Abstract
Purpose
Age-related macular degeneration (AMD) is a common disease in which both environmental and genetic factors have been implicated. Various single nucleotide polymorphisms (SNPs) have been correlated, through candidate gene association studies, with age-related diseases, including AMD. Recently we identified an association between AMD and two SNPs in CX3CR1, which encodes a chemokine receptor. This study investigates the Hemicentin-1 (an extracellular matrix protein) Q5345R, hOgg1 (DNA repair gene) S326C, and E-selectin (an adhesion molecule) S149R SNPs in association with AMD.
Methods
Genomic DNA was extracted from peripheral blood of 89 patients with advanced AMD, 97 age-matched controls without clinical AMD, and 170 random unscreened healthy volunteers. DNA was subjected to polymerase chain reaction amplification coupled with the restriction fragment length polymorphism assay.
Results
The distribution of the Hemicentin-1 Q5345R, hOgg1 S326C, and E-selectin S149R SNPs did not differ significantly (all P values > .05) between the AMD patients and controls. Hemincentin-1 5345R was not found in any subject. hOgg1 326C allele frequency was 21.35% (38 of 178) in the AMD group compared with 19.12% (65 of 340) in the random controls and 19.59% (38 of 194) in the age-matched controls. E-selectin 149R allele frequencies were 8.99% (16 of 178) in AMD cases, 9.41% (32 of 340) in random controls, and 10.82% (21 of 194) in age-matched controls.
Conclusions
We were not able to demonstrate an association between the Hemicentin-1, hOgg1, and E-selectin SNPs and AMD development in the currently available cases and controls. Further candidate genes, particularly those involved in extracellular matrix, oxidative stress, and immune system functions, are currently being screened in our laboratory.
INTRODUCTION
Age-related macular degeneration (AMD) is a chronic and progressive disease marked by degeneration of the photoreceptors, retinal pigment epithelium (RPE), Bruch’s membrane, and possibly the choriocapillaries in the macula.1–3 AMD is the third leading cause of visual impairment and blindness in the United States and the developed world among people aged 65 years and older.4;5 It has been projected that by the year 2020, approximately 7.3 million people in the United States alone will have developed at minimum the early stages of AMD in at least one eye.6 However, AMD prevalence has been rising across the globe. In 2002, an estimated 37 million people in the world were blind. Among these individuals, more than 82% were aged 50 years or older. Over recent years in the developed countries, the number of people over the age of 50 years has increased by 16%. In the developing countries excluding China, this increase was by 47%. China itself had a marked increase of 27% in their elderly population.7 As the average life span of humans continues to increase, particularly in the developed countries, the incidence of AMD is expected to nearly double within the next 25 years.
Despite remarkable disease prevalence, the etiology and pathogenesis of AMD remain unclear. AMD is a common and multifactorial disease in which both genetic and environmental factors have been implicated.8–10 Complex diseases such as AMD are marked by genetic heterogeneity, a low penetrance, a continuous phenotypic distribution, and a high susceptibility to nongenetic factors.11,12 There have been several controversial reports concerning potential risk factors for AMD development.13 To date, however, only age, smoking, exposure to light, and diet have been successfully identified.13–16
The strongest evidence of a genetic component in AMD development stems from the broad tendency for familial aggregation among cases, with roughly 20% of afflicted individuals reporting a positive family history.17,18 There is also a higher incidence of AMD among monozygotic twins as compared with their spouses or other first-degree relatives.19,20 It is very likely that in common complex diseases such as AMD, variations within several genes, each with a small overall contribution and relative risk, interact to create a genetic background that can be triggered by environmental factors.
Several types of genetic polymorphisms can be found within the human genome, such as repeat polymorphisms, insertions, and deletions. However, most DNA sequence variation in human populations is in the form of single nucleotide polymorphisms (SNPs).21 SNPs can be defined as persistent substitutions of a single base with a frequency of more than 1% in at least one population. Recently, investigators have begun to explore the potential role of SNPs in AMD development. Various SNPs have been correlated, through candidate gene association studies, with age-related diseases, including AMD.22,23
The candidate gene approach is a common method used in association analyses. This approach is based on generating hypotheses about, and selecting candidate genes involved with, plausible pathological pathways.12 This study investigates the Hemicentin-1 (an extracellular matrix protein identified through a genome-wide scan of extended families with AMD) Q5345R, hOgg1 (which is involved in oxidatively damaged DNA repair) S326C, and E-selectin (an adhesion molecule) S149R SNPs in association with AMD.
