Abstract
The respiratory NADH:quinone oxidoreductase (complex I) (NDH-1) is a multisubunit enzyme that translocates protons (or in some cases Na+) across energy-conserving membranes from bacteria or mitochondria. We studied the reaction of the Na+-translocating complex I from the enterobacterium Klebsiella pneumoniae with N,N′-dicyclohexylcarbodiimide (DCCD), with the aim of identifying a subunit critical for Na+ binding. At low Na+ concentrations (0.6 mM), DCCD inhibited both quinone reduction and Na+ transport by NDH-1 concurrent with the covalent modification of a 30-kDa polypeptide. In the presence of 50 mM Na+, NDH-1 was protected from inhibition by DCCD, and the modification of the 30-kDa polypeptide with [14C]DCCD was prevented, indicating that Na+ and DCCD competed for the binding to a critical carboxyl group in NDH-1. The 30-kDa polypeptide was assigned to NuoH, the homologue of the ND1 subunit from mitochondrial complex I. It is proposed that Na+ binds to the NuoH subunit during NADH-driven Na+ transport by NDH-1.
The production of energy in a living cell depends on electrochemical ion gradients established by primary pumps, which transport protons (or in some cases sodium ions) across energy-conserving membranes. In many organisms, energy conversion relies on the oxidation of NADH with O2, which drives the translocation of protons by respiratory complexes I, III, and IV across the inner membrane of bacteria or mitochondria (33). With 46 different subunits (5), NADH:quinone oxidoreductase (complex I) is the largest and most complicated redox pump in the mitochondrial respiratory chain. The low-resolution structure of complex I reveals a peripheral part that harbors flavin mononucleotide and several Fe-S clusters and a hydrophobic part embedded in the membrane (4, 35). The smaller complex I from bacteria (termed NDH-1) comprises 14 subunits (NuoA to NuoN), which are homologues of subunits found in the larger mitochondrial enzyme (12). These 14 subunits are considered to represent the functional core of mitochondrial complex I. Only 6 out of these 14 subunits, namely, homologues of subunits NuoBCDI in the peripheral part and NuoL and NuoH in the membrane part, build up the functional unit of energy-conserving hydrogenases from bacteria and archaea. These [NiFe] hydrogenases, which are phylogenetically related to NDH-1, couple the oxidation of ferredoxin with H+ as an electron acceptor to the transport of H+ or Na+ (16).
The coupling ion specificity (Na+ versus H+) of NDH-1 has important implications for the energy metabolism of bacteria and for the mechanism of cation transport by complex I. In 1999, we showed that the enterobacterium Klebsiella pneumoniae contains a Na+-translocating NADH dehydrogenase that belongs to the complex I family of respiratory enzymes (23). We subsequently determined the cofactor composition and Na+/electron transport stoichiometry of the complex (13) and showed that NADH-driven Na+ transport by NDH-1 is electrogenic (14). Our views have been challenged by Bertsova and Bogachev, who claimed that K. pneumoniae NDH-1 acts exclusively as a proton pump (3). Those authors proposed that NADH-driven Na+ transport in K. pneumoniae is catalyzed by another Na+-translocating NADH:quinone oxidoreductase (Na+-NQR) related to the enzyme found in Vibrio species (3).
Here, we reinvestigate NADH-driven Na+ translocation by NDH-1 from K. pneumoniae and further corroborate our previous conclusion (13) that Na+ transport is not catalyzed by a Na+-NQR present in our enzyme preparation. The binding of Na+ to NDH-1 is studied using the reagent N,N′-dicyclohexylcarbodiimide (DCCD), which selectively modifies carboxyl groups in hydrophobic environments (15). These carboxylates in the membrane-embedded part of a pump are likely ligands for Na+ in sodium ion-translocating enzymes (8). Using DCCD, a functional Na+ binding site on the NuoH subunit in the membrane-bound part of NDH-1 from K. pneumoniae is identified. The NuoH subunit is highly conserved in the complex I family of enzymes and in energy-conserving hydrogenases, supporting the notion that NuoH plays a prominent role during redox-driven cation transport. We discuss the physiological conditions of Na+ transport versus H+ transport by NDH-1 and speculate on the mechanism of Na+ translocation.
MATERIALS AND METHODS
Growth and subcellular fractionation.
One hundred liters of Klebsiella pneumoniae cells was grown anaerobically in a bioreactor (Bioengineering AG) at 37°C (7) in the presence of 46 mM glycerol as a carbon source and 50 mM fumarate as an electron acceptor. The addition of fumarate resulted in a 50% increase of the specific ubiquinone-1 (Q1) reduction activity in the membrane fraction. Cells were harvested after less than 3 h using a flowthrough separator (K25; Westfalia), resuspended in a solution containing HEPES-KOH, pH 7.0, 50 mM KCl, and 7% (vol/vol) dimethyl sulfoxide, and stored at −20°C. All subsequent steps were carried out in an anaerobic chamber (Coy) under an atmosphere of 90% N2 and 10% H2 or in sealed vessels flushed with N2. Membranes from K. pneumoniae cells were prepared as described previously (13). Crude extracts from Vibrio alginolyticus cells were obtained as described previously (41).
Solubilization of NDH-1 and precipitation with polyethylene glycol.
Membranes from K. pneumoniae (700 mg of protein) were solubilized at 4°C for 30 min with 20 ml buffer (10 mM HEPES-KOH, pH 7.5, 50 mM KCl, 10% [vol/vol] glycerol) containing 13 mM l-α-diheptanoylphospatidylcholine (DHPC). The detergent-to-protein ratio was 5:1. The membranes were resuspended by agitation every 5 min. The solubilized membrane proteins (180 mg in 14 ml) obtained by ultracentrifugation (150,000 × g, 60 min) were subjected to fractionated precipitation with polyethylene glycol (PEG) (PEG 6000). The solubilizate was mixed with 45 mM MgCl2 and 0.42 ml 50% PEG (final concentration, 1.5% PEG), and the suspension was stirred on ice for 15 min. Precipitated proteins were removed by centrifugation (36,000 × g, 15 min), and 2.24 ml 50% PEG was added to the supernatant (final concentration, 9.5% PEG). The suspension was stirred on ice for 15 min, and precipitated NDH-1 (30 to 50 mg) was collected by centrifugation (70,000 × g, 15 min). NDH-1 was resuspended in 3 ml 10 mM HEPES-KOH, pH 7.5, containing 1.5 mM DHPC and 1.7 mg ml−1 l-α-phosphatidylcholine (from soybean, type II-S; Sigma). NDH-1 obtained by fractionated PEG precipitation contained 18 nmol Fe and 8 nmol acid-labile sulfide mg−1 protein and catalyzed NADH-dependent Q1 reduction (0.3 μmol min−1 mg−1 protein) and Na+ transport (0.7 μmol min−1 mg−1 lipid).
Purification of NDH-1 by anionic-exchange chromatography.
Chromatographic steps were carried out in an anaerobic chamber (Coy) using a Jasco high-performance liquid chromatography system. Solubilized membranes (96 mg) were loaded onto a Fractogel TSK DEAE-650(S) Sepharose column (1.5 by 8 cm; Merck), and the column was washed with 15 ml buffer (10 mM HEPES-KOH, pH 7.5, 50 mM KCl, 10% glycerol). NDH-1 was eluted with a linear gradient (53 ml) from 250 to 1,000 mM KCl in buffer at a flow rate of 1.5 ml min−1. Fractions showing NADH-dependent Q1 reduction activity eluted between 445 and 740 mM KCl and were combined (13 ml). A total of 2.5 mM DHPC and 0.2 mg ml−1 l-α-phosphatidylcholine were added, and NDH-1 was concentrated by ultracentrifugation (150,000 × g) overnight. The concentrated NDH-1 was resuspended in 1.5 ml buffer and loaded onto a Source 15Q Superformance column (1.6 by 2 cm; Amersham). The column was washed with 2 volumes of buffer, and a linear gradient (53 ml) from 250 mM to 1,000 mM KCl was applied. NDH-1 eluted between 315 and 445 mM KCl and was concentrated by precipitation with 15% PEG 6000 in the presence of 45 mM MgCl2. NDH-1 (3 mg) was collected by centrifugation (70,000 × g, 15 min) and resuspended in 1 ml 10 mM HEPES-KOH, pH 7.5, containing 2.5 mM DHPC and 0.2 mg ml−1 l-α-phosphatidylcholine. The addition of solubilized lipids stabilized NDH-1 as judged by its Q reductase activity.
