Abstract
We describe a platform technology, termed the dock and lock method, which uses a natural binding between the regulatory subunits of cAMP-dependent protein kinase and the anchoring domains of A kinase anchor proteins for general application in constructing bioactive conjugates of different protein and nonprotein molecules from modular subunits on demand. This approach could allow quantitative and site-specific coupling of many different biological substances for diverse medical applications. The dock and lock method is validated herein by producing bispecific, trivalent-binding complexes composed of three stably linked Fab fragments capable of selective delivery of radiotracers to human cancer xenografts, resulting in rapid, significantly improved cancer targeting and imaging, providing tumor/blood ratios from 66 ± 5 at 1 h to 395 ± 26 at 24 h.
Keywords: bispecific antibody, fusion proteins, A kinase, protein engineering, site-specific conjugation
Innovative fusion proteins created by recombinant technologies may be built into more complex structures to gain additional attributes that are highly desirable, yet not technically attainable, in the individual engineered construct. Well known examples include antibodies armed with toxic drugs or radionuclides to enhance therapeutic effects or facilitate target detection (1), cytokines modified with polyethylene glycols to increase serum half-lives (2), biotinylated proteins to enable immobilization into microarrays (3), and protein-DNA chimeras to quantify specific molecules to which the protein binds (4). To date, these goals are commonly achieved with varied success by judicious application of conjugation chemistries, but there remains a need for a general method that would ensure facile, quantitative, and site-specific coupling of bioactive molecules to form a multimeric product that not only is of defined composition but also retains the original activity of each linked constituent. New strategies that are based on enzyme-substrate or enzyme-inhibitor binding to tether two or more moieties of distinct functions into covalent (5) or quasicovalent (6) assemblies have been reported; however, the complicated methods may limit their widespread uses.
We describe a versatile method of generating multifunctional binary complexes composed of stably tethered fusion proteins by using the specific protein/protein interactions between the regulatory subunit of cAMP-dependent protein kinase (PKA) and the anchoring domains (AD) of A kinase anchor proteins (AKAPs). PKA, which plays a central role in one of the best studied signal transduction pathways triggered by the binding of the second messenger cAMP to the regulatory subunit of PKA (R), was first reported in 1968 (7). The structure of the holoenzyme, consisting of two catalytic subunits that are held in an inactive form by an R subunit dimer, was elucidated in the mid-1970s (8). Binding of cAMP to the R subunits leads to the release of active catalytic subunits for a broad spectrum of kinase activities, which are regulated through compartmentalization of the holoenzyme via AKAPs (9). Currently, >50 AKAPs that localize to various subcellular sites, including plasma membrane, actin cytoskeleton, nucleus, mitochondria, and endoplasmic reticulum, have been identified with diverse structures in species ranging from yeast to humans (10). Two types of R subunits (RI and RII) are found in PKA and each has α and β isoforms. The R subunits have been isolated only as stable dimers, and for type II, the dimerization domain has been shown to consist of the 44 amino-terminal residues (11). The AD of AKAPs for PKA is an amphipathic helix of 14–18 residues (12). The amino acid sequences of the AD are quite varied among AKAPs, and the binding affinities for RII dimers range from 2 × 10−9 M to 9 × 10−8 M, whereas the binding affinities for RI dimers are ≈100-fold weaker (13). AKAPs will bind only to dimeric R subunits. For human RIIα, the AD binds to a hydrophobic surface formed by the 23 amino-terminal residues (14). Thus, the dimerization domain and AKAP binding domain of human RIIα both are located within the same N-terminal 44-aa sequence, which is termed the dimerization and docking domain (DDD).
We envisioned a platform technology, referred to as the dock and lock (DNL) method, for quantitatively generating an exclusive binary complex consisting of any two components, referred to hereafter as A and B, via specific interactions between the DDD and the AD peptide sequences described above. One component of the binary complex, A, would be produced by linking a DDD sequence to a precursor of A resulting in a first structure, hereafter referred to as a. Because the DDD sequence would effect the spontaneous formation of a dimer, A thus would be composed of a2. The other component of the binary complex, B, would be produced by linking an AD sequence to a precursor of B, resulting in a second structure hereafter referred to as b. The dimeric motif of DDD contained in a2 should create a docking site for binding to the AD sequence contained in b, thereby resulting in a ready association of a2 and b to form a binary complex composed of a2b. This binding event could be stabilized further with a subsequent reaction to covalently secure the two components of the assembly via disulfide bridges, which might occur very efficiently, because initial binding interactions would orient the reactive thiol groups to ligate site-specifically and, therefore, without compromising the original functions of the two precursors. The approach is modular in nature and potentially can be applied to link, site-specifically and covalently, a wide range of substances, including peptides, proteins, and nucleic acids.
We have previously demonstrated the advantage of using a chemically prepared bispecific antibody with bivalent, instead of monovalent, binding to the tumor antigen for enhancing the amount and retention of radiotracer bound to the tumor (15). A recombinant bispecific trivalent construct, referred to as hBS14, with bivalent carcinoembryonic antigen (CEA) binding was produced in myeloma cell culture and performed very well as a pretargeting agent (16, 17). However, the relatively low productivity of hBS14 prompted us to develop alternative structures that would be functionally similar to hBS14 yet could be produced with a higher yield. In this paper, we describe the use of the DNL method to generate bispecific, trivalent-binding antibody complexes composed of three stably linked Fab fragments for selective delivery of radiotracers to human cancer xenografts by a method of pretargeting, resulting in improved cancer targeting and imaging.
