Abstract
The transcription factor NFATc1 plays an essential role in transducing signals from RANKL in osteoclast differentiation. To date, however, the specific transcriptional targets of NFATc1 are unknown. Expression of the β3 integrin is required for normal osteoclast function. We therefore examined the role of NFATc1 in human β3 integrin expression in osteoclast differentiation. Analysis of the mouse and human β3 gene promoters revealed considerable sequence homology across a 1.3 kb region upstream of the transcription start site (TSS), with conserved NFAT binding elements present. The region −1242 to +29 (relative to the TSS) was cloned as a luciferase reporter construct (pB3-1.3) and a deletion construct removing to −997 (pB3-1) made. The deletion of 245 bp 5′ removed three conserved NFAT sites including a consensus NFAT:AP-1 site. The pB3-1.3 reporter construct was induced by treatment with RANKL in the range 2.5–40 ng/ml and dose-dependently induced by co-transfection with human NFATc1 in RAW264.7 cells. The pB3-1 deletion construct was minimally induced with RANKL treatment and unresponsive to co-transfected NFATc1. Direct NFAT binding to two of the consensus NFAT sites within this 245 bp 5′ region was demonstrated by EMSA and supershift with anti-NFAT antibodies. Mutation of two of the conserved NFAT sites in the −1242 to −997 fragment was required to prevent binding. The double NFAT mutant, in the context of the full-length promoter was unresponsive to RANKL treatment or co-transfected NFATc1. We generated cell-permeable TAT-dominant-negative (dn)NFATc1 fusion proteins to assess the effect of blockade of NFAT signaling. Transduction with dnNFAT inhibited RANKL induction of the human β3 integrin promoter. Involvement of the NFATc1-calcineurin pathway in regulating the human β3 integrin promoter was further confirmed using the calcineurin pathway inhibitory peptide 11R-VIVIT. Together these results establish the β3 gene as a direct target of NFATc1 in RANKL-dependent osteoclast formation.
Keywords: Transcriptional regulation, Beta 3, Bone, RANKL
Abbreviations: BLAST, basic local alignment search tool; mBMM, mouse bone marrow macrophage; bp, base pairs; CTR, calcitonin receptor; cath K, cathepsin K; dn, dominant negative; TBE, Tris Buffered EDTA; EMSAs, electrophoretic mobility shift assays; HA, hemagglutinin; IPTG, isopropyl-β-d-thiogalactopyranoside; luc, luciferase; NFAT, nuclear factor of activated T cells; OSCAR, osteoclast associated receptor; PBS, phosphate buffered saline; PMA, phorbol 12-myristate 13-acetate; RANKL, receptor activator NFκB ligand; S.D., standard deviation; TSS, transcription start site; WT, wild type
1. Introduction
Osteoclasts are multinucleated cells derived from hematopoietic progenitor cells of the monocyte/macrophage lineage that are unique in their capacity to resorb mineralized matrix (Baron, 1989). Studies have shown that receptor activator NFκB ligand (RANKL) (Kong et al., 1999), in the presence of M-CSF (Yoshida et al., 1990; Kodama et al., 1991; Tanaka et al., 1993), is the essential mediator of osteoclast differentiation. RANKL acts through its receptor RANK to initiate a signaling cascade that is crucial for osteoclast differentiation and activation. The transcription factor nuclear factor of activated T cells (NFATc1) is up-regulated by RANKL and has been identified as playing a crucial role in osteoclast differentiation and function (Ishida et al., 2002; Takayanagi et al., 2002; Hirotani et al., 2004).
NFATc1 is activated by the Ca2+/calmodulin-regulated phosphatase calcineurin (Macian et al., 2001). Over-expression of NFATc1 (Takayanagi et al., 2002) or ectopic expression of constitutively active NFATc1 (Hirotani et al., 2004) is able to bypass the requirement for RANKL in osteoclast differentiation. Selective inhibition of calcineurin-induced NFATc1 activation results in the impaired spreading of TRAP-positive cells and reduced bone-resorbing capacity (Hirotani et al., 2004). Pertinent to the current study are recent findings demonstrating the ability of NFATc1 to induce the expression of various osteoclast genes, including the β3 integrin (Hirotani et al., 2004).