METHODS
STUDY SUBJECTS
This protocol was approved by the National Eye Institute Institutional Review Board. Each participant included in this study signed the informed consent prior to participation. This multiple case-control study included an AMD patient group and two normal control groups. The patients and controls included in this study were all white of non-Hispanic descent residing in the surrounding greater Washington, DC, area.
Sporadic patients (n = 89) with advanced AMD and screened normal controls (n = 97) were enrolled in this study. A clinical diagnosis of advanced AMD was defined by geographic atrophy involving the center of the macula and/or choroidal neovascularization in the presence of drusen in at least one eye. Stereoscopic fundus photographs of the optic disc and the macula were taken for all AMD patients. The screened normal controls were spouses and friends of the patients as well as randomly recruited normal volunteers. These subjects were all older than 50 years of age. These clinically screened controls received dilated fundus examinations that showed an absence of drusen or less than five small drusen (<63 μm) in the center of the macula and an absence of all other retinal disease affecting the photoreceptors, outer retinal layers, or both, such as high myopia, retinal dystrophies, central serous retinopathy, vein occlusion, diabetic retinopathy, uveitis, and other retinal diseases.
Healthy blood donors (n = 170), who did not receive a clinical eye examination, served as another control group in order to obtain information on SNP frequencies in a population that has not yet reached the average age for AMD onset. Therefore, the average age of this population is much lower than that of the AMD cases and screened controls. This group is referred to as the random controls in this study. The demographics of the study groups are summarized in Table 1.
TABLE 1.
DEMOGRAPHICS OF THREE STUDY GROUPS: PATIENTS WITH AGE-RELATED MACULAR DEGENERATION (AMD) AND TWO CONTROL GROUPS (AGE-MATCHED SCREENED VOLUNTEERS AND RANDOM BLOOD BANK DONORS)
| GROUP | N | MALE | FEMALE | MEAN AGE (YEARS) |
|---|---|---|---|---|
| Screened control | 97 | 48 | 49 | 67.5 ± 11.7 |
| Random control | 170 | 113 | 5 | 45.3 ± 14.0 |
| AMD | 89 | 42 | 4 | 79.4 ± 7.0 |
DNA EXTRACTION
Venous whole blood (10 mL) was collected from the study subjects. Genomic DNA was extracted and isolated by using a QIAamp DNA Blood Maxi kit (catalog No. 51194; Qiagen, Valencia, California).
SNP TYPING
The SNPs Hemicentin-1 Gln5345Arg, hOgg1 Ser326Cys, and E-selectin Ser149Arg were detected by polymerase chain reaction (PCR) coupled with the restriction fragment length polymorphism (RFLP) method. For each gene, the PCR mixture included 1XJumpStart ReadyMix REDTaq (Sigma, St Louis, Missouri), 25 ng DNA, and 70 pmol of the respective primers. Following RFLP assay, the DNA fragments were separated on 15% TBE polyacrylamide gels and visualized after ethidium bromide staining. A negative (H2O) control and standards for both alleles and a heterozygous DNA control when available were included on each genotyping plate. Representative gel images used for SNP typing are shown in Figures 1 through 3.
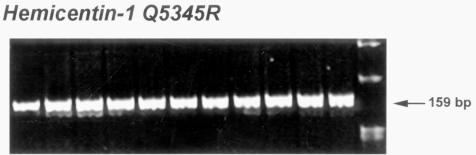
FIGURE 1.
Autoradiography of Hemicentin-1 Q5345R in patients with age-related macular degeneration and age-matched screened volunteers. Representative gel picture of Hemicentin-1 Q5345R shows the banding patterns that determined the respective single nucleotide polymorphism types. The restriction fragment length polymorphism pattern of Hemicentin-1 Q5345R shows a 159–base pair (bp) pattern representing the wild-type (Q/Q). No variant alleles were detected in either of the case or control populations.
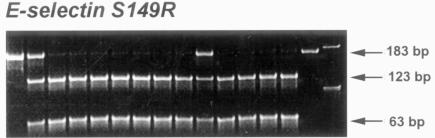
FIGURE 3.
Autoradiography of E-selectin S149R in patients with age-related macular degeneration and two control groups (age-matched screened volunteers and random blood bank donors). Representative gel pictures of E-selectin S149R showing the banding patterns that determined the respective single nucleotide polymorphism types. The restriction fragment length polymorphism pattern of E-selectin S149R shows bands at 123 bp and 63 bp in wild-types (S/S); 183 bp, 123bp, and 63 bp in heterozygotes (S/R); and 183 bp in variant homozygotes (R/R).