Modification of NDH-1 with DCCD and electron transfer activity.
DCCD reacts with protonated carboxyl groups of proteins with the formation of dicyclohexyl-O-isourea. In the absence of water or other nucleophiles, e.g., in membrane-embedded regions of proteins, a rearrangement occurs, which results in the stable binding of dicyclohexyl N-isourea into the protein (15). The reaction of NDH-1 with DCCD was performed at room temperature with the exclusion of O2 in the anaerobic chamber or in reaction tubes sealed with rubber stoppers. The reaction conditions varied with respect to pH and Na+ concentration. Aliquots of NDH-1 obtained by fractionated PEG precipitation (1 mg in 40 ml 10 mM HEPES-KOH, pH 7.5, 1.5 mM DHPC, 1.7 mg ml−1 l-α-phosphatidylcholine) were added to 40 μl buffer (50 mM MES [morpholineethanesulfonic acid], Tris, or MOPS [morpholinepropanesulfonic acid]) containing 50 mM KCl, 4% glycerol, and 0.6 or 50 mM Na+. The pH of the buffer was adjusted with KOH to 6.0, 6.5, 7.0, 7.5, or 8.0. The aliquots were mixed with 6 μl 10 mM DCCD in ethanol (final concentration, 698 μM). A control reaction mixture contained 6 μl ethanol. After 3 min, the oxidation of NADH and the reduction of Q1 by the enzyme specimens were determined using a solution containing 50 mM Tris-HCl, pH 8.0, and 50 mM KCl with the exclusion of O2. The Q reductase activity corresponding to the formation of ubiquinol-1 from ubiquinone-1 was determined from the difference in absorbance at the wavelength pair 248 and 268 nm (Δɛ at 248 to 268 nm of 7.8 mM−1 cm−1) (44).
Reconstitution of NDH-1 and Na+ transport.
NDH-1 was reconstituted into proteoliposomes by diluting a mixture of NDH-1, l-α-phosphatidylcholine, and DHPC with buffer (10 mM HEPES-KOH, pH 7.5, or 10 mM MES-KOH, pH 6.5) containing 50 mM KCl and 10% glycerol (13). The protein-to-lipid ratios were 1:1 (gram/gram) with NDH-1 enriched by fractionated PEG precipitation and 1:4 (gram/gram) with NDH-1 from the Source 15Q chromatographic step. The modification of reconstituted NDH-1 with DCCD and the Na+ uptake measurements were performed at 25°C. Aliquots of reconstituted NDH-1 (1.6 mg lipids in 270 μl) were mixed with 0, 100, 200, 500, or 800 μM DCCD. After 4 min, Na+ transport was initiated by the addition of 5 mM NaCl and 0.1 mM NADH. At different times, samples of 70 μl were applied to a 1-ml plastic syringe containing 0.6 ml Dowex 50 (K+) and eluted with 0.8 ml of deionized water. The eluate was collected in plastic tubes, and the amount of Na+ entrapped in the proteoliposomes was determined by atomic absorption spectroscopy (13). All experiments were repeated at least four times. Mean values or representative measurements performed with a single enzyme batch are presented.
H+ transport.
Purification of the H+-translocating F1Fo ATPase from Escherichia coli (17) and determination of ATP hydrolysis activity (26) were performed as described previously. The ATPase exhibited a specific ATP hydrolysis activity of 0.18 μmol min−1 mg−1. The ATPase from E. coli and the NDH-1 from K. pneumoniae were coreconstituted into proteoliposomes by adding the proteins in DHPC to a lipid film in a round-bottom flask. The protein-to-lipid ratio was 1:4 (gram/gram), respectively. The lipids were dispersed by vortexing for 1 min. The formation of proteoliposomes was accomplished by the dropwise addition of buffer (10 mM HEPES-KOH, pH 7.0, 50 mM KCl, 10% glycerol) to dilute the DHPC below the critical micellar concentration (13, 14). The modification of the membrane-embedded c subunit of the F1Fo ATPase with DCCD blocks proton transport through Fo and ATP hydrolysis catalyzed by F1 (49). In the presence of 0.5 mM DCCD, ATP hydrolysis by the ATPase decreased to 0.06 μmol min−1 mg−1, indicating that 66% of the hydrolytic activity was coupled to proton transport. Assuming a ratio of four protons translocated per ATP hydrolyzed (49), the calculated proton transport activity was as follows: 0.12 × 4 = 0.48 μmol min−1 mg−1. During ATP hydrolysis, a transmembrane voltage of at least 30 mV was generated (14), demonstrating that a proton gradient was maintained across the liposomal membrane. ATP-driven proton transport into proteoliposomes was followed by the quenching of 9-amino-6-chloro-2-methoxyacridin (ACMA) fluorescence (18) in 10 mM Tris-HCl, pH 7.0, containing 5 mM NaCl, 50 mM KCl, 10% glycerol, and 10 mM MgCl2.
Modification of NDH-1 with [14C]DCCD and SDS-PAGE.
The reaction of NDH-1 from the Source 15Q chromatographic step with radioactively labeled DCCD was performed in 30 mM HEPES-MES, pH 6.5, with or without added NaCl. Two aliquots of NDH-1 (each 30 μg in 40 μl buffer containing 50 or 0.6 mM Na+) were mixed with 100 μM [14C]DCCD (54 μCi/μmol; Amersham). After 1 h, 40 μl sample buffer containing 2.5% sodium dodecyl sulfate (SDS) was added, and the modified NDH-1 complexes were separated by SDS-polyacrylamide gel electrophoresis (PAGE) (36). Proteins were visualized with Coomassie blue 250-R. The gel was treated with NAMP 100V Amplify enhancer solution (Amersham) for 5 min, dried under a vacuum on Whatman 3MM paper, and exposed to a phosphor screen for 7 days. Radioactively labeled polypeptides were detected with a PhosphorImager (Molecular Dynamics) and quantified using the program ImageQuant 5.0.
Extraction of hydrophobic NDH-1 subunits with 2-butanol.
NDH-1 from the Source 15Q chromatographic step (1 to 2 mg in 0.5 ml) was extracted with 1 ml H2O and 2 ml of ice-cold 2-butanol according to a method described previously (48). The suspension was agitated for 5 min at 4°C and centrifuged for 15 min at 9,000 × g. The upper organic phase containing hydrophobic polypeptides was transferred into a reaction tube and mixed with 80 μl 2.5% SDS (final concentration, 0.1% SDS). The polypeptides were dried in a SpeedVac, resolved for 6 h in 50 μl sample buffer containing 2.5% SDS, and separated by SDS-PAGE (36). NDH-1 was also extracted after modification with [14C]DCCD. NDH-1 (2 mg) in 30 mM HEPES-MES, pH 6.5, containing 0.6 mM Na+ was incubated with 50 μM [14C]DCCD for 1 h and extracted with 2-butanol as described above.
Analytical procedures.
The amount of protein was determined by the bicinchoninic acid method using a reagent from Pierce (39). Bovine serum albumin served as a standard. Iron content was determined colorimetrically by the 5,5′(3-(2-pyridyl)-1,2,4-triazine-5,6 diyl)-bis-2-furansulfonic acid, disodium salt (Ferene) complex (1). For the determination of acid-labile sulfur, the methylene blue method (2) was applied. Tryptic in-gel digestion was performed as described previously (37). The digests were analyzed using a QTOF Ultima API mass spectrometer (Waters, United Kingdom), and proteins were identified using ProteinLynx Global Server software (Waters, United Kingdom). Transfer of polypeptides from SDS-PAGE gels to polyvinyl fluoride membranes was performed as described previously (9). Edman degradation was carried out using a G1005A protein sequencer (Agilent).
