Abstract
Escherichia coli microcin J25 (MccJ25) is a plasmid-encoded antibiotic peptide consisting of 21 l-amino acid residues (G1-G-A-G-H5-V-P-E-Y-F10-V-G-I-G-T15-P-I-S-F-Y20-G). E. coli RNA polymerase (RNAP) is the intracellular target of MccJ25. MccJ25 enters cells after binding to specific membrane transporters: FhuA in the outer membrane and SbmA in the inner membrane. Here, we studied MccJ25 mutants carrying a substitution of His5 by Lys, Arg, or Ala. The inhibitory effects on cellular growth and in vitro RNAP activity were determined for each mutant microcin. The results show that all mutants inhibited RNAP in vitro. However, the mutants were defective in their ability to inhibit cellular growth. Experiments in which the FhuA protein was bypassed showed that substitutions of MccJ25 His5 affected the SbmA-dependent transport. Our results thus suggest that MccJ25 His5 located in the lariat ring is involved, directly or indirectly, in specific interaction with SbmA and is not required for MccJ25 inhibition of RNAP.
Escherichia coli microcin J25 (MccJ25) is a plasmid-encoded, ribosomally synthesized peptide antibiotic consisting of 21 amino acid residues (5, 13). It is primarily active on gram-negative bacteria related to the producer strain.
Four plasmid genes, mcjA, mcjB, mcjC, and mcjD, are involved in MccJ25 production (16). The mcjA gene codes for a 58-amino-acid-long MccJ25 precursor. The processing enzymes McjB and McjC are required for production of active, mature MccJ25 from the McjA precursor. The MccJ25 immunity protein, McjD, is highly similar to membrane translocator proteins of the ABC exporter family. Thus, immunity conferred by McjD is probably mediated by active efflux of the peptide, which would keep its concentration inside the producing cell below a critical level and thus ensure survival. The E. coli outer membrane protein TolC may form an export complex with McjD and is thus also implicated in MccJ25 secretion (7).
MccJ25 uptake into E. coli cells is dependent on the outer membrane protein FhuA and the inner membrane proteins TonB, ExbD, ExbB, and SbmA (14, 15). The complex TonB-ExbB-ExbD uses the proton-motive force from the cytoplasmic membrane to transduce energy to the outer membrane, thus providing energy for transport. To accomplish energy transfer, TonB contacts the outer membrane receptor FhuA (10). SbmA transports MccJ25 through the inner membrane. Once inside, the peptide inhibits E. coli RNA polymerase (RNAP) (8, 19).
The tertiary structure of MccJ25 (G1-G-A-G-H5-V-P-E-Y-F10-V-G-I-G-T15-P-I-S-F-Y20-G) was recently elucidated (2, 12, 18). MccJ25 contains a lactam linkage between the α-amino group of Gly1 and the γ-carboxyl of Glu8, forming an eight-residue ring (Gly1 to Glu8) named the lariat ring. The “tail” (Tyr9 to Gly21) passes through the ring, with Phe19 and Tyr20 straddling each side of it, sterically trapping the tail within the ring. The MccJ25 amino acids F10-V-G-I-G-T-P16 form a β-hairpin structure comprising two β-strands (F10-V11 and T15-P16) and a β-turn (V11-G-I-G14). This region is involved in antibiotic uptake (4) by interacting with the outer membrane protein FhuA (9). MccJ25 carries only two charged residues: a positively charged histidine (His5) localized in the lariat ring and a negatively charged glycine (Gly21) at the carboxyl terminus of the molecule. The two charged groups are close in the three-dimensional structure (12) and were shown to be important for MccJ25 activity (3). Amidation of MccJ25 Gly21 specifically blocked RNAP inhibition but not transport inside the cell (17). On the other hand, carbethoxylation of the imidazole ring of MccJ25 His5 decreased the inhibitory effect of the antibiotic on both E. coli cell growth and in vivo RNA synthesis. The biological activity of carbethoxylated MccJ25 was completely recovered after treatment with hydroxylamine, which gives the native MccJ25. Thus, it appeared that the polar histidyl residue is required for MccJ25 transport into the cell, its extrusion outside the cell, or RNAP inhibition (3). In the present paper, we offer evidence that the His5 residue of the lariat ring is important for transport of the antibiotic into the cell through interaction with the inner membrane protein SbmA.
