Abstract
Heterosis is the phenomenon whereby the progeny of particular inbred lines have enhanced agronomic performance relative to both parents. Although several hypotheses have been proposed to explain this fundamental biological phenomenon, the responsible molecular mechanisms have not been determined. The maize inbred lines B73 and Mo17 produce a heterotic F1 hybrid. Global patterns of gene expression were compared in seedlings of these three genotypes by using a microarray that contains 13,999 cDNAs. Using an estimated 15% false discovery rate as a cutoff, 1,367 ESTs (9.8%) were identified as being significantly differentially expressed among genotypes. All possible modes of gene action were observed, including additivity, high- and low-parent dominance, underdominance, and overdominance. The largest proportion of the ESTs (78%; 1,062 of 1,367) exhibited expression patterns that are not statistically distinguishable from additivity. Even so, 22% of the differentially regulated ESTs exhibited nonadditive modes of gene expression. Classified on the basis of significant pairwise comparisons of genotype means, 181 of these 305 nonadditive ESTs exhibited high-parent dominance and 23 exhibited low-parent dominance. In addition, 44 ESTs exhibited underdominance or overdominance. These findings are consistent with the hypothesis that multiple molecular mechanisms, including overdominance, contribute to heterosis.
Keywords: global transcript profiling, heterosis, overdominance
The hybrid progeny of selected pairs of inbred lines exhibit enhanced agronomic performance relative to both parents (1), a phenomenon that is termed heterosis or hybrid vigor. Heterosis is widely exploited in applied plant breeding. For example, ≈95% of U.S. maize acreage is planted to hybrids. Duvick (2) estimates that maize hybrids exhibit a 15% yield advantage relative to superior open-pollinated varieties and that worldwide heterosis accounts for an additional 55 million metric tons of grain yield annually. Despite the fact that heterosis has been widely exploited by plant breeders to the benefit of agriculture and society, the molecular mechanisms responsible for this basic biological phenomenon are not well understood.
Multiple models have been proposed to explain heterosis (3). The two most commonly invoked are the dominance (or complementation) hypothesis and the overdominance hypothesis. The first hypothesis (4, 5) states that deleterious alleles at different loci in the two homozygous parental genomes are complemented in the heterozygous F1 hybrid. More recently, the special case that complementation of genes that differ in their presence and absence among maize lines may contribute to heterosis has been proposed (6). Complementation cannot by itself, however, explain heterosis because although the per se performance of inbred lines can be improved by purging them of detrimental alleles, doing so has little impact on heterosis (3). Additional evidence for this view comes from the findings that progressively more heterosis occurs in polyploids as the diversity of the component genomes increases and inbreeding depression in autotetraploids increases faster than homozygosity.
The overdominance hypothesis (1, 5, 7) states that the improved performance of an F1 hybrid relative to its inbred parents is a consequence of favorable allelic interactions at heterozygous loci that outperform either homozygous state. Although these classical hypotheses have provided guidance for experimentation (8–11), it is likely that heterosis depends on multiple mechanisms, including epigenetic phenomena. It is also possible that differential accumulation of allele-specific transcripts in hybrids may contribute to heterosis (12).
It has been hypothesized that differential gene expression in inbreds and hybrids may be responsible for heterosis (13, 14). For example, a hybrid could accumulate levels of transcript equal to the mid-parent (additivity), the high or low parent (high or low parent dominance), above the high parent (overdominance), or below the low parent (underdominance). Prior studies of gene expression in inbreds and their F1 hybrids have focused on relatively few genes.
Here, we apply global transcript profiling technology to examine the expression of thousands of genes in two inbred parents and their F1 hybrid to begin to understand the underlying mechanisms and complex regulatory network surrounding heterosis.
More than 1,300 ESTs exhibited significant differential expression patterns among the three genotypes at an estimated false discovery rate (FDR) of 15%. The most common mode of action was additivity, but several hundred genes exhibited high- or low-parent dominant, overdominant, or underdominant modes of gene action. The expression patterns of >90% of sampled genes were validated by using quantitative real-time PCR (qRT-PCR). The finding that all modes of gene action can be detected in inbreds and their F1 hybrid is consistent with the hypothesis that multiple molecular mechanisms, including overdominance, contribute to heterosis.
Results
The maize F1 hybrid generated by crossing the inbred lines B73 and Mo17 is taller, matures more quickly, and produces higher grain yields than both parents (15). We elected to analyze global patterns of gene expression in these three genotypes because this hybrid and its relatives are widely grown in the Corn Belt (16) and the genetic map of maize is based on recombinant inbreds developed from this hybrid.
Because heterosis affects most aspects of plant growth and development, one of the challenges in designing such an experiment is deciding which tissue to analyze. In making this decision, we sought a system in which we could tightly control environmental variability and that, therefore, would provide the statistical power to detect even subtle changes in gene expression that nevertheless may be biologically relevant. We elected to analyze seedlings because seedling dry weight exhibits a substantial degree of heterosis (Table 1), and seedlings can be grown under controlled conditions (see Methods). Although the B73 × Mo17 hybrid is used commercially, the Mo17 × B73 hybrid exhibits a greater degree of heterosis for seedling dry weight (Table 1) and, therefore, was selected for the profiling experiments. Above-ground seedling tissue was harvested from the three genotypes 14 days after planting. RNA extracted from these seedlings was reverse transcribed, labeled with fluorescent dyes, and hybridized to a cDNA array that contains 13,999 informative spots (see Methods). Nine biological replications were analyzed to provide a higher degree of statistical power.
