Abstract
Objective: To determine whether automated classification of document citations can be useful in reducing the time spent by experts reviewing journal articles for inclusion in updating systematic reviews of drug class efficacy for treatment of disease.
Design: A test collection was built using the annotated reference files from 15 systematic drug class reviews. A voting perceptron-based automated citation classification system was constructed to classify each article as containing high-quality, drug class–specific evidence or not. Cross-validation experiments were performed to evaluate performance.
Measurements: Precision, recall, and F-measure were evaluated at a range of sample weightings. Work saved over sampling at 95% recall was used as the measure of value to the review process.
Results: A reduction in the number of articles needing manual review was found for 11 of the 15 drug review topics studied. For three of the topics, the reduction was 50% or greater.
Conclusion: Automated document citation classification could be a useful tool in maintaining systematic reviews of the efficacy of drug therapy. Further work is needed to refine the classification system and determine the best manner to integrate the system into the production of systematic reviews.
The practice of evidence-based medicine (EBM) involves applying the best and most up-to-date evidence, in the form of published literature, to patient care decision making.1,2 While the original vision of EBM appeared to require physicians directly searching the primary literature for evidence applicable to their patients, the modern conception of EBM relies heavily on distillations of the primarily literature in the form of systematic reviews (also called evidence reports),3,4 such as those produced by the Cochrane Collaboration and the Evidence-based Practice Centers (EPCs) of the Agency for Healthcare Research and Quality (AHRQ).5 AHRQ reports are available to the public (http://www.ahrq.gov/clinic/epcindex.htm/) and abstracts of Cochrane reviews are also available (http://www.cochrane.org/reviews/).
The Department of Medical Informatics and Clinical Epidemiology at the Oregon Health and Science University is home to one of the AHRQ EPCs (http://www.ohsu.edu/epc/). The EPC has focused on providing comprehensive literature reviews comparing classes of drugs used for treating specific conditions. To date, the Oregon EPC, Southern California EPC, and Research Triangle Institute/University of North Carolina (RTI/UNC) EPC have completed and published 15 evidence reports, evaluating the efficacy of medications in drug classes such as oral opioids, skeletal muscle relaxants, and estrogen replacement.6,7,8
Making these evidence reports comprehensive and keeping them up to date is a labor-intensive process.9,10 Like any systematic review, those of drug classes identify thousands of articles that must be located, triaged, reviewed, and summarized. Potentially relevant articles are located using an iteratively refined query-based search of biomedical electronic databases, such as MEDLINE and EMBASE. These queries are developed by starting with the optimized clinical queries proposed and studied by Haynes et al.11,12,13 and refined based on the experience and knowledge of the EPC staff. For the 15 systematic drug reviews mentioned above, the staff created queries for randomized controlled trials by combining terms for health conditions and interventions with the Haynes et al. research methodology filters for therapy.
Articles are then triaged in a two-step process. First the abstract is reviewed, and, if the abstract meets the inclusion criteria, the entire article is read. If the full text article proves to meet the inclusion criteria, the evidence presented in the article is summarized and included in the EPC report. Advances in clinical evaluation and pharmacology require that EPC drug reviews be updated on a periodic basis. This inevitably leads to the workload of the center increasing over time as reviewers must both produce new reviews as well as monitor and update the old ones.
The process of creating these drug reviews is very methodical. Reviewers keep detailed records of their search methods, the articles for which they have reviewed the abstracts and read full text, and, finally, which articles include sufficient high-quality evidence to warrant inclusion in the systematic review. This process motivated our interest in using these data to train an automated classification system that would have the ability to predict which new articles were most likely to include evidence warranting inclusion in a review update. An automated classification system would function to triage (or filter) new articles that match the search criteria of the original study. This would be useful to the reviewers in several ways. First, by monitoring the number of new articles on a given topic containing high-quality evidence (as determined by the classification system), the reviewers would have a simple and clear indication when substantial new information exists on a topic and the report needs to be revised. Second, the classification system could decrease the number of articles that require manual review and therefore reduce one of the most time-consuming steps in preparing or updating a systematic review. Third, classifying the most likely documents to contain high-quality evidence can help reviewers prioritize which articles are read first and which are read only if there is sufficient time. This paper presents the investigators' application of a machine learning–based classification system to reduce the labor required and improve the efficiency of keeping drug reviews up to date.
We are unaware of any prior work that applies automated classification of literature for topic-specific evidence-based drug or therapy reviews similar to the work we present here. The closest research is the work of Aphinyanaphongs et al.,14 who have published work investigating the use of machine-learning algorithms to improve information retrieval of high-quality articles useful for evidence-based medicine in the nontopic-specific high-level categories of etiology, prognosis, diagnosis, and treatment. Their research focuses on improving performance over the clinical query filters first proposed by Haynes et al.,11,12,13 using all the articles published in ten journals from 1986 through 1991, and whether those articles were included in the ACP Journal Club as a gold standard.
There has been much more work in the related area of automated document classification to assist curator annotation of biomedical databases. Like the work of Aphinyanaphongs et al.,14 the goals of these tasks are to place articles into one of a few high-level, nontopic-specific categories. The results of these high-level automated tasks vary because the recall requirements are different and are difficult to compare to the topic-specific classification presented here. Dobrokhotov et al.15 used a combination of NLP and statistical classification techniques to achieve a recall of 0.4477 at a precision of 0.6716 for identifying articles containing information about human genetic diseases and polymorphisms, using a test set where 15% of the original articles were relevant. The TREC 2004 Genomics track included a task to identify articles containing information on mouse gene function for Gene Ontology (GO) annotation. With a utility function giving recall 20 times the importance of precision, the best results achieved were a normalized utility of 0.6512, and precision of 0.1579 at a recall of 0.8881, resulting in an F-measure of 0.2681.16
Methods
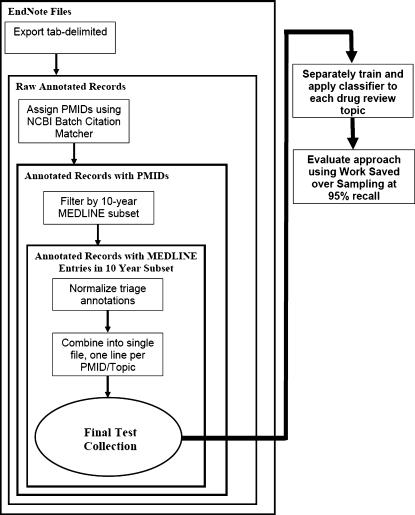
In order to build and test an automated classification system for these evidence reports, we proceeded in three phases. In the first phase, we built test collections for each of 15 review topics. In the second phase, we trained a machine learning–based classifier on the test collections. In the final phase, we evaluated the approach on each of the review topics. ▶ shows a diagram of the overall process.
Figure 1.
High-level diagram of the overall process.
Building the Text Collections
The initial data set consisted of 15 EndNote (http://www.endnote.com/) reference files with annotations (in the user-defined fields) made available to the investigators by the EPC. Each field in the reference file contained information on the article's title, authors, journal, and year of publication, as well as several user-defined fields. The user-defined fields included information entered by the EPC reviewers about whether each article passed the triage evaluation done by an expert reviewer at the abstract and article level. In addition, sometimes a free-text reason was coded as to why a paper was excluded. Therefore, these fields were “semicoded” in that the EPC used a consistent set of strings to encode triage decisions and may also have appended additional free text describing a reason for the assigning of a specific code. Occasional data entry typographical errors were also present.
In order to transform these data into a consistent set of information that could be used by a classification system, we processed the data in several steps. In the first step, we exported the data from EndNote using a tab-delimited, one record per line text format. In the second step, we extracted author, journal, title, year, and user-defined information and used the NCBI Batch Citation Matcher (http://www.ncbi.nlm.nih.gov/entrez/citmatch.cgi) to look up the corresponding PubMed identifiers (PMID). Then we produced a joined file that included the author, journal, title, year, user-defined information, and PMID for each article.
In order to make our work reproducible and comparable with work by others who apply different approaches to this task, we needed to have a fixed, static collection of MEDLINE records. We decided to use the TREC 2004 Genomics Track document corpus, a general MEDLINE subset that has been widely used in other experimental systems. The corpus consists of all MEDLINE records (not just genomics records, as might be inferred from the name) for the years 1994 through the end of 2003. Using this document corpus allows straightforward replicability of our results and allows other research groups to directly compare their results with our own.16
We needed to limit the test collection to articles for which the TREC 2004 Genomics Track document corpus had available MEDLINE records. Not all the articles included in the reference files were indexed in MEDLINE. For example, some were in non-English journals not indexed in MEDLINE. We filtered the test collection by the PMIDs present in the TREC corpus to ensure that we had a MEDLINE record for each article in the test collection.
In the next step, we normalized the EndNote user-defined fields that contained the EPC-coded information for the abstract and article triage decisions for each article. We used a set of simple regular expressions, customized for each drug review and determined by inspection of the data, to normalize the reason fields into a set of ten consistently coded values. The codes and meanings are shown in ▶. The investigators had extended discussions with the EPC reviewers to determine the correct regular expressions to map the user-defined fields into the coded values shown in ▶ and to detect and resolve any typographical errors. No articles were excluded from the test collection due to an inability to process the user-coded fields. In this study, we sought to distinguish articles included at the abstract and full text levels from all excluded ones; however, we chose to preserve the information provided in the exclusion reason codes for use in future work.