RESULTS
Purification of NDH-1.
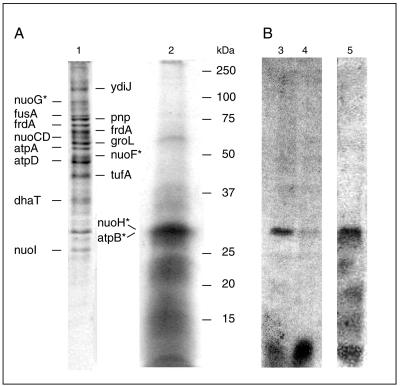
NDH-1 was purified by two anionic-exchange chromatographic steps using DHPC as a detergent. During the purification, the formation of quinol by NDH-1 was monitored to observe the electron transfer from NADH oxidized at the peripheral part to the substrate quinone interacting with the membrane-embedded part of the complex. The specific quinol formation activity increased from 0.22 μmol min−1 mg−1 in solubilized membranes to 0.77 μmol min−1 mg−1 after the Source 15Q chromatographic step. Comparable rates are reported for purified bovine complex I, which exhibits a quinol formation activity of 0.4 to 0.8 μmol min−1 mg−1 (30). The NADH dehydrogenase activity of K. pneumoniae NDH-1 was generally higher (2 to 4 μmol min−1 mg−1), indicating some leakage of electrons to exogenous oxidants. The content of nonheme iron increased from 1 nmol Fe mg−1 protein in the solubilized membranes to 28 to 37 nmol Fe mg−1 in NDH-1 after Source 15Q chromatography. The iron in NDH-1 from K. pneumoniae was predominantly associated with Fe-S clusters, as indicated by the presence of 26 to 35 nmol acid-labile sulfide mg−1 protein. For the related NDH-1 from Escherichia coli, an iron content of 38 to 45 nmol Fe mg−1 was reported previously (27). Bertsova and Bogachev recently proposed that K. pneumoniae NDH-1 exclusively acts as a proton pump. They concluded that the NADH-driven Na+ transport described in our previous studies (13, 14) was caused by a contamination of our NDH-1 with a Na+-NQR related to the enzyme found in Vibrio species (3). Silver ions specifically inhibit Na+-NQR but not NDH-1 or complex I (41, 47). Ag+ (10 μM) completely prevented NADH oxidation by crude extracts from Vibrio alginolyticus, whereas no inhibition was observed with our K. pneumoniae extracts in the presence of 40 μM Ag+. Annonin VI is a specific inhibitor of bacterial NDH-1 that is partially competitive with respect to Q (11). In the presence of 50 μM annonin VI, Na+ translocation by our NADH dehydrogenase purified by anionic-exchange chromatography was inhibited by 50%, indicating that the observed Na+ transport activity is catalyzed by NDH-1. We then searched for polypeptides of the putative Na+-NQR from K. pneumoniae in NDH-1 from the Source 15Q chromatographic step. NDH-1 was separated by SDS-PAGE (Fig. 1) and subjected to N-terminal sequencing or tryptic digestion and mass spectroscopic analyses. We did not detect polypeptides that would indicate the presence of a NADH dehydrogenase related to the Na+ pump found in Vibrio cholerae (Table 1). Notably, we did not detect a K. pneumoniae homologue of the 45-kDa subunit catalyzing the oxidation of NADH in the Na+-translocating NADH dehydrogenase from V. cholerae (46). On the other hand, the NuoF and NuoG subunits of the peripheral, NADH-oxidizing part of NDH-1 were unambiguously identified in the Na+-translocating NADH:quinone oxidoreductase purified from K. pneumoniae (13, 23). The subunits NuoCD and NuoI from the connecting fragment of NDH-1 were assigned by mass spectrometry of tryptic peptides (Fig. 1 and Table 1). In summary, these results demonstrate that NADH-driven Na+ transport is an intrinsic catalytic property of NDH-1 from K. pneumoniae and is not caused by a Na+-NQR present in our enzyme preparation. A major contaminant in NDH-1 from the Source 15Q chromatographic step is the proton-translocating F1Fo ATPase (20). Mass spectrometry of peptides obtained by tryptic digestion revealed that the 51- and 55-kDa polypeptides in the NDH-1 preparation represent the α and β subunits from the soluble F1 part of the ATPase (Fig. 1 and Table 1). Extraction of NDH-1 with 2-butanol yielded a prominent band at 30 kDa (Fig. 1), which represented a mixture of subunit a from the membrane-bound Fo part of the ATPase and the NuoH subunit of NDH-1 (see below).
FIG. 1.
Identification of proteins in NDH-1 and modification of subunit NuoH with [14C]DCCD. Panel A shows the SDS-PAGE of NDH-1 (100 μg) after Source 15Q chromatography (lane 1) and the 2-butanol extract from 2 mg NDH-1 (lane 2). The gels were stained with Coomassie. Polypeptides were identified by mass spectroscopy (see Table 1 for results) or N-terminal sequencing (indicated with an asterisk). Nuo and Atp denote subunits of NDH-1 and the F1Fo ATPase, respectively. Panel B shows the autoradiograms of NDH-1 after modification with [14C]DCCD and separation by SDS-PAGE. Two aliquots of NDH-1 (30 μg each) were mixed with [14C]DCCD in the presence of 0.6 mM Na+ (lane 3) or 50 mM Na+ (lane 4). Lane 5 shows the autoradiogram of the 2-butanol extract from 2 mg NDH-1 after modification with [14C]DCCD in the presence of 0.6 mM Na+.
TABLE 1.
Identification of proteins in NDH-1 from the source 15Q chromatographic stepa
| Swiss-Prot accession no. | Protein | Gene | Predicted mass (Da) | No. of peptide matches | Protein score |
|---|---|---|---|---|---|
| Q8FH47 | Hypothetical protein YdiJ | ydiJ | 113,164 | 2 | 39 |
| Q8ZLT3 | Polynucleotide phosporylase | pnp | 76,991 | 13 | 589 |
| P0A1H3 | Elongation factor G | fusA | 77,419 | 10 | 425 |
| Q8FAL6 | Fumarate reductase flavoprotein subunitb | frdA | 65,948 | 5 | 130 |
| Q8FAL6 | Fumarate reductase flavoprotein subunitb | frdA | 65,948 | 5 | 106 |
| P0A1Y7 | NADH dehydrogenase I, subunit NuoCD | nuoCD | 68,817 | 4 | 88 |
| O66026 | 60-kDa chaperonin | groL | 57,003 | 18 | 602 |
| Q8XG95 | ATP synthase alpha subunit | atpA | 55,079 | 15 | 644 |
| P0ABB4 | ATP synthase beta subunit | atpD | 50,163 | 19 | 671 |
| P0A6N1 | Elongation factor Tu | tufA | 43,286 | 6 | 201 |
| Q7WRJ3 | 1,3-Propanediol oxidoreductase | dhaT | 41,518 | 2 | 73 |
| Q8ZH40 | d-Methionine binding protein | metQ | 29,359 | 2 | 83 |
| Q83BR3 | NADH dehydrogenase I, subunit NuoI | nuoI | 18,985 | 1 | 29 |
Polypeptides subjected to tryptic digestion and mass spectroscopic analysis are indicated in the SDS-PAGE gel shown in Fig. 1. Results were compared with the UniRef100 database (http://www.ebi.ac.uk/uniref/) using the Mascot search engine (Matrix Science Ltd., United Kingdom). In cases where the identified K. pneumoniae protein is not deposited in the Swiss-Prot database, the accession number of the corresponding enterobacterial protein with the highest sequence similarity is given as a reference.
Both polypeptides separated by SDS-PAGE (Fig. 1) were assigned to frdA.
