain LT2 using λ-Red-mediated recombination (9). The tetRA element was amplified by PCR with primers flgE403tetR and flgE403tetA (Table 2). The PCR-amplified tetRA fragment is flanked at its 5′ and 3′ ends by 40 bases of DNA sequence identical to the 40 bases upstream of the flgE stop codon and the 40 bases downstream of the last amino acid codon of flgE. Selection for tetracycline resistance (Tcr) allows the isolation of the desired recombinants that have the tetRA element between the last amino acid codon of flgE and the stop codon. The tetRA element was replaced by λ-Red-mediated recombination with a PCR-amplified β-lactamase gene (bla) that was missing its N-terminal 23-amino-acid Sec-dependent secretion signal and flanked by the same 40-bp sequences identical to the upstream and downstream regions of the stop codon of flgE. Primers flgE5amp and flgE3amp (Table 2) were used to amplify the bla gene. The replacement of flgE::tetRA was achieved by electroporation of the PCR-amplified bla gene into the flgE::tetRA-containing strain on tetracycline-sensitive (Tcs) selection medium (26) to create strain TH9948 (flgE-bla). Deletion of the flagellar rod structural genes flgBC was accomplished by targeted deletion using λ-Red-mediated recombination with a tetRA fragment that was PCR amplified with primers flgBCtetR and flgBCtetA. The resulting DNA fragment contains the tetRA element flanked by 40 bases of sequence identical to the 40 bases upstream of the start codon of flgB and the 40 bases downstream of the flgC stop codon. Selection for Tcr by λ-Red-mediated recombination resulted in the construction of the ΔflgBC::tetRA allele. The ΔflgBC::tetRA element was replaced using λ-Red-mediated recombination with a PCR-amplified DNA fragment designed with the 40 bases upstream of the flgB start codon fused to the 168 bases downstream of flgC stop codon. Primers flgBCdelet and flgDrtR were used to amplify the PCR products.
TABLE 2.
DNA primers used in this study
| Primer | Sequence |
|---|---|
| 5SF | 5′-GCGAGAGTAGGGAACTGCCA-3′ |
| flgE403tetR | 5′-AACCCAGGACCAGATCCTCAATACGCTGGTTAACCTGCGCTTAAGACCCACTTTCACATT-3′ |
| flgE403tetA | 5′-TATAAATTGCGTGATCCATTAAGCTATCCCGTCAGGCGCTTACTAAGCACTTGTCTCCTG-3′ |
| flgE5amp | 5′-AACCCAGGACCAGATCCTCAATACGCTGGTTAACCTGCGCCACCCAGAAACGCTGGTGAA-3′ |
| flgE3amp | 5′-TATAAATTGCGTGATCCATTAAGCTATCCCGTCAGGCGCTTACCAATGCTTAATCAGTGA-3′ |
| flgBCtetR | 5′-AAGCTGTCGGCTGAATTTTGCCATTTGCGGAGGAGATATGTTAAGACCCACTTTCACATT-3′ |
| flgBCtetA | 5′-TCATTCATATTTACGGCAATAGACATACGCGCCTCCTTTACTAAGCACTTGTCTCCTG-3′ |
| flgBCdelet | 5′-TTTAAGCTGTCGGCTGAATTTTGCCATTTGCGGAGGAGATTAAAGGAGGCGCGTATGTCT-3′ |
| flgDrtR | 5′-ATGGGTTAGTCGGGTCCTGG-3′ |
| flgCtetR | 5′-AAAAAGCATGATGCTTAAAACGCTGACATTAGGCCAGTAATTAAGACCCACTTTCACATT-3′ |
| flgD-p-5tetRA | 5′-TTGAAGAACCAGGACCCGACTAACCCATTACAAAATAATGTTAAGACCCACTTTCACATT-3′ |
| flgD-p-3tetRA | 5′-GAGAAAAAGACATGACTGACTCCTGATAAGTGTAAGGGCTTACTAAGCACTTGTCTCCTG-3′ |
| flgBseq5 | 5′-CATTTTGCGTTTATTCCGGCG-3′ |
| flgER | 5′-CCACTTTGGAACCGGCGAAC-3′ |
| flgDF | 5′-CTCGCCATGGCAGCTATCAGGCAA-3′ |
| flgDR | 5′-TGCGGCTGCAGTAGAACACGGAA-3′ |
| flgERHindIII | 5′-TGGTTAAGCTTGCCCGGTCGGA-3′ |
| flgDdelet | 5′-TTAAAACGCTGACATTAGGCCAGTAAAGGAGGCGCGTATGTAAGCCCTTACACTTATCAG-3′ |
| flgDFRTdel | 5′-ATTGAAGAACCAGGACCCGACTAACCCATTACAAAATAATTAAGCCCTTACACTTATCAG-3′ |
| flgD5′deletI | 5′-CAGCGGATCGATGACCGGAAGCAACGCTGCCGATCTGCAATAAGCCCTTACACTTATCAG-3′ |
| flgD5′deletII | 5′-TATGTCTATTGCCGTAAATATGAATGACCCGACCAACACGTAAGCCCTTACACTTATCAG-3′ |
| flgD3′delet1 | 5′-TTAAAACGCTGACATTAGGCCAGTAAAGGAGGCGCGTATGGAGTTAACGACACAGTTGGC-3′ |
| flgD3′delet2 | 5′-TTAAAACGCTGACATTAGGCCAGTAAAGGAGGCGCGTATGGGTTCTTACAACATTGCGAT-3′ |
| flgD3′delet3 | 5′-TTAAAACGCTGACATTAGGCCAGTAAAGGAGGCGCGTATGGCTCTGGTACAGGGCGTGAC-3′ |