Table 1.
Standardized Coded Values for Abstract and Article Triage Decisions
| Abstract or Article Code | Meaning |
|---|---|
| I | Included at abstract or article level |
| E | Nonspecifically excluded |
| 1 | Excluded due to foreign language |
| 2 | Excluded due to wrong outcome |
| 3 | Excluded due to wrong drug |
| 4 | Excluded due to wrong population |
| 5 | Excluded due to wrong publication type |
| 6 | Excluded due to wrong study design |
| 7 | Excluded due to wrong study duration |
| 8 | Excluded due to background article |
| 9 | Excluded due to only abstract being available |
As a final step, we combined the information from all the studies into a single text file. Each line corresponds to an individual article triaged by the EPC staff for a specific drug review, and each field corresponds to the drug review name, a reference file identifier, the PMID, and the abstract and full text article triage fields encoded as shown in ▶. Example data records are shown in ▶. Note that even though the data set will only include citations and not full text articles, the expert EPC reviewers had access to the full text articles and made their triage decisions in two stages, the first based on the abstract and the second based on the full text article. It is this final triage decision that we would like to predict using a machine-learning algorithm using only the information contained within the article citation.
Table 2.
Example Drug Review Journal Citation Records
| Drug Class Review | EndNote ID | PMID | Abstract Triage | Article Triage | Year |
|---|---|---|---|---|---|
| ADHD | 1010 | 11483145 | 5 | E | 2001 |
| Antihistamines | 615 | 1342896 | I | E | 1992 |
| BetaBlockers | 211 | 10826501 | I | I | 2000 |
| CalciumChannelBlockers | 3139 | 11718496 | 2 | E | 2001 |
PMID = PubMed identifier; ADHD = attention-deficit/hyperactivity disorder.
Note that in the process just described, there are two steps where we exclude articles originally contained in the EndNote files. We lose articles when the Batch Citation Matcher cannot find a matching article (i.e., the reference is not indexed in MEDLINE) and when the article is not present in the ten-year MEDLINE subset. After processing, we were able to convert between 30% and 50% of the references into the test collection.
Descriptive statistics about the number of original articles reviewed for each study, the number of articles included in the text collection, and the percentage of positive articles in each text collection are shown in ▶. The first column lists the drug review name, the second the number of citations reviewed by the EPC and therefore the number of entries in the corresponding EndNote file, the third column gives the number of citations included in the test collection for each review, the fourth shows the percentage of the original citations in the EndNote file retained in the text collection, and the final column gives the percentage of articles retained in the test collection that were included in the final EPC systematic review.
Table 3.
Descriptive Statistics on the Number of Citations for Each Study
| Drug Review Name | No. of Citations and Articles Reviewed by EPC | No. of Citations Included in Test Collection | % Retained in Test Collection | % Retained Included in EPC Review |
|---|---|---|---|---|
| ACEInhibitors | 6,866 | 2,544 | 37.05 | 1.60 |
| ADHD | 2,191 | 851 | 38.84 | 2.40 |
| Antihistamines | 1,037 | 310 | 29.89 | 5.20 |
| AtypicalAntipsychotics | 2,947 | 1,120 | 38.00 | 13.0 |
| BetaBlockers | 5,437 | 2,072 | 38.11 | 2.00 |
| CalciumChannelBlockers | 3,717 | 1,218 | 32.77 | 8.20 |
| Estrogens | 718 | 368 | 51.25 | 21.7 |
| NSAIDs | 766 | 393 | 51.31 | 10.4 |
| Opioids | 4,996 | 1,915 | 38.33 | 0.80 |
| OralHypoglycemics | 1,460 | 503 | 34.45 | 27.0 |
| ProtonPumpInhibitors | 2,698 | 1,333 | 49.41 | 3.80 |
| SkeletalMuscleRelaxants | 5,460 | 1,643 | 30.09 | 0.50 |
| Statins | 7,922 | 3,465 | 43.74 | 2.50 |
| Triptans | 1,343 | 671 | 49.96 | 3.60 |
| UrinaryIncontinence | 809 | 327 | 40.42 | 12.2 |
EPC = Evidence-based Practice Center; ACE = angiotensin-converting enzyme; ADHD = attention-deficit/hyperactivity disorder; NSAIDs = nonsteroidal anti-inflammatory drug.
The percentage of articles in the test collection that were selected for inclusion in each of the drug reviews varied widely, from a low of 0.5% (for SkeletalMuscleRelaxants) to a high of 27% (for OralHypoglycemics). A low percentage of true positives is often typical for biomedical document classification tasks and requires a special approach to classification and evaluation to provide useful results.16,17,18 Unlike many typical automated document classification tasks where accuracy (percentage of correct predictions) or F-measure (defined later) may provide a useful metric for evaluating systems, in biomedical scenarios such as we have here, the goal is often to improve precision, but only if a very high level of recall can be sustained. For systematic reviews, the reviewers try to include every article relevant to the topic that provides high-quality evidence. While it is reasonable to assume that a small percentage of articles are missed, any automated classification system must maintain a high recall, or the work saved by improving the precision of the set of documents that require manual review is made irrelevant by the large number of relevant articles missed.
Classifier System
We used the MEDLINE records for each of the articles to generate the feature set as input to the machine learning system. Features included all the words from the title and abstract in a “bag-of-words” approach as well as the article's Medical Subject Headings (MeSH) and MEDLINE publication type. For MeSH-based features, we included the main headings, the headings with subheadings, the primary heading, and the subheadings by themselves. The MeSH and MEDLINE publication type features were pre-pended with “MESH_” and “PUBTYPE_” respectively to ensure that these features were treated distinct from words in the title or abstract. We counted the frequency of each feature appearing in positive and negative documents and applied the χ2 test with an α of 0.05 to use the statistically significant features as relevant features for input to the classifier system, using the 2 × 2 table shown in ▶. Our previous work on document classification has shown this value of α to produce good results.17 Each feature was treated as a binary quantity, either present in each document or absent, resulting in a feature vector for each document consisting of entirely ones and zeros.
Table 4.
Arrangement of 2 × 2 Table for Computing Feature Significance
| Feature is the one under test? |
||
|---|---|---|
| Training document is triage positive? | Yes | No |
| Yes | Number of times feature seen in positive documents | Number of times other features seen in positive documents |
| No | Number of times feature seen in negative documents | Number of times other features seen in negative documents |
We did not weight the features by term frequencies. Preliminary testing found that weighting features by intra-document frequency and/or inverse document frequency (TF, IDF, TFIDF) decreased the performance of the classifier system. We also tried applying the Porter stemming algorithm19 as well as a stop list of the 300 most common English words20 to the word features. ▶ shows the number and type of significant features for each drug class review. The first column gives the drug review name, and the second column gives the number of statistically significant features found in the training data for that review. The last three columns break the total number of features down into three distinct categories: number of word-based features, number of MeSH tag-based features, and number of MEDLINE publication type tag-based features.
Table 5.
Number of Significant Features for Each Study
| Drug Review Name | Total No. of Significant Features | Word Features | MeSH Features | PubType Features |
|---|---|---|---|---|
| ACEInhibitors | 210 | 165 | 40 | 5 |
| ADHD | 80 | 56 | 24 | 0 |
| Antihistamines | 29 | 19 | 9 | 1 |
| AtypicalAntipsychotics | 381 | 302 | 71 | 8 |
| BetaBlockers | 194 | 147 | 42 | 5 |
| CalciumChannelBlockers | 329 | 247 | 77 | 5 |
| Estrogens | 233 | 184 | 44 | 5 |
| NSAIDs | 242 | 186 | 51 | 5 |
| Opioids | 55 | 41 | 14 | 0 |
| OralHypoglycemics | 234 | 175 | 55 | 4 |
| ProtonPumpInhibitors | 206 | 146 | 54 | 6 |
| SkeletalMuscleRelaxants | 11 | 7 | 2 | 2 |
| Statins | 467 | 374 | 87 | 6 |
| Triptans | 121 | 96 | 22 | 3 |
| UrinaryIncontinence | 215 | 165 | 45 | 5 |
MeSH = medical subject headings; ACE = angiotensin-converting enzyme; ADHD = attention-deficit/hyperactivity disorder; NSAIDs = nonsteroidal anti-inflammatory drugs.
Successfully training a classifier to identify low-probability classes such as the articles included in these drug reviews can be challenging when using machine-learning algorithms without sufficient configuration options. We have previously had success in classifying document collections with highly asymmetric positive and negative sample frequency (e.g., a small number of positives in a collection with a large number of negatives) using a variation of the voting perceptron classifier first proposed by Freund and Schapire.17,21 The voting perceptron algorithm has very good performance, is quite fast, and is easy to implement. While the algorithm as published does not include a means of compensating for asymmetric false positive and negative penalties we have created a modification to the algorithm that does provide this flexibility. This allows the system to work well in situations where the cost of a recall error (false negative) differs significantly from the cost of a precision error (false positive).