NFATc1 is capable of inducing osteoclast precursors to differentiate into mature osteoclasts, however, the direct and key transcriptional target genes of NFATc1 have not been defined. Recent work has identified NFAT binding sites in the TRAP promoter, osteoclast-specific P3 promoter of the calcitonin receptor (CTR), the cathepsin K (cath K) promoter and the osteoclast associated receptor (OSCAR) promoter (Takayanagi et al., 2002; Matsumoto et al., 2004; Matsuo et al., 2004; Kim et al., 2005a) and demonstrated specific regulation of the promoters by NFATc1 (Anusaksathien et al., 2001; Matsuo et al., 2004; Kim et al., 2005b).
NFAT co-operatively binds with transcription factors of the AP-1 (Fos/Jun) family and AP-1 proteins to a number of functionally important sites in the promoters of numerous cytokine genes (Rao et al., 1997; Macian et al., 2001). For instance, the interaction between NFATc1 and c-Fos has been shown to be necessary in the regulation of the TRAP promoter in osteoclasts. In addition, in vitro promoter analyses identified nuclear factor of activated T-cells (NFAT)/AP-1 sites in the osteoclast-specific TRAP and CTR promoters (Matsuo et al., 2004). It is possible that c-Fos or c-Jun interaction with NFATc1 may also be involved in the regulation of the human β3 promoter.
The integrin αvβ3 is expressed on bone resorbing osteoclasts (Shinar et al., 1993) and evidence suggests that it is involved in the attachment of osteoclasts to bone (Horton et al., 1991). Blocking experiments have identified the αvβ3 integrin as a major functional adhesion receptor in osteoclasts (Horton et al., 1991; Engleman et al., 1997), where it appears to be crucial for cell spreading (Grano et al., 1994). The αvβ3 integrin is also involved in cell migration and maintenance of the sealing zone (Nakamura et al., 1999). These observations suggest that αvβ3 plays a major role in the function of osteoclasts, mediating aspects of cellular organization and function (Horton et al., 1991; Grano et al., 1994; Engleman et al., 1997; Nakamura et al., 1999).
The requirement for αvβ3 in normal osteoclast function is clearly demonstrated by genetic ablation of the β3 gene in mice (McHugh et al., 2000). Osteoclasts from mutant mice fail to spread in vitro (McHugh et al., 2000), supporting the results from blocking experiments using β3 antibodies (Horton et al., 1991). In vivo, osteoclasts in β3 knockout mice fail to form actin rings and do not form ruffled membranes, impeding them from resorbing bone effectively and resulting in an osteosclerotic phenotype (McHugh et al., 2000).
Expression of the αvβ3 integrin is regulated by expression of the β3 subunit during osteoclast differentiation (Kitazawa et al., 1995; Li et al., 1995; Zhu et al., 1996). We have previously shown that IL-4-dependent expression of the αvβ3 integrin, in osteoclast precursors is mediated by expression of the β3 gene (Kitazawa et al., 1995) and, using β3 gene promoter-reporter constructs, we have demonstrated that IL-4 dependent up-regulation is directly mediated by the transcription factor STAT-6 (McHugh et al., 2001). Thus far, little is known regarding the transcriptional mechanisms mediating β3 integrin expression during osteoclast differentiation. Therefore, we sought to understand the molecular mechanisms governing expression of the human β3 gene. In this study, we focused on the role of the calcineurin/NFATc1 pathway in the regulation of the human β3 integrin promoter during osteoclast differentiation. Using reporter constructs containing the promoter region the human β3 promoter, we demonstrated induction by RANKL and provide evidence that induction is mediated directly by NFATc1.