MATERIALS AND METHODS
Bacteria and culture media.
The bacteria and plasmids used in this work are listed in Table 1. Liquid media were Luria broth (LB) or M9 minimal medium supplemented with glucose (0.2%), thiamine (1 μg/ml), and casein enzymatic hydrolysate (0.2%). Solid media contained 1.2% agar. When required, antibiotics were added at the following concentrations: ampicillin, 50 μg/ml; chloramphenicol, 25 μg/ml.
TABLE 1.
E. coli strains and plasmidsa
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| MC4100 | F−araD139 Δ(argF-lac)205 λ−rpsL150 flbB5301 relA deoC1 pstF25 | CGSC |
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 supE44 hsdR17(rK− mK−) thi-1 gyrA (Nalr) | BRL |
| MG1655 | F−λ−rph-1 | CGSC |
| AB1133 | F−thr-1 ara-14 leuB6 lacY1 (gpt-proA)62 supE44 galK2 λ-rac hisG4 rfbD1 rpsL31 (smr) kdgK51 xyl-5 mtl-1 argE3 thi-1 | CGSC |
| XL1-Blue | recA1 endA1 gyrA96 thi hsdR17(rK− mK−) supE44 relA1 lac [F′proAB+lacIqZΔM15::Tn10 (Tetr)] | Stratagene |
| AB259 | HfrH supQ80 λ−relA1 spoT1 thi-1 | CGSC |
| EZE100 | MC4100 sbmA::Tn5 | This work |
| SBG303 | MC4100 fhuA::Tn5 | 14 |
| RYC1000 | F−araD139 ΔlacU169 Δrbs7 rpsL relA thiA recA56 gyrA | CGSC |
| Plasmids | ||
| pTUC342 | pBR322 mcjA, Apr | |
| pTUC341 | pBR322 mcjBCD, Apr | |
| pEVA100 | pTUC342 mcjA (His5xAla) | This work |
| pEVA101 | pTUC342 mcjA (His5xLys) | This work |
| pEVA102 | pTUC342 mcjA (His5xArg) | This work |
| pEVA103 | pTUC342 mcjA (Gly2xAla) | This work |
| pMM73.4 | pBR322 carrying sbmA+, Tcr | F. Moreno |
CGSC, E. coli Genetic Stock Center; BRL, Bethesda Research Laboratory; Apr, ampicillin resistant; Tcr, tetracycline resistant.
Mutagenesis and isolation of MccJ25 analogues.
Plasmid pTUC342, carrying the mcjA gene under its own promoter, was used in order to obtain the MccJ25 mutants. They were constructed with the QuikChange mutagenesis kit (Stratagene) according to the manufacturer's recommendations. Mutations were confirmed by DNA sequencing of the mutagenized plasmids. To produce and purify the microcin variants, the mutant plasmids were transformed into a strain carrying the compatible plasmid pTUC341, containing the mcjBCD genes required for synthesis and export of the antibiotic (16). Native MccJ25 and MccJ25 analogues were purified by high-performance liquid chromatography as described previously (3). Amino acid substitutions in mutant peptides were confirmed by electrospray ionization mass spectrometry.
MccJ25 sensitivity determination.
Antibiotic activities of native MccJ25 and each analogue were determined using a spot-on-lawn test. Doubling dilutions of each sample (1 mg/ml) were prepared in distilled water containing 0.01% Tween 80 to prevent peptide aggregation. Ten-microliter samples of each dilution were spotted on M9 medium. After the drops had dried on the plates, they were layered with 4 ml of soft agar containing approximately 107 cells of the indicator strain. The plates were incubated overnight at 37°C and examined for growth inhibition. The concentration (μM) of the last dilution giving a clear or turbid spot was taken as the MIC of MccJ25 and its analogues.