A perceptron is essentially an equation for a linear combination of the values of the feature set, represented as a vector. There is one term in the perceptron vector for each feature in the feature set plus an optional bias term. A document is classified by taking the dot product of a document's feature vector with the perceptron vector and adding in the bias term. If the result is greater than zero, then the document is classified as positive; if it less than or equal to zero, then the document is classified as negative.
Rosenblatt's22 original algorithm trained the perceptron by applying it to each sample in the training data. If the sample was incorrectly classified, the perceptron was modified by adding or subtracting the sample back into the perceptron, adding when the sample was a true positive and subtracting when the sample was a true negative. Over a large number of training samples, the perceptron converges on the solution that best approximates the separation between positive and negative documents in the training set.
Freund and Schapire improved the performance of the perceptron by modifying the algorithm to produce a series of perceptrons, each of which makes a prediction on the class of each document and gets a number of “votes” depending on how many documents that perceptron classified correctly in the training set. The class with the most votes is the predicted class assigned to the document. They also extended the perceptron algorithm to enable the use of kernels using more complex mathematical operations rather than the straightforward linear kernel that simply adds and subtracts samples as described above.
In our modified voting perceptron, we use a linear kernel and differentially adjust the learning rate of the perceptron for false negatives and false positives. While in the original implementation of Freund and Schapire, incorrectly classified samples are directly added or subtracted back into the perceptron, we first multiply the sample by a factor known as the learning rate. Furthermore, we use different learning rates for false positives and false negatives.
We fixed the false-positive learning rate at 1.0 and adjusted the false-negative learning rate (FNLR) to optimize performance. The frequency of positives differs widely across the 15 studies. In general, we have found that the FNLR should be proportional to the ratio of negative to positive samples in the data set. In order to process and analyze all the studies in a consistent manner, we created a normalized FNLR parameter, w. The parameter w is input to the learning algorithm and the actual FNLR used in training the classifier for each study is given by:
 |
(1) |
In our experiments, we vary w in a consistent manner across all reviews and compute FNLR for each topic. In general, the optimal value of w will be different for each classification task. As we will show, w can be determined by applying cross-validation to the training data and interpolating to solve for the value of w that results in the best value for the chosen scoring measure for each task.
We also tried a rule-based classifier, Slipper,23 with the same feature sets and document weights but found that it consistently underperformed the voting perceptron classifier. We did not explore the use of the currently very popular support vector machine (SVM)-based classifiers such as SVM-Light.24 The voting perceptron classifier divides the parameter space in a similar way to SVM and has been shown to perform similarly to SVM-based classifiers.21 Also, our prior experience with SVM-Light has found that the program settings are insufficient to handle classification tasks with very low rates of positive samples when high recall is required.17
Evaluating the Classifier
We wanted to evaluate how our classifier approach performs on identifying new articles for inclusion in future updates of the drug evidence reviews. In order to do that, we needed to decide on an appropriate metric as well as optimize the normalized FNLR weight w for each drug review. To make the most efficient use of the data sets and to get the best estimate of system performance on future data, we chose to use 5 × 2 cross-validation.
In 5 × 2 cross-validation, the data set is randomly split in half, and then one half is used to train the classifier, and the classifier is scored using the other half as a test set. Then the roles of the two half data sets are exchanged, with the second half used for training and the first half used for testing, with the results accumulated from both halves of the split. What makes 5 × 2 different from the ten-way cross-validation more commonly used is that the half-and-half split and score process is repeated five times. This approach uses each data sample five times for training and five times for testing among random splits and averages the results together for all runs. The 5 × 2 cross-validation approach is thought to give better estimates of actual performance than the ten-way cross-validation method, which frequently overestimates performance.25
It was challenging to determine an appropriate metric with which to evaluate our approach. The most commonly used measures, precision and recall, separately measure two things of importance to a classifier system, namely, how accurately the classifier performs when predicting items in the class of interest (precision) and how completely the classifier identifies the items of interest (recall). For classification tasks, precision (P) and recall (R) are defined as:
 |
(2) |
 |
(3) |
Since precision and recall are two separate numbers, comparing one system or the result of one set of parameters to another is difficult. Precision and recall are commonly combined into a weighted harmonic mean called F-measure, usually weighting both components equally and defined as 2*P*R/(P + R).
However, neither precision, recall, nor F-measure captures what we were interested in measuring for this task. For a document classification system to provide value for systematic reviews, the system has to save the reviewers the work of reading every paper. At the same time, the number of missed papers containing quality evidence has to be very low. For this study, we assumed that a recall of 0.95 or greater was required for the system to identify an adequate fraction of the positive papers. Precision should be as high as possible, as long as recall is at least 0.95.
Furthermore, the most important feature of the system to measure is how much future work the reviewers could save for each drug review. Rather than simply reporting the highest precision obtained at a recall at or above 0.95, we chose to use as our metric a measure of the work saved at a recall fixed at 0.95. We define the work saved as the percentage of papers that meet the original search criteria that the reviewers do not have to read (because they have been screened out by the classifier). A recall of 0.95 can be obtained with a 0.95 random sampling of the data, and this process would save the reviewers 5% of the work of reading the papers. Clearly, for the classifier system to provide an advantage, the work saved must be greater than the work saved by simple random sampling. Therefore, we measure the work saved over and above the work saved by simple sampling for a given level of recall. We define the work saved over sampling (WSS) as:
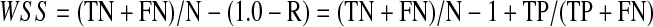 |
(4) |
where TP is the number of true positives identified by the classifier, TN is the number of true negatives identified by the classifier, FN is the number of false negatives identified by the classifier, R is recall and N is the total number of samples in the test set. For the present work, we have fixed recall at 0.95 and therefore work saved over sampling at 95% recall (WSS@95%) is
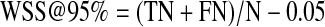 |
(5) |
This measure does make some simplifying assumptions. No specific consideration is given to variations in document length or the work expended during review of an article citation versus the review of a full text article. We assume the actual work saved by documents predicted to be negative by the classification system is on average the same as the work required by the average document retrieved during the literature search.
We applied the 5 × 2 cross-validation process to the data sets for each of the drug reviews multiple times, varying w between 0.25 and 8.00. This range of w used covered the useful range of FNLR for each of the review topics. At lower values of w (< 0.25), the recall was well below the 0.95 that we were targeting. For higher values of w (>8.00), recall was at or near 100%. Finally, we estimated WSS@95% by linearly interpolating the WSS for the values of w with recall immediately above and below 0.95. This allowed us to both determine the best value of w for training the classifier for a specific systematic review and to estimate the performance of the classifier on future articles that meet the search criteria of the original drug review.
Results
The results of our cross-validation experiments are presented in ▶. Precision, recall, F-measure, FNLR, and WSS for each drug review at eight different values of the parameter w, from 0.25 to 8.00 are shown. For each drug review, the value of w that leads to a recall closest to 0.95 is marked with an asterisk. This value varies between a low of 0.75 and a high of 4.00. For some topics, estimating WSS@95% recall required evaluating WSS at 8.0 to bracket the 95% recall value. Evaluating performance at values of w below 0.75 appears not to be necessary to determine the best value of w to optimize WSS@95%.
Table 6.