Na+ transport by NDH-1.
NDH-1 purified by two anionic-exchange chromatographic steps catalyzed NADH-driven Na+ transport with an initial rate of 3.6 μmol Na+ min−1 mg−1 protein or 0.8 μmol Na+ min−1 mg−1 lipid. The pH in the lumen of the proteoliposomes and in the assay buffer was 7.5. The Na+ transport activity of NDH-1 was significantly higher than rates observed with other Na+ pumps like oxaloacetate decarboxylase (10 nmol Na+ min−1 mg−1 lipid) (6) and the F1Fo ATPase (20 nmol Na+ min−1 mg−1 lipid) (50). We previously studied the Na+/electron stoichiometry of NDH-1 in proteoliposomes enriched with Q6 and Q8 and determined a ratio of two Na+ ions translocated per two electrons transferred (13). In the present study, the specific Q1 reductase activity of NDH-1 prior to reconstitution was 0.8 μmol QH2 formed min−1 mg−1 protein (or 1.6 μmol electrons transferred min−1 mg−1), resulting in a ratio of 4.5 Na+ ions/2 electrons. Note that the electron transfer activity of NDH-1 determined with the artificial substrate Q1 is roughly 50% of the activity observed with long-chain quinones like Q6 and Q8 (13).
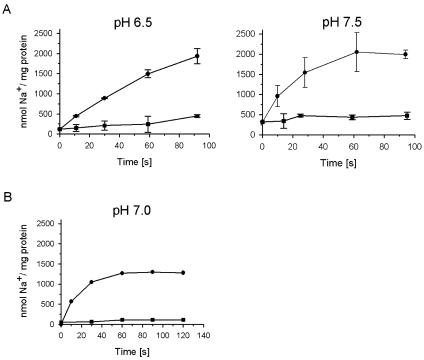
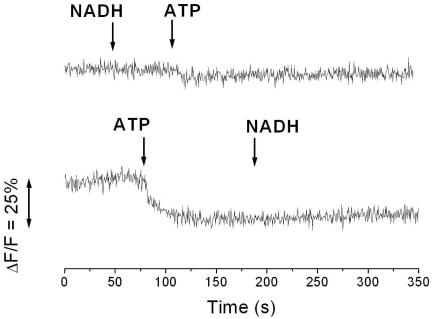
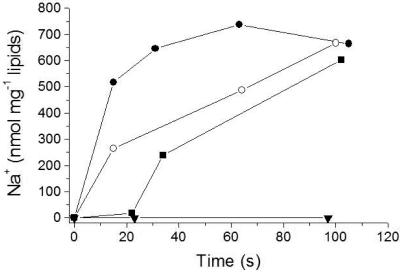
Lowering the internal and external pHs to 6.5 resulted in a decrease of Na+ transport by NDH-1 to 1.6 μmol Na+ min−1 mg−1 protein or 0.4 μmol Na+ min−1 mg−1 lipid. At pH 6.5 and pH 7.5, the lumen of the proteoliposomes contained 0.5 μmol Na+ mg−1 lipid at the end of the experiment (Fig. 2). Using K. pneumoniae cells grown anaerobically on glycerol and fumarate, Bertsova and Bogachev previously presented evidence for H+ translocation by NDH-1 at pH 6.5. The experiments were performed without added Na+ using cells deprived of internal Na+ by repeated washing, and the possibility that Na+ could replace H+ as the preferred coupling cation was not tested (3). We investigated whether reconstituted NDH-1 from K. pneumoniae catalyzed NADH-driven proton transport at pH 7.0 in the presence of 5 mM Na+. The proton-translocating F1Fo ATPase from Escherichia coli was coreconstituted together with NDH-1 into the same artificial membrane system. Proton transport was followed by the quenching of ACMA fluorescence. No change in fluorescence intensity was observed upon the addition of NADH, whereas ATP elicited quenching of the signal, demonstrating that ATP hydrolysis, but not NADH oxidation, was coupled to the translocation of protons into the lumen of the proteoliposomes (Fig. 3). Note that coreconstitution of the two complexes was performed according to a protocol previously established for NDH-1 (14), which is not optimal for ATPase. We previously demonstrated that oxidation of NADH by reconstituted NDH-1 results in the alkalization of the internal lumen of the proteoliposomes (14). If NDH-1 operates as a proton pump, acidification of the proteoliposomes is expected. We also confirmed NADH-driven Na+ transport by NDH-1 coreconstituted with the F1Fo ATPase under identical conditions (Fig. 2). The rate of Na+ transport (2 μmol min−1 mg−1 protein) was higher than the proton transport activity of the ATPase (0.5 μmol min−1 mg−1 protein). We therefore expect that NADH oxidation would elicit the quenching of ACMA fluorescence if NDH-1 catalyzes proton transport at rates comparable to its Na+ transport activity. If NADH was added prior to the addition of ATP, proton transport by the ATPase was drastically diminished (Fig. 3). Most likely, the transmembrane voltage (inside positive) generated during NADH-driven Na+ transport by NDH-1 (14) prevented the translocation of protons by the ATPase into the lumen of the proteoliposomes.
FIG. 2.
Influence of pH on NADH-driven Na+ transport by NDH-1. NADH-driven Na+ uptake (•) by NDH-1 purified by anionic-exchange chromatography in the buffer used for the reconstitution of the complex into proteoliposomes (10 mM HEPES-KOH, 50 mM KCl, 10% glycerol) was monitored. The protein-to-lipid ratio was 1:4. At time zero, 0.1 mM NADH and 5 mM NaCl were added, and the Na+ content of the proteoliposomes was determined at the indicated times. In the control reactions, NADH was omitted (▪). (A) Proteoliposomes containing NDH-1. Na+ transport at pH 6.5 or pH 7.5 was monitored. (B) Proteoliposomes containing NDH-1 and the H+-translocating ATPase. Na+ transport at pH 7.0 was monitored.
FIG. 3.
Proton transport into proteoliposomes containing NDH-1 from Klebsiella pneumoniae and the F1Fo ATPase from Escherichia coli. Proton transport was followed by the quenching of ACMA fluorescence in 10 mM Tris-HCl, pH 7.0, containing 10 mM MgCl2, 50 mM KCl, 10% glycerol, and 5 mM NaCl. The arrows indicate the addition of 0.1 mM NADH or 2.5 mM ATP.
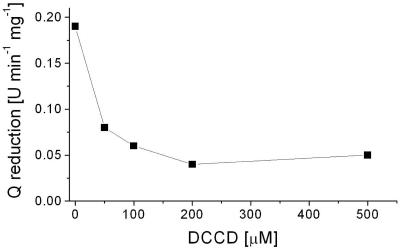
The quinone reduction activity of NDH-1 decreased from 0.19 μmol min−1 mg−1 protein at pH 7.5 to 0.04 μmol min−1 mg−1 protein at pH 6.5 (Fig. 4), concomitant with a drop of the Na+ transport activities from 3.6 μmol Na+ min−1 mg−1 protein (pH 7.5) to 1.6 μmol Na+ min−1 mg−1 protein (pH 6.5). Most likely, the lower rates of electron transfer at pH 6.5 resulted in decreased transport activities, but a switch from Na+ to H+ as the preferred coupling cation should also be considered. In summary, sodium ion translocation by NDH-1 was observed at a pH range from 6.5 to 7.5 and in the presence of 5 mM Na+. These in vitro conditions are reminiscent of the situation in the bacterial cytoplasm, which is regulated to pH 7.6 to 7.8 and contains 4 to 25 mM Na+ (32). Bertsova and Bogachev previously reported that NDH-1 did not contribute to Na+ transport in K. pneumoniae cells grown aerobically on LB. This negative result was taken as further evidence for proton transport by NDH-1 (3). The failure of Bertsova and Bogachev to observe Na+ translocation by NDH-1 during aerobic respiration is not surprising, as electron flow in enterobacteria is not directed through NDH-1 under these growth conditions (45). In anaerobically grown E. coli cells (42) or K. pneumoniae cells (23), NADH is predominantly oxidized by NDH-1, which acts as a Na+ pump. During transport, Na+ is proposed to bind to a critical carboxylic group in the membrane-bound NuoH subunit of NDH-1 from K. pneumoniae, as will be outlined below.