Antimicrobial activity tests with various cell concentrations in liquid medium.
Antimicrobial activity tests in liquid cultures with different cell concentrations were carried out as follows. Serial 10-fold dilutions in LB from an overnight culture were incubated in the absence and in the presence of 5 μM MccJ25 or 45 μM MccJ25 analogues. After 2 h at 37°C, samples were removed, serially diluted into sterile LB medium, and plated on LB agar. After 24 h of incubation at 37°C, the viable count was used to establish the survival curves.
Mild osmotic shock.
For outer membrane permeabilization, the washing procedure of Cavard and Lazdunski (6) was used. MC4100 cells from a stationary-phase culture in LB medium were harvested by centrifugation, washed three times with 10 mM sodium phosphate buffer, pH 6.8, and resuspended to the desired concentration (about 2,000 cells/ml) in the same buffer. Cell survival was evaluated in the absence or in the presence of either 38 μM native MccJ25 or MccJ25 analogues. After 2 h of incubation at 37°C, 100-μl aliquots were plated in duplicate onto LB medium. After overnight incubation at 37°C, the number of CFU was recorded. The control experiments without microcin showed that the washing treatment did not affect cell viability.
RNA polymerase activity.
An abortive initiation assay was performed in 10 μl transcription buffer (20 mM Tris-HCl, pH 7.9, 40 mM KCl, 10 mM MgCl2) containing 20 nM RNAP, 200 nM DNA fragment containing the T7A1 promoter, 100 μM CpA primer, 40 μM [α-32P]UTP (10 Ci/mmol), and different concentrations of MccJ25 and its mutant derivatives. Reactions were allowed to proceed for 10 min at 37°C and stopped with urea-containing buffer. Products were resolved by urea-polyacrylamide gel electrophoresis (7 M urea, 23% acrylamide) and then revealed by autoradiography. The resulting image was analyzed, and the amounts of transcripts were quantitated using ImageQuant software.
RESULTS AND DISCUSSION
MccJ25 His mutants and antimicrobial activity.
Site-specific mutations in plasmid-borne mcjA were designed to test the function of the unique histidine residue of MccJ25. Substitution of His5 for Lys or Arg (His5xLys or His5xArg) maintained the positive charge, whereas the change of His5 to Ala (His5xAla) resulted in removal of the side chain beyond the β-carbon and the loss of charge. Cells harboring plasmids with mutant mcjA and wild-type mcjBCD produced mature, processed MccJ25 mutants as judged by the appearance of characteristic peaks during reverse-phase HPLC of culture medium (data not shown) and matrix-assisted laser desorption ionization-time-of-flight mass spectrometry analysis of the peaks. The m/z spectra showed major ions with m/z values of 2,098, 2,126, and 2,041 for MccJ25(His5xLys), MccJ25(His5xArg), and MccJ25(His5xAla), respectively. No contamination with native MccJ25 could be detected. Further fragmentation led to tandem mass spectrometry spectra that showed ions with m/z values of 678, 706, and 621, corresponding to the eight-residue ring (Gly1 to Glu8) for MccJ25(His5xLys), MccJ25(His5xArg), and MccJ25(His5xAla), respectively. These results confirm that the integrity of the lariat ring is not affected by the substitutions. Table 2 shows a comparison of inhibitory activities of MccJ25 mutants and wild-type MccJ25 in spot tests on several E. coli strains. As can be seen, wild-type MccJ25 showed significantly higher activity than either of the mutants with all strains tested. However, mutant microcins had various inhibitory effects on different E. coli strains. The Lys mutant was significantly more active than the Ala and Arg mutants. The positive charge of the arginine side chain appears to be unimportant, since the Arg and Ala mutants were similarly defective except in the MG1655 strain, for which the Arg mutant shows only an 18-fold defect versus a 73-fold defect of the Ala mutant. The greater effect of the Arg mutant compared with His and Lys (all positively charged amino acids) may reflect the larger size of this amino acid side chain (seven C or N atoms, versus six and five for His and Lys, respectively). In total, the data suggest that either positive charge or size may be important in the observed effects.