2. Materials and methods
2.1. Phylogenic foot printing
The mouse and human genomic sequences encompassing the human β3 gene were identified by basic local alignment search tool (BLAST) (NCBI; www.ncbi.nlm.nih.gov/BLAST). VISTA (www.gsd.lbl.gov/vista/index.shtml) was used to identify regions of high identity and conserved regulatory elements (comparing the human promoter to the mouse genome). Putative transcription factor binding sites in the −1242 to +29 bp region of the human β3 gene, were identified using MATCH based on the TRANSFAC4.0 matrices (transfac.gbf.de/TRANSFAC/) (Wingender et al., 2001), using a core match of 1. Sequence manipulation and alignment of 15 kb of the mouse and human promoter regions were completed using Pustell matrix (MacVector 7.2.2). NFAT:AP1 composite sites were identified using Model Inspector (Genomatix software GmbH) (Klingenhoff et al., 1999).
2.2. Plasmid construction
A 1.3 kb region of the human β3 promoter was a kind gift from P. Bray (pSLuc125) (Villa-Garcia et al., 1994; Wilhide et al., 1997). The β3 fragment was subcloned into the luciferase reporter vector pGL3 basic (Promega Corporation, Madison, WI), as described below to generate pGL3-B3 1.3.
The region −1242 to +29 bp relative to the TSS of the human β3 promoter was excised from the pSLuc125 construct by digestion with SpeI, blunt-ended with DeepVent (exo−) polymerase (New England Biolabs) and digested with BglII. The pGL3 basic vector was prepared by digestion with BglII and SmaI. The insert was ligated into the prepared vector with T4 ligase. The resulting promoter–reporter construct is designated pGL3B3-1.3. Plasmids were grown in JM109 cells (Promega) and were purified with the Endofree Maxiprep kit (QIAgen). A 5′ deletion construct pB3-1 was created by digestion with BstXI, followed by digestion with KpnI. The linearized construct was blunt-ended with DeepVent (exo+) polymerase (New England Biolabs) and recircularized with T4 DNA ligase (New England Biolabs). The human NFATc1 expression plasmid (pSH102) was a generous gift from G.R. Crabtree (Stanford University).
2.3. PCR mutagenesis
Mutations were introduced into two NFAT sites in the 5′ region of the pB3-1.3 reporter construct by a two-step PCR strategy. For the first step, overlapping PCR products containing the mutations were generated using the wild type construct as a template and the forward primer, containing mutations (5′-AGTTTCCAAAGGAATTCCAATCAGTCAGATAGG-3′) (mutations of NFAT sites underlined) and a reverse primer (5′-AAATAGTGGTAGTCCTGATGGGTAA-3′), complementary to nucleotide −899 to −770 of the wild type promoter. The second set of primers was a forward primer from the pGL3 vector, and the reverse complement containing the mutations (5′-ACCTTAAAAGGTTCAAAGGTTTCCTTAAGGTTA-3′). Overlapping, PCR products were isolated and combined as template for the second round of PCR with the flanking primers alone. The final PCR product was digested with KpnI plus BstXI and subcloned into the pB3-1.3 construct, digested with the same enzymes, producing pB3-1.3M1 and M2. All plasmid constructs were confirmed by DNA sequencing.