For Hemicentin-1, a 159–base pair (bp) DNA fragment was PCR amplified with use of the following primers: 5′-CAA GTG TAT CTG TCC ACC AGG TC-3′ and 5′ TGT CTG TAA TGC TGT TGA GGT TG-3′. The PCR program was run at 2 minutes at 94°C, followed by 34 cycles of 30 seconds denaturation at 94°C, 40 seconds annealing at 59°C, and 55 seconds extension at 72°C. RFLP analysis was conducted by using the restriction endonuclease SalI. The digestion patterns for each SNP type are as follows: wild-type (Gln/Gln), 159 bp; heterozygous variant (Gln/Arg), 159 bp and 137 bp; and homozygous variant (Arg/Arg), 137 bp.
For hOgg1, a 200-bp DNA fragment was PCR amplified by using the following primers: 5′-ACT GTC ACT AGT CTC ACC AG-3′ and 5′-TGA ATT CGG AAG GTG CTT GGG GAA T-3′. The PCR conditions were 2 minutes at 94°C, followed by 34 cycles of 30 seconds denaturation at 94°C, 40 seconds annealing at 61°C, and 55 seconds extension at 72°C. RFLP analysis was conducted by using the restriction endonuclease Fnu4H2. The digestion patterns for each SNP type are as follows: wild-type (Ser/Ser), 200 bp; heterozygous variant (Ser/Cys), 200 bp and 100 bp; and homozygous variant (Cys/Cys), 100 bp.
For E-selectin, a 186-bp DNA fragment was PCR amplified with use of the following primers: 5′-AGT AAT AGT CCT CCT CAT CAT G-3′ and 5′-ACC ATC TCA AGT GAA GAA AGA G-3′. The program was run at 2 minutes at 94°C, followed by 34 cycles of 30 seconds denaturation at 94°C, 40 seconds annealing at 59°C, and 55 seconds extension at 72°C. RFLP analysis was conducted by using the restriction endonuclease Pst1. The digestion patterns for each SNP type are as follows: wild-type (Ser/Ser), 123 bp and 63 bp; heterozygous variant (Ser/Arg), 183 bp, 123 bp, and 63 bp; and homozygous variant (Arg/Arg), 183 bp.
STATISTICAL ANALYSIS
The chi-square test was performed in order to compare the carrier and allele frequencies of the cases and controls. Hardy-Weinberg equilibrium was also tested by using the chi-square test within 1 degree of freedom. A P value of less than .05 was considered to be significant. Odds ratios were calculated and an estimation of confidence intervals was made on the basis of the unmatched case-control design.24
RESULTS
The 89 AMD patients and 97 screened controls were matched as closely as possible for both gender and race (Table 1). Although the screened patients and controls were roughly matched for gender, the random control population was largely male. Of the 89 AMD patients, 57 had the neovascular, or “wet,” form of the disease.
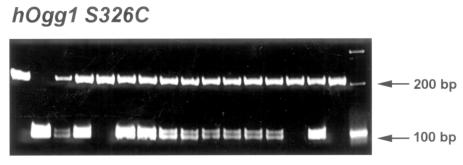
DNA was successfully extracted from all enrolled patients and controls. The distribution of the Hemicentin-1 Q5345R, hOgg1 S326C, and E-selectin S149R polymorphisms did not deviate from Hardy-Weinberg equilibrium in each group included in this study (all P values < .05). The distribution of the Hemicentin-1 5345R, hOgg1 326C, and E-selectin 149R variant alleles did not differ significantly (all P values > .05) between the AMD patients and control populations (Tables 2 through 4). All gels were unable to detect a positive Hemicentin-1 5345R variant band in the screened patients and controls (Figure 1). hOgg1 326C allele frequency was 21.35% (38 of 178) in the AMD group compared with 19.12% (65 of 340) in the random controls and 19.59% (38 of 194) in the age-matched controls (Table 3). Representative autoradiography showed positive hOgg1 SNP banding patterns in several cases (Figure 2). E-selectin 149R allele frequencies were 8.99% (16 of 178) in AMD cases, 9.41% (32 of 340) in random controls, and 10.82% (21 of 194) in age-matched controls (Table 4).
TABLE 2.
ALLELE FREQUENCIES OF HEMICENTIN-1 Q5345R IN PATIENTS WITH AGE-RELATED MACULAR DEGENERATION (AMD) AND AGE-MATCHED SCREENED VOLUNTEERS
|
HEMICENTIN-1Q5345R |
|||||
|---|---|---|---|---|---|
| GROUP | ALLELES N | Q | R (%) | R OR | χ2/Pvalue |
| Screened control (n = 81) | 162 | 162 | 0 (0) | 0 | No SNP |
| AMD (n = 88) | 176 | 176 | 0 (0) | 0 | No SNP |
OR = odds ratio; SNP = single nucleotide polymorphism.