Results
Bispecific Trivalent Structures Composed of Three Stably Linked Fab Fragments.
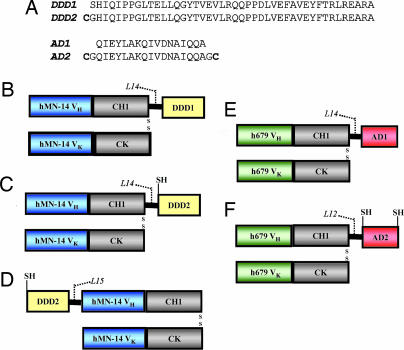
To prove the concept, we used the DNL method to assemble highly stable bispecific a2b complexes, comprising three Fab fragments. Five fusion proteins were generated (Fig. 1), for which detailed descriptions of the design, engineering, and stable transfection of each vector, as well as production, purification, and biochemical characterization of each fusion protein, are provided as Supporting Text, which is published as supporting information on the PNAS web site. Three A components were made recombinantly by using as a precursor the Fab fragment of the humanized monoclonal antibody, hMN-14 (18), which has binding specificity for human CEA. The first A, C-DDD1-hMN-14, was generated by linking the DDD1 peptide sequence, which is composed of amino acids 1–44 of human RIIα, to the carboxyl-terminal end of the Fd chain via a 14-residue flexible peptide linker (Fig. 1B). This construct was modified by incorporation of a cysteine residue adjacent to the amino-terminal end of DDD1 to create C-DDD2-hMN-14 (Fig. 1C). The DDD2 sequence was moved to the amino-terminal end of the Fd to generate N-DDD2-hMN-14 (Fig. 1D). Two B components were generated recombinantly by using as a precursor the Fab fragment of the humanized monoclonal antibody h679 (19), which has binding specificity for histamine-succinyl-glycine (HSG). The first B, h679-AD1, was generated by linking the AD1 sequence to the carboxyl-terminal end of the Fd chain via a 15-residue flexible peptide linker (Fig. 1E). AD1 is a 17-residue amino acid sequence derived from AKAP-IS, a synthetic peptide optimized for RII-selective binding with a reported dissociation constant (Kd) of 4 × 10−10 M (13). A second B, h679-AD2, was generated in the same fashion as h679-AD1 except with the addition of cysteine residues to both the amino- and carboxyl-terminal ends of AD1 (Fig. 1F).
Fig. 1.
Amino acid sequences of DDD and AD peptides (A) and schematic diagrams of C-DDD1-hMN-14 (B), C-DDD2-hMN-14 (C), N-DDD2-hMN-14 (D), h679-AD1 (E), and h679-AD2 (F). The heavy-chain constant domain 1 (CH1) and the light-chain constant domain (CK) are shown in gray. Variable domains of the heavy (VH) and light (VK) chains of hMN-14 and h679 are shown in blue and green, respectively. DDD and AD are shown in yellow and red, respectively. Peptide linker sequences (L14, L15 and L12) consisting of GGGGS repeats indicate the number of amino acids in each linker. Disulfide bridges and free sulfhydryl groups are indicated as SS and SH, respectively.
As expected, C-DDD1-hMN-14 and h679-AD1 were purified from culture media exclusively in the a2 form (as a homodimer of Fab) and b form (as a monomer of Fab), respectively (Fig. 5, which is published as supporting information on the PNAS web site). When C-DDD1-hMN-14 was combined with h679-AD1, the formation of an a2b complex was readily demonstrated by size-exclusion HPLC (SE-HPLC) and biacore (Fig. 6, which is published as supporting information on the PNAS web site). Equilibrium gel filtration analysis (20) further determined the Kd for the interaction between the a2 of C-DDD1-hMN-14 and the b of h679-AD1 to be ≈8 × 10−9 M (Fig. 7, which is published as supporting information on the PNAS web site), which is likely too weak of an affinity to keep the a2b complex intact at concentrations typical (<1 μg/ml) for in vivo applications.