TABLE 2.
Effect of MccJ25 mutants on several E. coli strainsa
| Strain | Sensitivity by spot-on-lawn test (microcin MIC, μM)
|
|||
|---|---|---|---|---|
| MccJ25 | MccJ25 (His5xLys) | MccJ25 (His5xArg) | MccJ25 (His5xAla) | |
| MC4100 | 0.9b | 7.3b | Rc | R |
| AB259 | 0.9b | 3.6b | R | R |
| RYC1000 | 0.9b | 3.6b | R | R |
| DH5α | 0.9 | 3.6b | 29.3b | 29.3 |
| XL1-Blue | 0.9 | 7.3b | 29.3b | 29.3 |
| AB1133 | 0.4 | 3.6b | 7.3b | 14.6 |
| MG1655 | 0.4 | 3.6b | 7.3b | 29.3 |
Representative results of one of several independent experiments are shown.
Turbid spot.
R, no inhibition was noted in the presence of 100 μM microcin.
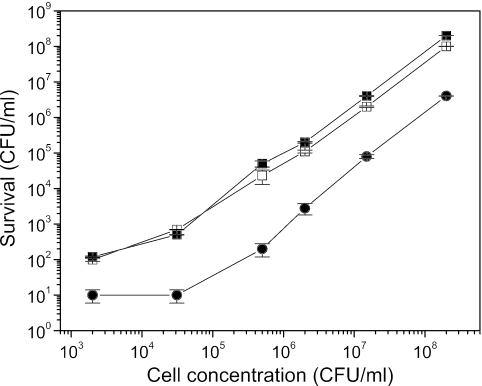
Figure 1 shows the sensitivity of E. coli MC4100 to wild-type MccJ25 and two of the mutants, MccJ25(His5xAla) and MccJ25(His5xArg). As can be seen, the viable cell count decreased, irrespective of the initial cell count, by 2 to 3 orders of magnitude in the presence of 5 μM wild-type MccJ25. Under identical conditions, 5 μM of microcin mutants had no effect on viable cell counts (data not shown), while 45 μM MccJ25(His5xAla) or MccJ25(His5xArg) decreased viable cell counts only by 1 order of magnitude.
FIG. 1.
Effects of 5 μM MccJ25 (•), 45 μM MccJ25(His5xAla) (▪), or MccJ25(His5xArg) (□) on survival of E. coli MC4100 at different cell concentrations. The results are expressed as mean values obtained from three independent experiments, and standard deviations are shown.
Effect of MccJ25 mutants on RNAP activity.
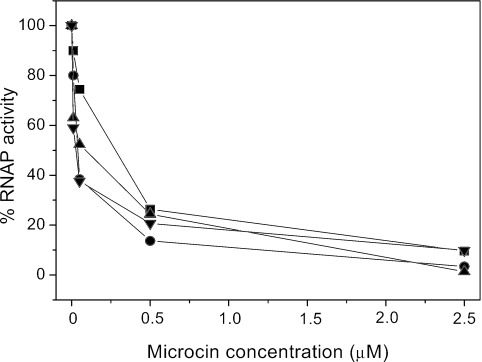
MccJ25 inhibits transcription by binding to and occluding the RNAP secondary channel, which is thought to conduct nucleotide substrates to the RNAP catalytic center (1, 11). We reported that E. coli cells harboring a mutant RNAP were able to grow on selective medium containing MccJ25. The mutation, designated sjmA1, changed Thr931 to Ile in the largest (β′) subunit of E. coli RNAP (8, 19). Thr931 is exposed on the surface of the secondary channel, and the T931I substitution decreases MccJ25 interaction with RNAP (19). Figure 2 shows the degree of inhibition by the wild-type MccJ25 or MccJ25 variants of transcription by wild-type RNAP. Transcription was essentially abolished by the addition of 2.5 μM of either mutant or wild-type MccJ25. As expected, even in the presence of a high concentration (50 μM) of MccJ25 and its mutants, only partial inhibition of the T931I RNAP was observed with MccJ25 (20%), MccJ25(His5xLys) (16%), or MccJ25(His5xAla) (24%) at the identical assay conditions of Fig. 2, while MccJ25(His5xArg) was totally ineffective.