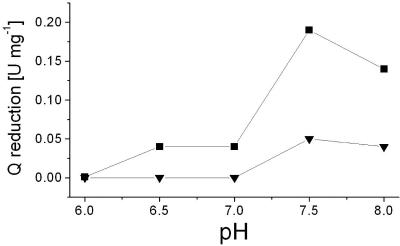
FIG. 4.
pH optimum for the modification of NDH-1 by DCCD. NDH-1 obtained by PEG precipitation was adjusted to the desired pH with Good buffers and incubated with 698 μM DCCD for 3 min. The control mixtures contained 6 μl ethanol. The residual Na+ concentration was 0.6 mM. The Q reductase activity of NDH-1 modified with DCCD (▾) or without DCCD added (▪) was determined at pH 8.0.
DCCD inhibits quinone reduction and Na+ transport by NDH-1.
The carboxyl group-modifying reagent DCCD specifically inhibits mitochondrial complex I and the related NDH-1 from bacteria (51). In bovine complex I, DCCD modifies the membrane-spanning ND1 subunit (NuoH homologue in NDH-1) and inhibits both electron transfer and proton transport (52). We investigated how the modification of NDH-1 from K. pneumoniae by DCCD at low (0.6 mM) Na+ concentrations was influenced by the pH of the reaction buffer. The extent of modification with the carbodiimide was estimated from the inhibition of the quinone reductase activity of NDH-1. At pH 7.5 and in the presence of 0.6 mM Na+, quinone reduction by NDH-1 was inhibited by 75% after modification with 200 μM DCCD for 4 min (Fig. 5). The pH dependency of DCCD inactivation was studied after incubation of NDH-1 with 698 μM DCCD for 3 min. We confirmed that inhibition was maximal after 3 min under these conditions (see Fig. S1 in the supplemental material). It is noteworthy that in the absence of DCCD, NDH-1 was completely inactivated at pH 6.0. At pH 6.5 and 7.0, NDH-1 exhibited only 25% of the maximum activity observed at pH 7.5 (Fig. 4). Incubation of NDH-1 with DCCD resulted in the inhibition of Q reductase activity, which was markedly influenced by the pH of the reaction medium. At pH 6.5 and 7.0, the modification of NDH-1 with DCCD led to complete inactivation. At pH 7.5 and 8.0, the electron transfer activity of NDH-1 modified with DCCD decreased to approximately 25% of the activity observed in the absence of the carbodiimide (Fig. 4). We now asked whether DCCD also inhibited NADH-driven Na+ transport by NDH-1. NDH-1 was reconstituted into proteoliposomes at pH 7.5 and incubated with DCCD in the presence of 0.6 mM Na+. The initial rates of Na+ transport by NDH-1 decreased from 1.8 μmol Na+ min−1 mg−1 lipid in the absence of DCCD to zero in the presence of 800 μM DCCD (Fig. 6). In the presence of 200 μM DCCD, the initial rate of Na+ transport was decreased by 50%.
FIG. 5.
DCCD inhibition of quinone reduction by NDH-1. Aliquots of NDH-1 obtained by PEG precipitation (0.3 mg in 50 μl 50 mM HEPES-KOH, pH 7.5, 50 mM KCl, and 4% glycerol) were incubated with 0, 100, 200, or 500 μM DCCD for 4 min, and the quinone reductase activity was monitored at pH 8.0.
FIG. 6.
DCCD inhibition of Na+ transport by NDH-1. NDH-1 obtained by PEG precipitation was reconstituted into proteoliposomes at pH 7.5 and incubated with 200 μM (○), 500 μM (▪), or 800 μM (▾) DCCD or without DCCD (•). The protein-to-lipid ratio was 1:1. Na+ transport was initiated by the addition of 5 mM NaCl and 0.1 mM NADH. The data were corrected by subtracting the Na+ content of proteoliposomes from a control reaction without NADH added (40 to 80 nmol Na+ mg−1 lipid) (Fig. 2).
It is concluded that DCCD inhibited both electron transfer and Na+ transport by NDH-1 from K. pneumoniae. The modification with DCCD was less effective at an alkaline pH, indicating that the DCCD-reactive residue of NDH-1 must be protonated in order to be modified. A likely target for DCCD modification is a carboxylic group located in the membrane-embedded part of the complex. A hydrophobic environment shifts the pK of a carboxylic group to the alkaline range, which facilitates protonation and enhances its reactivity with the carbodiimide (21). In the Na+-translocating F1Fo ATPase from Propionigenium modestum, DCCD modifies the protonated carboxylic group of glutamate 65 in subunit c from the membrane-bound Fo part. Glutamate 65 is protected by Na+ from the reaction with DCCD and represents a ligand for the coupling cation, which can be either a proton or a sodium ion (8). We hypothesized that the DCCD-reactive carboxylic group in NDH-1 could ligate a sodium ion and investigated the effect of Na+ on the modification of NDH-1 with DCCD.
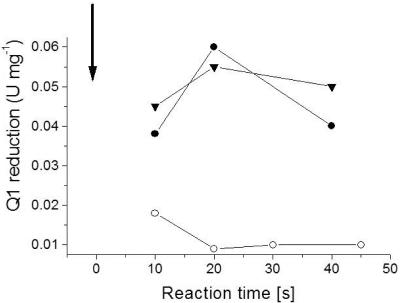
Na+ protects NDH-1 from modification with DCCD.
Figure 7 shows the time course of the reaction of NDH-1 with DCCD at pH 6.5 in the presence of 0.6 or 50 mM Na+. The extent of modification with the carbodiimide was estimated from the Q reductase activity of NDH-1 after different incubation periods. In the presence of 0.6 mM Na+, we observed a rapid inactivation of NDH-1 by DCCD. NDH-1 activity was decreased to 25% of the initial activity within the first 20 s. If 50 mM Na+ was added to NDH-1 prior to the addition of DCCD, the inactivation of NDH-1 was completely prevented, indicating full protection of NDH-1 from the modification with the carbodiimide. Other cations like K+ (Fig. 5) or Li+ (50 mM) did not protect NDH-1 from DCCD modification. If the inhibition of NDH-1 by DCCD is due to the specific modification of a carboxylic group, the loss of Q reductase activity should correlate with the incorporation of radioactivity from [14C]DCCD into a subunit of NDH-1. Figure 1 shows the autoradiogram of NDH-1, which was modified with [14C]DCCD at pH 6.5 in the presence of 0.6 or 50 mM Na+. At 0.6 mM Na+, two radioactive bands at apparent molecular masses of 30 and 7 kDa were detected (Fig. 1, lane 3). Raising the Na+ concentration in the reaction mixture to 50 mM decreased the radioactivity incorporated into the 30-kDa polypeptide to less than 30% of the radioactivity detected at 0.6 mM Na+. In contrast, radioactivity associated with the 7-kDa polypeptide slightly increased in the presence of 50 mM Na+ (Fig. 1, lane 4). It is concluded that Na+ prevented the reaction of the 30-kDa polypeptide with [14C]DCCD. This protection from modification by [14C]DCCD correlated with the protection of NDH-1 from DCCD inactivation by Na+. In a parallel experiment performed under identical conditions, the Q reduction activity of NDH-1 decreased from an initial value of 0.1 μmol min−1 mg−1 to zero after incubation with 100 μM DCCD for 1 h. By adding 50 mM Na+ prior to the addition of DCCD, NDH-1 was protected from inhibition. We hypothesized that the 30-kDa band represented a membrane-embedded polypeptide, since the stable modification of a protein by DCCD preferentially occurs in hydrophobic environments. After extraction of NDH-1 with 2-butanol, a prominent polypeptide with an apparent molecular mass of 30 kDa and several additional polypeptides were recovered in the organic phase (Fig. 1, lane 2). We did not detect flavins in the 2-butanol extract from NDH-1 by fluorescence spectroscopy (see Fig. S2 in the supplemental material) and conclude that the extract did not contain polypeptides with covalently attached flavins like subunits NqrB and NqrC from Na+-NQR (31). Hydrophobic extraction of NDH-1 modified with [14C]DCCD yielded radioactively labeled polypeptides with apparent molecular masses between 30 and 7 kDa (Fig. 1, lane 5). The 30-kDa polypeptide was subjected to N-terminal sequencing by Edman degradation. We detected two prominent amino acids during each degradation cycle, indicating the presence of two distinct polypeptides, and obtained the mixed sequence A/S, S/X, E/L, N/T, M/P, T/D, P/L (X, not identified). A major contamination of our NDH-1 preparation is the proton-pumping F1Fo ATPase (Fig. 1). Treatment with 2-butanol will extract hydrophobic subunits of the membrane-embedded part of NDH-1 together with subunits a (30.1 kDa) and c (8.1 kDa) from the Fo part of the ATPase. The N-terminal sequence of subunit a from K. pneumoniae ATPase is ASENMTP (20). Assuming that subunit a of the ATPase is present in the organic extract of NDH-1, the second polypeptide detected in the 30-kDa band with the sequence SXLTPD is the NuoH subunit (ND1 homologue) from NDH-1, which has the N-terminal sequence SWLTPD, deduced from the nuoH gene of K. pneumoniae (http://genome.wustl.edu/projects/bacterial/kpneumoniae/). The DCCD-reactive polypeptide with an apparent mass of 7 kDa detected in NDH-1 and in the 2-butanol extract from NDH-1 (Fig. 1B) could represent subunit c from the proton-translocating F1Fo ATPase or another hydrophobic subunit from NDH-1, like NuoA (19). Note that multiple bands of the organic extract from NDH-1 in the SDS-PAGE gel could represent a single polypeptide that binds various amounts of lipids, as observed with subunit c of ATPase (22). The detergent-solubilized NDH-1 might also contain additional [14C]DCCD-reactive subunits that escape detection due to insufficient separation by SDS-PAGE.