Table 1.
Heterosis for seedling dry weight
| Genotype | Mean seedling dry weight, g |
|---|---|
| B73 | 0.351 ± 0.092 |
| Mo17 | 0.310 ± 0.082 |
| B73 × Mo17 | 0.392 ± 0.085 |
| Mo17 × B73 | 0.517 ± 0.078 |
Seedlings of each genotype were grown under the conditions described in Methods and harvested 14 days after planting. B73 × Mo17 and Mo17 × B73 designate reciprocal hybrids in which the female parent is B73 or Mo17, respectively. Dry weights were determined for 36 individual seedlings per genotype.
Statistical Analysis of Microarray Hybridization Data.
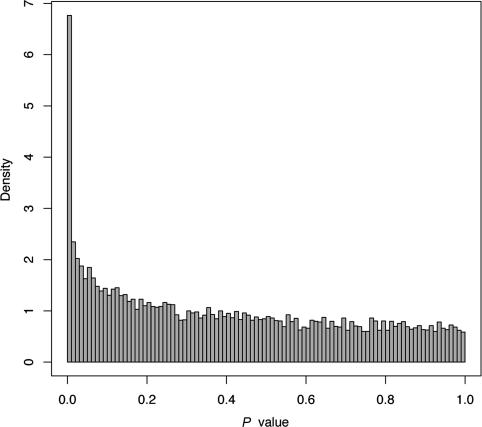
Data normalization and transformation were performed to reduce nonbiological variation, make signal intensities comparable across arrays, and achieve approximate normality and constant variance for statistical modeling (see Methods). A mixed model analysis of the data revealed genes having significant differences in gene expression levels in at least one of the three genotypes. The distribution of P values generated from the tests for genotype (Fig. 1) was used to detect genes with significant differential expression across genotypes. Multiple significance thresholds were investigated, providing significance lists of lengths 280 (1% FDR), 460 (3%), 990 (10%), and 1,367 (15%) genes. The estimated 15% FDR level was chosen to sample a large pool of significant genes for further analyses. At this threshold, 9.8% (1,367 of 13,999) of informative cDNAs on the microarray were differentially expressed among the genotypes. As a group, these genes are involved in a wide variety of cellular processes. The statistical power of this experiment made it possible to detect even small changes in gene expression. For example, the significant fold changes between the low- and high-expressing genotypes for the 1,367 differentially expressed ESTs ranged from 1.2 to 88 (Fig. 3, which is published as supporting information on the PNAS web site).
Fig. 1.
Distribution of P values for the 13,999 gene-specific hypothesis tests for equality of means across genotypes.
Analysis of Gene Action.
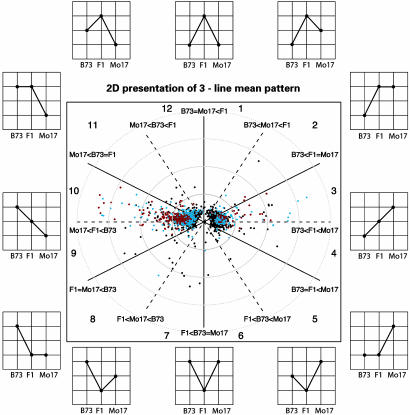
The 1,367 ESTs identified as differentially expressed at the 15% FDR level were further investigated to determine their modes of gene action (Table 4, which is published as supporting information on the PNAS web site). The estimated genotype means from the mixed model analyses were used to determine the expression pattern for each EST. To visualize the patterns of gene expression, a 2D polar coordinate plot of the 3D line mean patterns was implemented (Fig. 2). The radius at which a gene is plotted represents the log2 fold change between the highest and lowest expression levels among the three genotypes. The angle at which a gene is plotted represents the relationships among the means of the three genotypes. A gene plotted on the horizontal line exhibits pure additivity. A gene plotted on the vertical line in the top or bottom half of the figure exhibits overdominance or underdominance, respectively. A gene plotted on a solid line exhibits the same expression level in two genotypes, and a different expression level in a third genotype (see plot labels for direction of differential expression). A gene plotted between lines exhibits a mode of gene action that is intermediate to that indicated by the two nearest lines.
Fig. 2.
Two-dimensional presentation of gene action and fold changes of ESTs that are differentially expressed in B73, Mo17, and their F1 hybrid. ESTs falling directly on a dashed line between 1 and 7 o’clock, 3 and 9 o’clock, or 5 and 11 o’clock exhibited differences in expression from the low to the middle expressing genotype that is equivalent to the change from the middle to the high expressing genotype. ESTs falling on the horizontal and vertical lines exhibited pure additivity and over- (or under-) dominance, respectively (see plot labels). The radius at which an EST is plotted represents the log2 of the fold change between the high- and low-expressing genotypes. ESTs associated with a FDR of 1%, 5%, and 15% are shown in red, blue, and black, respectively. To provide better resolution for those of the 1,367 differentially expressed ESTs with smaller fold changes, only the 1,361 ESTs that exhibited changes of <16-fold are plotted. The remaining six ESTs are listed in Table 4.
