Quorum sensing is a process by which bacteria release and subsequently respond to signal molecules, as a mechanism for sensing population density (4). Acylated homoserine lactones (AHLs) are well-studied quorum-sensing signals among proteobacteria and are most commonly synthesized by enzymes of the LuxI family (3). AHLs are usually recognized by members of the LuxR family of transcription factors, often encoded adjacent to their corresponding LuxI-type AHL synthases. Proteobacterial genome sequencing has revealed the existence of many more LuxR homologues than were known or suspected to exist, frequently in significant excess over the number of recognized AHL synthases in the genome. Many of these regulatory genes are orphans, retaining key attributes of better-studied luxR homologues but not linked to or associated with an AHL synthase gene. Some of these orphans are substantially different in size from characterized LuxR homologues, with large truncations or additional sequences. Of those typically sized LuxR-type proteins with end-to-end similarity, many lack one or more conserved amino acid residues known to be critical to the function of most LuxR-type proteins (3). These imperfect LuxR homologues might function to recognize AHLs by an alternate mechanism, provide ligand-independent activity, act as dominant-negative inhibitors, or even detect alternate small molecules. In other cases, all or most of the critical residues are present. These more highly conserved LuxR-type orphans may respond to endogenously synthesized AHL(s), generated by an otherwise unassociated AHL synthase of the same microbe or possibly signals from different microbes. A study from Lequette et al., published in this issue of the Journal of Bacteriology, establishes the role of an intriguing orphan LuxR homologue called QscR (quorum-sensing control repressor) in a previously unrecognized regulatory pathway within the larger AHL quorum-sensing network of the opportunistic human pathogen Pseudomonas aeruginosa (12). This work provides important new insights into the activity of the orphan QscR protein within this complex control system and also illustrates an impressive application of DNA microarray technology to address specific hypotheses at a genomic scale.
Quorum sensing in Pseudomonas aeruginosa.
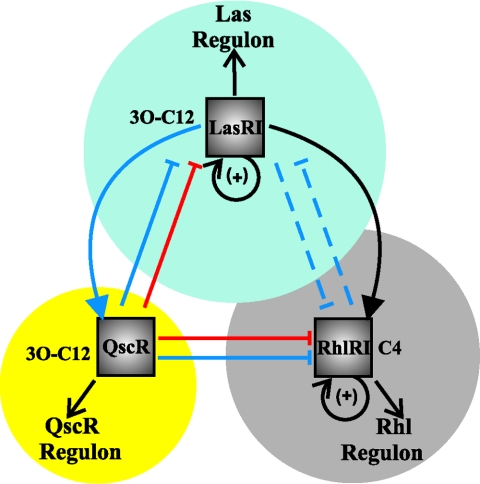
AHL quorum sensing in P. aeruginosa is a complex, multisignal, global regulatory network with control over diverse target functions including virulence factors, exoenzymes, motility, nutrient acquisition, and biofilm formation (9). Two LuxI-type proteins encoded at separate sites within the P. aeruginosa PAO1 genome, LasI and RhlI, direct the synthesis of N-3-oxo-dodecanoyl-l-homoserine lactone (3O-C12-HSL) and N-butyryl-l-homoserine lactone (C4-HSL), respectively (15, 16). LasR is a LuxR-type transcription factor encoded adjacent to lasI and in response to 3O-C12-HSL controls many target functions such as elastases, toxins, and other virulence factors. LasR also activates expression of the rhlR gene (Fig. 1) (see reference 5). RhlR is a second LuxR-type protein, encoded adjacent to rhlI and responsive to C4-HSL (14). RhlR also has numerous genomic targets, including rhamnolipid biosynthesis and siderophore production, many of which overlap to various degrees with the LasR regulon but others of which are clearly discrete (19, 20). LasR and RhlR are also linked to wider signaling pathways, including synthesis of the Pseudomonas quinolone signal and related hydroxyalkylquinolones (22). Transcription profiling suggests that the Las and Rhl pathways influence the expression of 3 to 11% of the P. aeruginosa genome (160 to 650 genes) (see references 7, 19, and 23).
FIG. 1.
Model of P. aeruginosa quorum-sensing network. Black arrows indicate direct transcriptional control (positive/negative or as indicated), blue arrows indicate protein-AHL interactions, and red arrows are protein-protein interactions. Solid arrows are well-supported mechanisms of regulation, while dashed arrows are more tentative. Looped arrows indicate positive feedback on cognate AHL synthesis via LasR and RhlR. Underlying circles represent genes under direct transcriptional control of each LuxR-type protein.