FIG. 7.
Protection of NDH-1 from DCCD inhibition by Na+. NDH-1 obtained by PEG precipitation (0.14 mg in 20 μl 1 M MES, pH 6.5) was mixed with 698 μM DCCD, and the quinone reduction activity was monitored after different reaction times (final DCCD concentration in the assay buffer, 14 μM). The arrow indicates the addition of DCCD at time zero. ○, with DCCD added; •, without DCCD added; ▾, 50 mM NaCl added prior to DCCD addition.
In summary, the results show that K. pneumoniae NDH-1 is inhibited by DCCD. This inhibition, which was promoted by more-acidic pH values, was accompanied by the modification of a 30-kDa polypeptide by [14C]DCCD. Sodium ions prevented the modification and inactivation of NDH-1 by DCCD. The DCCD-reactive 30-kDa polypeptide most likely represents the NuoH subunit of the membrane-embedded part of NDH-1.
DISCUSSION
During the oxidation of NADH with quinone, NDH-1 generates an electrochemical potential that drives ATP synthesis (45), but how charge separation across the inner bacterial membrane is achieved remains enigmatic. NDH-1 could establish transmembrane potential by the transport of electrons, protons, or another coupling ion. The results presented here corroborate our previous conclusion that the bacterial homologue of complex I (NDH-1) from Klebsiella pneumoniae catalyzes electrogenic Na+ transport. Two Na+ ions are translocated per NADH oxidized (13), and a transmembrane potential of at least 30 mV is generated (14). NDH-1 is activated by Na+ in a positively cooperative manner, suggesting that at least two sodium ions bind to the complex (13). Prompted by the findings of Bertsova and Bogachev (3), we reinvestigated the cation selectivity of reconstituted NDH-1 and confirmed Na+ transport at a pH range of 6.5 to 7.5. In addition, we found no evidence for H+ transport at pH 7.0 in the presence of 5 mM Na+. We previously discussed the possibility that NDH-1 from K. pneumoniae could act as a proton pump at low Na+ concentrations and an acidic pH but preferentially translocates Na+ under physiological conditions (13), e.g., in the presence of 4 to 25 mM Na+ (32). Recent studies with NDH-1 from E. coli and with the energy-converting hydrogenase from Carboxydothermus hydrogenoformans support the idea that either H+ or Na+ can be used as a coupling cation in these phylogenetically related ion pumps. The Coo hydrogenase from C. hydrogenoformans catalyzes CO-dependent H2 production and comprises six subunits that are homologues of the peripheral NuoBCDI and the membrane-bound NuoH and NuoL subunits of NDH-1 (16). Hedderich and coworkers previously observed that the Coo hydrogenase translocated protons at pH 5.9 but switched to Na+ transport at pH 7.0. Those authors discussed the possibility that the Coo hydrogenase is a Na+ pump that can use H+ at a low pH (10a). Using reconstituted E. coli NDH-1, Stolpe and Friedrich previously observed an effect of Na+ on NADH-driven proton transport and speculated that the redox reaction of NDH-1 is coupled with some type of Na+/H+ antiporter (43).
In the Na+-translocating F1Fo ATPase from Propionigenium modestum, the coupling cation is either a proton or a sodium ion, which is ligated by a DCCD-reactive carboxylic group in the membrane-bound Fo part (8). A similar interaction between Na+ and the membrane-embedded regions of the complex can be expected for NDH-1. The high-resolution structures of the rotor rings from Na+-translocating ATPases provide insight into a Na+ binding site and show how the positive charge of the sodium ion is shielded by the negative charge of a carboxylate in the membrane-spanning part of the Fo ring (28) and the K ring (29). At an acidic pH and a low Na+ concentration, this carboxylate located in subunit c of the F1Fo ATPase becomes protonated and specifically reacts with DCCD (8). In NDH-1 from K. pneumoniae, Na+ prevents the inactivation of the complex by DCCD and protects the NuoH subunit from modification with the carbodiimide. There is evidence that Na+ also protects the Coo hydrogenase from inhibition by DCCD (10a). The NuoH subunit has eight transmembrane helices and is one of the most conserved subunits in NDH-1 or complex I (34). Likely targets for DCCD are conserved carboxylic groups in membrane-embedded regions of NuoH, but the possibility that DCCD modifies a carboxylic group from a peripheral loop that extends into a hydrophobic environment should also be considered. An attractive candidate for DCCD modification is the conserved Glu247 in Nqo8 (NuoH homologue), which seems to be important for in vivo activity of NDH-1 from Paracoccus denitrificans (25).
In a direct mechanism of NDH-1 inactivation, DCCD modifies a protonated carboxylic group in a sodium ion binding site, but this reaction is prevented if the proton is replaced by Na+. DCCD could also indirectly inhibit Na+ transport by blocking other functions of NDH-1 like substrate binding or electron transfer. In this scenario, the protective effect of Na+ reflects a decreased accessibility of DCCD to NDH-1 at elevated Na+ concentrations. An indirect mechanism of NDH-1 inactivation seems unlikely, because the electron flow from NADH to the Fe-S centers in bovine complex I is not affected after modification with DCCD (24). We therefore propose a direct mechanism of DCCD inhibition where Na+ and DCCD compete for the binding to a critical carboxylic group in the NuoH subunit of NDH-1 from K. pneumoniae.