A large proportion of the ESTs (78%, 1,062 of 1,367) exhibited expression patterns that are not statistically distinguishable from additivity. The 305 ESTs that exhibited nonadditive mode of gene expression were further classified based on significant pairwise comparisons (P < 0.05). Of these 305 ESTs, 181 exhibited high-parent dominance and 23 exhibited low-parent dominance. Twelve ESTs exhibited modes of gene action intermediate between additivity and dominance. In addition, 34 ESTs exhibited clear overdominance, and 10 exhibited clear underdominance. Although it was possible to conclude that the remaining 45 ESTs exhibited nonadditive gene action, there was insufficient statistical power to assign them to one of the above categories. Genes that exhibited overdominance and underdominance exhibited near parent fold changes of 1.2–2.0 and 1.3–4.7, respectively, and are involved in multiple cellular processes (Table 2).
Table 2.
The 44 ESTs that exhibited overdominant or underdominant gene action in the microarray analysis
| GenBank accession no.* | blast results (e-value)† | Significant pattern‡ | NP P value§ | NP fold change¶ |
|---|---|---|---|---|
| Overdominant gene action | ||||
| BG841554 | Unknown protein (6 e−41) | B≈M<F | 0.0313 | 2.01 |
| CB334498 | ns | B≈M<F | 0.0060 | 0.63 |
| DV550679 | ns | B≈M<F | 0.0140 | 1.42 |
| DV489625 | Putative serine-threonine protein kinase (1 e−42) | B≈M<F | 0.0063 | 1.40 |
| DV496092 | Putative peroxidase (2 e−14) | B≈M<F | 0.0163 | 1.40 |
| CD661986 | Putative jasmonate-induced protein (2 e−19) | B≈M<F | 0.0185 | 1.35 |
| CD484960 | β-glucosidase aggregating factor precursor (2 e−37) | M<B<F | 0.0007 | 1.33 |
| DV494333 | Pre-mRNA splicing factor (3 e−15) | B≈M<F | 0.0036 | 1.29 |
| DV549450 | Putative SSR α subunit (9 e−75) | B≈M<F | 0.0156 | 1.26 |
| CD651121 | β-glucosidase aggregating factor precursor (3 e−35) | B≈M<F | 0.0496 | 1.26 |
| DV549419 | Single-strand DNA endonuclease (4 e−6) | B≈M<F | 0.0271 | 1.25 |
| DV550393 | Unknown protein (2 e−11) | B≈M<F | 0.0393 | 1.25 |
| CB331016 | Putative glucose-1-phosphate adenyltransferase (6 e−13) | B≈M<F | 0.0311 | 1.24 |
| BM379680 | Superoxide dismutase (1 e−43) | B≈M<F | 0.0104 | 1.24 |
| CB280807 | Proteasome component (2 e−18) | B≈M<F | 0.0034 | 1.24 |
| DV491691 | ns | B≈M<F | 0.0383 | 1.23 |
| CB603924 | Putative jasmonate-induced protein (2 e−19) | B≈M<F | 0.0284 | 1.23 |
| DV943290 | Calcineurin B protein (3 e−51) | B≈M<F | 0.0205 | 1.23 |
| BM080645 | Unknown protein (1 e−10) | M<B<F | 0.0433 | 1.22 |
| CB381444 | Pollen 2-phosphoglycerate dehydrogenase 2 precursor (6 e−92) | B≈M<F | 0.0103 | 1.22 |
| DV622265 | ADP-ribosylation factor (5 e−81) | B≈M<F | 0.0349 | 1.21 |
| DV489865 | Expressed protein (2 e−39) | B≈M<F | 0.0154 | 1.21 |
| CB617229 | Superoxide dismutase (2 e−84) | B≈M<F | 0.0107 | 1.20 |
| CD651750 | β-glucosidase aggregating factor precursor (4 e−22) | B≈M<F | 0.0430 | 1.20 |
| BM073302 | Superoxide dismutase (7 e−74) | B≈M<F | 0.0182 | 1.19 |
| CB381307 | Putative 20-kDa chaperonin (7 e−47) | B≈M<F | 0.0286 | 1.19 |
| CB331033 | Putative translation elongation factor (2 e−30) | B≈M<F | 0.0495 | 1.18 |
| CB329753 | Superoxide dismutase (4e−77) | B≈M<F | 0.0330 | 1.18 |
| CB380870 | Putative jasmonate-induced protein (6 e−19) | M<B<F | 0.0240 | 1.17 |
| CB605313 | β-glucosidase aggregating factor precursor (6 e−22) | B≈M<F | 0.0427 | 1.17 |
| CB886104 | Histone H2B (4 e−35) | B≈M<F | 0.0315 | 1.17 |
| CD001350 | Nucleosome chromatin assembly factor group D protein (7 e−12) | B≈M<F | 0.0200 | 1.16 |
| CD485186 | DNA-binding protein (1 e−22) | B≈M<F | 0.0344 | 1.16 |
| DV492982 | Putative translation elongation factor (5 e−40) | B≈M<F | 0.0496 | 1.16 |
| Underdominant gene action | ||||
| DV493742 | Zein (2 e−67) | F<B≈M | 0.0137 | 4.27 |
| CB281986 | Putative nucleoside diphosphate kinase (7 e−31) | F<B≈M | 0.0201 | 2.71 |
| BM337640 | 15-kDa beta zein (8 e−44) | F<B≈M | 0.0074 | 3.85 |
| AI612441 | ns | F<B≈M | 0.0162 | 3.45 |
| BM351629 | ns | F<B≈M | 0.0433 | 2.46 |
| BM073434 | Pathogenesis-related protein (5 e−66) | F<B≈M | 0.0002 | 2.00 |
| DV489785 | Multidrug resistance protein (6 e−19) | F<B≈M | 0.0047 | 1.45 |
| DV942972 | ns | F<B≈M | 0.0068 | 1.61 |
| BM073507 | ns | F<B≈M | 0.0252 | 1.58 |
| DV622486 | Branched silkless 1 (2 e−9) | F<B≈M | 0.0341 | 1.24 |
The P value for equality of means among the three genotypes was <0.0212 for all listed ESTs. The ESTs are listed in decreasing order of near-parent fold changes.