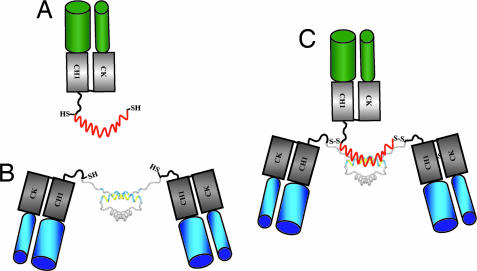
To prevent the dissociation of the noncovalent complex formed from C-DDD1-hMN-14 and h679-AD1 at lower concentrations, cysteine residues were introduced into the DDD and AD sequences of the A (N-DDD2-hMN-14 and C-DDD2-hMN-14) and B (h679-AD2) components, respectively. We anticipated that upon mixing of the cysteine-modified components, an a2b complex would promptly form, which could be further stabilized by the formation of disulfide bridges. Such stably tethered trivalent bispecific structures (Fig. 2), referred to as TF1 for N-DDD2-hMN-14/h679-AD2 and TF2 for C-DDD2-hMN-14/h679-AD2, were generated as follows. Because the analysis of the cysteine-modified fusion proteins revealed that disulfide reduction of each component is required for DDD2/AD2 binding (Supporting Text; see also Figs. 8–10, which are published as supporting information on the PNAS web site), h679-AD2 was mixed with a molar excess of N-DDD2-hMN-14 or C-DDD2-hMN-14 in the presence of the thiol-reducing agent Tris [2-carboxyethyl] phosphine hydrochloride at room temperature for 1 h. The Tris [2-carboxyethyl] phosphine hydrochloride was removed by hydrophobic interaction chromatography and DMSO was added to the solution to facilitate oxidative disulfide bond formation. The resulting disulfide-linked a2b structures (TF1 or TF2) were purified to homogeneity by HSG-based affinity chromatography (16). The events of the process at the stage of mixing, reduction, and final purification were monitored by SE-HPLC, as shown for the production of TF2 in Fig. 11, which is published as supporting information on the PNAS web site. A more detailed description and analysis of this process is given as Supporting Text.
Fig. 2.
Schematic diagrams of h679-AD2 as b (A), C-DDD2-hMN-14 as a2 (B); and TF2 as disulfide-linked a2b (C). The AD2 peptide is shown in red. The DDD dimer is shown in gray with the docking region shown in blue and yellow. Free sulfhydryl groups and disulfide bridges are shown as SH and S-S, respectively.
Biochemical Characterization of TF2.
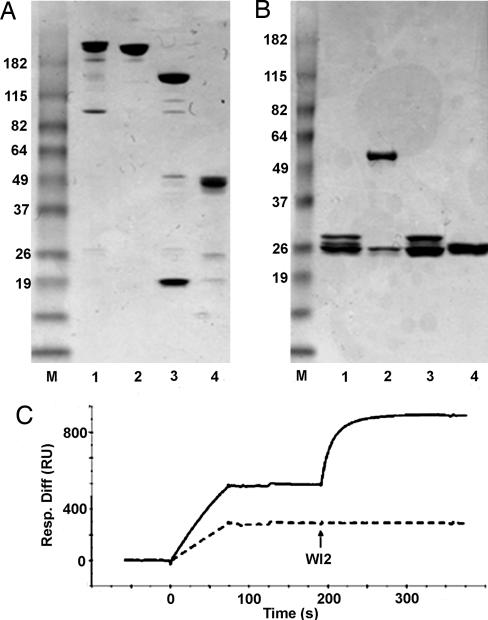
Both TF1 and TF2 were characterized in the same way, and the results for TF1 were similar to those discussed below for TF2, unless indicated otherwise. Ideally, TF2 should be a fully covalent complex composed of C-DDD2-hMN-14 and h679-AD2 having two disulfide bonds formed between the four cysteine residues introduced into DDD2 and AD2. However, complete disulfide formation between all reduced cysteines (including those from the inadvertently split Fd and light chains) may not be necessary to have a highly stable complex, provided that there is at least one disulfide bridge formed between AD2 and DDD2 to prevent the dissociation of a2 from b. Nonreducing SDS/PAGE analysis demonstrates that the majority of TF2 exists as an intact molecule (Fig. 3A, lane 1) with a relative mobility near that of IgG (Fig. 3A, lane 2). The additional minor bands (Fig. 3A, lane 1) reveal that disulfide formation is not complete under the conditions used for generating TF2, because only bands representing the constituent polypeptides of TF2 are evident by reducing SDS/PAGE (Fig. 3B). MALDI-TOF MS identified a single peak indicating a molecular mass of 156,434 Da, which is within 0.5% of the calculated molecular mass (157,319 Da) of the deduced amino acid sequences comprising TF2.
Fig. 3.
Characterization of TF2. Nonreducing (A) and reducing (B) SDS/PAGE analysis of TF2. Lanes: 1, TF2; 2, hMN-14 IgG; 3, C-DDD2-hMN-14; 4, h679-AD2. The positions of molecular mass markers (lane M) are indicated. (C) biacore analysis of TF2 by using an HSG-coupled sensorchip. TF2 (solid line) or a noncovalent a2b composed of C-DDD1-hMN-14 and h679-AD1 (dashed line) was diluted to 1 μg/ml total protein, and 50 μl was injected. A subsequent 100-μl injection of 20 μg/ml WI2 IgG (indicated by arrow) measured CEA binding.
The bispecificity of TF2 to bind simultaneously to both HSG and CEA was evaluated by biacore with an HSG-coupled sensorchip (Fig. 3C). Samples containing TF2 or an equimolar mixture of C-DDD1-hMN-14 and h679-AD1 (representing noncovalent a2b) were diluted to 1 μg/ml of total protein before Biacore injection. Because the noncovalent complex is injected at a concentration below the Kd determined for the a2b association by equilibrium gel filtration, the DDD1/AD1 complex falls apart during and immediately after injection, resulting in ≈50% decrease in the response units (RU) obtained from binding to the immobilized HSG when compared with that of TF2. Subsequent injections of WI2 IgG, a rat antiidiotype to hMN-14 (21), demonstrated that only TF2 contained the hMN-14 component that was tightly associated with the h679 component, as indicated by an additional increase in RU. If the WI2 binding is allowed to approach saturation, the stoichiometry of the binding between TF2 and WI2 can be determined by using the formula (RUWI2/RUTF2) × (MWTF2/MWWI2). The calculated stoichiometry was 1:1, as expected for a divalent TF2 reacting with a divalent WI2 IgG.