The quinone reductase activity of NDH-1 from K. pneumoniae is stimulated by Na+, and the activation profile is consistent with the cooperative binding of at least two sodium ions to NDH-1 (13). The membrane-bound NuoL subunit is likely to interact with Na+, since a truncated version of NuoL from E. coli NDH-1 exhibits Na+ transport activity (40). The results described here suggest that NuoH could also bind Na+, but whether NuoH and NuoL together participate in Na+ binding or whether they offer two distinct Na+ binding sites cannot be answered yet. Inhibitor studies suggest that the ND1 subunit of mitochondrial complex I (NuoH homologue) does not necessarily bind quinone but is located close to a quinone binding site (10, 38). The complex I inhibitor rotenone, which acts at a quinone reduction site, does not prevent the modification of the bovine ND1 subunit (NuoH homologue) with DCCD (52). While NuoH does not seem to directly bind quinone, it might provide a ligand for Na+ during transport by NDH-1. The modification with DCCD is a promising tool to characterize the Na+ binding site in the NuoH subunit of NDH-1 in future studies.
Supplementary Material
Acknowledgments
We thank Po-Chi Lin for providing the F1Fo ATPase from E. coli. We also thank Reiner Hedderich and Lucia Forzi, Max Planck Institut für terrestrische Mikrobiologie, Marburg, for in-depth discussions. We are indebted to Peter Hunziker, Universität Zürich, for carrying out the tryptic digests and mass spectroscopic analyses of proteins.
This work was supported by the Swiss National Science Foundation and the Vontobel Stiftung.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Dedicated to Peter Dimroth on the occasion of his 65th birthday.
REFERENCES
- 1.Beinert, H. 1978. Micro methods for the quantitative determination of iron and copper in biological material. Methods Enzymol. 54:435-445. [DOI] [PubMed] [Google Scholar]
- 2.Beinert, H. 1983. Semi-micro methods for analysis of labile sulfide and of labile sulfide plus sulfane sulfur in unusually stable iron-sulfur proteins. Anal. Biochem. 131:373-378. [DOI] [PubMed] [Google Scholar]
- 3.Bertsova, Y. V., and A. V. Bogachev. 2004. The origin of the sodium-dependent NADH oxidation by the respiratory chain of Klebsiella pneumoniae. FEBS Lett. 563:207-212. [DOI] [PubMed] [Google Scholar]
- 4.Böttcher, B., D. Scheide, M. Hesterberg, L. Nagel-Steger, and T. Friedrich. 2002. A novel, enzymatically active conformation of the Escherichia coli NADH:ubiquinone oxidoreductase (complex I). J. Biol. Chem. 277:17970-17977. [DOI] [PubMed] [Google Scholar]
- 5.Carroll, J., I. M. Fearnley, J. M. Skehel, M. J. Runswick, R. J. Shannon, J. Hirst, and J. E. Walker. 2005. The post-translational modifications of the nuclear encoded subunits of complex I from bovine heart mitochondria. Mol. Cell. Proteomics 4:693-699. [DOI] [PubMed] [Google Scholar]
- 6.DiBerardino, M., and P. Dimroth. 1996. Aspartate 203 of the oxaloacetate decarboxylase β-subunit catalyses both the chemical and vectorial reaction of the Na+ pump. EMBO J. 15:1842-1849. [PMC free article] [PubMed] [Google Scholar]
- 7.Dimroth, P. 1986. Preparation, characterization and reconstitution of oxaloacetate decarboxylase from Klebsiella aerogenes, a sodium pump. Methods Enzymol. 125:530-540. [DOI] [PubMed] [Google Scholar]
- 8.Dimroth, P. 1996. Sodium ion coupled F1Fo ATPases, p. 21-46. In W. N. Konings, H. R. Kaback, and J. S. Lolkema (ed.), Handbook of biological physics. Elsevier Science B.V., Amsterdam, The Netherlands.
- 9.Dunn, S. D. 1986. Effects of the modification of transfer buffer composition and the renaturation of proteins in gels on the recognition of proteins on Western blots by monoclonal antibodies. Anal. Biochem. 157:144-153. [DOI] [PubMed] [Google Scholar]
- 10.Earley, F. G. P., S. D. Patel, C. I. Ragan, and G. Attardi. 1987. Photolabelling of a mitochondrially encoded subunit of NADH dehydrogenase with [3H]dihydrorotenone. FEBS Lett. 219:108-113. [DOI] [PubMed] [Google Scholar]
- 10a.Forzi, L. 2005. Ph.D. thesis. University of Marburg, Marburg, Germany.
- 11.Friedrich, T., P. van Heek, H. Leif, T. Ohnishi, E. Forche, B. Kunze, R. Jansen, W. Trowitzsch-Kienast, G. Höfle, H. Reichenbach, and H. Weiss. 1994. Two binding sites of inhibitors in NADH:ubiquinone oxidoreductase (complex I): relationship of one site with the ubiquinone-binding site of bacterial glucose:ubiquinone oxidoreductase. Eur. J. Biochem. 219:691-698. [DOI] [PubMed] [Google Scholar]
- 12.Gabaldon, T., D. Rainey, and M. A. Huynen. 2005. Tracing the evolution of a large protein complex in the eukaryotes, NADH:ubiquinone oxidoreductase (complex I). J. Mol. Biol. 348:857-870. [DOI] [PubMed] [Google Scholar]
- 13.Gemperli, A. C., P. Dimroth, and J. Steuber. 2002. The respiratory complex I (NDH I) from Klebsiella pneumoniae, a sodium pump. J. Biol. Chem. 277:33811-33817. [DOI] [PubMed] [Google Scholar]
- 14.Gemperli, A. C., P. Dimroth, and J. Steuber. 2003. Sodium ion cycling mediates energy coupling between complex I and ATP synthase. Proc. Natl. Acad. Sci. USA 100:839-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hassinen, E. I., and P. T. Vuokila. 1993. Reaction of dicyclohexylcarbodiimide with mitochondrial proteins. Biochim. Biophys. Acta 1114:107-124. [DOI] [PubMed] [Google Scholar]
- 16.Hedderich, R. 2004. Energy-converting [NiFe] hydrogenases from archaea and extremophiles: ancestors of complex I. J. Bioenerg. Biomembr. 36:65-75. [DOI] [PubMed] [Google Scholar]
- 17.Ishmukhametov, R. R., M. A. Galkin, and S. B. Vik. 2005. Ultrafast purification and reconstitution of His-tagged cysteine-less Escherichia coli F1Fo ATP synthase. Biochim. Biophys. Acta 1706:110-116. [DOI] [PubMed] [Google Scholar]
- 18.Kaim, G., and P. Dimroth. 1993. Formation of a functionally active sodium-translocating hybrid F1Fo ATPase in Escherichia coli by homologous recombination. Eur. J. Biochem. 218:937-944. [DOI] [PubMed] [Google Scholar]
- 19.Kao, M. C., S. Di Bernardo, M. Perego, E. Nakamaru-Ogiso, A. Matsuno-Yagi, and T. Yagi. 2004. Functional roles of four conserved charged residues in the membrane domain subunit NuoA of the proton-translocating NADH-quinone oxidoreductase from Escherichia coli. J. Biol. Chem. 279:32360-32366. [DOI] [PubMed] [Google Scholar]
- 20.Kauffer, S., R. Schmid, K. Steffens, G. Deckers-Hebestreit, and K. Altendorf. 1987. Evolutionary relationship between Enterobacteriaceae: comparison of the ATP synthases (F1F0) of Escherichia coli and Klebsiella pneumoniae. Arch. Microbiol. 148:187-192. [DOI] [PubMed] [Google Scholar]
- 21.Kluge, C., and P. Dimroth. 1993. Kinetics of inactivation of the F1FO ATPase of Propionigenium modestum by dicyclohexylcarbodiimide in relationship to H+ and Na+ concentration: probing the binding site for the coupling ions. Biochemistry 32:10378-10386. [DOI] [PubMed] [Google Scholar]