TABLE 4.
ALLELE FREQUENCIES OF E-SELECTIN S149R IN PATIENTS WITH AGE-RELATED MACULAR DEGENERATION (AMD) AND TWO CONTROL GROUPS (AGE-MATCHED SCREENED VOLUNTEERS AND RANDOM BLOOD BANK DONORS)
| E-SELECTINS149R | |||||
|---|---|---|---|---|---|
| GROUP | ALLELES N | S | R (%) | R OR | χ2/PVALUE |
| Screened control (n = 97) | 194 | 173 | 21 (10.82) | 0.81 | .35/.55 |
| Random control (n = 170) | 340 | 308 | 32 (9.41) | 0.95 | .02/.87 |
| AMD (n = 89) | 178 | 162 | 16 (8.99) | — | — |
OR = odds ratio.
TABLE 3.
ALLELE FREQUENCIES OF HOGG1 S326C IN PATIENTS WITH AGE-RELATED MACULAR DEGENERATION (AMD) AND TWO CONTROL GROUPS (AGE-MATCHED SCREENED VOLUNTEERS AND RANDOM BLOOD BANK DONORS)
|
HOGG1S326C |
|||||
|---|---|---|---|---|---|
| GROUP | ALLELES N | S | C (%) | C OR | χ2/PVALUE |
| Screened control (n = 97) | 194 | 156 | 38 (19.59) | 1.114 | .18/.67 |
| Random control (n = 170) | 340 | 275 | 65 (19.12) | 1.148 | .36/.55 |
| AMD (n = 89) | 178 | 140 | 38 (21.35) | — | — |
OR = odds ratio.
FIGURE 2.
Autoradiography of hOgg1 S326C in patients with age-related macular degeneration and two control groups (age-matched screened volunteers and random blood bank donors). Representative gel picture of hOgg1 S326C shows the banding patterns that determined the respective single nucleotide polymorphism types. The restriction fragment length polymorphism pattern of hOgg1 S326C shows a 200–base pair (bp) pattern representing the wild-type (S/S), 200 and 100 bp representing the heterozygote (S/C), and 100 bp representing the variant homozygote (C/C).
A slightly lowered prevalence of the E-selectin 149R variant was found in the AMD patients as compared with the two control groups. However, this difference was not significant. Figure 3 illustrates a representative gel image of the E-selectin banding patterns. These results do not demonstrate an association between the Hemicentin-1 5345R, hOgg1 326C, and E-selectin 149R alleles and AMD in this small study.
DISCUSSION
Statistically significant differences in variant allele frequencies were not obtained between the AMD case and control groups after screening for the Hemicentin-1 Q5345R, hOgg1 S326C, and E-selectin S149R SNPs. Our results suggest that these polymorphisms may not be associated with AMD.
Genome-wide scans and genetic linkage analyses of extended families with AMD are two of the approaches taken to locate candidate AMD loci. ARMD1, an identified disease-related locus, was previously mapped to 1q25-31 in a large family with AMD.25,26 Recently, a gene within this region was found to be significantly associated with AMD risk. The Y402H polymorphism within the gene encoding the complement factor H protein has been reported in significant association with AMD by several independent groups.27–29 Complement factor H (CFH) is located on chromosome 1q31. It was reported that the 402H variant allele may account for up to one half of all AMD cases and that individuals homozygous for this variant allele have a 7.4-fold increased risk of developing AMD.29 We also found that a SNP in a CFH intron, which is strongly haplotyped with CFH 402, was significantly higher in our AMD patients compared with the controls (data not shown).
Hemicentin-1 was identified and mapped to 1q25.30-1q31.1 after further paring down of the ARMD1 locus.26 Several groups have screened this gene for potential AMD associated variants; however, no evidence for any significant allele associations has been generated thus far. Schultz and colleagues26 previously screened for the Hemicentin-1 5345R variant in sporadic AMD cases and found a very low frequency in both patients and controls. Iyengar and associates30 recently screened exon 104 where Hemicentin-1 is located and found no evidence of mutations in this region. Abecasis and colleagues31 were also unable to detect this variation in a total of 620 patients and 237 controls.
In the present study, no variant allele was detected in either the screened patients or controls. It may be possible that this gene’s significant involvement in disease development is specific to the particular multigenerational AMD-affected pedigrees previously studied. In other words, Hemicentin-1 may not be relevant to most isolated or sporadic AMD cases or even necessarily to other family pedigrees that may have more complex underlying genetic factors. Furthermore, successfully identifying an association between a particular allele and a given disease phenotype is greatly influenced by the variant allele frequency in the general population. It is highly possible also that the rarity of the Hemicentin-1 5345R allele in the general population renders the small sample populations included in this current study incapable of detecting or determining this variation’s involvement in AMD. Lastly, there may be additional variations within or in very close proximity to this gene that may account for the risk associated with this region other than Q5345R. Larger sample sizes are needed to elucidate to what degree and by which mechanisms this gene is involved in AMD development.