2.4. Transient transfection of RAW264.7 Cells
Clonal RAW264.7 cells (McHugh, 2001: ASBMR abstract) were cultured in α-modified Eagle’s medium α-MEM (Cellgro) containing 10% heat inactivated fetal bovine serum (HIFBS) (Hyclone) at 3 × 105 cells/well in 6 well plates (Nunc) in or 1.5 × 105 cells/well in 12 well plates (Nunc) for 24 h before transfection. Where indicated, cells were treated with RANKL 20 ng/ml (R and D Systems) on day 0 (when seeded) and day 3. Cells were transfected with constructs with the Transit-LT transfection reagent, (Mirus Corporation, WI), according to manufacturer’s instructions. At 48–72 h following transfection, cells were washed with phosphate buffered saline (PBS), and then lysed with the addition of reporter lysis buffer (Promega). Following a freeze (−80 °C)/thaw cycle, cells were lysed with Luciferase Assay Buffer (Promega) and luciferase activity of the clarified supernatant was determined with an Auto Luminat LB953 EG and G Luminometer (Berthold). Each experiment was repeated 2–3 times in triplicate and the results of a representative experiment are shown. One preparation was used across the range of concentrations. Results were normalized to protein concentration in the extracts as assessed by bicinchoninic acid protein assay (BCA assay) (Pierce) and the average of the data+standard deviation (S.D.) is presented. The pB3-1.3 reporter construct was assessed for dose response to RANKL by treatment with 2.5–40 ng/ml RANKL. In co-transfection experiments, cells were transfected with 0.001–0.5 μg of NFATc1 expression constructs. Expression vector, pBJ5, lacking an insert was used in control wells or in combination with NFATc1 expression constructs such that all wells were transfected with equal amount of DNA (to 0.5 μg total). For dose–response experiments the data is presented as the fold induction, which was calculated as the activity of each replicate divided by the untreated empty vector and then averaged (+S.D.).
2.5. Nuclear extract preparation
Jurkat cells obtained from ATCC were cultured in RPMI (Cellgro) with 10% HIFCS (Hyclone) overnight before stimulation with 30 μM phorbol 12-myristate 13-acetate (PMA) (Sigma) and 100 nM ionomycin (Fisher) for 30 min. Cells were washed in PBS then resuspended in 1 ml of ice-cold hypotonic buffer (10 mM HEPES-KOH, pH 7.9; 10 mM KCL; 1.5 mM MgCl2; 10 μl/ml protease inhibitor cocktail (Sigma) and 0.5 mM DTT) and placed on ice for 15 min. Cells were lysed by the addition of 64 μl of 10% NP-40 and incubated on ice for 10 min. Nuclei were pelleted then resuspended in 100 μl of high salt extraction buffer (20 mM HEPES–KOH, pH 7.9, 420 mM KCl, 1.2 mM MgCl2, 0.5 mM DTT, 0.2 mM EDTA, 25% glycerol) and 10 μl/ml protease inhibitor cocktail (Sigma) and incubated on ice for 20 min. The concentration of protein in clarified supernatants was determined using the BCA protocol (Pierce). Nuclear extracts were snap-frozen and stored at −80 °C.
2.6. Electrophoretic mobility shift assays (EMSAs)
Complementary oligonucleotides, containing two conserved NFAT sites of the human wild type (WT) β3 promoter were annealed (the sequences are listed in Table 1). Oligonucleotides from the mouse IL-2 promoter containing the NFAT consensus site or a mutant were obtained from Santa Cruz. Double-stranded oligonucleotides were end-labeled with T4 polynucleotide kinase (Promega) and [γ32P]-ATP (PerkinElmer) and purified on Sephadex G-25 Quick Spin columns (Boehringer Mannheim). Binding reactions (20 μl) were prepared with 2.5 μg of nuclear extract, 1 μg poly dI/dC (Pharmacia) in binding buffer [(10 mM Tris HCl (pH 7.5), 100 mM NaCl, 1 mM EDTA, 10% glycerol], 1 mM DTT, and 5×104 cpm of labeled oligonucleotide probe and incubated 30 min at room temperature. Nucleoprotein–oligonucleotide complexes were resolved by electrophoresis on 5% polyacrylamide gels (acrylamide/bisacrylamide at 40:1) in 0.25× Tris Buffered EDTA (TBE) buffer containing 5% glycerol at 200 V for 2 1/2 h at 4 °C. Where indicated, antibodies to NFATc1 (7A6) and NFATc2 (G1-D10) (Santa Cruz) (1 μl each) were added to binding reactions. For competition assays, unlabeled wild-type and mutant probes were added at 50× the concentration of the labeled probe. The gels were dried and autoradiographed for 16–36 h at −80 °C with intensifying screens.
Table 1.