The results of immunoreactivity experiments by using SE-HPLC corroborated the interpretation of the biacore data, demonstrating that TF2 has two functional CEA binding sites (Fig. 12, which is published as supporting information on the PNAS web site). Competitive ELISA provided further evidence that TF2 binds to CEA divalently with the same avidity as the parental hMN-14 IgG, whereas the avidity of TF1 appeared to be 2-fold lower (Fig. 12). We speculate that the lower affinity of TF1 for CEA may be due to the close proximity of the two hMN-14 VH domains to each other as well as to the dimerized DDD2, resulting in the observed reduction in binding avidity.
Serum Stability.
TF1 and TF2 were designed to be stably tethered structures that should maintain full functionality when substantially diluted and could resist degradation in serum. The stability in human sera was assessed by diluting TF2 to 0.1 mg/ml in pooled fresh human serum, followed by incubation at 37°C under 5% CO2 for 7 days. Daily samples upon dilution 25-fold were analyzed by biacore with the same HSG-coupled sensorchip as described above. An injection of WI2 IgG was followed to quantify the amount of intact and fully active TF2. Serum samples were compared with control samples that were diluted directly from stock solutions. TF2 is stable in serum, retaining 98% of its bispecific binding activity after 7 days (Fig. 13, which is published as supporting information on the PNAS web site).
In Vivo Validation by Pretargeted Human Cancer Imaging in Mice.
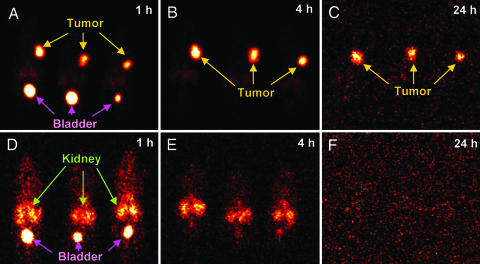
TF2 was evaluated in a pretargeting radioimmunodetection experiment by using nude mice bearing CEA-expressing, human colorectal adenocarcinoma xenografts (LS-174T). Pretargeting is a multistep process originally developed to resolve the slow blood clearance of directly targeting antibodies, which contributes to undesirable toxicity to normal tissues, in particular, bone marrow. With pretargeting, a radionuclide or other therapeutic agent is attached to a small compound that is cleared within minutes from the blood. The pretargeting agent, which is capable of recognizing the small radiolabeled compound in addition to the target antigen, is administered first, and the radiolabeled compound is administered at a later time when the pretargeting agent is sufficiently cleared from the blood. Superior uptake of a pretargeted 99mTc di-HSG peptide (99mTc-radiotracer) in a xenograft model, when compared with that of a directly radiolabeled anti-CEA Fab′, had been reported (17) recently by using hBS14, which comprises the same binding domains for CEA and HSG as TF2 but is different in structural design. Our previous findings (22, 23) indicated that the optimal time interval between administration of a bispecific antibody and the radiotracer is one that allows the former to clear the blood to <1% of the injected dose per gram (% ID/g). Preliminary biodistribution experiments demonstrated that by 16 h after injection, the amount of TF2 was <1% ID/g in the blood and all other normal tissues, whereas tumor uptake was >5% ID/g (Table 1, which is published as supporting information on the PNAS web site). Thus, 16 h after i.v. injection of TF2, the 99mTc-radiotracer was given, with animals being imaged and necropsied at various times after the radiotracer injection. The 99mTc-radiotracer uptake in tumor and normal tissues is summarized in Table 2, which is published as supporting information on the PNAS web site). At 1 h after injection, tumor uptake was 30.1 ± 13.7% ID/g, whereas the levels in blood and normal tissues, except kidney (5% ± 0.4% ID/g), were <1% ID/g. Tumor uptake remained high (16.3 ± 2.9% ID/g) over the 24-h study. Exceptional tumor-to-nontumor ratios were achieved as early as 0.5 h and improved over time (Table 3, which is published as supporting information on the PNAS web site). For example, the tumor-to-blood ratio was 13 ± 2, 66 ± 5, 237 ± 36, and 395 ± 26 at 0.5, 1, 4, and 24 h, respectively. Scintigraphic imaging confirmed the highly specific TF2-mediated tumor targeting of the 99mTc-radiotracer (Fig. 4). Images taken at 1, 4, and 24 h after injection of the radiotracer all showed an intense tumor-specific signal. Other than tumor, only the bladder at 1 h had a strong signal, indicating elimination of the radiotracer in the urine. These data demonstrate that TF2 is highly stable for in vivo applications, and we predict that other similarly constructed stably tethered structures will be as well.
Fig. 4.