Results of 5 × 2 Cross-validation on Data Set for Each Drug Review
| Drug Review | WSS@95% | w | FNLR | P | R | F | WSS |
|---|---|---|---|---|---|---|---|
| ACEInhibitors | 56.61% | 0.25 | 15.2620 | 0.1211 | 0.6293 | 0.2031 | 54.55% |
| 0.5 | 30.5240 | 0.0949 | 0.8098 | 0.1699 | 67.23% | ||
| 0.75 | 45.7870 | 0.0761 | 0.8732 | 0.1400 | 68.82% | ||
| 1 | 61.0490 | 0.0642 | 0.8780 | 0.1197 | 65.77% | ||
| *2 | 122.0980 | 0.0387 | 0.9561 | 0.0745 | 55.84% | ||
| 4 | 244.1950 | 0.0238 | 0.9854 | 0.0465 | 31.81% | ||
| 8 | 488.3900 | 0.0183 | 1.0000 | 0.0359 | 11.75% | ||
| ADHD | 67.95% | 0.25 | 10.3870 | 0.1379 | 0.6300 | 0.2262 | 52.26% |
| 0.5 | 20.7750 | 0.0998 | 0.8500 | 0.1786 | 64.98% | ||
| *0.75 | 31.1630 | 0.0945 | 0.9200 | 0.1713 | 69.11% | ||
| 1 | 41.5500 | 0.0738 | 0.9800 | 0.1373 | 66.79% | ||
| 2 | 83.1000 | 0.0436 | 1.0000 | 0.0835 | 46.09% | ||
| 4 | 166.2000 | 0.0292 | 1.0000 | 0.0568 | 19.53% | ||
| 8 | 332.4000 | 0.0249 | 1.0000 | 0.0486 | 5.57% | ||
| Antihistamines | 0.00% | 0.25 | 4.5940 | 0.0894 | 0.1375 | 0.1084 | 5.81% |
| 0.5 | 9.1880 | 0.0798 | 0.2125 | 0.1160 | 7.51% | ||
| 0.75 | 13.7810 | 0.0737 | 0.3125 | 0.1193 | 9.38% | ||
| 1 | 18.3750 | 0.0714 | 0.3625 | 0.1193 | 10.06% | ||
| 2 | 36.7500 | 0.0600 | 0.5125 | 0.1075 | 7.19% | ||
| 4 | 73.5000 | 0.0502 | 0.8500 | 0.0948 | −2.35% | ||
| 8 | 147.0000 | 0.0488 | 0.8500 | 0.0923 | −4.87% | ||
| AtypicalAntipsychotics | 14.11% | 0.25 | 1.6680 | 0.3890 | 0.2712 | 0.3196 | 18.03% |
| 0.5 | 3.3360 | 0.3239 | 0.5014 | 0.3935 | 29.96% | ||
| 0.75 | 5.0030 | 0.2701 | 0.6027 | 0.3730 | 31.18% | ||
| 1 | 6.6710 | 0.2527 | 0.7014 | 0.3716 | 33.96% | ||
| 2 | 13.3420 | 0.1968 | 0.8479 | 0.3194 | 28.62% | ||
| *4 | 26.6850 | 0.1534 | 0.9493 | 0.2642 | 14.27% | ||
| 8 | 53.3700 | 0.1362 | 0.9932 | 0.2396 | 4.28% | ||
| BetaBlockers | 28.44% | 0.25 | 12.0830 | 0.1253 | 0.4905 | 0.1996 | 41.11% |
| 0.5 | 24.1670 | 0.0799 | 0.6429 | 0.1422 | 47.98% | ||
| 0.75 | 36.2500 | 0.0621 | 0.7381 | 0.1146 | 49.72% | ||
| 1 | 48.3330 | 0.0503 | 0.8190 | 0.0948 | 48.91% | ||
| *2 | 96.6670 | 0.0334 | 0.9286 | 0.0644 | 36.47% | ||
| 4 | 193.3330 | 0.0257 | 0.9714 | 0.0500 | 20.42% | ||
| 8 | 386.6670 | 0.0226 | 0.9952 | 0.0443 | 10.39% | ||
| CalciumChannelBlockers | 12.21% | 0.25 | 2.7950 | 0.2756 | 0.3440 | 0.3060 | 24.15% |
| 0.5 | 5.5900 | 0.2259 | 0.5620 | 0.3222 | 35.77% | ||
| 0.75 | 8.3850 | 0.1968 | 0.6460 | 0.3017 | 37.65% | ||
| 1 | 11.1800 | 0.1694 | 0.6760 | 0.2709 | 34.84% | ||
| 2 | 22.3600 | 0.1248 | 0.8840 | 0.2188 | 30.26% | ||
| *4 | 44.7200 | 0.0952 | 0.9460 | 0.1730 | 13.02% | ||
| 8 | 89.4400 | 0.0845 | 0.9960 | 0.1558 | 2.83% | ||
| Estrogens | 18.34% | 0.25 | 0.9000 | 0.4953 | 0.2625 | 0.3431 | 14.73% |
| 0.5 | 1.8000 | 0.4648 | 0.5275 | 0.4941 | 28.08% | ||
| 0.75 | 2.7000 | 0.4288 | 0.6775 | 0.5252 | 33.40% | ||
| 1 | 3.6000 | 0.3968 | 0.7500 | 0.5190 | 33.91% | ||
| 2 | 7.2000 | 0.3213 | 0.8900 | 0.4721 | 28.78% | ||
| *4 | 14.4000 | 0.2552 | 0.9725 | 0.4044 | 14.42% | ||
| 8 | 28.8000 | 0.2263 | 0.9975 | 0.3689 | 3.93% | ||
| NSAIDs | 49.67% | 0.25 | 2.1460 | 0.3720 | 0.3756 | 0.3738 | 27.03% |
| 0.5 | 4.2930 | 0.3453 | 0.7024 | 0.4630 | 49.02% | ||
| 0.75 | 6.4390 | 0.2831 | 0.8146 | 0.4201 | 51.44% | ||
| *1 | 8.5850 | 0.2631 | 0.9317 | 0.4103 | 56.22% | ||
| 2 | 17.1710 | 0.1620 | 0.9902 | 0.2785 | 35.26% | ||
| 4 | 34.3410 | 0.1161 | 1.0000 | 0.2081 | 10.18% | ||
| 8 | 68.6830 | 0.1080 | 1.0000 | 0.1950 | 3.41% | ||
| Opioids | 13.32% | 0.25 | 31.6670 | 0.0347 | 0.2533 | 0.0610 | 19.61% |
| 0.5 | 63.3330 | 0.0187 | 0.4667 | 0.0359 | 27.09% | ||
| 0.75 | 95.0000 | 0.0128 | 0.4533 | 0.0249 | 17.57% | ||
| 1 | 126.6670 | 0.0109 | 0.6933 | 0.0214 | 19.44% | ||
| *2 | 253.3330 | 0.0092 | 0.9467 | 0.0182 | 14.19% | ||
| 4 | 506.6670 | 0.0082 | 0.9867 | 0.0162 | 3.85% | ||
| 8 | 1013.3338 | 0.0078 | 1.0000 | 0.0156 | 0.05% | ||
| OralHypoglycemics | 8.96% | 0.25 | 0.6750 | 0.5437 | 0.2471 | 0.3397 | 12.42% |
| 0.5 | 1.3490 | 0.4840 | 0.4882 | 0.4861 | 21.55% | ||
| 0.75 | 2.0240 | 0.4284 | 0.5941 | 0.4978 | 21.92% | ||
| 1 | 2.6990 | 0.3962 | 0.6735 | 0.4989 | 21.39% | ||
| 2 | 5.3970 | 0.3375 | 0.8382 | 0.4812 | 16.67% | ||
| *4 | 10.7940 | 0.3004 | 0.9471 | 0.4561 | 9.46% | ||
| 8 | 21.5880 | 0.2797 | 0.9838 | 0.4355 | 3.27% | ||
| ProtonPumpInhibitors | 27.68% | 0.25 | 6.2840 | 0.2335 | 0.4706 | 0.3121 | 39.35% |
| 0.5 | 12.5690 | 0.1522 | 0.6588 | 0.2472 | 49.32% | ||
| 0.75 | 18.8530 | 0.1162 | 0.7059 | 0.1996 | 47.35% | ||
| 1 | 25.1370 | 0.0879 | 0.7882 | 0.1582 | 44.52% | ||
| *2 | 50.2750 | 0.0602 | 0.9373 | 0.1132 | 34.18% | ||
| 4 | 100.5490 | 0.0471 | 0.9686 | 0.0898 | 18.18% | ||
| 8 | 201.0980 | 0.0412 | 0.9961 | 0.0792 | 7.17% | ||
| SkeletalMuscleRelaxants | 0.00% | 0.25 | 45.3890 | 0.0042 | 0.0222 | 0.0070 | −0.70% |
| 0.5 | 90.7780 | 0.0038 | 0.0222 | 0.0064 | −1.02% | ||
| 0.75 | 136.1670 | 0.0044 | 0.3556 | 0.0088 | −8.34% | ||
| 1 | 181.5560 | 0.0050 | 0.4000 | 0.0099 | −3.90% | ||
| 2 | 363.1110 | 0.0051 | 0.8444 | 0.0102 | −5.56% | ||
| 4 | 726.2220 | 0.0055 | 1.0000 | 0.0109 | 0.00% | ||
| 8 | 1452.4440 | 0.0055 | 1.0000 | 0.0109 | 0.00% | ||
| Statins | 24.71% | 0.25 | 9.9410 | 0.1707 | 0.5106 | 0.2559 | 43.72% |
| 0.5 | 19.8820 | 0.1154 | 0.6612 | 0.1965 | 52.06% | ||
| 0.75 | 29.8240 | 0.0883 | 0.7600 | 0.1583 | 54.89% | ||
| 1 | 39.7650 | 0.0702 | 0.8071 | 0.1291 | 52.49% | ||
| 2 | 79.5290 | 0.0469 | 0.8894 | 0.0891 | 42.44% | ||
| *4 | 159.0590 | 0.0311 | 0.9647 | 0.0603 | 20.41% | ||
| 8 | 318.1180 | 0.0263 | 0.9906 | 0.0512 | 6.55% | ||
| Triptans | 3.37% | 0.25 | 6.7400 | 0.1389 | 0.2500 | 0.1786 | 18.56% |
| 0.5 | 13.4790 | 0.0883 | 0.5583 | 0.1524 | 33.21% | ||
| 0.75 | 20.2190 | 0.0711 | 0.6917 | 0.1290 | 34.38% | ||
| 1 | 29.9580 | 0.0639 | 0.7583 | 0.1178 | 33.36% | ||
| 2 | 53.9170 | 0.0449 | 0.8667 | 0.0854 | 17.64% | ||
| *4 | 107.8330 | 0.0365 | 0.9583 | 0.0703 | 1.94% | ||
| 8 | 215.6670 | 0.0359 | 1.0000 | 0.0693 | 0.39% | ||
| UrinaryIncontinence | 26.14% | 0.25 | 1.7940 | 0.4509 | 0.5050 | 0.4764 | 36.80% |
| 0.5 | 3.5870 | 0.3434 | 0.7400 | 0.4691 | 47.64% | ||
| 0.75 | 5.3810 | 0.2824 | 0.7400 | 0.4088 | 41.95% | ||
| 1 | 7.1750 | 0.2523 | 0.8400 | 0.3880 | 43.27% | ||
| 2 | 14.3500 | 0.1880 | 0.9100 | 0.3116 | 31.80% | ||
| *4 | 28.7000 | 0.1559 | 0.9850 | 0.2691 | 21.19% | ||
| 8 | 57.4000 | 0.1325 | 1.0000 | 0.2341 | 7.71% |
WSS = work saved over sampling; w = normalized FNLR parameter; FNLR = false-negative learning rate; P = precision; R = recall; F = F1-measure; ACE = angiotensin-converting enzyme; ADHD = attention-deficit/hyperactivity disorder; NSAIDs = nonsteroidal anti-inflammatory drugs; * = recall closest to 0.95.