*Based on sequence similarity, the following groups of ESTs appear to correspond to single genes: CD661986, CB603924, CD651750, CB380870, and CB605313; CD484960 and CD651121; BM379680, CB617229, and BM073302; and CD001350 and CD485186.
†Individual ESTs or the corresponding EST contigs (if available) were screened against a copy of the NCBI nr database downloaded February 8, 2006 by using blastx. ns indicates no significant blast hits by using an e-value cutoff of 1 e−5.
‡≈ indicates a failure to reject the null hypothesis that the values of the indicated genotypes are identical at P < 0.05. B, B73; M, Mo17; F, F1.
§Near-parent P value from equality of means test between F1 and parent with the nearest level of gene expression.
¶Fold change between F1 and parent with the nearest level of gene expression shown as a ratio. For overdominant genes, the ratio of F1 to near parent expression level is listed; for underdominant genes, the ratio of the near parent to F1 expression level is listed.
Validation of Modes of Gene Action via qRT-PCR.
A sample of 45 genes identified in the mixed model analysis as having significant differences in gene expression across genotypes was selected for validation via qRT-PCR (Table 3). Genes that exhibited a variety of modes of gene action (i.e., from all 12 sectors of Fig. 2) were chosen for validation. Selected genes exhibited changes in gene expression from the low- to high-expressing genotype of 1.4- to 88-fold.
Table 3.
The 35 genes identified as being differentially regulated among genotypes in the microarray analysis that had significant qRT-PCR validation results
| GenBank accession no. | blast results (e-value)* | Microarray results |
qRT-PCR results |
|||
|---|---|---|---|---|---|---|
| Fold change† | Sector‡ | Significant pattern§ | P value | Significant pattern | ||
| Equivalent gene action observed in microarray and qRT-PCR experiments | ||||||
| BM073941 | β-amyrin synthase (2 e−48) | 87.98 | 10 | M<B≈F | <0.0001 | M<B≈F |
| BM340381 | ns | 10.73 | 10 | M<B≈F | 0.0008 | M<B≈F |
| BM338817 | ns | 9.79 | 10 | M<B≈F | 0.0014 | M<B≈F |
| BM074072 | Heme A:farnesyltransferase (1 e−14) | 9.37 | 10 | M<F<B | <0.0001 | M<F<B |
| BM334691 | ns | 6.64 | 3 | B<M≈F | 0.0053 | B<M≈F |
| BM073390 | ns | 6.27 | 3 | B<F<M | <0.0001 | B<F<M |
| BM337350 | ns | 5.06 | 10 | M<B≈F | <0.0001 | M<B≈F |
| BM079864 | Circulin B (8 e−07) | 4.31 | 3 | B<F<M | <0.0001 | B<F<M |
| DV942972 | ns | 3.86 | 9 | M<F<B | 0.0002 | M<F<B |
| DV550757 | Unknown protein (2 e−59) | 3.47 | 3 | B<F<M | <0.0001 | B<F<M |
| DV549373 | Putative chloroplast 50S ribosomal protein (4 e−30) | 2.40 | 11 | M<B≈F | <0.0001 | M<B≈F |
| BM080645 | Unknown protein (1 e−10) | 2.08 | 11 | M<B<F | 0.0002 | M<B<F |
| BM073434 | Pathogenesis-related protein (5 e−66) | 2.05 | 6 | F<B≈M | 0.0035 | F<B≈M |
| BM072868 | Unknown protein (2 e−12) | 2.01 | 11 | M<B≈F | 0.0001 | M<B<F |
| DV490892 | ns | 1.76 | 2 | B<M≈F | <0.0001 | B<M≈F |
| Detectable gene action patterns are consistent in microarray and qRT-PCR experiments | ||||||
| BM348583 | Putative cytochrome P450 (2 e−6) | 26.00 | 10 | M<B≈F | <0.0001 | M<F<B |
| BM073611 | ns | 22.77 | 10 | M<B≈F | <0.0001 | M<F<B |
| BG841239 | Unknown protein (4 e−76) | 17.07 | 10 | M<F<B | 0.0260 | M<B≈F |
| BM073284 | Circulin B (7 e−7) | 16.57 | 3 | B<M≈F | <0.0001 | B<F<M |
| BM073916 | ns | 5.01 | 9 | F≈M<B | <0.0001 | M<F<B |
| BM336730 | ns | 4.18 | 9 | F≈M<B | <0.0001 | F≈M<B |
| BM337359 | Putative wound-inducive gene (1 e−14) | 3.42 | 8 | F<B | <0.0001 | M<F<B |
| BM337880 | ns | 3.26 | 8 | F≈M<B | <0.0001 | M<F<B |
| DV489988 | Cytosolic aldehyde dehydrogenase RF2D (2 e−49) | 2.26 | 2 | B<M≈F | <0.0001 | B<F<M |
| DV489868 | Putative MYB29 protein (2 e−48) | 2.24 | 4 | F≈B<M | <0.0001 | B<F<M |
| AI861151 | Putative Xa1-like protein (2 e−68) | 2.22 | 4 | B<F<M | 0.0107 | F≈B<M |
| DV942867 | ns | 2.20 | 8 | F≈M<B | <0.0001 | M<F<B |
| BG842276 | Unknown protein (1 e−10) | 2.05 | 11 | M<B≈F | <0.0001 | M<B<F |
| BG841156 | Carboxypeptidase D (7 e−74) | 2.02 | 2 | B<M≈F | <0.0001 | B<F<M |
| CB334498 | ns | 1.65 | 1 | B≈M<F | 0.0423 | M<F |
| CD661986 | Putative 32.7-kDa jasmonate-induced protein (2 e−19) | 1.59 | 12 | B≈M<F | 0.0385 | B<F |
| DV489625 | Putative serine-threonine protein kinase (1 e−42) | 1.41 | 12 | B≈M<F | 0.0411 | B<F |
| Different modes of gene action detected in microarray and qRT-PCR experiments | ||||||
| BG841472 | Putative cystatin (3 e−61) | 4.66 | 3 | B<M≈F | <0.0001 | M<B≈F |
| BM073340 | Nonspecific lipid transfer protein (5 e−23) | 2.92 | 3 | B<M≈F | <0.0001 | M<B≈F |
| BM268642 | Transcription factor MADS57 (1 e−23) | 2.14 | 11 | M<B≈F | <0.0001 | B<F<M |
The 10 genes with nonsignificant patterns in the qRT-PCR experiment are listed in Table 5.