| flgD3′delet4 | 5′-TTAAAACGCTGACATTAGGCCAGTAAAGGAGGCGCGTATGGGCACCACCACACTCGACGA-3′ |
| flgD3′delet5 | 5′-TTAAAACGCTGACATTAGGCCAGTAAAGGAGGCGCGTATGCTCGACGAAGTTCGGCAAAT-3′ |
| flgD3′delet6 | 5′-TTAAAACGCTGACATTAGGCCAGTAAAGGAGGCGCGTATGCGGCAAATAATCTAAGCCCT-3′ |
| flgD3′delet8 | 5′-TTAAAACGCTGACATTAGGCCAGTAAAGGAGGCGCGTATGATCTAAGCCCTTACACTTAT-3′ |
| flgD3′delet11 | 5′-TTAAAACGCTGACATTAGGCCAGTAAAGGAGGCGCGTATGGTTCGGCAAATAATCTAAGC-3′ |
| flgDtetdelet | 5′-TTAAAACGCTGACATTAGGCCAGTAAAGGAGGCGCGTATGATTGGCCCATTACTGTTTAC-3′ |
| flgE3′RT-PCR | 5′-CCACTTTGGAACCGGCGAAC-3′ |
| flgFrtR | 5′-TGGTTAACTGCCCGGTCGGA-3′ |
Site-directed mutagenesis.
The ΔflgD(Δaa2-232) (missing amino acids 2 through 232) G828T (TH9945) and ΔflgD(Δaa2-232) G827A (TH9946) alleles were obtained by first constructing a deletion/insertion of the region between the two stop codons of flgC and flgD in LT2 with a tetRA element as described above using λ-Red-mediated recombination. The tetRA element was amplified by PCR with primers flgCtetR and flgD-p-3tetRA. The PCR-amplified tetRA fragment is flanked at its 5′ and 3′ ends by 40 bases of DNA sequence identical to the 40 bases upstream of the flgC stop codon and the 40 bases downstream of the flgD stop codon. The tetRA element was replaced using λ-Red-mediated recombination and Tcs medium with PCR products that contain the flgBC sequence and 140 bp of 5′ flgE sequence from TH9936 and TH9938, respectively, as described above. The PCR products were amplified using primers flgBSeq5 and flgER.
Construction of plasmids pflgD and pflgDE.
The flgD region of LT2 was amplified by PCR using primers flgDF and flgDR (Table 2). The PCR products were digested with NcoI and PstI and ligated into pTrc99A, digested with NcoI and PstI, to make pflgD. The flgDE region of LT2 was also amplified by PCR using primers flgDF and flgERHindIII (Table 2). The PCR products were digested with NcoI and HindIII and ligated into pTrc99A, digested with NcoI and HindIII, to make pflgDE.
Construction of flgD deletion strains.
The flgD gene was replaced with a tetRA element in LT2 using λ-Red-mediated recombination as described above. The tetRA element was amplified by PCR with primers flgD-p-5tetRA and flgD-p-3tetRA (Table 2). The tetRA fragment is flanked at its 5′ and 3′ ends by 40 bases of DNA sequence identical to the 40 bases upstream of the 56th amino acid codon of flgD and the 40 bases downstream of the flgD stop codon. To construct flgD deletion strains, the ΔflgD::tetRA element was replaced using λ-Red-mediated recombination and Tcs medium with PCR products that contain the relevant flgD regions. Primers flgDdelet, flgDFRTdel, flgD5′deletI, flgD5′deletII, flgD3′delet1, flgD3′delet2, flgD3′delet3, flgD3′delet4, flgD3′delet5, flgD3′delet11, flgD3′delet6, and flgD3′delet8 (Table 2) were used to make TH8547, TH9924, TH9926, TH9927, TH9929, TH9930, TH9931, TH9932, TH9933, TH9943, TH9934, and TH9935, respectively. Each of the 5′ 40 bases of these primers are identical to each of the 40 bases upstream of the deleted regions, and each of the 3′ 20 bases of the primers are identical to the 20 bases downstream of the deleted regions. Primer flgE3′RT-PCR was used as a reverse primer that is complementary to the region 20 bases upstream of base 146 of the flgE coding sequence. The genomic DNA of LT2 was used as a PCR template. Primer flgDtetdelet, whose 5′ 40 bases are identical to the 40 bases upstream of second amino acid codon of flgD and whose 3′ 20 bases are identical to the 20 bases downstream of the 1,041st base of tetA, and primer flgE3′RT-PCR were used to make TH9944. The genomic DNA of the ΔflgD::tetRA strain was used as a PCR template.
Assay for FlgE-β-lactamase activity.