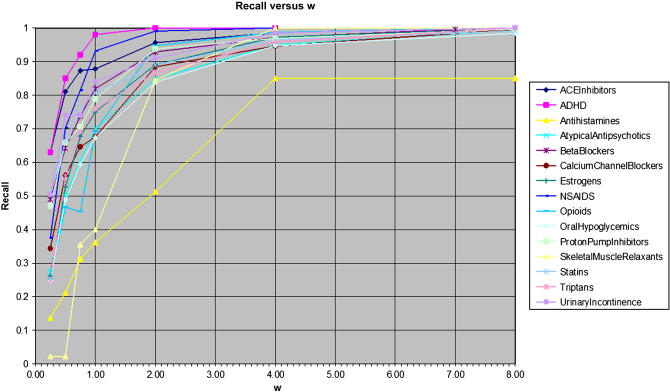
▶ presents the relationship between w and recall for each of the 15 drug reviews. While recall monotonically increases very smoothly with w for all 15 reviews, the value of w for which recall reaches 0.95 varies widely. Furthermore, the curves flatten out substantially as w was raised above 2.0, making 4.0 a practical upper limit for w.
Figure 2.
Recall versus w for each of the 15 drug review topics.
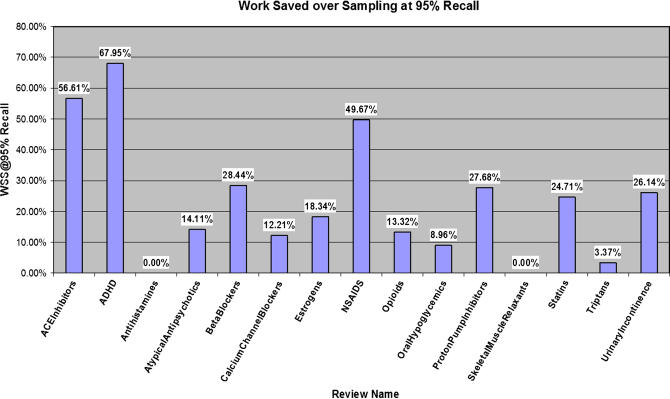
▶ presents estimates of the potential work saved for each of the 15 reviews. For 13 of the 15 topics, the WSS@95% was positive, meaning that the automated classification process is predicted to save the reviewers work in the future. For three of these topics, ACEInhibitors, ADHD, and NSAIDs, the predicted work savings was very large, about ≥50%. For these tasks the classification system is clearly successful at producing a significant work savings.
Figure 3.
Work saved over sampling at 95% recall for each of the 15 drug review topics.
The work savings for the 15 tasks varied between 0% for Antihistamines and SkeletalMuscleRelaxants and 67.95% for ADHD. To fully evaluate our method, it was necessary to set a WSS@95% threshold for success. We determined a success threshold in the following manner. EPC staff estimates that performing a thorough literature database search, review of abstracts, full text procurement of identified documents, and review of those documents takes on average about 332 hours of a total 3,648-hour timeline to produce a systematic review or to update a review. WSS@95% measures the work saved over sampling at 95% recall, that is, the work saved over and above if one just took a 95% sample of the documents. When computing the actual time saved, this additional 5% (1.0 − 0.95 = 0.05) has to be added back in to arrive at an estimate of the total time saved by using the system, as opposed to just the benefit over sampling.
At a WSS@95% of 10.0%, actually 15% of the 332 hours spent on the tasks listed above is saved. This comes to about 50 hours of decreased work or about one week less time spent on each report. The EPC staff thought that saving more than a week's worth of labor was significant and that the saved time could be put to good use in writing a better review, spending more time synthesizing the evidence, or conducting further analyses. Therefore, a WSS@95% of 10% is a reasonable threshold for distinguishing the review topics where our methods provide a significant savings from those topics that do not derive a significant savings.
For 11 of the 15 review topics, the predicted WSS@95% savings was above 10%, and for seven of the 15 topics, the predicted savings was greater than 20%. For only four of the topics was the predicted savings less than 10%. The overall average WSS@95% for the 15 topics was 23.43%.
For two topics, Antihistamines and SkeletalMuscleRelaxants, the classification process did not provide any savings. For the Antihistamine review, a recall of 95% was unable to be achieved, and increasing w from 4.00 to 8.00 did not increase the recall at all. For the SkeletalMuscleRelaxants review, a recall of 95% was achievable, but the proportion of positives in the set chosen by the classifier was lower than in the original sample. For the Triptans review, the savings was positive (3.37%) , but smaller than our required threshold.
The last column in ▶, the percentage retained included in EPC review, can be interpreted as the precision of the queries used in the literature search, as applied to the articles in our text collection. These numbers can be compared to the precision shown in ▶. ▶ presents these results also showing in the last column the factor by which the classification process multiplies precision at approximately 95% recall, as compared to the original expert-created clinical query. The precision and recall figures are chosen for the recall from ▶ that are closest to 0.95. The precision improvement factor varies widely, from a very modest 1.0139 for Triptans to a very large 3.9375 for ADHD. Since the classification process was unable to achieve any improvement for Antihistamines and SkeletalMuscleRelaxants, results for these two reviews are labeled “NA” (not applicable) in the table. The last row of ▶ provides an overall average for the 13 topics for which the classifier system was successful.
Table 7.
Comparison of Precision with Original Query
| Drug Review Name | Query Precision | Classifier Precision | Classifier Recall | Precision Improvement |
|---|---|---|---|---|
| ACEInhibitors | 0.0160 | 0.0387 | 0.9561 | 2.4188 |
| ADHD | 0.0240 | 0.0945 | 0.9200 | 3.9375 |
| Antihistamines | 0.0520 | NA | NA | NA |
| AtypicalAntipsychotics | 0.1300 | 0.1534 | 0.9493 | 1.1800 |
| BetaBlockers | 0.0200 | 0.0334 | 0.9286 | 1.6700 |
| CalciumChannelBlockers | 0.0820 | 0.0952 | 0.9460 | 1.1610 |
| Estrogens | 0.2170 | 0.2552 | 0.9725 | 1.1760 |
| NSAIDs | 0.1040 | 0.2631 | 0.9317 | 2.5298 |
| Opioids | 0.0080 | 0.0092 | 0.9467 | 1.1500 |
| OralHypoglycemics | 0.2700 | 0.3004 | 0.9471 | 1.1126 |
| ProtonPumpInhibitors | 0.0380 | 0.0602 | 0.9373 | 1.5842 |
| SkeletalMuscleRelaxants | 0.0050 | NA | NA | NA |
| Statins | 0.0250 | 0.0311 | 0.9647 | 1.2440 |
| Triptans | 0.0360 | 0.0365 | 0.9583 | 1.0139 |
| UrinaryIncontinence | 0.1220 | 0.1559 | 0.9850 | 1.2779 |
| Mean for not NA | 0.0840 | 0.1174 | 0.9495 | 1.6504 |
NA = Not applicable.
To ensure that our results were not confounded by the sizes or other descriptive statistics of the samples, we explored possible connections to classifier performance by computing linear correlation between WSS@95% and sample size, percentage of positive samples, and the number of relevant features. These calculations showed a very low level of nonstatistically significant correlation between WSS@95% and sample size (R2 = 0.025, p = 0.576), between WSS@95% and percentage of positive samples (R2 = 0.036, p = 0.498), and between WSS@95% and the number of features (R2 = 0.013, p = 0.689).
Results obtained applying the 300-word stop list and the Porter stemming in addition to the previously described classification algorithm are shown in ▶. Overall stemming and stopping increased the range of best to worst scores, but the average effect was small. The results for NSAIDs decreased markedly, but the score for CalciumChannelBlockers almost doubled. Since stemming and stopping did not provide any clear and consistent benefit and added to the overall computational complexity, we decided to base our results and analysis on the baseline classification system without stemming or stopping.
Table 8.