Sequences of oligonucleoprobes used for EMSAs
| hβ3 WT | 5′ AGTGGAAAAAGGAAGGAAAATCAGTCAGATAGG 3′ |
| hβ3 MT1 | 5′ AGTGGAAAAAGGAATTCCAATCAGTCAGATAGG 3′ |
| hβ3 MT2 | 5′ AGTTTCCAAAGGAAGGAAAATCAGTCAGATAGG 3′ |
| hβ3 MT1 & 2 | 5′ AGTTTCCAAAGGAATTCCAATCAGTCAGATAGG 3′ |
| IL-2 NFAT cons | 5′ CGCCCAAAGAGGAAAATTTGTTTCATA 3′ |
| IL-2 NFAT MT | 5′ CGCCCAAAGCTTAAAATTTGTTTCATA 3′ |
The sequences represent the top strand of double-stranded probes. The hB3 wild type (WT) probe contains NFAT sites located −1098 to −1092 and −1109 to −1103 (relative to the TSS) in the human β3 promoter. Mutant sequences are indicated by bold text. NFAT binding sites are underlined.
2.7. Generation of TAT–HA dominant negative (dn) NFATc1 proteins
Human NFATc1 was amplified by PCR from a construct containing NFATc1 (cDNA provided by G. Crabtree) using primers containing Age-1 and Mfe1 restriction sites (5′-CGGACCGGTATGCCAAGCACCAGCTTTCCAGTCC-3′) and ( 5 ′ - CGGCAATTGTTAGAAAAAGCACCCCACGCGCTC-3′). The cDNA was cloned into the Age1/Eco R1 sites of the pTAT–HA vector provided by S. Dowdy (St. Louis) (Nagahara et al., 1998). In the current study, we generated pTAT-dominant negative (dn) NFATc1 constructs by deleting the DNA binding domain of NFATc1 from the construct by restriction digest with PvuII, as described (Northrop et al., 1994).
The TAT-dnNFATc1 expression plasmids were transformed into Rosetta 2 cells (Novagen, Madison WI). Cells were cultured in 25 ml of LB medium containing 50 μg/ml of ampicillin and 50 μg/ml chloramphenicol overnight at 30 °C then diluted in 300 ml of LB (no antibiotics) and cultured to log phase for a further 2 1/2 h at 37 °C. Expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (Bio-Rad, CA) for 3 h at 37 °C. Cells were sonicated and proteins were denatured using 6 μM guanidine followed by purification on a Ni-NTA nickel columns (Pierce) and eluted by stepping from 10 mM to 1 M imidazole in the presence of 8 M urea (Sigma-Aldrich, MI), as described (Dolgilevich et al., 2002). Fractions were desalted on PD-10 columns (Amersham, Pharmacia) or D-salt Dextran columns (Pierce). Fusion proteins were snap frozen in 10% glycerol, as suggested (Becker-Hapak et al., 2001). Samples were run on SDS-PAGE gels (Bio-Rad) to determine size and yield. TAT–HA proteins without inserts were isolated and used as a control. The identity of the fusion proteins was verified by immunoblot using a mouse monoclonal anti-HA antibody (clone 12CA5) (Roche, IN) and NFATc1 (7A6) (Santa Cruz) (data not shown) (Abu-Amer et al., 2001). Successful transduction of the cells with TAT proteins was verified by detection of the HA tag on pTAT constructs by immunohistochemistry (data not shown).
2.8. Inhibition of the NFAT/calcineurin pathway
To inhibit the NFAT/calcineurin pathway with dnNFATc1 or 11R-VIVIT, RAW264.7 cells were seeded as above for transfection. Where indicated, cells were treated with 200 nM of TAT-dn NFATc1 or the TAT–HA protein when cells were seeded and 20 ng/ml RANKL added. Cells were transfected on day 1. Results were confirmed by treatment with the cell-permeable peptide, 11R-VIVIT (2 μM) (Calbiochem) (Noguchi et al., 2004), or an equal volume of DMSO. An additional 1 ml of media was replaced after 48 h post-transfection and 20 ng/ml RANKL was added in relevant wells. Where indicated, 200 nM of TAT proteins was added 24 h post-transfection, as suggested by Becker-Hapak et al. (2001). 11R-VIVIT was also replenished as it has been shown to have a half-life of 30 h (Noguchi et al., 2004). Cells were lysed with Luciferase Assay Buffer (Promega) 48 h post-transfection.