One-milliliter cultures in LB medium were grown overnight at 37°C with aeration. The cells were diluted 100-fold into 3 ml LB medium and grown at 37°C with aeration to mid-log phase (optical density at 600 nm [OD600] of 0.5 to 0.7). The cells were diluted 50-fold in LB medium containing variable concentrations of ampicillin (Ap) (twofold serially diluted from 800 μg/ml to 3.125 μg/ml). After 6 h of growth at 37°C, the OD600 of each sample was measured. If the OD600 was lower than 0.05, the Ap concentration in LB was considered as a MIC.
Preparation of RNA.
Three-milliliter cultures grown to about 5 × 108 cells/ml were pelleted and resuspended in 50 μl protoplasting buffer (15 mM Tris-HCl [pH 8], 0.45 M sucrose, 8 mM EDTA). A 5-μl portion of Lysozyme (10 mg/ml) was added, and the mixture was incubated for 5 min at 25°C. A 500-μl portion of RNAwiz (Ambion Inc., Austin, TX) was added, and the sample was mixed thoroughly by vortexing vigorously for 10 s and incubated for 5 min at 25°C. After the addition of 100 μl of chloroform (Sigma), the sample was mixed by vortexing vigorously for 20 s and incubated for 10 min at 25°C. The sample was centrifuged at 10,000 × g for 15 min at 4°C. The 250-μl aqueous-phase sample was extracted and added to 250 μl of water. After the addition of 500 μl of isopropyl alcohol (Sigma), the sample was mixed and incubated for 10 min at 25°C. The precipitate was collected by centrifugation at 10,000 × g for 15 min at 4°C and washed with 500 μl of 75% cold ethanol. The precipitate was then dissolved in 50 μl of RNA storage buffer (Ambion Inc., Austin, TX). The concentration of RNA was determined by measuring the absorbance at 260 nm.
RACE.
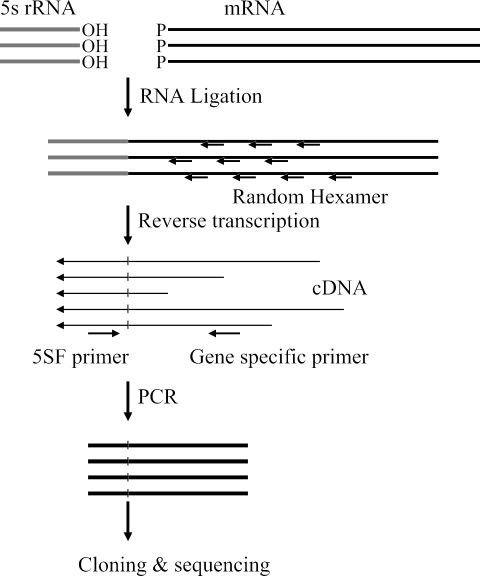
Cells were grown at 37°C in LB medium. The RNA was extracted as described above, and when required, 5′ phosphate groups were removed by treatment with tobacco acid pyrophosphate (Epicenter). RNA ligation was performed at 37°C for 3 h in a 25-μl volume containing 2.5 μg of total RNA, 10 units of T4 RNA ligase (Ambion Inc., Austin, TX), and 20 units of RNasin (Promega). The ligated RNA was cleaned up using a G-50 column (Amersham Biosciences), and 1 μg of RNA was reverse transcribed at 37°C for 1 h in a 20-μl reaction volume containing 4 units of Ominiscript reverse transcriptase (QIAGEN), 0.5 mM each deoxynucleoside triphosphate, 0.5 mM random hexamer (Invitrogen), and 10 units of RNasin. PCR amplification was performed with a gene-specific primer and primer 5SF (5′-GCGAGAGTAGGGAACTGCCA-3′), complementary to the 5S rRNA gene, using Hot Start Taq polymerase (QIAGEN). The PCR products were purified by using a PCR purification kit (QIAGEN) and separated by agarose gel electrophoresis. The desired band was eluted, cloned into the pGEM-T-Easy vector (Promega), and sequenced (Fig. 1).
FIG. 1.
Flow diagram of 5′ RACE. Total RNAs were randomly ligated and then reverse transcribed using a random primer. PCR amplification was performed with 5SF and gene-specific primers. The PCR products were cloned and sequenced.
Complementation tests.
Complementation tests were performed using a motility plate assay. The pflgD or pflgDE plasmid was introduced into flgD mutants, and the motility activity of those strains was measured on motility agar plates (3 g Difco agar, 10 g Bacto tryptone, and 5 g NaCl per liter) without IPTG (isopropyl-β-d-thiogalactopyranoside). Motility plates were illuminated with a dark-field colony counter (Reichert) and photographed using a digital camera (Nikon E990).
Isolation and characterization of motile revertants from strain TH9947 [pflgD/ΔflgD(aa2-232)].