Results of 5 × 2 Cross-validation with Stemming and Stop List on Data Set for Each Review
| Drug Review | WSS@95% | w | FNLR | P | R | F | WSS |
|---|---|---|---|---|---|---|---|
| ACEInhibitors | 60.95% | 0.25 | 15.2620 | 0.128 | 0.737 | 0.218 | 64.38% |
| 0.5 | 30.5240 | 0.092 | 0.854 | 0.167 | 70.49% | ||
| 0.75 | 45.7870 | 0.075 | 0.898 | 0.139 | 70.50% | ||
| 1 | 61.0490 | 0.058 | 0.907 | 0.109 | 65.49% | ||
| 2 | 122.0980 | 0.042 | 0.966 | 0.080 | 59.26% | ||
| 4 | 244.1950 | 0.026 | 0.966 | 0.051 | 37.36% | ||
| 8 | 488.3900 | 0.020 | 0.985 | 0.039 | 19.27% | ||
| ADHD | 67.60% | 0.25 | 10.3870 | 0.130 | 0.680 | 0.218 | 55.69% |
| 0.5 | 20.7750 | 0.104 | 0.800 | 0.184 | 61.93% | ||
| 0.75 | 31.1630 | 0.091 | 0.890 | 0.165 | 65.97% | ||
| 1 | 41.5500 | 0.078 | 0.980 | 0.144 | 68.41% | ||
| 2 | 83.1000 | 0.049 | 0.990 | 0.093 | 51.36% | ||
| 4 | 166.2000 | 0.031 | 1.000 | 0.059 | 22.96% | ||
| 8 | 332.4000 | 0.025 | 1.000 | 0.049 | 6.93% | ||
| Antihistamines | 0.00% | 0.25 | 4.5940 | 0.097 | 0.175 | 0.125 | 8.21% |
| 0.5 | 9.1880 | 0.083 | 0.275 | 0.128 | 10.40% | ||
| 0.75 | 13.7810 | 0.061 | 0.275 | 0.100 | 4.27% | ||
| 1 | 18.3750 | 0.073 | 0.413 | 0.124 | 11.96% | ||
| 2 | 36.7500 | 0.054 | 0.488 | 0.098 | 2.30% | ||
| 4 | 73.5000 | 0.050 | 0.850 | 0.094 | −3.52% | ||
| 8 | 147.0000 | 0.049 | 0.850 | 0.092 | −5.00% | ||
| AtypicalAntipsychotics | 15.09% | 0.25 | 1.6680 | 0.370 | 0.297 | 0.330 | 19.26% |
| 0.5 | 3.3360 | 0.298 | 0.521 | 0.379 | 29.30% | ||
| 0.75 | 5.0030 | 0.263 | 0.663 | 0.377 | 33.46% | ||
| 1 | 6.6710 | 0.238 | 0.725 | 0.359 | 32.84% | ||
| 2 | 13.3420 | 0.186 | 0.881 | 0.307 | 26.33% | ||
| 4 | 26.6850 | 0.155 | 0.951 | 0.266 | 14.98% | ||
| 8 | 53.3700 | 0.138 | 0.989 | 0.242 | 5.33% | ||
| BetaBlockers | 34.14% | 0.25 | 12.0830 | 0.123 | 0.514 | 0.199 | 42.95% |
| 0.5 | 24.1670 | 0.076 | 0.695 | 0.137 | 50.90% | ||
| 0.75 | 36.2500 | 0.054 | 0.805 | 0.102 | 50.50% | ||
| 1 | 48.3330 | 0.055 | 0.824 | 0.103 | 52.01% | ||
| 2 | 96.6670 | 0.032 | 0.948 | 0.062 | 35.24% | ||
| 4 | 193.3330 | 0.026 | 0.976 | 0.051 | 22.03% | ||
| 8 | 386.6670 | 0.023 | 0.995 | 0.044 | 10.68% | ||
| CalciumChannelBlockers | 23.83% | 0.25 | 2.7950 | 0.236 | 0.312 | 0.269 | 20.35% |
| 0.5 | 5.5900 | 0.216 | 0.484 | 0.298 | 29.98% | ||
| 0.75 | 8.3850 | 0.187 | 0.664 | 0.292 | 37.25% | ||
| 1 | 11.1800 | 0.164 | 0.754 | 0.269 | 37.60% | ||
| 2 | 22.3600 | 0.125 | 0.930 | 0.221 | 32.08% | ||
| 4 | 44.7200 | 0.096 | 0.974 | 0.174 | 13.94% | ||
| 8 | 89.4400 | 0.085 | 1.000 | 0.157 | 3.40% | ||
| Estrogens | 14.03% | 0.25 | 0.9000 | 0.506 | 0.303 | 0.379 | 17.26% |
| 0.5 | 1.8000 | 0.468 | 0.560 | 0.510 | 29.97% | ||
| 0.75 | 2.7000 | 0.418 | 0.670 | 0.515 | 32.16% | ||
| 1 | 3.6000 | 0.398 | 0.780 | 0.527 | 35.39% | ||
| 2 | 7.2000 | 0.319 | 0.890 | 0.469 | 28.29% | ||
| 4 | 14.4000 | 0.251 | 0.955 | 0.398 | 12.84% | ||
| 8 | 28.8000 | 0.226 | 0.990 | 0.369 | 3.95% | ||
| NSAIDs | 29.29% | 0.25 | 2.1460 | 0.353 | 0.371 | 0.362 | 26.13% |
| 0.5 | 4.2930 | 0.301 | 0.629 | 0.407 | 41.09% | ||
| 0.75 | 6.4390 | 0.259 | 0.815 | 0.393 | 48.69% | ||
| 1 | 8.5850 | 0.203 | 0.912 | 0.333 | 44.45% | ||
| 2 | 17.1710 | 0.155 | 0.946 | 0.266 | 30.77% | ||
| 4 | 34.3410 | 0.115 | 1.000 | 0.206 | 9.06% | ||
| 8 | 68.6830 | 0.108 | 1.000 | 0.195 | 3.21% | ||
| Opioids | 16.23% | 0.25 | 31.6670 | 0.029 | 0.267 | 0.053 | 19.51% |
| 0.5 | 63.3330 | 0.018 | 0.360 | 0.035 | 20.66% | ||
| 0.75 | 95.0000 | 0.016 | 0.507 | 0.031 | 25.71% | ||
| 1 | 126.6670 | 0.011 | 0.760 | 0.022 | 22.54% | ||
| 2 | 253.3330 | 0.009 | 0.973 | 0.018 | 15.45% | ||
| 4 | 506.6670 | 0.008 | 1.000 | 0.016 | 4.76% | ||
| 8 | 1013.3338 | 0.008 | 1.000 | 0.016 | 0.01% | ||
| OralHypoglycemics | 6.96% | 0.25 | 0.6750 | 0.502 | 0.240 | 0.324 | 11.05% |
| 0.5 | 1.3490 | 0.446 | 0.457 | 0.451 | 17.98% | ||
| 0.75 | 2.0240 | 0.419 | 0.578 | 0.486 | 20.50% | ||
| 1 | 2.6990 | 0.394 | 0.646 | 0.489 | 20.26% | ||
| 2 | 5.3970 | 0.340 | 0.846 | 0.485 | 17.28% | ||
| 4 | 10.7940 | 0.298 | 0.934 | 0.452 | 8.65% | ||
| 8 | 21.5880 | 0.280 | 0.984 | 0.436 | 3.43% | ||
| ProtonPumpInhibitors | 28.47% | 0.25 | 6.2840 | 0.217 | 0.463 | 0.295 | 38.10% |
| 0.5 | 12.5690 | 0.135 | 0.643 | 0.223 | 46.04% | ||
| 0.75 | 18.8530 | 0.110 | 0.722 | 0.190 | 46.98% | ||
| 1 | 25.1370 | 0.085 | 0.792 | 0.153 | 43.36% | ||
| 2 | 50.2750 | 0.059 | 0.941 | 0.112 | 33.53% | ||
| 4 | 100.5490 | 0.047 | 0.969 | 0.089 | 17.78% | ||
| 8 | 201.0980 | 0.041 | 0.996 | 0.078 | 5.74% | ||
| SkeletalMuscleRelaxants | 0.00% | 0.25 | 45.3890 | 0.006 | 0.022 | 0.010 | 0.25% |
| 0.5 | 90.7780 | 0.006 | 0.044 | 0.010 | 0.31% | ||
| 0.75 | 136.1670 | 0.005 | 0.378 | 0.009 | −6.67% | ||
| 1 | 181.5560 | 0.005 | 0.422 | 0.010 | −2.71% | ||
| 2 | 363.1110 | 0.005 | 0.844 | 0.010 | −5.56% | ||
| 4 | 726.2220 | 0.005 | 1.000 | 0.011 | 0.00% | ||
| 8 | 1452.4440 | 0.005 | 1.000 | 0.011 | 0.00% | ||
| Statins | 27.41% | 0.25 | 9.9410 | 0.137 | 0.494 | 0.214 | 40.56% |
| 0.5 | 19.8820 | 0.088 | 0.654 | 0.156 | 47.28% | ||
| 0.75 | 29.8240 | 0.072 | 0.762 | 0.131 | 50.16% | ||
| 1 | 39.7650 | 0.062 | 0.805 | 0.115 | 48.63% | ||
| 2 | 79.5290 | 0.040 | 0.911 | 0.077 | 35.83% | ||
| 4 | 159.0590 | 0.031 | 0.984 | 0.060 | 20.25% | ||
| 8 | 318.1180 | 0.028 | 0.988 | 0.055 | 12.33% | ||
| Triptans | 2.38% | 0.25 | 6.7400 | 0.132 | 0.325 | 0.188 | 23.71% |
| 0.5 | 13.4790 | 0.077 | 0.500 | 0.134 | 26.81% | ||
| 0.75 | 20.2190 | 0.069 | 0.775 | 0.127 | 37.38% | ||
| 1 | 29.9580 | 0.060 | 0.742 | 0.111 | 29.90% | ||
| 2 | 53.9170 | 0.045 | 0.883 | 0.086 | 18.44% | ||
| 4 | 107.8330 | 0.037 | 0.933 | 0.071 | 2.87% | ||
| 8 | 215.6670 | 0.036 | 1.000 | 0.070 | 0.89% | ||
| UrinaryIncontinence | 25.74% | 0.25 | 1.7940 | 0.375 | 0.495 | 0.427 | 33.35% |
| 0.5 | 3.5870 | 0.305 | 0.715 | 0.428 | 42.81% | ||
| 0.75 | 5.3810 | 0.255 | 0.810 | 0.388 | 42.16% | ||
| 1 | 7.1750 | 0.224 | 0.870 | 0.357 | 39.54% | ||
| 2 | 14.3500 | 0.184 | 0.930 | 0.307 | 31.17% | ||
| 4 | 28.7000 | 0.152 | 0.975 | 0.263 | 18.97% | ||
| 8 | 57.4000 | 0.130 | 0.995 | 0.230 | 5.98% |
WSS = work saved over sampling; w = normalized FNLR parameter; FNLR = false-negative learning rate; P = precision; R = recall; F = F1- measure; ACE = angiotensin-converting enzyme; ADHD = attention-deficit/hyperactivity disorder; NSAIDs = nonsteroidal anti-inflammatory drugs.