- 22.Kluge, C., and P. Dimroth. 1993. Specific protection by Na+ or Li+ of the F1Fo-ATPase of Propionigenium modestum from the reaction with dicyclohexylcarbodiimide. J. Biol. Chem. 268:14557-14560. [PubMed] [Google Scholar]
- 23.Krebs, W., J. Steuber, A. C. Gemperli, and P. Dimroth. 1999. Na+ translocation by the NADH:ubiquinone oxidoreductase (complex I) from Klebsiella pneumoniae. Mol. Microbiol. 33:590-598. [DOI] [PubMed] [Google Scholar]
- 24.Krishnamoorthy, G., and P. C. Hinkle. 1988. Studies on the electron transfer pathway, topography of iron-sulfur clusters, and site of coupling in NADH-Q oxidoreductase. J. Biol. Chem. 263:17566-17575. [PubMed] [Google Scholar]
- 25.Kurki, S., V. Zickermann, M. Kervinen, I. Hassinen, and M. Finel. 2000. Mutagenesis of three conserved Glu residues in a bacterial homologue of the ND1 subunit of complex I affects ubiquinone reduction kinetics but not inhibition by dicyclohexylcarbodiimide. Biochemistry 39:13496-13502. [DOI] [PubMed] [Google Scholar]
- 26.Laubinger, W., and P. Dimroth. 1987. Characterization of the Na+-stimulated ATPase of Propionigenium modestum as an enzyme of the F1F0 type. Eur. J. Biochem. 168:475-480. [DOI] [PubMed] [Google Scholar]
- 27.Leif, H., V. D. Sled, T. Ohnishi, H. Weiss, and T. Friedrich. 1995. Isolation and characterization of the proton-translocating NADH:ubiquinone oxidoreductase from Escherichia coli. Eur. J. Biochem. 230:538-548. [DOI] [PubMed] [Google Scholar]
- 28.Meier, T., P. Polzer, K. Diederichs, W. Welte, and P. Dimroth. 2005. Structure of the rotor ring of F-type Na+-ATPase from Ilyobacter tartaricus. Science 308:659-662. [DOI] [PubMed] [Google Scholar]
- 29.Murata, T., I. Yamato, Y. Kakinuma, A. G. Leslie, and J. E. Walker. 2005. Structure of the rotor of the V-type Na+-ATPase from Enterococcus hirae. Science 308:654-659. [DOI] [PubMed] [Google Scholar]
- 30.Nakashima, Y., K. Shinzawa-Itoh, K. Watanabe, K. Naoki, N. Hano, and S. Yoshikawa. 2002. Steady-state kinetics of NADH:coenzyme Q oxidoreductase isolated from bovine heart mitochondria. J. Bioenerg. Biomembr. 34:11-19. [DOI] [PubMed] [Google Scholar]
- 31.Nakayama, Y., M. Yasui, K. Sugahara, M. Hayashi, and T. Unemoto. 2000. Covalently bound flavin in the NqrB and NqrC subunits of Na+-translocating NADH-quinone reductase from Vibrio alginolyticus. FEBS Lett. 474:165-168. [DOI] [PubMed] [Google Scholar]
- 32.Padan, E., and S. Schuldiner. 1992. Na+ transport systems in prokaryotes, p. 3-24. In E. Bakker (ed.), Alkali cation transport systems in prokaryotes. CRC Press, Boca Raton, Fla.
- 33.Rich, P. R. 2003. The molecular machinery of Keilin's respiratory chain. Biochem. Soc. Trans. 31:1095-1105. [DOI] [PubMed] [Google Scholar]
- 34.Roth, R., and C. Hägerhäll. 2001. Transmembrane orientation and topology of the NADH:quinone oxidoreductase putative quinone binding subunit NuoH. Biochim. Biophys. Acta 1504:352-362. [DOI] [PubMed] [Google Scholar]
- 35.Sazanov, L. A., and J. E. Walker. 2000. Cryo-electron crystallography of two sub-complexes of bovine complex I reveals the relationship between the membrane and peripheral arms. J. Mol. Biol. 302:455-464. [DOI] [PubMed] [Google Scholar]
- 36.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 37.Schrimpf, S. P., H. Langen, A. V. Gomes, and C. Wahlestedt. 2001. A two-dimensional protein map of Caenorhabditis elegans. Electrophoresis 22:1224-1232. [DOI] [PubMed] [Google Scholar]
- 38.Schuler, F., and J. E. Casida. 2001. Functional coupling of PSST and ND1 subunits in NADH:ubiquinone oxidoreductase established by photoaffinity labeling. Biochim. Biophys. Acta 1506:79-87. [DOI] [PubMed] [Google Scholar]
- 39.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 40.Steuber, J. 2003. The C-terminally truncated NuoL subunit (ND5 homologue) of the Na+-dependent complex I from Escherichia coli transports Na+. J. Biol. Chem. 278:26817-26822. [DOI] [PubMed] [Google Scholar]
- 41.Steuber, J., W. Krebs, and P. Dimroth. 1997. The Na+-translocating NADH:ubiquinone oxidoreductase from Vibrio alginolyticus: redox states of the FAD prosthetic group and mechanism of Ag+ inhibition. Eur. J. Biochem. 249:770-776. [DOI] [PubMed] [Google Scholar]
- 42.Steuber, J., C. Schmid, M. Rufibach, and P. Dimroth. 2000. Na+ translocation by complex I (NADH:quinone oxidoreductase) of Escherichia coli. Mol. Microbiol. 35:428-434. [DOI] [PubMed] [Google Scholar]
- 43.Stolpe, S., and T. Friedrich. 2004. The Escherichia coli NADH:ubiquinone oxidoreductase (complex I) is a primary proton pump but may be capable of secondary sodium antiport. J. Biol. Chem. 279:18377-18383. [DOI] [PubMed] [Google Scholar]
- 44.Tokuda, H., and K. Kogure. 1989. Generalized distribution and common properties of Na+-dependent NADH:quinone oxidoreductases in gram-negative marine bacteria. J. Gen. Microbiol. 135:703-709. [Google Scholar]
- 45.Tran, Q. H., J. Bongaerts, D. Vlad, and G. Unden. 1997. Requirement for the proton-pumping NADH dehydrogenase I of Escherichia coli in respiration of NADH to fumarate and its bioenergetic implication. Eur. J. Biochem. 244:155-160. [DOI] [PubMed] [Google Scholar]
- 46.Türk, K., A. Puhar, F. Neese, E. Bill, G. Fritz, and J. Steuber. 2004. NADH oxidation by the Na+-translocating NADH:quinone oxidoreductase from Vibrio cholerae: functional role of the NqrF subunit. J. Biol. Chem. 279:21349-21355. [DOI] [PubMed] [Google Scholar]
- 47.Unemoto, T., T. Ogura, and M. Hayashi. 1993. Modifications by Na+ and K+, and the site of Ag+ inhibition in the Na+-translocating NADH-quinone reductase from a marine Vibrio alginolyticus. Biochim. Biophys. Acta 1183:201-205. [Google Scholar]
- 48.Walker, J. E., J. M. Skehel, and S. K. Buchanan. 1995. Structural analysis of NADH:ubiquinone oxidoreductase from bovine heart mitochondria. Methods Enzymol. 260:14-34. [DOI] [PubMed] [Google Scholar]
- 49.Weber, J., and A. E. Senior. 2003. ATP synthesis driven by proton transport in F1F0-ATP synthase. FEBS Lett. 545:61-70. [DOI] [PubMed] [Google Scholar]
- 50.Wehrle, F., G. Kaim, and P. Dimroth. 2002. Molecular mechanism of the ATP synthase's Fo motor probed by mutational analyses of subunit a. J. Mol. Biol. 322:369-381. [DOI] [PubMed] [Google Scholar]
- 51.Yagi, T. 1987. Inhibition of NADH-ubiquinone reductase activity by N,N′-dicyclohexylcarbodiimide and correlation of this inhibition with the occurrence of energy-coupling site 1 in various organisms. Biochemistry 26:2822-2828. [DOI] [PubMed] [Google Scholar]
- 52.Yagi, T., and Y. Hatefi. 1988. Identification of the dicyclohexylcarbodiimide-binding subunit of NADH-ubiquinone oxidoreductase (complex I). J. Biol. Chem. 263:16150-16155. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.