Motile revertants of strain TH9947 (pflgD/ΔflgD) were isolated by transferring cells from isolated colonies with a round toothpick into motility plates by poking the cells through the agar followed by incubation at 30°C until revertant motility flares appeared (up to 3 days). Motile (Mot+) revertants were single-colony isolated two times on an LB agar plate for purification. The Mot+ phenotypes were mapped to the flg operon by cotransduction with the pyrC locus. A P22 transducing phage grown on the motile revertants was used to infect pflgD/TH9925 [ΔpyrC::FRT-Kan-FRT(FKF) ΔflgD::tetRA], and the mixture was incubated on minimal glucose plates to select pyrC+ transductants. One-hundred pyrC+ Tcs colonies were screened for motility. The screening of only the pyrC+ Tcs transductants ensured that the region of the flg operon between pyrC and flgD was replaced (although a low level of double recombination events that separately replaced the pyrC and flgD regions of the recipient DNA [a few percent] was also possible). If the Mot+ revertant allele was upstream of flgD, we expected nearly 100% of the pyrC+ Tcs transductants to be Mot+. If the Mot+ revertant allele was downstream of the ΔflgD::tetRA allele, we expected significantly less than 100% of the pyrC+ Tcs transductants to be Mot+. The percentage of pyrC+ Tcs transductants was determined for each cross, and revertants predicted to be upstream of the ΔflgD::tetRA allele were subjected to DNA sequence analysis of the region upstream of flgD.
RESULTS
Analysis of mRNA processing in the flgB-L operon.
The finding that FlgE was posttranscriptionally regulated in response to flagellar assembly led us to examine a possible role of mRNA processing in the expression of the 9.9-kbp class 2 flgBCDEFGHIJKL operon. Also, cleavage of the flgBCDEFGHIJ operon has been implicated in posttranscriptional regulation of the flgE gene in Rhodobacter sphaeroides (4, 18).
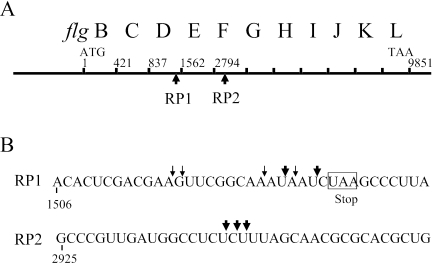
The mRNA processing sites in the upstream and downstream regions of the flgE gene of S. enterica serovar Typhimurium were determined by the technique of RACE (Fig. 1) (3). Following the ligation of genomic RNA to the 5s rRNA gene, two primers, flgE3′RT-PCR and flgFrtR, which are complementary to flgE and flgF, respectively, were used as gene-specific primers for performing PCR of the region. The PCR products showed discrete DNA bands by agarose gel electrophoresis (date not shown). The DNA bands were eluted, cloned, and analyzed by DNA sequencing. We analyzed 20 samples of each PCR product to determine the mRNA processing sites that were 5′ and 3′ of the flgE coding sequence (Fig. 2). The mRNA processing sites in the flgD and flgF genes were located just upstream of the flgD stop codon and 250 bases into the flgF coding region. Major (more than 5 out of 20 samples) and minor (less than 5 out of 20 samples) mRNA processing sites were mapped in each of the mRNA processing sites (Fig. 2B).
FIG. 2.
RNA processing sites in the flgB-L operon. (A) Organization of genes in the flgB-L operon. The mRNA processing sites are indicated. (B) Major (large arrows) and minor (small arrows) mRNA processing sites were mapped. The stop codon of flgD is indicated in RP1. RP, RNA processing.
Deletion of the flgD coding sequence inhibits flgE gene expression.
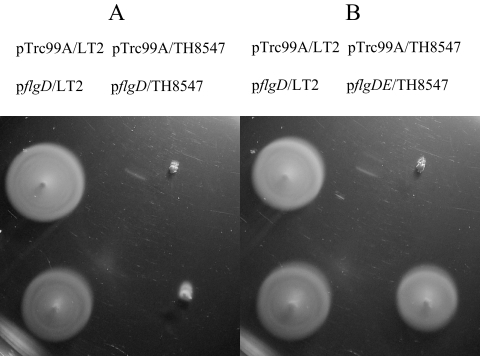
As described above, we detected mRNA processing sites at the end of the flgD coding sequence. To test whether the upstream flgD sequence might affect the production of FlgE, the flgD gene was deleted, leaving just the start and stop codons (strain TH8547). Our rationale was that if mRNA processing of the flgD region is necessary for the expression of flgE, then the ΔflgD allele would not be complemented with a plasmid expressing flgD alone but would be complemented with a plasmid expressing both flgD and flgE. This was found to be the case. The flgD and flgDE genes were cloned separately into the pTrc99A expression plasmid, resulting in the pflgD and pflgDE expression plasmids, respectively. Plasmid pflgD or pflgDE was introduced into the ΔflgD strain and screened for motility on tryptone soft-agar plates. Plasmid pflgD did not restore motility, while the pflgDE plasmid did (Fig. 3). This result suggested that the loss of the flgD coding region has a negative effect on expression of the flgE gene, perhaps due to the loss of the mRNA processing site.
FIG. 3.
Motility assay of flgD-deficient strain TH8547 with pflgD (A) or pflgDE (B). pTrc99A (vector control), pflgD, or pflgDE was transformed into the wild type (LT2) or TH8547. pTrc99A and pflgD did not affect the motility of the wild type. TH8547 was not complemented with pflgD (A) but was complemented with pflgDE (B).
The effects of flgD deletions on downstream flgE gene expression.