Discussion
For more than 70% (11/15) of the drug reviews, the automated classification process showed a significant savings of reviewer effort at the 95% recall level. At this high level of recall, for 20% (3/15) of the reviews this savings was very large, about ≥50%. Clearly, a savings of ≥50% of the effort needed to review journal articles is substantial and attractive.
Our research demonstrates a means of training an automated document classification system to select the papers with the highest likelihood of containing high-quality evidence. We have shown that automated document classification has strong potential for aiding the labor-intensive literature review process for systematic treatment reviews and other similar studies. Of course, the work savings could be greater if a lower level of recall were found acceptable.
Furthermore, our research demonstrates how to determine which review topics the automated classifier will provide benefit for and an estimate of the expected labor reduction. The 5 × 2 cross-validation method should provide an accurate estimate of the classification performance on future articles that meet the given search criteria. This allows us to apply the automated classification system to only the review topics that gain sufficient value in the review process. While further work is necessary to determine the requirements of systematic reviewers before adding document classification to the drug review work process, we have shown a robust means of estimating the benefit that an automated document classification approach could provide.
We found that the performance of the classifier system, while most often providing at least some benefit, varied widely. In general, it is difficult to determine why a machine-learning algorithm such as the voting perceptron performs well on one task and not on another. Unlike with rules-based classifiers (e.g., Cohen et al.23), the decision processes of a margin-based learning algorithm such as ours are rather opaque. Nevertheless, some observations can be made from the data in ▶ and ▶ about why the classification system performed very well on some review topics and poorly on others.
Statistical analysis showed essentially no correlation between performance and the sample size or fraction of positive samples. The two zero scoring topics, Antihistamines and SkeletalMuscleRelaxants, had the lowest number of significant features, meaning that the classifier system had fewer dimensions with which to separate positives from negatives. It appears that 30 or fewer significant features are not enough to adequately model the triage process. However, the highest scoring topics did not necessarily have the highest number of significant features and the correlation between number of significant features and WSS was not statistically significant. The best performance occurred on ACEInhibitors, ADHD, and NSAIDs topics, but these topics had significant feature counts in the middle of the range. The tasks with the highest number of significant feature counts, AtypicalAntipsychotics and Statins, scored at the low and high extremes of the moderate performing group of tasks. We examined the data for correlations between performance and descriptive measures about the set of significant features such as number of individual features above various recall and precision thresholds and were unable to find any correlation.
Our previous work in biomedical text mining has shown that examining the single most predictive feature can provide insight into the performance of text classification tasks and at times can even dominate the performance,16 especially when human annotated features such as MeSH are available. We therefore conducted an analysis looking at the most strongly predictive feature for each of the 15 tasks. ▶ presents the single best feature for each of the review tasks, as determined by the F-measure prediction score on the individual features. There is no statistically significant correlation of WSS@95% performance with the F-measure of the single best feature. Even when removing the Triptans review topic by assuming that it is an outlier, the correlation is very weak (R2 = 0.079) and still not significant (p = 0.329).
Table 9.
Best Single Feature and F-Measure of That Feature for Each Task
| Review | WSS@95% | Best Feature | F-measure of Feature |
|---|---|---|---|
| ACEInhibitors | 0.5661 | MESH_Myocardial_Infarction/mortality | 0.3206 |
| ADHD | 0.6795 | adults | 0.3098 |
| Antihistamines | 0.0000 | scale | 0.2641 |
| AtypicalAntipsychotics | 0.1411 | PUBTYPE_Multicenter_Study | 0.3464 |
| BetaBlockers | 0.2844 | hospitalization | 0.2429 |
| CalciumChannelBlockers | 0.1221 | CI (abbreviation for Confidence Interval) | 0.3128 |
| Estrogens | 0.1834 | years | 0.5242 |
| NSAIDs | 0.4967 | PUBTYPE_Multicenter_Study | 0.5217 |
| Opioids | 0.1332 | safety | 0.0965 |
| OralHypoglycemics | 0.0896 | study | 0.4743 |
| ProtonPumpInhibitors | 0.2768 | superior | 0.2620 |
| SkeletalMuscleRelaxants | 0.0000 | MESH_MAIN_Research_Support_Non-U | 0.0437 |
| Statins | 0.2471 | PUBTYPE_Multicenter_Study | 0.2255 |
| Triptans | 0.0337 | MESH_PRIMARY_Indoles/*therapeutic_use | 0.3157 |
| UrinaryIncontinence | 0.2614 | weeks | 0.5192 |
Note, however, that the best features are most commonly words, as this is the case for nine of the 15 topics. Since the discriminatory power of the publication type and MeSH is one of the primary means by which the original expert-created queries were constructed, there have to be other useful features in order for the classifier system to outperform the original query. Therefore, it is reasonable to infer that for this to be possible there must exist effective classification features that are not as immediately obvious (to the human experts constructing search queries) as MeSH and publication types are. For example, for the highest scoring topic, ADHD, the word “adults” was the most discriminating classifier feature, although this term does not appear in the original expert-created query (the report inclusion criteria encompassed both pediatric and adult populations). Similarly, the terms for the single best features for BetaBlockers (hospitalization), ProtonPumpInhibitors (superior), and UrinaryIncontinence (weeks) do not appear in the search strategy. For ACEInhibitors, the MeSH Myocardial Infarction does appear in the original search strategy, but not with the mortality subheading included in the single best feature. In fact, the word mortality does not appear in any form in the original search strategy. Most interesting is the best feature confidence interval (CI) for the topic CalciumChannelBlockers. While this term, nor any obviously overlapping terms, does not appear in the original search strategy, one can infer that the presence of explicit confidence intervals was one characteristic that reviewers were looking for when deciding whether to include a given paper in the systematic review.
Conversely, for the Triptans topic, while the MeSH Indoles/*therapeutic use does not appear explicitly in the expert search strategy, the same concepts are largely covered by the inclusion of each of the triptans of interest explicitly (sumatriptan, almotriptan, etc.) in the search strategy along with the MeSH Migraine/Drug Therapy. Therefore, there may not be enough strong classification concepts for the classifier to use in addition to those already expressed in the original search strategy. This may be one reason why the performance on the Triptans topic is so poor, despite the presence of many statistically significant features. For the OralHypoglycemics, another low-scoring topic, the most predictive feature was the word study. This concept was certainly covered by the inclusion of expansions for Retrospective and Comparative Study in the original search strategy.
The previously described work of Aphinyanaphongs et al. focused on improving sensitivity and specificity as measured by the area under the receiver-operating curve, and not on labor reduction as we have done here. Nevertheless, at the highest levels of recall (sensitivity) such as we use in our work, the improvement in precision of the Aphinyanaphongs et al. approach is roughly similar to ours, varying from just over unity to above 2.0. It is interesting that the improvement in precision is similar considering the difference in sample sizes and topic specificity, as the topic-specific nature of our work leads to our sample sizes being much smaller and the inclusion requirements for each drug review study are much more specific.