FIG. 2.
Effects of different concentrations of MccJ25 (▪), MccJ25(His5xLys) (▾), MccJ25(His5xAla) (•), and MccJ25(His5xArg) (▴) on RNAP activity. The transcription activity was plotted as a percentage of activity observed in the absence of MccJ25.
We conclude that different in vivo inhibitory activities of MccJ25 and its mutants (Table 2 and Fig. 1) are not due to the altered ability to inhibit RNAP and could thus be due to differences in the uptake into cells and/or intracellular stability of MccJ25 mutants.
Low inhibitory activity of MccJ25 mutants is due to defective transport by the SbmA protein.
To find its way to its target, RNAP, MccJ25 must be transported through E. coli membranes by the outer membrane protein FhuA and the inner membrane protein SbmA (14, 15). The following experiments were performed to clarify which of these proteins is responsible for the differences between the wild-type and mutant microcins shown in Table 2. Cavard and Lazdunski (6) reported that a mild osmotic shock allows a bypass of the receptors that are normally required for penetration of the E. coli outer membrane bilayer by colicin A. We adapted the assay to bypass the FhuA-dependent step in MccJ25 uptake. Table 3 shows the results obtained when MC4100 cells were subjected to osmotic shock by washing them three times with 10 mM sodium phosphate buffer, pH 6.8, followed by a 2-h incubation at 37°C with MccJ25, MccJ25(His5xAla), or MccJ25(His5xArg) and plating on medium without MccJ25. Incubation with MccJ25 mutants had no effect on colony formation by washed cells; in contrast, little or no viable cells were detected after exposure of washed (or not washed) cells to wild-type MccJ25 (Table 3, rows 1 and 2). These results suggest that the barrier for passage of MccJ25 mutants to the cell interior is not at the outer membrane level. This notion was independently confirmed using E. coli SBG103 (MC4100 fhuA::Tn5) cells. As can be seen from Table 3 (compare rows 3 and 4), the fhuA mutation caused MccJ25 resistance, as expected (14), and this resistance was overcome by osmotic shock, but only in the case of wild-type MccJ25. On the other hand, E. coli EZE100 (MC4100 sbmA::Tn5) cells were resistant to both wild-type and mutant MccJ25, and washing had no effect on the resistance level (Table 3, rows 5 and 6).
TABLE 3.
Sensitivities of washed E. coli MC4100 cells to MccJ25 analoguesa
| Strain | No. of CFU with MccJ25 mutation
|
||
|---|---|---|---|
| None | His5xAla | His5xArg | |
| MC4100 | 30 | 1,930 | 1,920 |
| MC4100, washed | 0 | 1,910 | 1,950 |
| SBG103 (4100 fhuA::Tn5) | 1,930 | 1,970 | 1,930 |
| SBG103 (4100 fhuA::Tn5), washed | 0 | 1,580 | 1,950 |
| EZE100 (MC4100 sbmA::Tn5) | 1,930 | 1,950 | 1,980 |
| EZE100 (MC4100 sbmA::Tn5), washed | 1,900 | 1,970 | 1,800 |
The assay was performed by treating about 2 × 103 cells/ml with 38 μM of MccJ25 or its mutant derivatives. Representative results of one of several independent experiments are shown.