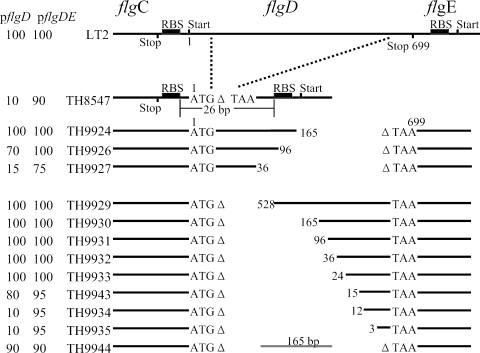
The deletion of the flgD coding region from amino acid 2 to the stop codon failed to express the downstream flgE gene. Deletions of flgD that left different amounts of the 5′ or 3′ coding sequences were constructed to determine which sequences of flgD are required for flgE gene expression (Fig. 4). Deletions of the 5′ coding sequence of flgD, leaving 15 to 528 bases of the 3′ coding sequence, exhibited normal complementation by pflgD alone, indicating normal expression of flgE. The deletions leaving 12 or 3 bases of the 3′ coding sequence did not express flgE. There was a very specific cutoff in flgE expression going from 15 bases at the 3′ end to 12 bases. This corresponds exactly to the very 3′ mRNA processing site for the flgE gene located by RACE in the 5′ end of the flgD coding sequence. Deletions of the 3′ coding sequence of flgD appeared to have a gradient effect on flgE expression. Deletions leaving 36, 96, or 165 bases of the flgD 5′ coding sequence showed weak, intermediate, and normal levels of flgE expression, respectively. Also, a deletion of 5′ and 3′ flgD coding sequences that left 165 bases from the middle of the flgD gene showed normal flgE expression. These results suggest that the 3′ 15 bases of the flgD coding sequence are critical (depending on the context) for normal flgE expression, but this can be suppressed if there is a sufficient coding sequence of any kind (between 96 and 165 bases) upstream of the flgD stop codon.
FIG. 4.
Construction of flgD strains and motility assay of flgD-deficient strains with pflgD or pflgDE. The motility activities (percentage of wild-type motility with pflgD or pflgDE) of flgD strains are indicated in the left two columns.
The flgD RBS prevents flgE gene expression.
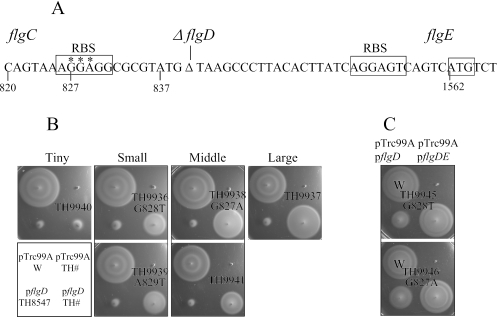
The complete deletion of the flgD coding sequence left 26 bp of DNA sequence between the putative flgD and flgE ribosome-binding sites (RBSs) (Fig. 5A). It was possible that the flgD RBS was interfering with the ability of ribosomes to initiate at the flgE RBS unless a sufficient amount of flgD coding sequence (between 96 and 165 bases) was translated. In an attempt to identify bases that interfered with flgE gene translation due to ΔflgD mutation, motile revertants of the pflgD/ΔflgD6219 strain were isolated and characterized. Motile revertants were isolated from 16 independent cultures and divided into four groups according to the degree of motility observed (Fig. 5B). The motile revertants were screened for linkage to the flgD region (see Materials and Methods). Some revertants showed 100% cotransduction with the pyrC-flgD region. Other revertants showed variable linkages, ranging from 10% to 60% cotransduction with the pyrC-flgD region. We expected that the revertants showing 100% cotransduction with the pyrC-flgD region were mutated in the flgBC region. Thus, we sequenced flgBC and 140 bp of the 5′ flgE region from three independent mutants (TH9936, TH9938, and TH9939) (Fig. 5A and B). Interestingly, all three mutants had different base substitution mutations in the RBS of flgD (G828T, G827A, and A829T, numbered relative to the flgB start codon in the wild type) (Fig. 5A and B). We constructed mutants with either the G828T (TH9945) or G827A (TH9946) substitution mutations by using site-directed mutagenesis to confirm that the phenotype observed was specific for these mutations. These strains were motile, suggesting that the flgD RBS was interfering with the ability of ribosomes to initiate at the flgE RBS in the ΔflgD mutant (Fig. 5C). We also sequenced the flgBC genes and 140 bp of the 5′ flgE region of three other motile revertants (TH9937, TH9940, and TH9941), which were less than 100% linked to the pyrC-flgD region. As expected, there were no mutations present in flgBC and 140 bp of the 5′ flgE region of these revertants. The cotransduction frequencies suggest that these mutations reside in the flgG-L operon downstream of the flgE gene and may affect the mRNA secondary structure in a manner that inhibits the ability of ribosomes to recognize the flgD RBS.
FIG. 5.
Isolation and characterization of motile revertants of pflgD/TH8547. (A) The sequences of two RBSs are shown. The sites of the mutations that restored the motility activity in TH9936, TH9938, and TH9939, respectively, are indicated by asterisks. The numbers are relative to the flgB start codon, ATG. (B) Motility assay of motile revertants. Three point mutants are indicated. (C) TH9945 and TH9946 were made by site-directed mutagenesis. Their motility activities are similar to those of TH9936 and TH9938, respectively.