Our current methods have several limitations. We are only using words from the title and abstract, MeSH, and MEDLINE publication types as potential classification features. Additional work needs to be done to explore possible performance improvements by including additional features such as full document text, N-grams, parts-of-speech, and conceptual relations derived from natural language- processing (NLP) techniques. Furthermore, while we have had consistently encouraging results using the modified voting perceptron classifier with a linear kernel, other classifiers or higher order kernels may improve performance.
Many classifier algorithms such as our voting perceptron produce, in addition to a predicted class, a score that can be used as a confidence measure. This confidence measure can be used to rank the individual documents returned by a search query leading to an information retrieval approach, in contrast with the binary document classification presented here. For the purposes of aiding the process of updating systematic treatment reviews, where the reviewers want to identify the best set of documents to review, query reranking may not be as useful as document classification. However, the ranking approach has been shown to have value in tasks that continuously rerun the same queries to determine the order in which query-matching publications are reviewed for database annotation, such as that done by the Swiss-Prot curators when annotating the relationship between genetic diseases and polymorphisms.15
Our data set included 15 drug reviews, all of which were available at the time this work was performed. While this number of topics is adequate to show that automated document classification can provide benefit in the drug review process, this sample size is small and our computation of average WSS@95% and precision improvement have to be interpreted in this context. Additional drug review data sets would help to further characterize the expected frequency and size of the work reduction for a typical drug review study.
Extending the system to process full text articles and using full text classification methods may make significant improvements in classifier performance possible. For example, Porter stemming may have a more consistent and beneficial effect on full text. However, it is not clear that all sources of full text, including MEDLINE-indexed journal articles, Cochrane reviews, and non-English journal articles should be processed together for classification. Further work is necessary to determine the best means of including the different types of full text articles. Full text may also allow us to include some of the reports that were removed during the initial data preparation process, which required the document to have an entry within MEDLINE.
An automated document classification system such as ours could be integrated into a full system that would automatically rerun the drug review queries on PubMed or Ovid, classify the resulting documents, and notify the reviewers when new positive documents were discovered for each drug review. It would be straightforward for this notification to be either an automated e-mail or from a news-feed (RSS) server. These notifications would be useful both for focusing the reviewer's time on the most likely articles to include high-quality topic-specific evidence, as well as to alert the review team when a sufficient amount of new evidence had accumulated to warrant an update of a given drug class review.
Conclusions
We have demonstrated that automated document classification systems can provide value to the process of preparing systematic evidence reviews. We have shown one method of constructing and training a classifier system to accomplish this task and have presented a means of estimating the performance of such a system and using that estimate to decide for which topics such a system would be most useful. For the vast majority of topics studied, automated document classification can provide some value in reducing the labor of manual review, and for about 20% of topics, the reduction is very large, approaching ≥50%.
Future work will focus on improving the classifier system and investigating the incorporation of the system into systematic review workflow. The results shown here are encouraging enough to warrant building a system that automatically reruns the queries and notifies reviewers of papers classified as positive for one of the 13 topics where a net gain was shown. This will allow collection of prospective data to determine the actual effect on the workload of revising systematic drug evidence reviews. It will also help determine the required level of recall and labor savings and how these parameters may change based on the topic, schedule, and other environmental factors. Finally, additional classifier features and classification algorithms should be investigated, such as making use of the nine specific exclusion codes.
We encourage others to investigate applying text mining and classification approaches to practical real-world problems in biomedical informatics such as presented here. Furthermore, we encourage other investigators to use publicly available collections whenever feasible. Therefore, we are publicly releasing the text collection and data set built and used in this work. The files are available for download off of the first author's home page at http://medir.ohsu.edu/∼cohenaa.
This work was funded in part by National Science Foundation grant ITR-0325160 and by NIH 2 T15 LM07088-11 from the National Library of Medicine.
The authors thank Mark Helfand, MD, MPH, and Marian McDonagh, PharmD, of the Oregon EPC, for their review and helpful suggestions during the writing of this article. They also acknowledge the Oregon EPC for providing the EndNote files.
References
- 1.Sackett DL, Haynes RB, Tugwell P. Clinical epidemiology: a basic science for clinical medicine. Boston: Little Brown, 1985.
- 2.Sackett DL, Rosenberg WM, Gray JA, Haynes RB, Richardson WS. Evidence based medicine: what it is and what it isn't. BMJ. 1996;312:71–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen AM, Stavri PZ, Hersh WR. A categorization and analysis of the criticisms of evidence-based medicine. Int J Med Inf. 2004;73:35–43. [DOI] [PubMed] [Google Scholar]
- 4.Hersh W. “A world of knowledge at your fingertips”: the promise, reality, and future directions of on-line information retrieval. Acad Med. 1999;74:240–3. [DOI] [PubMed] [Google Scholar]
- 5.Haynes RB. What kind of evidence is it that evidence-based medicine advocates want health care providers and consumers to pay attention to? BMC Health Serv Res. 2002;2:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chou R, Clark E, Helfand M. Comparative efficacy and safety of long-acting oral opioids for chronic non-cancer pain: a systematic review. J Pain Symptom Manage. 2003;26:1026–48. [DOI] [PubMed] [Google Scholar]
- 7.Chou R, Peterson K, Helfand M. Comparative efficacy and safety of skeletal muscle relaxants for spasticity and musculoskeletal conditions: a systematic review. J Pain Symptom Manage. 2004;28:140–75. [DOI] [PubMed] [Google Scholar]
- 8.Nelson HD, Humphrey LL, Nygren P, Teutsch SM, Allan JD. Postmenopausal hormone replacement therapy: scientific review. JAMA. 2002;288:872–81. [DOI] [PubMed] [Google Scholar]
- 9.Oregon Health & Science Universy, Drug Effectiveness Review Project Methods, 2003. Available from: http://www.ohsu.edu/drugeffectiveness/methods/index.htm/. Accessed 11/03/05.
- 10.Mulrow C, Cook D. Systematic reviews: synthesis of best evidence for health care decisions. Philadelphia: The American College of Physicians, 1998.
- 11.Haynes RB, Wilczynski N, McKibbon KA, Walker CJ, Sinclair JC. Developing optimal search strategies for detecting clinically sound studies in MEDLINE. J Am Med Inform Assoc. 1994;1:447–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilczynski NL, Haynes RB. Developing optimal search strategies for detecting clinically sound causation studies in MEDLINE. AMIA Annu Symp Proc. 2003:719–23. [PMC free article] [PubMed]
- 13.Wong SS, Wilczynski NL, Haynes RB, Ramkissoonsingh R. Developing optimal search strategies for detecting sound clinical prediction studies in MEDLINE. AMIA Annu Symp Proc. 2003:728–32. [PMC free article] [PubMed]
- 14.Aphinyanaphongs Y, Tsamardinos I, Statnikov A, Hardin D, Aliferis CF. Text categorization models for high-quality article retrieval in internal medicine. J Am Med Inform Assoc. 2005;12:207–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dobrokhotov PB, Goutte C, Veuthey AL, Gaussier E. Assisting medical annotation in Swiss-Prot using statistical classifiers. Int J Med Inform. 2005;74:317–24. [DOI] [PubMed] [Google Scholar]
- 16.Hersh WR, Bhupatiraju RT, Ross L, Johnson P, Cohen AM, Kraemer DF. TREC 2004 Genomics Track Overview. In: Proceedings of the Thirteenth Text Retrieval Conference–TREC 2004. Gaithersburg, MD. Available at http://trec.nist.gov/pubs/trec13/papers/GEO.OVERVIEW.pdf.
- 17.Cohen AM, Hersh WR, Bhupatiraju RT. Feature generation, feature selection, classifiers, and conceptual drift for biomedical document triage. In: Proceedings of the Thirteenth Text Retrieval Conference–TREC 2004. Gaithersburg, MD. Available at http://trec.nist.gov/pubs/trec13/papers/ohsu_hersh.geo.pdf.
- 18.Blaschke C, Leon EA, Krallinger M, Valencia A. Evaluation of BioCreAtIvE assessment of task 2. BMC Bioinformatics. 2005:, 6 Suppl 1:S16. [DOI] [PMC free article] [PubMed]
- 19.Porter MF. An algorithm for suffix stripping. Program. 1980;14:127–30. [Google Scholar]
- 20.Carroll JB, Davies P, Richman B. The American Heritage word frequency book. Boston: Houghton Mifflin, 1971.
- 21.Freund Y, Schapire RE. Large margin classification using the perceptron algorithm. Machine Learn. 1999;37:277–96. [Google Scholar]
- 22.Rosenblatt F. The perceptron: a probabilistic model for information storage and organization in the brain. Psychol Rev. 1958:386–407. [DOI] [PubMed]
- 23.Cohen WW, Singer Y. A Simple, fast, and effective rule learner. In: Proceedings of the Annual Conference of the American Association for Artificial Intelligence (AAAI). 1999; 335–42.
- 24.Joachims T. SVM-Light Support Vector Machine, 2004. Available from: http://svmlight.joachims.org/. Accessed 07/20/05.
- 25.Dietterich TG. Approximate statistical tests for comparing supervised classification learning algorithms. Neural Comput. 1998;10:1895–924. [DOI] [PubMed] [Google Scholar]