The results presented above suggest that the mutant microcins cannot pass through the inner membrane, i.e., they do not properly interact with the SbmA transporter. To obtain further support for the notion of defective recognition of MccJ25 mutants by SbmA, the MccJ25 sensitivity of MC4100 cells harboring the sbmA expression plasmid pMM73.4 was determined. Table 4 shows that the overexpression of sbmA appears to sensitize cells equally well to both the wild-type and mutant microcins. For wild-type MccJ25 and MccJ25(His5xLys), MICs decreased ∼8-fold upon sbmA overexpression. For the other two MccJ25 mutants, no quantification was possible, since they had no effect on the growth of control cells. Overexpression of sbmA made cells sensitive to these mutant microcins, though compared to the wild-type, ∼8.0 [MccJ25(His5xLys)], ∼60 [MccJ25(His5xArg)], and ∼130 [MccJ25(His5xAla)] times higher concentrations were required for growth inhibition (Table 4). The reduced antibiotic activity of the MccJ25 mutants with these cells is consistent with the idea that MccJ25 substitutions of His5 in the MccJ25 lariat ring interfere with recognition by the inner membrane receptor SbmA. However, this raises a question of whether these cells would also be more sensitive to any other functionally defective microcin derivatives, such as those carrying substitutions in the lariat ring other than His5. Preliminary results indicate that the sensitization effect of SbmA overexpression also occurs with a defective mutant carrying another substitution in the lariat ring (Gly2xAla) (data not shown), suggesting that this region could be crucial for MccJ25 import through the SbmA protein. Further studies are under way in our laboratories to confirm this hypothesis.
TABLE 4.
MICs of microcins for E. coli MC4100 transformed with pMM73.4
| Microcin | MIC (μM) for strain
|
|
|---|---|---|
| MC4100 | MC4100(pMM73.4) | |
| MccJ25 | 0.9 | 0.1 (8.1)c |
| MccJ25(His5xLys) | 7.3a | 0.9 (8.1)c; (8.3)d |
| MccJ25(His5xArg) | Rb | 7.3 (66.7)d |
| MccJ25(His5xAla) | R | 14.6 (132.7)d |
Turbid spot.
R, no inhibition was noted in the presence of 100 μM microcin.
In parentheses: MIC for MC4100/MIC for MC4100(pMM73.4).
In parentheses: MIC of MccJ25/MIC of MccJ25 mutant.
Acknowledgments
FONCYT (grant PICT 7231), CIUNT (grant 26/D235). and NIH FIRCA (R03 grant TW006828) funded this work. R.E.D.C. is the recipient of a CONICET fellowship, and P.A.V., R.A.S., and R.N.F. are career investigators of CONICET.
REFERENCES
- 1.Adelman, K., J. Yuzenkova, A. LaPorta, N. Zenkin, J. Lee, J. T. Lis, S. Borukhov, M. D. Wang, and K. Severinov. 2004. Molecular mechanism of transcription inhibition by peptide antibiotic microcin J25. Mol. Cell 14:753-762. [DOI] [PubMed] [Google Scholar]
- 2.Bayro, M. J., J. Mukhopadhyay, G. V. T. Swapna, J. Y. Huang, L.-C. Ma, E. Sineva, P. E. Dawson, G. T. Montelione, and R. H. Ebright. 2003. Structure of antibacterial peptide microcin J25: a 21-residue lariat protoknot. J. Am. Chem. Soc. 125:12382-12383. [DOI] [PubMed] [Google Scholar]
- 3.Bellomio, A., M. R. Rintoul, and R. D. Morero. 2003. Chemical modification of microcin J25 with diethylpyrocarbonate and carbodiimide: evidence for essential histidyl and carboxyl residues. Biochem. Biophys. Res. Commun. 303:458-462. [DOI] [PubMed] [Google Scholar]
- 4.Bellomio, A., P. A. Vincent, B. F. de Arcuri, R. A. Salomón, R. D. Morero, and R. N. Farías. 2004. The microcin J25 β-hairpin region is important for antibiotic uptake but not for RNA polymerase and respiration inhibition. Biochem. Biophys. Res. Commun. 325:1454-1458. [DOI] [PubMed] [Google Scholar]