Definition of QscR as a quorum-sensing inhibitor.
The P. aeruginosa PAO1 sequence revealed the existence of multiple LuxR-type proteins, beyond LasR and RhlR (www.pseudomonas.com). Only one of these translation products, defined as QscR (quorum-sensing control repressor, PA1898), exhibits full conservation with AHL-responsive LuxR homologues. Initial genetic studies of QscR suggested that this protein functions to modulate the activity of the Las and Rhl regulons (2). The qscR gene is not adjacent to an AHL synthase gene but is immediately upstream of phenazine pigment biosynthetic genes (phzA2 to -G2). Null mutants in qscR form blue-pigmented colonies, indicative of phenazine overproduction, and aberrantly express two separate phenazine biosynthetic operons (phzA to -G and phzA2 to -G2) (2, 10, 13). Additional quorum-sensing-controlled genes were also expressed early and more strongly in the qscR mutant. Consistent with this finding, there were elevated levels of the 3O-C12-HSL and C4-HSL signals in early-stage cultures and early expression of lasI and rhlI (2). It seemed that QscR was limiting the activity of the LasR and RhlR regulators, although the mechanism(s) by which this occurred was unclear.
Studies of QscR expressed in Escherichia coli contributed to the impression of this protein as a Las-Rhl antagonist. Coexpression of QscR with either LasR or RhlR in the absence of AHL, followed by in vivo chemical cross-linking, identified apparent QscR-LasR and QscR-RhlR heterodimers (10). Fluorescence anisotropy analysis of these E. coli cells supported this interpretation. Furthermore, this same approach suggested that in E. coli QscR exists as an oligomer that is destabilized by addition of either 3O-C12-HSL or C4-HSL. This work suggested a potent mechanism for QscR-dependent repression through sequestration of LasR and RhlR monomers and perhaps through binding to each of their inducing ligands (Fig. 1).
Using arrays to resolve the disarray.
The current work from Lequette et al., using P. aeruginosa PAO1 DNA microarrays to profile genes under QscR control, provides a strikingly different view from the previous studies. In addition to its presumptive role as a negative effector of Las and Rhl, these new findings suggest that QscR also controls its own discrete regulon. Comparison of expression profiles of a qscR null mutant and wild-type PAO1 at different stages of culture growth revealed that more than 400 different genes were differentially expressed more than 2.5-fold and could be categorized into five different groups based on their expression pattern over the growth curve. Many of the regulated genes were repressed by QscR (329 genes), some of which had not been identified in the previous, rather exhaustive expression analyses of lasR and rhlR mutants (6, 19, 23, 25). Even more striking, a smaller set of 76 genes declined in expression in the qscR mutant, suggesting a positive function for QscR.
DNA microarrays are becoming readily available for analysis of genome-wide gene expression patterns in many different microbial systems. As with other transcriptional profiling studies, Lequette et al. present extensive tables of QscR-regulated genes, with many presumptive products that may provide clues as to the cellular logic that underlies QscR function. In this study, however, the authors have focused most of their attention on the QscR mechanism of action and the structure of the P. aeruginosa quorum-sensing network. The first experiments suggested that QscR might control a separate, perhaps overlapping, set of genes relative to LasR and RhlR. The simplest explanation was that QscR associates with DNA sequences proximal to specific target promoters to regulate gene expression, as with many other LuxR homologues. It was clear from the preceding work that QscR could indirectly influence gene expression, through mechanisms including formation of heterodimers with LasR and RhlR and by sequestration of the AHL signal molecules. How could those differences in expression due to less direct mechanisms be distinguished from those that were the result of QscR-mediated transcriptional regulation, given the extensive interdigitation of the quorum-sensing pathways in P. aeruginosa? To address this question on the genomic scale, Lequette et al. devised a clever DNA microarray experiment (12). The indirect mechanisms through which QscR is likely to act, heterodimer formation and AHL binding, are independent of the ability of this protein to associate with DNA. In contrast, those P. aeruginosa genes under direct QscR control would inherently require QscR DNA binding activity. A qscR deletion derivative in which the carboxy-terminal DNA binding domain was ablated (qscR-Δdbd) was integrated into the PAO1 chromosome under the control of an inducible promoter. A similar construct was created with full-length qscR, and P. aeruginosa PAO1 strains harboring these constructs were transcriptionally profiled under inducing conditions. A set of 38 QscR-regulated genes identified in the previous microarray experiment, distributed throughout the PAO1 genome, also required the QscR DNA binding domain. Some of these genes were activated and some were repressed by elevated expression of full-length qscR, again consistent with the differential expression of these genes resulting either immediately from or as a consequence of transcriptional regulation via QscR.