*Individual ESTs or the corresponding EST contigs (if available) were screened against a copy of the NCBI nr database downloaded February 8, 2006 by using blastx. ns indicates no significant blast hits by using an e-value cutoff of 1 e−5.
†Fold changes were calculated between highest- and lowest-expressing genotypes.
‡Sector location in Fig. 2.
§≈ indicates a failure to reject the null hypothesis that the values of the indicated genotypes are identical at P < 0.05. B, B73; M, Mo17; F, F1.
Primers were designed to specifically amplify each of the 45 genes (see Methods). These primers were used to conduct qRT-PCR on seven biological replications of RNA from the three genotypes. Using threshold cycle (Ct) values generated from qRT-PCR experiments (see Methods), the null hypothesis of equal expression across the B73, Mo17, and F1 genotypes was tested. For 10 genes, there was not sufficient statistical power in the qRT-PCR experiments to determine a mode of gene action (Table 5, which is published as supporting information on the PNAS web site). There was sufficient statistical power to make conclusions regarding modes of gene action for the remaining 35 genes. For 15 of these genes, the modes of gene action detected in the qRT-PCR experiments were indistinguishable from the modes based on the microarray experiment (P < 0.05). For 17 of the genes, the mode of gene action obtained via the qRT-PCR experiments were at least consistent with the modes obtained from the microarray experiment. Hence, the qRT-PCR experiments either validated or were consistent with the modes of gene action exhibited by 91% (32 of 35) of the genes in the microarray experiment.
Discussion
Despite its critical importance to agriculture, a mechanistic understanding of heterosis has not been achieved. As a step toward generating data needed to test existing hypotheses, prior studies have analyzed modes of gene action in small sets of maize genes (13, 17). Even so, a global understanding of the behavior of gene expression in inbreds and their F1 hybrids is lacking. The current study used microarray technology to characterize the modes of gene action for 13,999 cDNAs.
Approximately 9.8% (1,367 of 13,999) of the ESTs assayed in this experiment exhibited differential expression among the three genotypes. The majority of these ESTs (n = 1,062) exhibited modes of gene action that could not be distinguished from additivity. The expression of these genes could be controlled by cis-acting regulatory elements and/or dosage-dependent trans-acting factors. The large number of genes that exhibited additive gene action is consistent with the complementation hypothesis of heterosis.
Approximately 22% (305 of 1,367) of the differentially regulated ESTs detected in this study exhibited nonadditive modes of gene expression. Most of these genes exhibited high-parent dominance (n = 181). However, low-parent dominance (n = 23), underdominance (n = 10), and overdominance (n = 34) also were observed. It was possible to validate via qRT-PCR the modes of gene action exhibited by 91% (32 of 35) of a sample of the differentially expressed genes.
Overall, 2.2% (305 of 13,999) of the ESTs analyzed in this survey of nearly 14,000 cDNAs exhibited nonadditive modes of gene action. These results differ substantially from a prior study of a smaller set of genes. Auger et al. (17) reported that 19 of the 30 genes (63%) exhibited nonadditive gene action. Although these two studies were conducted by using the same genotypes, Auger et al. (17) used gel blots to analyze RNA extracted from leaves of adult field-grown plants, whereas we used microarrays to analyze gene expression in seedlings grown under highly controlled environmental conditions. One explanation for the different rates of nonadditive gene expression observed in the two studies is differential sampling of the maize gene space. The 30 genes studied by Auger et al. (17) may be a less random sample than the 13,999 cDNAs present on our microarray. Alternatively (or in addition), the percentage of genes that exhibit nonadditive gene action may differ during development even though both stages of development analyzed in these studies exhibit heterosis. The nonadditive expression of these genes could be explained by dominant allelic and nonallelic epistatic control of transcript accumulation.