Variation within genes involved in the proposed protective pathways against the effects of oxygen toxicity within the eye has been a focus of many AMD association studies. The eye is particularly vulnerable to oxidative damage because oxygen consumption is much higher in the retina than in other human tissues.32 Thus, failure of the normal protective enzymes and mechanisms needed to defend the macula against the oxidative stresses imposed by reactive oxygen species may result in the development of AMD and other general ailments of the aging eye. Three SNPs (MnSOD, MEHE, Paraoxonase) related to oxidative stress have previously been reported in association with AMD in Japanese populations.33,34
DNA base modifications resulting in mutations and genetic instability are a major consequence of oxidative stress.35 Efficient DNA repair mechanisms are necessary to counterbalance the damaging effects of oxidizing species. DNA repair mechanisms are also vital in maintaining cellular integrity after oxidative damage has occurred.36 The disruption of DNA repair genes has been associated with the other degenerative and earlier-onset age-related diseases, such as the Werner and Bloom syndromes, as well as with the pathogenesis of aging.37–39
The human Ogg1 (hOgg1) gene encodes a DNA glycosylase that is involved in the base excision repair of 8-hydroxy-2′ deoxyguanin (8-OH-dG) from oxidatively damaged DNA.37 Defects in 8-OH-dG repair have been implicated in the development of several diseases, including cancer.37 8-OH-dG has been found to be highly mutagenic, leading to a mutator phenotype characterized by an increase in GC to TA transversions. These transversions are frequently observed in several oncogenes and tumor suppressor genes.37
Although a statistically significant association was not established between hOgg1 and AMD in this study, this avenue of exploration has yielded promising findings. The Cockayne syndrome B (CSB) gene, also called ERCC6, collaborates with hOgg1 to carry out preferential DNA repair in eukaryotes. This gene also plays a role in the maintenance of efficient hOgg1 expression.35,40–42 Recently, we have found an association between SNPs in the promoter region of this gene and AMD (Tuo J, et al, FASEB meeting, 2005, Abstract). We have found that there is an increased prevalence of variant alleles in AMD cases as compared with the control populations. These findings suggest that an increased risk of AMD development is associated with these polymorphisms.
Investigators have recently begun looking into the potentially critical immunologic mechanisms hypothesized to be involved in the development and pathogenesis of AMD. Macrophage chemoattractant CCL2 (alternate name MCP-1, a CC chemokine) or its cognate receptor-2 (CCR2) knockout mice have been shown to spontaneously develop hallmark features of AMD in their senescent stage.43 Recently, we reported an association between AMD and two SNPs within CX3CRI, a CX3C chemokine receptor of fraktalkine/CX3CL1.44 These SNPs have been associated with a decreased number of CX3CR1/CX3CL1 binding sites as well as with decreased binding affinity.45,46 We have hypothesized that the risk introduced in AMD development is due to inefficient and/or insufficient macrophage recruitment to Bruch’s membrane.44 Efficient macrophage recruitment is needed in order to aid in the clearing of the lipid and protein retinal deposits that later accumulate and develop into soft drusen, the early hallmark of AMD. With such promising initial findings, further investigation of the genes involved in the immunologic mechanisms implicated in AMD development and pathogenesis has become an interesting avenue of research for future association studies.
Adhesion of leukocytes to endothelial cells is an essential step in their migration, rolling, strong adhesion, and diapedesis from the circulation to sites of inflammation.47 E-selectin, a cellular adhesion molecule, mediates leukocyte infiltration and lymphocyte trafficking. Polymorphisms within this gene have been associated with risk of atherosclerosis.47,48 Although we did not establish a significant association between this particular SNP in E-selectin and AMD, we observed a slightly lowered prevalence of the variant allele in our AMD cases compared with our two control populations. This difference is slightly greater when comparing the AMD cases with the screened control group alone. Although not necessarily of high importance in this specific instance, it is worth pointing out that some of the individuals in the random control group with the younger mean age may go on to develop some degree of AMD later on in life. This trend may be attributed simply to the age-related nature of the disease. Therefore, we would expect the AMD cases and the screened controls to have allele frequencies that lie on opposite ends of the spectrum, with the allele frequency of the random control group falling somewhere in between. Again, although these differences are not significant, the allele frequency patterns observed in this study align with this theory. Additional genes involved in chemoattraction, cellular adhesion, and inflammation should be explored in larger sample populations.