One of the more strongly activated genes (PA1897, the phzA2 promoter adjacent to qscR) was subsequently introduced into an E. coli strain expressing QscR and was strongly activated in the presence of the Las signal 3O-C12-HSL but was unaffected by the Rhl signal C4-HSL. This AHL-dependent activation required several amino acid residues known to be important for LuxR protein function. The authors were therefore able to validate the prediction of QscR functionality suggested by their DNA microarray experiments. In a parallel publication, the authors have also determined that QscR binds the PA1897 promoter and that this binding requires AHL (11).
A new model for the P. aeruginosa quorum-sensing network.
In contrast to a dispersed and somewhat opaque role for QscR in P. aeruginosa quorum sensing, we are instead provided with evidence of a relatively simple core for the P. aeruginosa quorum-sensing network. LasR, RhlR, and QscR each control the transcription of specific sets of target genes, some of which may overlap due to promoter cross-recognition (Fig. 1). There is a clear regulatory hierarchy by which both QscR and RhlR are subjugated to the LasR regulator, as rhlR expression is activated by LasR, and QscR activity requires the LasI-synthesized AHL, production of which is under positive feedback control from LasR. In addition to this core regulatory cascade, these systems may feed back on one another in significant ways. QscR has a negative influence on Rhl and Las systems, as validated by Lequette et al. and in earlier studies, at least partially independent of transcription control (Fig. 1). There is also evidence for weak cross-recognition of AHL signals between the Las and Rhl systems (17). More recently, the LasI-directed signal 3O-C12-HSL was reported to disrupt dimers of RhlR (21). Which of these more baroque feedback mechanisms are physiologically significant remains to be determined.
QscR joins several other orphan LuxR-type proteins which can respond to AHL signals generated via a different R-I regulatory system(s) within the same cell. An example is ExpR from Sinorhizobium meliloti, an orphan LuxR-type protein which has the dominant role in regulating the quorum-sensing network in this microbe and responds to AHL(s) synthesized by the sinRI regulatory pair (8, 18). In cases such as QscR and ExpR, the additional AHL receptor may allow for expansion of the regulatory network or integration of additional environmental controls. Lequette et al. demonstrate that QscR responds strongly to 3O-C12-HSL synthesized by LasI (12). In a parallel study, this same group demonstrates that QscR may be even more sensitive to N-3-oxo-decanoyl-l-homoserine lactone (3O-C10-HSL) than to the LasI-generated signal, whereas LasR is much more sensitive to 3O-C12-HSL (11). SdiA from E. coli and Salmonella enterica is a LuxR-type protein that apparently enables these bacteria, which do not synthesize AHLs, to respond to signals produced by neighboring cells (1, 24). Similarly, it seems plausible that QscR could also function to respond to cohabiting microbes that produce C10-HSL derivatives, acting to integrate monospecies and multispecies signaling.
Acknowledgments
I wish to thank E. P. Greenberg and colleagues for sharing findings prior to publication and helpful discussions on this commentary. Thanks also to T. G. Platt for critiquing the manuscript.
Research on quorum sensing in the Fuqua lab is supported by the National Science Foundation (MCB 0223724 and MCB 0238515) and the Indiana University MetaCyt Program.