The existence of overdominant gene action has important implications for evolutionary theory, in particular the maintenance of genetic variability. The evidence for overdominant gene action however, has been limited so far. Some, but not all, experiments conducted by using Drosophila (18), and more recently by using C. elegans, (19) have uncovered evidence for overdominance. Although the results of the current study in maize are consistent with prevailing views that most loci exhibit additive, or less frequently, dominant gene action, the identification of 34 ESTs that exhibited overdominance suggests that hypothesized genetic processes, including heterosis, that invoke overdominance cannot be excluded from consideration. Although it is not possible to exclude the possibility that the overdominant gene action observed in this study is the result of “pseudooverdominance,” caused by the combined action of linked loci, such blocks of genes would have similar effects on genetic processes as overdominant loci.
Analysis of the microarray experiment resulted in the identification of 44 ESTs that exhibit overdominance or underdominance. qRT-PCR experiments validated these modes of gene action for five of eight tested genes. The existence of genes that exhibit overdominant or underdominant modes of gene action in B73, Mo17, and their F1 hybrid is at least consistent with the overdominance hypothesis of heterosis. Although heterosis is controlled by many genes, only a small fraction of all genes are involved (3). Hence, it is at least possible that some of these genes may contribute to heterosis. Consistent with this hypothesis, genes that exhibit overdominance include those that potentially affect a wide variety of regulatory steps, including splicing, translation, protein folding, modification, and degradation (Table 2); others are involved in stress response. All of these functions could contribute to posttranscriptional regulatory cascades contributing to heterosis.
Among other mechanisms, one attractive hypothesis for the existence of underdominant and overdominant gene action invokes the action of small interfering RNAs (siRNAs). siRNAs are typically derived from transposons and repeats, although some genes and other sequences can generate siRNAs (20). siRNAs can regulate gene expression by cleaving target mRNAs (21) and via transcriptional silencing (22). Maize inbreds differ radically in transposon and repeat content (6, 23, 24). In addition, in this study, at least two transposons exhibited >4-fold differences in expression between B73 and Mo17 (Table 4). Hence, inbreds are likely to differ in their complement of siRNAs. If siRNAs from one inbred do not match genes (e.g., repetitive sequences in 3′ UTRs) from the other inbred, the resulting hybrid could exhibit novel patterns of gene expression, including overdominance or underdominance. Consistent with this hypothesis, we observe profound differences in the accumulation of antisense RNAs in B73 and Mo17 (Y.J., R.A.S.-W., S. J. Emrich, R.D., L. Guo, Y. Fu, D. A. Ashlock, D.N., and P.S.S., unpublished data). Overall, our results are consistent with the hypothesis that multiple molecular mechanisms contribute to heterosis.
Methods
Genetic Stocks and Experimental Design.
The inbreds B73 (Schnable laboratory accession no. 660) and Mo17 (Schnable laboratory accession no. 2618) used in this study were derived by self-pollination from stocks originally obtained from Donald Robertson and Mike Lee (Iowa State University), respectively. Mo17 was crossed as a female by B73 to generate the F1. Kernels from three different seed sources (ears) per genotype were used in the experimental design. Individual genotypes within a replication, however, were all derived from the same source. Before microarray analyses, genotypes were confirmed by using codominant IDP genetic markers that distinguish B73 from Mo17 (Y. Fu, T. J. Wen, Y. I. Ronin, D. I. Mester, Y. Yang, M. Lee, A. B. Korol, D. A. Ashlock, and P.S.S., unpublished data). Ten biological replications of B73, Mo17, and their F1 (Mo17 × B73) were grown under highly controlled conditions in a randomized complete block design. For each replication, the B73, Mo17, and F1 samples were hybridized to three two-color cDNA microarrays by using a loop design such that each loop included all pairwise comparisons between genotypes. RNA pools for each genotype were alternately labeled providing dye balance within each loop. After hybridization, one biological replicate was removed because of poor quality. The final analysis incorporated 27 microarray slides (three slides for each of nine high-quality biological replicates).
Plant Growth and RNA Isolation.
Kernels were planted in SB 300 Universal soil (Sun Gro Horticulture, Bellevue, WA) within a PGW-40 (Percival Scientific, Perry, IA) growth chamber that provided 15 h of light (25°C) and 9 h of dark (20°C). Light intensity was ≈650–800 μmol·m−2·s−1. Seedlings were watered as needed by using a 0.7 M calcium nitrate solution. Fourteen days after planting, six random healthy plants were harvested as a pool for each genotype-replication. All aboveground tissue was separated from root tissue and immediately submerged in liquid nitrogen. After separately grinding each genotype-replication pool in liquid nitrogen, RNA was extracted from ≈10 g of frozen tissue by using TRIzol reagent (Invitrogen) as per the manufacturer’s instructions, with slight modifications. RNA integrity was confirmed via gel electrophoresis. The OligoTex mRNA midi kit (Qiagen, Valencia, CA) was used to extract mRNA from 500 μg of RNA by using the manufacturer’s protocol, with slight modifications. mRNA yields were typically between 0.75 and 1.5% of the starting RNA.
Microarray Printing.
The SAM1.1 cDNA array was printed on the UltraGAPs slide (Corning, Corning, NY) by using a PixSYS 5500 Arrayer (Cartesian Technologies, Irvine, CA). The GEO platform file for this chip is posted at www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi?acc = GPL2613.
Reverse Transcription, Labeling, and Microarray Hybridization.