Only advanced AMD cases were included in this study in order to minimize the possible bias introduced by subjective and/or poorly defined phenotyping criteria. Association studies are strengthened by the incorporation of larger, more carefully age-matched patient and selected control populations. However, because AMD is an age-dependent disease of high incidence, multiple control groups with different average ages can be used to elucidate a possible quantitative correlation and potential gene-dosage effect of the tested genetic risk. We utilized a study design that aims to compensate for our small sample sizes.
In conclusion, we were not able to demonstrate an association between the Hemicentin-1 Q5345R, hOgg1 S326C, and E-selectin S149R SNPs and advanced AMD. Common multifactorial diseases such as AMD are thought to arise from a complex combination of genetic and environmental risk factors. Therefore, it is highly unlikely that a single gene variant is solely responsible for disease development. Furthermore, it is probable that the contribution of each associated gene is relatively small. Larger sample sizes are needed to adequately study gene-gene and gene-environment interactions as well as to more accurately determine relative risk. We are continuing to recruit larger patient and control populations in order to investigate the potential role of further genetic variants in AMD development and pathogenesis.
ACKNOWLEDGMENTS
We would like to thank Katherine Shimel, RN, and Young Kim, RN, from the National Eye Institute, National Institutes of Health, Bethesda, Maryland, for patient and control recruitment and care.
Footnotes
This research was supported by the Intramural Research Program of the National Eye Institute, National Institutes of Health.
REFERENCES
- 1.Fine SL, Berger JW, Maguire MG, et al. Age-related macular degeneration. N Engl J Med. 2000;342:483–492. doi: 10.1056/NEJM200002173420707. [DOI] [PubMed] [Google Scholar]
- 2.Lutty G, Grunwald J, Majji AB, et al. Changes in choriocapillaris and retinal pigment epithelium in age-related macular degeneration. Mol Vis. 1999;5:35. [PubMed] [Google Scholar]
- 3.Ambati J, Ambati BK, Yoo SH, et al. Age-related macular degeneration: etiology, pathogenesis, and therapeutic strategies. Surv Ophthalmol. 2003;48:257–293. doi: 10.1016/s0039-6257(03)00030-4. [DOI] [PubMed] [Google Scholar]
- 4.Gottlieb JL. Age-related macular degeneration. JAMA. 2002;288:2233–2236. doi: 10.1001/jama.288.18.2233. [DOI] [PubMed] [Google Scholar]
- 5.Resnikoff S, Pascolini D, Etya’ale D, et al. Global data on visual impairment in the year 2002. Bull World Health Organ. 2004;82:844–851. [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman DS, O’Colmain BJ, Munoz B, et al. the Eye Diseases Prevalence Research Group. Prevalence of age-related macular degeneration in the United States. Arch Ophthalmol. 2004;122:564–572. doi: 10.1001/archopht.122.4.564. [DOI] [PubMed] [Google Scholar]
- 7.Kingman S. Glaucoma is second leading cause of blindness globally. Bull World Health Organ. 2004;82:887–888. [PMC free article] [PubMed] [Google Scholar]
- 8.Tuo J, Bojanowski CM, Chan CC. Genetic factors of age-related macular degeneration. Prog Retin Eye Res. 2004;23:229–249. doi: 10.1016/j.preteyeres.2004.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silvestri G. Age-related macular degeneration: genetics and implications for detection and treatment. Mol Med Today. 1997;3:84–91. doi: 10.1016/S1357-4310(96)10057-5. [DOI] [PubMed] [Google Scholar]
- 10.Gorin MB, Breitner JC, de Jong PT, et al. The genetics of age-related macular degeneration. Mol Vis. 1999;5:29. [PubMed] [Google Scholar]
- 11.Lander ES, Schork NJ. Genetic dissection of complex traits. Science. 1994;265:2037–2048. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- 12.Tabor HK, Risch NJ, Myers RM. Candidate-gene approaches for studying complex genetic traits: practical considerations [opinion] Nat Rev Genet. 2002;3:391–397. doi: 10.1038/nrg796. [DOI] [PubMed] [Google Scholar]