- 5.Blond, A., J. Péduzzi, C. Goulard, M. J. Chiuchiolo, M. Barthélémy, Y. Prigent, R. A. Salomón, R. N. Farías, F. Moreno, and S. Rebuffat. 1999. The cyclic structure of microcin J25, a 21-residue peptide antibiotic from Escherichia coli. Eur. J. Biochem. 259:747-755. [DOI] [PubMed] [Google Scholar]
- 6.Cavard, D., and C. Lazdunski. 1981. Involvement of Btub and OmpF proteins in binding and uptake of colicin A. FEMS Microbiol. Lett. 12:311-316. [Google Scholar]
- 7.Delgado, M. A., J. O. Solbiati, M. J. Chiuchiolo, R. N. Farías, and R. A. Salomón. 1999. Escherichia coli outer membrane protein TolC is involved in the production of the peptide antibiotic microcin J25. J. Bacteriol. 181:1968-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delgado, M. A., M. R. Rintoul, R. N. Farías, and R. A. Salomón. 2001. Escherichia coli RNA polymerase is the target of the cyclopeptide antibiotic microcin J25. J. Bacteriol. 183:4543-4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Destoumieux-Garzón, D., S. Duquesne, J. Peduzzi, C. Goulard, M. Desmadril, L. Letellier, S. Rebuffat, and P. Boulanger. 2005. The iron-siderophore transporter FhuA is the receptor for microcin J25. Role of the microcin Val11-Pro16 β-hairpin region in the recognition mechanism. Biochem. J. 389:869-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Killmann, H., C. Herrmann, A. Torun, G. Jung, and V. Braun. 2002. TonB of Escherichia coli activates FhuA through interaction with the beta-barrel. Microbiology 148:3497-3509. [DOI] [PubMed] [Google Scholar]
- 11.Mukhopadhyay, J., E. Sineva, J. Knight, R. M. Levy, and R. H. Ebright. 2004. Antibacterial peptide microcin J25 inhibits transcription by binding within and obstructing the RNA polymerase secondary channel. Mol. Cell 14:739-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosengren, K. J., R. J. Clark, N. L. Daly, U. Goransson, A. Jones, and D. J. Craik. 2003. Microcin J25 has a threaded sidechain-to-backbone ring structure and not a head-to-tail cyclized backbone. J. Am. Chem. Soc. 125:12464-12474. [DOI] [PubMed] [Google Scholar]
- 13.Salomón, R. A., and R. N. Farías. 1992. Microcin 25, a novel antimicrobial peptide produced by Escherichia coli. J. Bacteriol. 174:7428-7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salomón, R. A., and R. N. Farías. 1993. The FhuA protein is involved in microcin 25 uptake. J. Bacteriol. 175:7741-7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salomón, R. A., and R. N. Farías. 1995. The peptide antibiotic microcin 25 is imported through the TonB pathway and the SbmA protein. J. Bacteriol. 177:3323-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Solbiati, J. O., M. Ciaccio, R. N. Farías, and R. A. Salomón. 1996. Genetic analysis of plasmid determinants for microcin 25 production and immunity. J. Bacteriol. 178:3661-3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vincent, P. A., A. Bellomio, B. F. de Arcuri, R. N. Farías, and R. D. Morero. 2005. MccJ25 C-terminal is involved in RNA-polymerase inhibition but not in respiration inhibition. Biochem. Biophys. Res. Commun. 331:549-551. [DOI] [PubMed] [Google Scholar]
- 18.Wilson, K. A., M. Kalkum, J. Ottesen, J. Yuzenkova, B. T. Chait, R. Landick, T. Muir, K. Severinov, and S. A. Darst. 2003. Structure of microcin J25, a peptide inhibitor of bacterial RNA polymerase, is a lassoed tail. J. Am. Chem. Soc. 125:12475-12483. [DOI] [PubMed] [Google Scholar]
- 19.Yuzenkova, J., M. Delgado, S. Nechaev, D. Savalia, V. Epshtein, I. Artsimovitch, R. A. Mooney, R. Landick, R. N. Farias, R. Salomon, and K. Severinov. 2002. Mutations of bacterial RNA polymerase leading to resistance to microcin J25. J. Biol. Chem. 277:50867-50875. [DOI] [PubMed] [Google Scholar]