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
REFERENCES
- 1.Ahmer, B. M. 2004. Cell-to-cell signalling in Escherichia coli and Salmonella enterica. Mol. Microbiol. 52:933-945. [DOI] [PubMed] [Google Scholar]
- 2.Chugani, S. A., M. Whiteley, K. M. Lee, D. D'Argenio, C. Manoil, and E. P. Greenberg. 2001. QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 98:2752-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuqua, C., and E. P. Greenberg. 2002. Listening in on bacteria: acylhomoserine lactone signalling. Nat. Rev. Mol. Cell Biol. 3:685-695. [DOI] [PubMed] [Google Scholar]
- 4.Fuqua, W. C., S. C. Winans, and E. P. Greenberg. 1994. Quorum sensing in bacteria: the LuxR/LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gambello, M. J., and B. H. Iglewski. 1991. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J. Bacteriol. 173:3000-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hentzer, M., K. Riedel, T. B. Rasmussen, A. Heydorn, J. B. Andersen, M. R. Parsek, S. A. Rice, L. Eberl, S. Molin, N. Hoiby, S. Kjelleberg, and M. Givskov. 2002. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology 148:87-102. [DOI] [PubMed] [Google Scholar]
- 7.Hentzer, M., H. Wu, J. B. Andersen, K. Riedel, T. B. Rasmussen, N. Bagge, N. Kumar, M. A. Schembri, Z. Song, P. Kristoffersen, M. Manefield, J. W. Costerton, S. Molin, L. Eberl, P. Steinberg, S. Kjelleberg, N. Hoiby, and M. Givskov. 2003. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 22:3803-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoang, H. H., A. Becker, and J. E. Gonzalez. 2004. The LuxR homolog ExpR, in combination with the Sin quorum sensing system, plays a central role in Sinorhizobium meliloti gene expression. J. Bacteriol. 186:5460-5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juhas, M., L. Eberl, and B. Tummler. 2005. Quorum sensing: the power of cooperation in the world of Pseudomonas. Environ. Microbiol. 7:459-471. [DOI] [PubMed] [Google Scholar]
- 10.Ledgham, F., I. Ventre, C. Soscia, M. Foglino, J. N. Sturgis, and A. Lazdunski. 2003. Interactions of the quorum sensing regulator QscR: interaction with itself and the other regulators of Pseudomonas aeruginosa LasR and RhlR. Mol. Microbiol. 48:199-210. [DOI] [PubMed] [Google Scholar]
- 11.Lee, J.-H., Y. Lequette, and E. P. Greenberg. 2006. Activity of purified QscR, a Pseudomonas aeruginosa orphan quorum-sensing transcription factor. Mol. Microbiol. 59:602-609. [DOI] [PubMed] [Google Scholar]
- 12.Lequette, Y., J.-H. Lee, F. Ledgham, A. Lazdunski, and E. P. Greenberg. 2006. A distinct QscR regulon in the Pseudomonas aeruginosa quorum-sensing circuit. J. Bacteriol. 188:3365-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mavrodi, D. V., R. F. Bonsall, S. M. Delaney, M. J. Soule, G. Phillips, and L. S. Thomashow. 2001. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J. Bacteriol. 183:6454-6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ochsner, U. A., and J. Reiser. 1995. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 92:6424-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pearson, J. P., K. M. Gray, L. Passador, K. D. Tucker, A. Eberhard, B. H. Iglewski, and E. P. Greenberg. 1994. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc. Natl. Acad. Sci. USA 91:197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pearson, J. P., L. Passador, B. H. Iglewski, and E. P. Greenberg. 1995. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 92:1490-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pearson, J. P., E. C. Pesci, and B. H. Iglewski. 1997. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in the control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 179:5756-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pellock, B. J., M. Teplitski, R. P. Boinay, W. D. Bauer, and G. C. Walker. 2002. A LuxR homolog controls production of symbiotically active extracellular polysaccharide II by Sinorhizobium meliloti. J. Bacteriol. 184:5067-5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schuster, M., C. P. Lostroh, T. Ogi, and E. P. Greenberg. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185:2066-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schuster, M., M. L. Urbanowski, and E. P. Greenberg. 2004. Promoter specificity in Pseudomonas aeruginosa quorum sensing revealed by DNA binding of purified LasR. Proc. Natl. Acad. Sci. USA 101:15833-15839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ventre, I., F. Ledgham, V. Prima, A. Lazdunski, M. Foglino, and J. N. Sturgis. 2003. Dimerization of the quorum sensing regulator RhlR: development of a method using EGFP fluorescence anisotropy. Mol. Microbiol. 48:187-198. [DOI] [PubMed] [Google Scholar]
- 22.Wade, D. S., M. W. Calfee, E. R. Rocha, E. A. Ling, E. Engstrom, J. P. Coleman, and E. C. Pesci. 2005. Regulation of Pseudomonas quinolone signal synthesis in Pseudomonas aeruginosa. J. Bacteriol. 187:4372-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wagner, V. E., D. Bushnell, L. Passador, A. I. Brooks, and B. H. Iglewski. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185:2080-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang, X., P. A. J. de Boer, and L. I. Rothfield. 1991. A factor that positively regulates cell division by activating transcription of the major cluster of essential cell division genes in Escherichia coli. EMBO J. 10:3363-3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whiteley, M., K. M. Lee, and E. P. Greenberg. 1999. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:13904-13909. [DOI] [PMC free article] [PubMed] [Google Scholar]