Cancer imaging of mice bearing human colorectal adenocarcinoma xenografts. Nude mice bearing s.c. LS-174T tumors were injected i.v. with 125I-TF2 16 h before administering a 99mTc-radiotracer. Three mice were imaged on a γ-camera at 1 h (A), 4 h (B), and 24 h (C) after injection of the 99mTc-radiotracer. As a control, three additional mice received only the 99mTc-radiotracer (no pretargeting) and were imaged at 1 h (D), 4 h (E), and 24 h (F) after injection. Yellow, magenta, and green arrows indicate the locations of tumor, bladder, and kidney, respectively.
Discussion
By means of the DNL method, we assembled bispecific trivalent Fab constructs, TF1 and TF2, which were stable in serum for 7 days and showed superior localization in a human colonic carcinoma model expressing the target antigen, CEA. Already at 1 h after injection, tumor accretion was 30.1 ± 13.7% ID/g of tumor, and the tumor uptake remained high (>16% ID/g) over 24 h. Exceptionally high tumor/blood ratios were observed, ranging from 13 at 0.5 h to 395 at 24 h, which are comparable with those achieved with hBS14, a recombinant bispecific, trivalent construct with similar binding properties to TF2 (17). Accordingly, the use of the DNL technology to produce such stable Fab-fusion proteins for pretargeted cancer localization now provides a rapid and convenient method of production for clinical applications of both imaging and therapy.
Although the examples presented involve the facile union of two recombinant antibody-based fusion proteins, the DNL method is certainly not limited to the creation of multivalent binding structures composed of Fab or other variant forms of antibodies as the constituent subunits. In fact, TF1 and TF2 may be among the most complicated a2b structures to build. Besides the two disulfide bridges formed between the DDD2 and AD2, such Tri-Fabs possess three additional disulfide bridges that link the three pairs of cognate Fd and light chains and 12 more that reside in the respective VH, VL, CH1, and CL domains; all of these bridges must be properly assembled to retain the binding specificity and affinity. Even though the total assembly of the stably tethered structures requires both cleavage and reformation of disulfide bonds, these reactions proceed readily, efficiently, and quantitatively with no formation of undesirable side products, achieving 90% yield of TF1 or TF2 after purification. Therefore, we do not expect any difficulty with the creation of less complex a2b structures, such as those involving only three polypeptide chains with few or no additional disulfide bridges.
We also anticipate that the DNL method is useful for creating complex structures consisting of protein and nonprotein components, because derivatives based on AD2, which consists of only 21 amino acids, can be made synthetically with suitable reactive groups for conjugation to nonprotein precursors, such as nucleic acids or a variety of effectors, including drugs, dyes, chelators, radionuclides, and fluorescent molecules. Such nonprotein B components may be tethered to any A component to confer added functions, allowing novel applications. The findings that TF1 and TF2 both form stably tethered and fully functional a2b structures further attest to the innate flexibility of the DNL method. The A components of TF1 and TF2 have DDD2 positioned at the amino terminal end (N-DDD2-hMN-14) and the carboxyl-terminal end (C-DDD2-hMN-14) of the Fd polypeptide, respectively. In nature, DDD is located at the amino terminus of RII. However, our results suggest that its position is not restricted to the amino terminus to effect both dimerization and docking functions. The PKA-tethering domains (AD) are found at a wide variety of positions among the >50 identified AKAPs. Thus, for B component fusion proteins, a desirable AD sequence may be positioned internally or at either end of the polypeptide. For increased functionality, an AD sequence could be used as a linker peptide to couple two or more functional domains within the same B component, which would then be tethered to any A component.
We have demonstrated here that the A and B components can be produced as secreted fusion proteins by mammalian cell culture and combined subsequently or on demand. The individual components are stable upon storage at 4°C in PBS for at least 6 months, as shown for C-DDD2-hMN-14 and h679-AD2 by SE-HPLC, SDS/PAGE, and biacore analyses. Neither component exhibited any loss of protein mass or binding activity. There was no evidence of aggregation, precipitation, or degradation. Other A and B components generated from different antibody-binding domains also showed similar stability upon storage (unpublished results).
The high yield (>100 mg/liter) of the Fab-based components was comparable to that of its precursor; therefore, fusion of either a DDD or AD sequence to a precursor is not likely to diminish productivity. Fusion proteins containing an AD or DDD and intact RII subunits have been expressed recombinantly in E. coli with good yield (24). Thus, it does not appear that the choice of expression systems for an A or B component will be restricted by the DDD or AD group, respectively. Rather, the mode of production should be determined by the host cell requirements of the precursor.
Among the numerous strategies that have been used for engineering multivalent, multifunctional, fusion proteins, the one using the barnase-barstar module, as described by Deyev et al. (6), is particularly worth comparing with the DNL method. These researchers used the high-affinity interaction of the prokaryotic ribonuclease barnase and its natural inhibitor, barstar, to generate fusion proteins that could be purified and kept separately, and then mixed to form dimeric structures. Although this system can be useful for the modular assembly of binary complexes, there are some notable drawbacks. Because barnase is lethal to the host cell, barstar must be coexpressed with barnase fusion proteins, which require denaturation and chromatography under denaturing conditions for barstar removal and subsequent refolding. Utilization of barnase (110 amino acids) and barstar (89 amino acids) as a heterodimerization pair adds significant protein mass (22 kDa) that, combined with their bacterial origin, will likely render the complexes immunogenic. Other approaches shared some of the features of the DNL method, such as the selection of protein domains (25, 26) or peptide motifs (27, 28) for dimerization or oligomerization and the use of disulfide bridges for enhanced stability (27, 29). However, fusions with these domains or peptides usually produce more than one form of the desired product that, at least, makes downstream purification impractical. These methods may be useful for the generation of certain multimeric constructs, but in general, they are inefficient for the exclusive assembly of multimeric structures with defined compositions and unaltered functions.