An FlgE-β-lactamase fusion is secreted into the periplasm in rod mutant strains.
Previous work from our laboratory showed that the FlgE (flagellar hook subunit) protein was regulated in response to the stage of flagellum assembly independent of transcriptional regulation (6). Mutants early in flagellar assembly and late in flagellar assembly had cell-associated FlgE levels comparable to those observed in wild-type cells. Mutants defective in rod assembly (flgB, flgC, flgF, flgG, and flgJ mutants), with the exception of a fliE mutant, had little or no detectable FlgE. Mutants defective in the stages between rod assembly and hook polymerization, including the hook-scaffold gene, flgD, or P- and L-ring assembly genes flgA, flgH, and flgI, had between 15 and 30% of wild-type levels of FlgE protein. In all these flagellar mutant backgrounds, flgE mRNA levels were similar to those seen in the wild-type strain. It was previously shown that the hook-capping protein, FlgD, could be detected in the periplasm in rod and ring mutant strains (except in a fliE mutant) but not in strains defective in the flagellar type III secretion apparatus (27). It is possible that both FlgD and FlgE are secreted into the periplasm of rod mutants, but FlgE is much less stable. This would be consistent with the results reported previously by Bonifield and Hughes, which showed that FlgE was barely detectable or absent in the rod mutants but present at low levels in the ring mutants (6). In ring mutants, hook polymerization initiates in the absence of the ring structures (19), so some FlgE protein is stabilized by polymerization into nascent hook structures. Our hypothesis is that both FlgD and FlgE proteins are secreted into the periplasm of rod mutants but that FlgE is more sensitive to proteolysis and is rapidly degraded.
To test for FlgE secretion into the periplasm, we took advantage of the recent finding that the fusion of β-lactamase lacking its type II secretion signal (Bla) to the C terminus of FlgM had the dual benefit of preventing FlgM proteolysis in the periplasm and providing a direct assay for FlgM-Bla secretion by conferring an ampicillin-resistant (Apr) phenotype to the cells but only if the fusion was secreted into the periplasm (2). Retention of FlgM-Bla in the cytoplasm or complete secretion from the cell resulted in an Aps phenotype. C-terminal fusions of Bla to FlgE were constructed and assayed for secretion of FlgE-Bla into the periplasm in different flagellar mutant strains. The results are presented in Table 3. Wild-type strain LT2 is the Aps control with a MIC of <3 μg/ml for Ap. Strain TH6733, the positive Apr control that has wild-type β-lactamase with its intact secretion signal expressed from a MudA transposon insertion (11), has a MIC of 400 μg/ml for Ap. A chromosomally expressed flgE-bla fusion in which the N-terminal 23 amino acids of the β-lactamase protein, the type II secretion signal (14), was removed and the rest of the bla coding region was fused to the very C-terminal codon of flgE was constructed. The resulting FlgE-Bla fusion confers resistance to high levels of Ap in a strain deleted for the proximal rod genes flgB and flgC (TH9949). The level of Apr (MIC of 400 μg/ml) was the same level observed in cells expressing the wild-type bla+ gene, whose protein product has its intact N-terminal type II secretion signal (TH6733). An intermediate Apr level (MIC of 100 μg/ml) was observed in a strain with intact flagellar rods, and the only flagellar mutation present was the fusion of Bla to the C terminus of FlgE (TH9948). The Apr observed in the Rod+ and rod mutant strains was dependent on both flagellar gene expression (TH9951) and the presence of a flagellar secretion apparatus (TH9950). The assembly of the flagellar type III secretion system is completely blocked in strain TH9950, which contains a transposon insertion in the fliF gene. The FliF protein constitutes the MS ring on which the flagellar type III secretion apparatus is constructed (24, 25).
TABLE 3.
MICs of ampicillin for the flgE-bla mutants in S. enterica serovar Typhimurium
| Strain | Relevent genotype | MIC (μg/ml) |
|---|---|---|
| LT2 | Wild type | <3 |
| TH6733a | flgE5315::MudA | 400 |
| TH9948 | flgE-bla | 100 |
| TH9949 | flgE-bla ΔflgBC | 400 |
| TH9950 | flgE-bla ΔflgBC fliF5036::Tn10dCm | 3 |
| TH9951 | flgE-bla ΔflgBC flhD5704::Tn10dTc | <3 |
| TH9952 | flgE-bla ΔflgBC ΔfliE::tetRA | 50 |
We used TH6733, which has a promoter and an intact bla gene (complete with type II secretion signal sequence) in MudA, as a positive control.
We also tested the effect of the rod protein FliE on the secretion of FlgE-Bla in a strain deleted for flgB and flgC and the rod gene, fliE (strain TH9952) (Table 3). FlgE was present in cell extracts from the fliE mutant background, but no FlgE was detectable in extracts from the other rod-specific mutant strains (flgB, flgC, flgF, flgG, and flgJ mutants) (6). Also, in separate studies, the secretion of FlgD was dependent on FliE (12, 27), and FliE was shown to interact with the proximal rod protein FlgB (28). Taken together, these data suggest that FliE is both a part of the flagellar type III secretion apparatus and a rod component.