- 13.Hyman L, Neborsky R. Risk factors for age-related macular degeneration: an update. Curr Opin Ophthalmol. 2002;13:171–175. doi: 10.1097/00055735-200206000-00007. [DOI] [PubMed] [Google Scholar]
- 14.Age-Related Eye Disease Study Research Group. A randomized, placebo-controlled, clinical trial of high-dose supplementation with vitamins C and E, beta carotene, and zinc for age-related macular degeneration and vision loss: AREDS report no. 8. Arch Ophthalmol. 2001;119:1417–1436. doi: 10.1001/archopht.119.10.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Husain D, Ambati B, Adamis AP, et al. Mechanisms of age-related macular degeneration. Ophthalmol Clin North Am. 2002;15:87–91. doi: 10.1016/s0896-1549(01)00009-8. [DOI] [PubMed] [Google Scholar]
- 16.Age-Related Eye Disease Study Research Group. Risk factors associated with age-related macular degeneration. A case-control study in the age-related eye disease study: Age-Related Eye Disease Study report number 3. Ophthalmology. 2000;107:2224–2232. doi: 10.1016/s0161-6420(00)00409-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Jong PT, Klaver CC, Wolfs RC, et al. Familial aggregation of age-related maculopathy. Am J Ophthalmol. 1997;124:862–863. doi: 10.1016/s0002-9394(14)71715-5. [DOI] [PubMed] [Google Scholar]
- 18.Klaver CC, Wolfs RC, Assink JJ, et al. Genetic risk of age-related maculopathy. Population-based familial aggregation study. Arch Ophthalmol. 1998;116:1646–1651. doi: 10.1001/archopht.116.12.1646. [DOI] [PubMed] [Google Scholar]
- 19.Meyers SM, Greene T, Gutman FA. A twin study of age-related macular degeneration. Am J Ophthalmol. 1995;120:757–766. doi: 10.1016/s0002-9394(14)72729-1. [DOI] [PubMed] [Google Scholar]
- 20.Seddon JM, Cote J, Page WF, et al. The US twin study of age-related macular degeneration: relative roles of genetic and environmental influences. Arch Ophthalmol. 2005;123:321–327. doi: 10.1001/archopht.123.3.321. [DOI] [PubMed] [Google Scholar]
- 21.Cargill M, Daley GQ. Mining for SNPs: putting the common variants—common disease hypothesis to the test. Pharmacogenomics. 2000;1:27–37. doi: 10.1517/14622416.1.1.27. [DOI] [PubMed] [Google Scholar]
- 22.Klaver CC, Allikmets R. Genetics of macular dystrophies and implications for age-related macular degeneration. Dev Ophthalmol. 2003;37:155–169. doi: 10.1159/000072045. [DOI] [PubMed] [Google Scholar]
- 23.Zack DJ, Dean M, Molday RS, et al. What can we learn about age-related macular degeneration from other retinal diseases? Mol Vis. 1999;5:30. [PubMed] [Google Scholar]
- 24.Bland JM, Altman DG. Statistics notes: the odds ratio. Br Med J. 2000;320:1468. doi: 10.1136/bmj.320.7247.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein ML, Schultz DW, Edwards A, et al. Age-related macular degeneration: clinical features in a large family and linkage to chromosome 1q. Arch Ophthalmol. 1998;116:1082–1088. doi: 10.1001/archopht.116.8.1082. [DOI] [PubMed] [Google Scholar]
- 26.Schultz DW, Klein ML, Humpert AJ, et al. Analysis of the ARMD1 locus: evidence that a mutation in HEMICENTIN-1 is associated with age-related macular degeneration in a large family. Hum Mol Genet. 2003;12:3315–3323. doi: 10.1093/hmg/ddg348. [DOI] [PubMed] [Google Scholar]
- 27.Klein RJ, Zeiss C, Chew EY, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edwards AO, Ritter IR, Abel KJ, et al. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 29.Haines JL, Hauser MA, Schmidt S, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 30.Iyengar SK, Song D, Klein BE, et al. Dissection of genomewide-scan data in extended families reveals a major locus and oligogenic susceptibility for age-related macular degeneration. Am J Hum Genet. 2004;74:20–39. doi: 10.1086/380912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abecasis GR, Yashar BM, Zhao Y, et al. Age-related macular degeneration: a high-resolution genome scan for susceptibility loci in a population enriched for late-stage disease. Am J Hum Genet. 2004;74:482–494. doi: 10.1086/382786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beatty S, Koh H, Phil M, et al. The role of oxidative stress in the pathogenesis of age-related macular degeneration. Surv Ophthalmol. 2000;45:115–134. doi: 10.1016/s0039-6257(00)00140-5. [DOI] [PubMed] [Google Scholar]