3. Results
3.1. Homologous regions in the proximal promoters of mouse and human β3 integrin genes
Alignment of the 5′ end of the mouse and human β3 integrin gene sequences revealed regions of greater than 80% sequence homology across a 1.3 kb region upstream of the TSS (Fig. 1). Conservation between mouse and humans suggests that this region contains key regulatory elements (Fig. 1B). Furthermore, multiple conserved transcription factor binding elements are present within the region, as determined using the TRANSFAC transcription factor data base (Wingender et al., 2001) and confirmed with Genomatix software GmbH. Since NFATc1 is implicated in osteoclast differentiation and gene expression, we screened the 1271 bp fragment of the human β3 gene, consisting −1242 to +29 bp (relative to the TSS), for putative NFAT transcription factor binding sites with TRANSFAC. Within this region, 8 putative NFAT binding sites were identified (Fig. 1B). When the 1271 bp fragment of the human β3 gene was aligned to 15 kb of the mouse β3 gene (Fig. 1A), 3 of the NFAT binding sites in the 5′ promoter region and one site immediately upstream of the 5′ UTR were spatially conserved, as shown in Fig. 1C. Notably, a single NFAT:AP-1 consensus site was identified in the 5′ promoter region using Genomatix software GmbH. Other conserved elements identified in the 5′ promoter region included 3 Ets sites, 2 of which overlapped the NFATc1. Two GATA sites with core matches of 1 were also identified. The homology of the mouse and human β3 promoter regions, from −1242 to +29 relative to TSS, that includes consensus sites for NFAT and AP-1 is presented in Fig. 1.
Fig. 1.
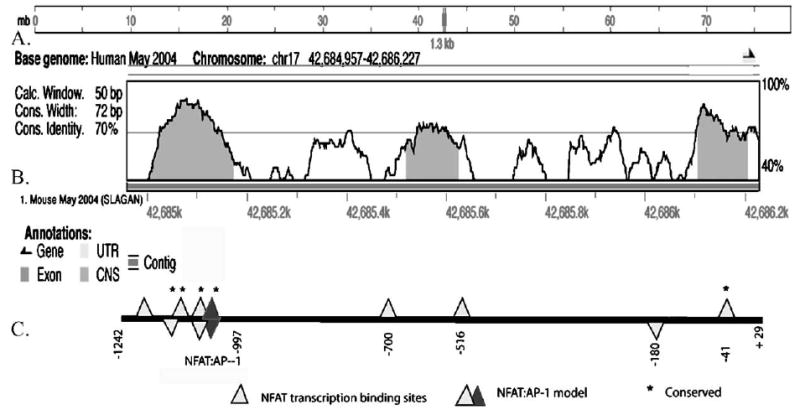
Identity between the mouse β3 promoter region and the 1.3 human β3 promoter. (A) 15 kb of the mouse β3 promoter region was aligned to the human promoter region in a Pustell matrix using MacVector to identify regions of high identity. The position of the human β3 promoter sequence within the human chromosome is indicated. (B) The human β3 integrin promoter sequence was subjected to analysis with VISTA (settings: window size=50 bp, conservation cut width=72% and the conserved identify of 70%) to predict sequences of high conservation with the mouse orthologue. The region −1242 to +29/pB3-1.3 (relative to the transcription start site, TSS) of the human β3 integrin is aligned to the mouse genome (May 2004 Mouse Genome Assembly > MLAGAN). The transcriptional start of the gene is indicated by the black arrow. (C) The pB3-1.3 full-length construct, −1242 to +29 (relative to the TSS) of the human β3 gene used in transfection studies is also shown above. Predicted binding sites for NFAT are displayed (light triangles) (core match of 1.0 as identified by MATCH/and genomatix). Sites on the top of line indicate sense and antisense orientation. The AP1 site, of the NFAT:AP-1 conserved composite site, is indicated (dark triangle). Transcription binding sites conserved between mouse and human are designated with*.