Two μg of mRNA were labeled according to Nakazono et al. (25), with slight modifications. Specific cDNA samples were labeled with Cy3 and Cy5 fluorescent dyes in accordance with the experimental design.
Microarray Data Acquisition, Normalization, and Analysis.
Replications 1–5 were scanned by using a ScanArray 5000 (Packard). Replications 6–9 were scanned by using a PerkinElmer Pro Scan Array HT. A minimum of six scans for each dye channel were completed at increasing photomultipler tube gain and laser power settings. Only one of these scans was selected for analysis for each channel per slide. A set of scans was selected that had similar natural logged median intensity values for the Cy3 and Cy5 channels of each individual slide and across all slides.
The lowess normalization method was applied to the log of background-corrected raw signal intensities to remove signal-intensity-dependent dye effects on each slide (26, 27). Normalization was conducted separately for each slide to avoid introducing dependencies among biological replications. After lowess normalization, the normalized data for each slide/dye combination were median-centered so that expression measures would be comparable across slides. Median centering involves subtracting the median value for a particular slide/dye combination from each individual value associated with the particular slide/dye combination. Thus, negative (positive) values indicate that a particular transcript was expressed below (above) the median for a particular slide/dye combination.
The SAM1.1 maize cDNA array chip contains 19,200 spots. Before statistical analysis 897 “empty” and “bad-PCR” spots were removed from the data set. For each of the remaining 18,303 spots on the microarray, a mixed linear model analysis (28) of the normalized, log-scale signal intensities was conducted to identify transcripts whose expression differed significantly among genotypes. The mixed linear model included fixed effects of genotype and dye and random effects related to the experimental design. The P values generated from the tests for line effect (testing for equality of the three genotype means) were used to determine significance of differential expression. The estimated means from the mixed model for each genotype were used to identify the mode of gene action for all significant genes. Each gene was classified into a significant pattern category by using pairwise comparison tests (P < 0.05). P values from the linear-in-genotype contrasts (testing for F1 genotype mean equal to the average of the two parental line means) from the mixed models were used to classify significant genes into the categories of not distinguishable from additivity (P > 0.05) and distinguishable from additivity (P < 0.05). Genes in the latter group were further classified into more specific nonadditive categories by using the aforementioned significant patterns. Genes with an F1 genotype mean not significantly different from one parent and significantly larger (smaller) than the other parent were said to exhibit high-parent (low-parent) dominance. Genes with an F1 genotype mean that was significantly larger (smaller) than both B73 and Mo17 were said to exhibit “clear” overdominance (underdominance).
By following the statistical analysis, an additional 4,112 spots were removed from the data set because of concerns regarding the quality of the associated DNA sequences and 192 exogenous spots also were removed. As a result, this study reports the gene expression patterns of 13,999 “informative” spots.
Validation of Gene Expression via qRT-PCR.
Primers were designed to amplify a sample of genes that exhibited statistically significant genotype effects in the analysis of the microarray data. Individual ESTs or EST contigs (if available) were compared to the MAGI 4.0 database (29) of assembled maize genomic sequences by using blast (30). The primers were designed by using primer3 (31). The design parameters were used as follows: Tm, 58°C to 61°C, no difference >2°C between the primers in a pair; primer length, 19–24 bp; GC content, 45–55%; amplicon length, 100–200 bp. Whenever possible, primers were designed to span introns. Only primer pairs having high scoring matches to a single MAGI were synthesized (Integrated DNA Technologies, Coralville, IA). Only primers yielding a single product in conventional PCR and qRT-PCR were used in the validation experiment.
RNA samples from seven biological replications of B73, Mo17, and the F1 were treated with RNase-free DNase I (Stratagene), extracted with 1:1 phenol:chloroform, and purified with the RNeasy Mini Kit (Qiagen). qRT-PCR was conducted by using an Mx4000 multiplex quantitative PCR system (Stratagene). A human gene (GenBank accession no. AA418251) was spiked into each reaction as an external reference for data normalization.
qRT-PCR data were initially analyzed by using mx4000 analysis software. Genotype-specific Ct values for each gene and control were calculated by using baseline-corrected, ROX-normalized parameters. Three technical replicates were included in each plate, and the average Ct value for each genotype was normalized within a plate to the human control gene by computing ΔCt,genotype = Ct,genotype –Ct,genotype(control) (32). The ΔCt,genotype values from the seven biological replicates were analyzed with sas statistical software (SAS, Cary, NC) by using a mixed linear model that included the fixed effect of genotype and random effects relevant to the experimental design. The fixed effect of genotype (B73, Mo17, and F1) was tested for significance (P < 0.05) and genes were classified into significant patterns by using pairwise comparison tests (P < 0.05).
Supplementary Material
Acknowledgments
We thank Nathan Springer, Jim Birchler, and Rob Martienssen for fruitful discussions; Karthik Viswanathan, Pengcheng Lu, and Cheng-Ting Yeh for computational support; Sang-Duck Seo for implementing the design of Fig. 2; and an anonymous reviewer for useful suggestions regarding this figure. This research was funded in part by the Iowa State University Plant Sciences Institute, Hatch Act, and State of Iowa funds.
Abbreviations
- FDR
false discovery rate
- qRT-PCR
quantitative real-time PCR
- siRNA
small interfering RNA.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The microarray data have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE3733).