The introduction of the fliE deletion into the ΔflgBC flgE-bla strain lowered the Ap MIC from 400 μg/ml to 50 μg/ml, suggesting a strong defect in secretion, but secretion could still occur, compared to the complete absence of the flagellar secretion system (Ap MIC of 3 μg/ml for TH9950 [fliF]). Thus, FliE is not essential for flagellar type III secretion but stimulates secretion eightfold by our assay.
DISCUSSION
In a previous study, we determined that the level of cellular FlgE protein was affected by a mutation in the flagellar assembly pathway, while flgE mRNA levels were unaffected (6). This led us to examine the possibility that the flgE gene, encoding the flagellar hook structural subunit protein, might be regulated by a posttranscriptional mechanism in a manner that couples flgE gene expression to FlgE assembly into the flagellum.
In this study, we determined the location of mRNA processing sites that flank the flgE gene of S. enterica serovar Typhimurium. Processing occurred within the last 15 bases of the flgD coding region and 250 bases into the coding region for the flgF gene. By examining the effect of flgD deletions on downstream flgE expression, we also determined that translation of flgD could have a negative effect on downstream flgE translation. The negative effect of flgD translation on flgE gene expression did not occur if enough flgD sequence was present (at least 100 bases) or if the last 15 bases upstream of the flgD stop codon were present. Somehow, the presence of the last 15 bases of the flgD coding sequence had the same effect as the presence of 165 bases of the N terminus on the internal flgD coding sequence. There are 27 bases between the flgD stop and flgE start codons. The 15 bases upstream of the flgD stop codon might play a role in the secondary structure of the flg mRNA that allows the binding of ribosomes to the flgE RBS. The identification of mRNA processing sites in this 15-bp region is consistent with the possibility that this sequence is needed for flgE processing and that the processing of the flgE gene from the flgB-L mRNA transcript might be critical for normal flgE expression. This might also be part of a mechanism that prevents FlgE secretion until the rod structure is completed. Thus, the processing of the flgE gene from the flgB-L mRNA transcript may play a role in the coupling of flgE translation to assembly as a mechanism to increase the efficiency of the assembly process.
In this study, we also present evidence to support the hypothesis that the absence of FlgE in rod mutant strains reported previously (6) was due to the export of FlgE into the periplasm, where it was degraded. The fusion of β-lactamase to FlgE (FlgE-Bla) conferred a strong Apr phenotype that is consistent with the export of FlgE-Bla into the periplasm, and the fusion of Bla to the C terminus of FlgE appears to stabilize periplasmic FlgE. It is not known if rod and hook proteins are exported in the order they are assembled. Our results suggest that rod and hook subunits can be exported simultaneously and that specific protein subunits that are exported before they are needed for assembly, such as a hook subunit secreted before the rod is completed, are simply degraded in the periplasm.
Acknowledgments
We thank the members of the Hughes laboratory for helpful discussions and comments on the manuscript.
This work was supported by PHS grant GM62206 from the National Institutes of Health to K.T.H. and grant KRF-2004-037-C00234 (to H.J.L.) from the Korea Research Foundation.
REFERENCES
- 1.Aizawa, S.-I. 1996. Flagellar assembly in Salmonella typhimurium. Mol. Microbiol. 20:1-4. [DOI] [PubMed] [Google Scholar]
- 2.Aldridge, P., J. E. Karlinsey, E. Becker, F. F. V. Chevance, and K. T. Hughes. Flk prevents premature secretion of the anti-σ factor FlgM into the periplasm. Mol. Microbiol., in press. [DOI] [PMC free article] [PubMed]
- 3.Argaman, L., R. Hershberg, J. Vogel, G. Bejerano, E. G. Wagner, H. Margalit, and S. Altuvia. 2001. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 11:941-950. [DOI] [PubMed] [Google Scholar]
- 4.Ballado, T., L. Camarena, B. Gonzalez-Pedrajo, E. Silva-Herzog, and G. Dreyfus. 2001. The hook gene (flgE) is expressed from the flgBCDEF operon in Rhodobacter sphaeroides: study of an flgE mutant. J. Bacteriol. 183:1680-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonifield, H. R., and K. T. Hughes. 2003. Flagellar phase variation in Salmonella enterica is mediated by a posttranscriptional control mechanism. J. Bacteriol. 185:3567-3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonifield, H. R., S. Yamaguchi, and K. T. Hughes. 2000. The flagellar hook protein, FlgE, of Salmonella enterica serovar Typhimurium is posttranscriptionally regulated in response to the stage of flagellar assembly. J. Bacteriol. 182:4044-4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chadsey, M. S. 1998. Regulation of the flagellar specific sigma factor, sigma 28, of Salmonella typhimurium by the anti-sigma factor FlgM. Ph.D. thesis. University of Washington, Seattle, Wash.