- 33.Kimura K, Isashiki Y, Sonoda S, et al. Genetic association of manganese superoxide dismutase with exudative age-related macular degeneration. Am J Ophthalmol. 2000;130:769–773. doi: 10.1016/s0002-9394(00)00552-3. [DOI] [PubMed] [Google Scholar]
- 34.Ikeda T, Obayashi H, Hasegawa G, et al. Paraoxonase gene polymorphisms and plasma oxidized low-density lipoprotein level as possible risk factors for exudative age-related macular degeneration. Am J Ophthalmol. 2001;132:191–195. doi: 10.1016/s0002-9394(01)00975-8. [DOI] [PubMed] [Google Scholar]
- 35.Tuo J, Chen C, Zeng X, et al. Functional crosstalk between hOgg1 and the helicase domain of Cockayne syndrome group B protein. DNA Repair (Amst) 2002;1:913–927. doi: 10.1016/s1568-7864(02)00116-7. [DOI] [PubMed] [Google Scholar]
- 36.Liang FQ, Godley BF. Oxidative stress-induced mitochondrial DNA damage in human retinal pigment epithelial cells: a possible mechanism for RPE aging and age-related macular degeneration. Exp Eye Res. 2003;76:397–403. doi: 10.1016/s0014-4835(03)00023-x. [DOI] [PubMed] [Google Scholar]
- 37.Boiteux S, Radicella JP. The human OGG1 gene: structure, functions, and its implication in the process of carcinogenesis. Arch Biochem Biophys. 2000;377:1–8. doi: 10.1006/abbi.2000.1773. [DOI] [PubMed] [Google Scholar]
- 38.Cai J, Nelson KC, Wu M, et al. Oxidative damage and protection of the RPE. Prog Ret Eye Res. 2000;19:205–221. doi: 10.1016/s1350-9462(99)00009-9. [DOI] [PubMed] [Google Scholar]
- 39.van Brabant AJ, Stan R, Ellis NA. DNA helicases, genomic instability, and human genetic disease. Ann Rev Gen Hum Genet. 2000;1:409–459. doi: 10.1146/annurev.genom.1.1.409. [DOI] [PubMed] [Google Scholar]
- 40.Tuo J, Jaruga P, Rodriguez H, et al. The Cockayne syndrome group B gene product is involved in cellular repair of 8-hydroxyadenine in DNA. J Biol Chem. 2002;277:30832–30837. doi: 10.1074/jbc.M204814200. [DOI] [PubMed] [Google Scholar]
- 41.Tuo J, Muftuoglu M, Chen C, et al. The Cockayne syndrome group B gene product is involved in general genome base excision repair of 8-hydroxyguanine in DNA. J Biol Chem. 2001;276:45772–45779. doi: 10.1074/jbc.M107888200. [DOI] [PubMed] [Google Scholar]
- 42.Tuo J, Jaruga P, Rodriguez H, et al. Primary fibroblasts of Cockayne syndrome patients are defective in cellular repair of 8-hydroxyguanine and 8-hydroxyadenine resulting from oxidative stress. FASEB J. 2003;17:668–674. doi: 10.1096/fj.02-0851com. [DOI] [PubMed] [Google Scholar]
- 43.Ambati J, Anand A, Fernandez S, et al. An animal model of age-related macular degeneration in senescent Ccl-2- or Ccr-2-deficient mice. Nat Med. 2003;9:1390–1397. doi: 10.1038/nm950. [DOI] [PubMed] [Google Scholar]
- 44.Tuo J, Smith B, Bojanowski CM, et al. The involvement of sequence variation and expression of CX3CR1 in the pathogenesis of age-related macular degeneration. FASEB J. 2004;18:1297–1299. doi: 10.1096/fj.04-1862fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McDermott DH, Fong AM, Yang Q, et al. Chemokine receptor mutant CX3CR1-M280 has impaired adhesive function and correlates with protection from cardiovascular disease in humans. J Clin Invest. 2003;111:1241–1250. doi: 10.1172/JCI16790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moatti D, Faure S, Fumeron F, et al. Polymorphism in the fractalkine receptor CX3CR1 as a genetic risk factor for coronary artery disease. Blood. 2001;97:1925–1928. doi: 10.1182/blood.v97.7.1925. [DOI] [PubMed] [Google Scholar]
- 47.Gbadegesin RA, Watson CJ, Cotton SA, et al. A PCR-RFLP typing method for adhesion molecule gene polymorphisms and allele frequencies in a normal UK population. Eur J Immunogenet. 2002;29:109–111. doi: 10.1046/j.1365-2370.2002.00277.x. [DOI] [PubMed] [Google Scholar]
- 48.Zheng F, Chevalier JA, Zhang LQ, et al. An HphI polymorphism in the E-selectin gene is associated with premature coronary artery disease. Clin Genet. 2001;59:58–64. doi: 10.1034/j.1399-0004.2001.590110.x. [DOI] [PubMed] [Google Scholar]