In conclusion, although molecular engineering has revolutionized many areas of research, allowing the invention and production of a wide variety of recombinant proteins, the existing methods for producing multivalent, multifunctional constructs have not been satisfactory for clinical or commercial development. Therefore, the general approach described here, which enables efficient and quantitative formation of stably tethered structures with defined composition and multiple functionalities, appears to solve this problem.
Materials and Methods
Molecular Biology, Cell Culture, and Protein Purification.
A detailed description of the cloning procedures involved in generating the five mammalian expression vectors from the pdHL2 plasmid (30), vector transfection, screening and selection of productive clones, cell culture, and purification of each fusion protein is provided as Supporting Text.
Generation of TF1 and TF2.
A typical example of preparing TF2 is provided below. TF1 was prepared similarly, and the procedures have been repeated successfully at different scales. A 25% molar excess of C-DDD2-hMN-14 (150 mg) was mixed with affinity-purified h679-AD2 (60 mg) at 2 mg/ml total protein in 1 mM EDTA/PBS. The mixture was reduced for 1 h at room temperature with 5 mM Tris [2-carboxyethyl] phosphine hydrochloride and then adjusted to 0.75 M ammonium sulfate before loading onto a 20-ml butyl fast flow hydrophobic interaction chromatography column (Amersham Pharmacia). The column was washed with 0.75 M ammonium sulfate/1 mM EDTA/PBS (pH 7.4) to remove Tris [2-carboxyethyl] phosphine hydrochloride and then eluted with 1 mM EDTA/PBS. The eluate was made to 10% DMSO and allowed to react for 2 days at 25°C. The solution was loaded onto a 30-ml HSG-based affinity column, which was washed to baseline with PBS and eluted with ≈10 column volumes of elution buffer (1 M imidazole/1 mM EDTA/0.1 M NaAc, pH 4.5). The final product was dialyzed against several changes of PBS.
Molecular Size.
SE-HPLC was performed on a Beckman System Gold Model 116 with a Bio-Sil SEC 250 column (Bio-Rad). For some experiments requiring increased resolution, two Bio-Sil SEC 250 columns were connected in tandem. The following standards were used to calibrate the column: hMN-14 IgG (≈150 kDa), hMN-14 Fab′ (≈50 kDa) modified with N-ethyl maleimide, 679 Fab′ (≈50 kDa) modified with N-ethyl maleimide, and hMN-14 F(ab′)2 (≈100 kDa).
biacore Analysis.
The bispecific binding properties were analyzed by using a biacore x system (Biacore) with a high-density HSG-coupled biosensor chip (16). A flow rate of 40 μl/min was used for each experiment. Culture media or purified samples were diluted in Biacore running buffer (0.15 M NaCl/1 mM EDTA/0.1 M Hepes, pH 7.4) before injection.
SDS/PAGE.
Reducing and nonreducing SDS/PAGE analyses were performed by using 4–20% gradient Tris-Glycine gels (Cambrex Bio Science Rockland, Rockland, ME). Samples were diluted to 0.1 mg/ml in sample buffer (2% SDS/5% glycerol/62.5 mM Tris·HCl, pH 6.8) and heated to 95°C before 1 μg of total protein was loaded per lane. For reducing gels, 5% of 2-mercaptoethanol was included in the sample buffer. Gels were stained with Coomassie blue to visualize protein bands.
MALDI-TOF MS.
MALDI-TOF MS was performed in a sinapinic acid matrix by The Scripps Center for Mass Spectrometry (La Jolla, CA).
ELISA for CEA Binding.
A competitive ELISA was performed to compare the CEA binding of TF1, TF2, and hMN-14 IgG. Microplate wells were coated with a fusion protein (0.5 μg per well) containing the A3B3 domain of CEA, which is recognized by hMN-14 (31), and blocked with 2% BSA-PBS. Two-fold serial dilutions of TF1, TF2, and hMN-14 IgG were made in quadruplicate and added to the wells along with horseradish peroxidase-conjugated hMN-14 IgG (1 nM). The plates were developed with o-phenylenediamine dihydrochloride and read at 490 nm, after stopping the reaction by adding 1.5 N H2SO4.
Serum Stability.
The stability of TF2 in fresh human serum was assayed by biacore. TF2 was diluted to 0.1 mg/ml in fresh human serum, which was pooled from four donors, and incubated at 37°C under 5% CO2 for 7 days. Daily samples were diluted to 4 μg/ml (25-fold) and injected (50 μl) on Biacore by using an HSG-coupled sensorchip. A subsequent injection (100 μl) of WI2 IgG (20 μg/ml) quantified the amount of intact and fully active TF2. The WI2 binding response of the daily serum samples were compared with that of control samples diluted directly from stock solutions. The stability of TF1 in fresh human serum was determined in a similar fashion.