- 8.Chilcott, G. S., and K. T. Hughes. 2000. The coupling of flagellar gene expression to flagellar assembly in Salmonella typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64:694-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gillen, K. L., and K. T. Hughes. 1991. Molecular characterization of flgM, a gene encoding a negative regulator of flagellin synthesis in Salmonella typhimurium. J. Bacteriol. 173:6453-6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Groisman, E. A. 1991. In vivo genetic engineering with bacteriophage Mu. Methods Enzymol. 204:180-212. [DOI] [PubMed] [Google Scholar]
- 12.Hirano, T., T. Minamino, K. Namba, and R. M. Macnab. 2003. Substrate specificity classes and the recognition signal for Salmonella type III flagellar export. J. Bacteriol. 185:2485-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes, K. T., K. L. Gillen, M. J. Semon, and J. E. Karlinsey. 1993. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science 262:1277-1280. [DOI] [PubMed] [Google Scholar]
- 14.Kadonaga, J. T., A. E. Gautier, D. R. Straus, A. D. Charles, M. D. Edge, and J. R. Knowles. 1984. The role of the beta-lactamase signal sequence in the secretion of proteins by Escherichia coli. J. Biol. Chem. 259:2149-2154. [PubMed] [Google Scholar]
- 15.Karlinsey, J. E., and K. T. Hughes. 2006. Genetic transplantation: Salmonella enterica serovar Typhimurium as a host to study sigma factor and anti-sigma factor interactions in genetically intractable systems. J. Bacteriol. 188:103-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karlinsey, J. E., S. S. Tanaka, V. Bettenworth, S. Yamaguchi, W. Boos, S. I. Aizawa, and K. T. Hughes. 2000. Completion of the hook-basal body of the Salmonella typhimurium flagellum is coupled to FlgM secretion and fliC transcription. Mol. Microbiol. 37:1220-1231. [DOI] [PubMed] [Google Scholar]
- 17.Kleckner, N. 1983. Transposon Tn10, p. 261-298. In J. A. Shapiro (ed.), Mobile genetic elements. Academic Press, New York, N.Y.
- 18.Kobayashi, K., T. Saitoh, D. S. Shah, K. Ohnishi, I. G. Goodfellow, R. E. Sockett, and S. I. Aizawa. 2003. Purification and characterization of the flagellar basal body of Rhodobacter sphaeroides. J. Bacteriol. 185:5295-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kubori, T., N. Shimamoto, S. Yamaguchi, K. Namba, and S. Aizawa. 1992. Morphological pathway of flagellar assembly in Salmonella typhimurium. J. Mol. Biol. 226:433-446. [DOI] [PubMed] [Google Scholar]
- 20.Kutsukake, K. 1997. Autogenous and global control of the flagellar master operon, flhD, in Salmonella typhimurium. Mol. Gen. Genet. 254:440-448. [DOI] [PubMed] [Google Scholar]
- 21.Kutsukake, K. 1994. Excretion of the anti-sigma factor through a flagellar substructure couples flagellar gene expression with flagellar assembly in Salmonella typhimurium. Mol. Gen. Genet. 243:605-612. [DOI] [PubMed] [Google Scholar]
- 22.Liu, X., and P. Matsumura. 1994. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J. Bacteriol. 176:7345-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macnab, R. M. 1996. Flagella and motility, p. 123-145. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 24.Macnab, R. M. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 57:77-100. [DOI] [PubMed] [Google Scholar]
- 25.Macnab, R. M. 2004. Type III flagellar protein export and flagellar assembly. Biochim. Biophys. Acta 1694:207-217. [DOI] [PubMed] [Google Scholar]
- 26.Maloy, S. R. 1990. Experimental techniques in bacterial genetics. Jones and Bartlett, Boston, Mass.
- 27.Minamino, T., and R. M. Macnab. 1999. Components of the Salmonella flagellar export apparatus and classification of export substrates. J. Bacteriol. 181:1388-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minamino, T., S. Yamaguchi, and R. M. Macnab. 2000. Interaction between FliE and FlgB, a proximal rod component of the flagellar basal body of Salmonella. J. Bacteriol. 182:3029-3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Motaleb, M. A., M. S. Sal, and N. W. Charon. 2004. The decrease in FlaA observed in a flaB mutant of Borrelia burgdorferi occurs posttranscriptionally. J. Bacteriol. 186:3703-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohnishi, K., K. Kutsukake, H. Suzuki, and T. Iino. 1992. A novel transcriptional regulatory mechanism in the flagellar regulon of Salmonella typhimurium: an anti sigma factor inhibits the activity of the flagellum-specific sigma factor, σF. Mol. Microbiol. 6:3149-3157. [DOI] [PubMed] [Google Scholar]
- 31.Ohnishi, K., K. Kutsukake, H. Suzuki, and T. Iino. 1990. Gene fliA encodes an alternative sigma factor specific for flagellar operons in Salmonella typhimurium. Mol. Gen. Genet. 221:139-147. [DOI] [PubMed] [Google Scholar]
- 32.Xie, H., N. Kozlova, and R. J. Lamont. 2004. Porphyromonas gingivalis genes involved in fimA regulation. Infect. Immun. 72:651-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yanagihara, S., S. Iyoda, K. Ohnishi, T. Iino, and K. Kutsukake. 1999. Structure and transcriptional control of the flagellar master operon of Salmonella typhimurium. Genes Genet. Syst. 74:105-111. [DOI] [PubMed] [Google Scholar]