3.2. The human β3 gene promoter is activated by RANKL
A −1242 to +29 bp (relative to the TSS) human β3 genomic fragment was subcloned into a luciferase reporter vector with the resulting construct designated pB3-1.3. The pB3-1.3 reporter was transfected into osteoclast-forming RAW264.7 cells, which were cultured under osteoclast inducing conditions. After 72 h, extracts were assayed for luciferase activity. As shown in Fig. 2A, the pB3-1.3 reporter construct responded to RANKL and was induced between 2.5 ng and 40 ng/ml. Maximal induction of greater than 3-fold over the untreated cells was observed at 5 ng/ml RANKL (29-fold over the untreated empty vector). A RANKL concentration of 20 ng/ml was chosen for the following studies, as it effectively induced TRAP-positive multinucleated cells.
Fig. 2.
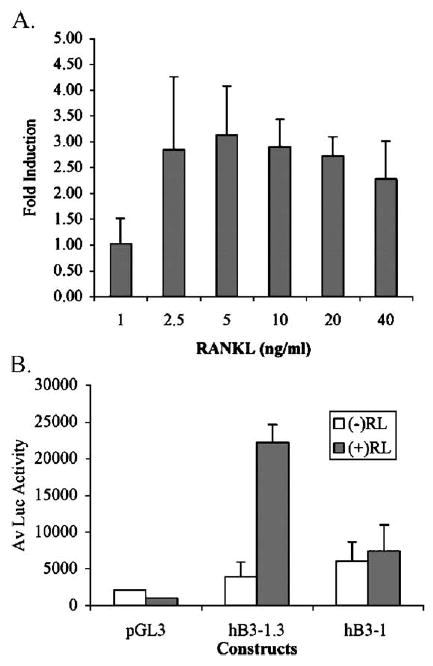
RANKL induction of the human β3 promoter. (A) RAW264.7 cells were treated with RANKL (0, 1, 2.5, 5, 10, 20 and 40 ng). Cells were transiently transfected on day 1 with the human β3 integrin promoter constructs, pB3-1.3 containing −1242 bp to +29 bp (relative to the TSS). Luciferase (luc) activity was assayed 72 h post-transfection and normalized to protein. Experiments were performed in triplicate and the fold induction of pB3-1.3 over the untreated vector was calculated for each replicate. Data represents the average fold induction (+S.D.) and results are representative of 3 independent experiments. (B) RAW264.7 cells were seeded and treated with +20 ng/ml RANKL. Cells were transfected, with the pGL3 Basic luciferase constructs, full-length pB3-1.3 or pB3-1 reporter gene constructs and assayed for Luc activity 72 h post-transfection. The experiment was performed in triplicate and the data are the average of the Luc activity normalized to protein (+S.D.). Results are representative of 2 independent experiments.
3.3. Identification of the RANKL responsive region within the human β3 integrin promoter
To identify the region of the β3 promoter responsible for RANKL induction, we prepared a 5′ deletion of the pB3-1.3 promoter construct. The resulting promoter fragment, −997 to +29 bp (relative to the TSS), was designated pB3-1. This 245 bp 5′ truncation removed the upstream region of >85% identity between the mouse and human promoter which contains numerous conserved transcription factor binding sites including NFAT, Ets, GATA-1, as well as the conserved NFAT: AP1 composite site. As shown in Fig. 2B, deletion of 245 bp 5′ of the 1.3 kb promoter markedly blunted induction by RANKL.