References
- 1.Shull G. H. Am. Breeders Assoc. 1908;4:296–301. [Google Scholar]
- 2.Duvick D. N. In: The Genetics and Exploitation of Heterosis in Crops. Coors J. G., Pandey S., editors. Madison, WI: Crop Sci. Soc. Amer.; 1999. pp. 19–29. [Google Scholar]
- 3.Birchler J. A., Auger D. L, Riddle N. C. Plant Cell. 2003;15:2236–2239. doi: 10.1105/tpc.151030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davenport C. B. Science. 1908;28:454–455. doi: 10.1126/science.28.718.454-b. [DOI] [PubMed] [Google Scholar]
- 5.Crow J. F. In: Heterosis. Gowen J. W., editor. Ames: Iowa State College Press; 1952. pp. 282–297. [Google Scholar]
- 6.Fu H., Dooner H. K. Proc. Natl. Acad. Sci. USA. 2002;99:9573–9580. doi: 10.1073/pnas.132259199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.East E. M. Conn. Agric. Exp. Stn. 1907:419–428. [Google Scholar]
- 8.Stuber C. W., Lincoln S. E., Wolff D. W., Helentjaris T., Lander E. S. Genetics. 1992;132:823–839. doi: 10.1093/genetics/132.3.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu H., Romero-Severson J., Bernardo R. Theor. Appl. Genet. 2003;107:494–502. doi: 10.1007/s00122-003-1271-7. [DOI] [PubMed] [Google Scholar]
- 10.Gibson G., Riley-Berger R., Harshman L., Kopp A., Vacha S., Nuzhdin S., Wayne M. Genetics. 2004;167:1791–1799. doi: 10.1534/genetics.104.026583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao J., Li J., Yuan L., Tanksley S. D. Genetics. 1995;140:745–754. doi: 10.1093/genetics/140.2.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo M., Rupe M. A., Danilevskaya O. N., Yang X., Hu Z. Plant J. 2003;36:30–44. doi: 10.1046/j.1365-313x.2003.01852.x. [DOI] [PubMed] [Google Scholar]
- 13.Song R., Messing J. Proc. Natl. Acad. Sci. USA. 2003;100:9055–9060. doi: 10.1073/pnas.1032999100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hubner N., Wallace C. A., Zimdahl H., Petretto E., Schulz H., Maciver F., Mueller M., Hummel O., Monti J., Zidek V., et al. Genetics. 2005;37:243–253. doi: 10.1038/ng1522. [DOI] [PubMed] [Google Scholar]
- 15.Hallauer A. R., Miranda J. B. in Quantitative Genetics in Maize Breeding. Ames: Iowa State Univ. Press; 1981. [Google Scholar]
- 16.Lu H., Bernardo R. Theor. Appl. Genet. 2001;103:613–617. [Google Scholar]
- 17.Auger D. L., Gray A. D., Ream T. S., Kato A., Coe E. H., Jr., Birchler J. A. Genetics. 2005;169:389–397. doi: 10.1534/genetics.104.032987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewontin R. C. in The Genetic Basis of Evolutionary Change. New York: Columbia Univ. Press; 1974. [Google Scholar]
- 19.Peters A. D., Halligan D. L., Whitlock M. C., Keightley P. D. Genetics. 2003;165:1031–1032. doi: 10.1093/genetics/165.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaughn M. W., Martienssen R. A. Science. 2005;309:1525–1526. doi: 10.1126/science.1117805. [DOI] [PubMed] [Google Scholar]
- 21.Hammond S. M. FEBS Lett. 2005;579:5822–5829. doi: 10.1016/j.febslet.2005.08.079. [DOI] [PubMed] [Google Scholar]
- 22.Lippman Z., Martienssen R. A. Nature. 2004;431:364–370. doi: 10.1038/nature02875. [DOI] [PubMed] [Google Scholar]
- 23.Yao H., Zhou Q., Li J., Smith H., Yandeau M., Nikolau B. J., Schnable P. S. Proc. Natl. Acad. Sci. USA. 2002;99:6157–6162. doi: 10.1073/pnas.082562199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morgante M., Brunner S., Pea G., Fengler K., Zuccolo A., Rafalski A. Nat. Genet. 2005;37:997–1002. doi: 10.1038/ng1615. [DOI] [PubMed] [Google Scholar]
- 25.Nakazono M., Qiu F., Borsuk L. A., Schnable P. S. Plant Cell. 2003;15:583–596. doi: 10.1105/tpc.008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dudoit S., Yang Y. H., Callow M. J., Speed T. P. Statistica. Sinica. 2002;12:111–140. [Google Scholar]
- 27.Cui X., Kerr M. K., Churchill G. A. Stat. Appl. Genet. Mol. Biol. 2003;2 doi: 10.2202/1544-6115.1009. article 4. [DOI] [PubMed] [Google Scholar]
- 28.Wolfinger R. D., Gibson G., Wolfinger E. D., Bennet L., Hamadeh H., Bushel P., Afshari C., Paules R. S. J. Comput. Biol. 2001;8:625–637. doi: 10.1089/106652701753307520. [DOI] [PubMed] [Google Scholar]
- 29.Fu Y., Emrich S. J., Guo L., Wen T.-J., Ashlock D. A., Aluru S., Schnable P. S. Proc. Natl. Acad. Sci. USA. 2005;102:12282–12287. doi: 10.1073/pnas.0503394102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altschul S. F. J. Mol. Biol. 1991;219:555–565. doi: 10.1016/0022-2836(91)90193-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rozen S., Skaletsky H. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 32.Livak K. J., Schmittgen T. D. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.