In Vivo Targeting and Imaging Studies.
In vivo pretargeting of CEA-expressing tumors was examined in female athymic nude mice (8 weeks old when transplanted; Taconic, Germantown, NY) bearing s.c. human colorectal adenocarcinoma xenografts (LS-174T). Tumor cells were expanded in tissue culture, and mice were injected s.c. with 1 × 107 cells per mouse. After 1 week, tumors were measured, and mice were assigned to groups of five per time point. The mean tumor size at the start of this study was 0.105 ± 0.068 cm3. One group of 20 mice was injected with 80 μg of 125I-TF2 (500 pmol, 2 μCi; 1 Ci = 37 GBq). The di-HSG peptide (IMP-245) was radiolabeled with 99mTc-pertechnetate (Mallinckrodt) as described in ref. 32. Mice were administered the 99mTc-di-HSG peptide (99mTc-radiotracer; 40 μCi, 92 ng, 50 pmol) 16 h after injection of 125I-TF2. The mice were killed and necropsied at 0.5, 1, 4, and 24 h after injection of the 99mTc-radiotracer. Tumor and various tissues were removed, weighed, and placed in a γ-counter to determine % ID/g of tissue at each time point. In addition, three mice from the 24 h time-point group were imaged at 1, 4, and 24 h after injection. As a control, three additional mice received only 99mTc-radiotracer (no pretargeting) and were imaged at 1, 4, and 24 h after injection, before being necropsied after the 24-h imaging session. Anesthetized mice were placed supine on the surface of a high-efficiency, low-energy collimator on an ADAC Solus gamma scintillation camera (ADAC Laboratories, Milpitas, CA). The images were adjusted to the same background, but with the brightness increased until a portion of the pixels in any given region were at their maximal intensity.
Supplementary Material
Acknowledgments
We thank Diane Nordstrom and John Kopinski for excellent technical assistance.
Abbreviations
- AD
anchoring domain
- AKAP
A-kinase anchor protein
- CEA
carcinoembryonic antigen
- DDD
dimerization and docking domain
- DNL
dock and lock
- HSG
histamine-succinyl-glycine
- % ID/g
percent of the injected dose per gram
- PKA
cAMP-dependent protein kinase
- R
regulatory subunit of PKA
- SE-HPLC
size-exclusion HPLC.
Footnotes
Conflict of interest statement: E.A.R., D.M.G., T.M.C., W.J.M., and C.-H.C. are employees of IBC Pharmaceuticals, Inc., or Immunomedics, Inc. D.M.G. is Chairman of the Board of Directors of both IBC Pharmaceuticals and Immunomedics.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Wu A. M., Senter P. D. Nat. Biotechnol. 2005;23:1137–1146. doi: 10.1038/nbt1141. [DOI] [PubMed] [Google Scholar]
- 2.Pepinsky R. B., LePage D. J., Gill A., Chakraborty A., Vaidyanathan S., Green M., Baker D. P., Whalley E., Hochman P. S., Martin P. J. Pharmacol. Exp. Ther. 2001;297:1059–1066. [PubMed] [Google Scholar]
- 3.Tan L.-P., Lue R. Y. P., Chen G. Y. J., Yao S. Q. Bioorg. Med. Chem. Lett. 2004;14:6067–6070. doi: 10.1016/j.bmcl.2004.09.083. [DOI] [PubMed] [Google Scholar]
- 4.Burbulis I., Yamaguchi K., Gordon A., Carlson R., Brent R. Nat. Methods. 2005;2:31–37. doi: 10.1038/nmeth729. [DOI] [PubMed] [Google Scholar]
- 5.Hodneland C. D., Lee Y.-S., Min D.-H., Mrksich M. Proc. Natl. Acad. Sci. USA. 2002;99:5048–5052. doi: 10.1073/pnas.072685299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deyev S. M., Waibel R., Lebedenko E. N., Schubuger A. P., Pluckthun A. Nat. Biotechnol. 2003;21:1486–1492. doi: 10.1038/nbt916. [DOI] [PubMed] [Google Scholar]
- 7.Walsh D. A., Perkins J. P., Krebs E. G. J. Biol. Chem. 1968;243:3763–3765. [PubMed] [Google Scholar]
- 8.Corbin J. D., Soderling T. R., Park C. R. J. Biol. Chem. 1973;248:1813–1821. [PubMed] [Google Scholar]
- 9.Scott J. D., Stofko R. E., McDonald J. R., Comer J. D., Vitalis E. A., Mangili J. A. J. Biol. Chem. 1990;265:21561–21566. [PubMed] [Google Scholar]
- 10.Wong W., Scott J. D. Nat. Rev. Mol. Cell. Biol. 2004;12:959–970. doi: 10.1038/nrm1527. [DOI] [PubMed] [Google Scholar]
- 11.Hausken Z. E., Dell’Acqua M. L., Coghlan V. M., Scott J. D. J. Biol. Chem. 1996;271:29016–29022. doi: 10.1074/jbc.271.46.29016. [DOI] [PubMed] [Google Scholar]
- 12.Carr D. W., Stofko-Hahn R. E., Fraser I. D., Bishop S. M., Acott T. S., Brennan R. G., Scott J. D. J. Biol. Chem. 1991;266:14188–14192. [PubMed] [Google Scholar]
- 13.Alto N. M., Soderling S. H., Hoshi N., Langeberg L. K., Fayos R., Jennings P. A., Scott J. D. Proc. Natl. Acad. Sci. USA. 2003;100:4445–4450. doi: 10.1073/pnas.0330734100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colledge M., Scott J. D. Trends Cell Biol. 1999;6:216–222. doi: 10.1016/s0962-8924(99)01558-5. [DOI] [PubMed] [Google Scholar]
- 15.Karacay H., Sharkey R. M., McBride W. J., Griffiths G. L., Qu Z., Chang K., Hansen H. J., Goldenberg D. M. Bioconjugate Chem. 2002;13:1054–1070. doi: 10.1021/bc0200172. [DOI] [PubMed] [Google Scholar]
- 16.Rossi E. A., Chang C.-H., Losman M. J., Sharkey R. M., Karacay H., McBride W., Cardillo T. M., Hansen H. J., Qu Z., Horak I. D., et al. Clin. Cancer Res. 2005;11:7122s–7129s. doi: 10.1158/1078-0432.CCR-1004-0020. [DOI] [PubMed] [Google Scholar]
- 17.Sharkey R. M., Cardillo T. M., Rossi E. A., Chang C.-H., Karacay H., McBride W. J., Hansen H. J., Horak I. D., Goldenberg D. M. Nat. Med. 2005;11:1250–1255. doi: 10.1038/nm1322. [DOI] [PubMed] [Google Scholar]
- 18.Sharkey R. M., Juweid M., Shevitz J., Behr T., Dunn R., Swayne L. C., Wong G. Y., Blumenthal R. D., Griffiths G. L., Siegel J. A., et al. Cancer Res. 1995;55:5935s–5945s. [PubMed] [Google Scholar]
- 19.Rossi E. A., Sharkey R. M., McBride W., Karacay H., Zeng L., Hansen H. J., Goldenberg D. M., Chang C.-H. Clin. Cancer Res. 2003;9:3886s–3896s. [PubMed] [Google Scholar]
- 20.Gegner J. A., Dahlquist F. W. Proc. Natl. Acad. Sci. USA. 1991;88:750–754. doi: 10.1073/pnas.88.3.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Losman M. J., Novick K. E., Goldenberg D. M., Monestier M. Int. J. Cancer. 1994;56:580–584. doi: 10.1002/ijc.2910560419. [DOI] [PubMed] [Google Scholar]
- 22.Sharkey R. M., McBride W. J., Karacay H., Chang K., Griffiths G. L., Hansen H. J., Goldenberg D. M. Cancer Res. 2003;63:354–363. [PubMed] [Google Scholar]
- 23.Sharkey R. M., Karacay H., Richel H., McBride W. J., Rossi E. A., Chang K., Yeldell D., Griffiths G. L., Hansen H. J., Goldenberg D. M. Clin. Cancer Res. 2003;9:3897s–3913s. [PubMed] [Google Scholar]
- 24.Herberg F. W., Maleszka A., Eide T., Vossebein L., Tasken K. J. Mol. Biol. 2000;298:329–339. doi: 10.1006/jmbi.2000.3662. [DOI] [PubMed] [Google Scholar]
- 25.Terskikh A. V., Le Doussal J.-M., Crameri R., Fisch I., Mach J.-P., Kajava A. V. Proc. Natl. Acad. Sci. USA. 1997;94:1663–1668. doi: 10.1073/pnas.94.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muller K. M., Arndt K. M., Strittmatter W., Pluckthun A. FEBS Lett. 1998;422:259–264. doi: 10.1016/s0014-5793(98)00021-0. [DOI] [PubMed] [Google Scholar]
- 27.de Kruif J., Logtenberg T. J. Biol. Chem. 1996;271:7630–7634. doi: 10.1074/jbc.271.13.7630. [DOI] [PubMed] [Google Scholar]
- 28.Muller K. M., Arndt K. M., Pluckthun A. FEBS Lett. 1998;432:45–49. doi: 10.1016/s0014-5793(98)00829-1. [DOI] [PubMed] [Google Scholar]
- 29.Schmiedl A., Breitling F., Dubel S. Protein Eng. 2000;13:725–734. doi: 10.1093/protein/13.10.725. [DOI] [PubMed] [Google Scholar]
- 30.Gillies S. D., Lo K. M., Wesolowski J. J. Immunol. Methods. 1989;125:191–202. doi: 10.1016/0022-1759(89)90093-8. [DOI] [PubMed] [Google Scholar]
- 31.Blumenthal R. D., Hansen H. J., Goldenberg D. M. Cancer Res. 2005;65:8809–8817. doi: 10.1158/0008-5472.CAN-05-0420. [DOI] [PubMed] [Google Scholar]
- 32.Hansen H. J., Jones A. L., Sharkey R. M., Grebenau R., Blazejewski N., Kunz A., Buckley M. J., Newman E. S., Ostella F., Goldenberg D. M. Cancer Res. 1990;50:794s–798s. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.