3.4. Dose-dependent transactivation of the human β3 integrin promoter by NFATc1
Due to the presence of several putative NFAT binding elements in the 1.3 kb human β3 promoter (Fig. 1C) and the essential role that NFATc1 has in osteoclast differentiation and activation (Takayanagi et al., 2002; Hirotani et al., 2004), we sought to determine if the human β3 gene is trans-activated by NFATc1. Human β3 pB3-1.3 reporter constructs were co-transfected with a range of human NFATc1 concentrations (0.001–0.5 μg) to test for trans-activation. DNA concentrations were maintained at equal levels with the addition of empty expression vector (pBJ5). Co-transfection of the human β3 pB3-1.3 reporter with human NFATc1 expression constructs resulted in dose-dependent trans-activation of the β3 promoter, as shown in Fig. 3A. Promoter induction was evident when co-transfected with the NFATc1 expression construct at greater than 0.005 μg, and was highest with 0.5 μg of the NFATc1 expression construct (60 fold induction over the untreated construct). At 0.5 μg, induction was 168 fold over the empty pGL3 basic luciferase vector (not shown). Trans-activation of the β3 promoter by NFATc1 is consistent with the presence of functional NFAT binding sites in the human β3 promoter.
Fig. 3.
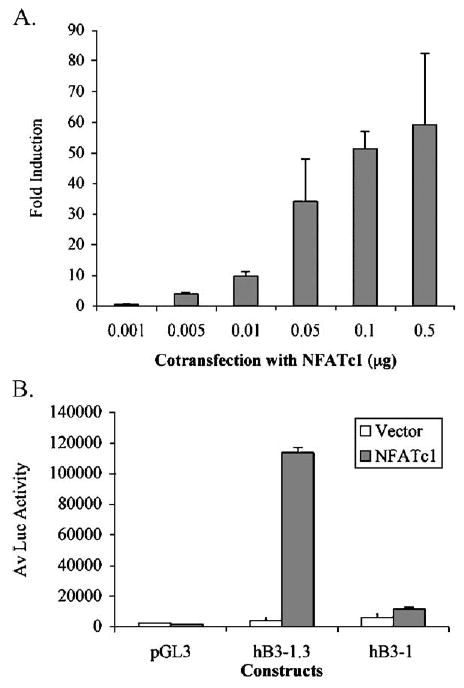
Trans-activation of the human β3 promoter by NFATc1. (A) RAW264.7 cells were transfected with pB3-1.3 reporter gene constructs (containing −1242 to +29 of the human β3 gene). Cells were co-transfected with 0.001–0.5 μg of NFATc1 in pBJ5 vectors (or the pBJ5 control vector) and assayed for luciferase (Luc) activity. Experiments were performed in triplicate and normalized to protein; the fold induction for each pB3-1.3 replicate over the average of the pB3-1.3 construct (with pBJ5 control vector) was determined. A fold induction was determined using the average of the induction calculated for each replicate (+SD). (B) RAW264.7 cells were transfected with human β3 reporter gene constructs pB3-1.3 or pB3-1 (containing portions −1242 and −997+29 bp (relative to the TSS)) and co-transfected with 0.5 μg of NFATc1 in pBJ5 vectors or pBJ5 (control). Experiments were performed in triplicate and Luc activity normalized to protein; the average+S.D. is shown and is representative of 2 independent experiments.
3.5. Identification of the NFAT-responsive region within the human β3 integrin promoter
The region −1242 to −997 (relative to the TSS) of the pB3-1.3 construct contains 3 conserved NFAT sites, including an NFAT:AP-1 site (Fig. 1C) and deletion of this region results in loss of RANKL induction (Fig. 2B). We therefore tested NFATc1 trans-activation of the deletion construct (pB3-1) to determine if this region of the human β3 promoter is responsible for NFAT, as well as RANKL induction. Deletion of 245 bp 5′ of the β3 promoter abolished the response to NFATc1 co-transfection, as shown in Fig. 3B. This indicates that the putative NFAT binding sites within the −1242 to −997 bp region (relative to the TSS) are the predominant sites responsible for NFAT induction of the β3 promoter. Notably, this region was found to be responsible for a large proportion of the RANKL response, as shown in Fig. 2B.
3.6. NFAT binds to consensus sites within the −1242 to −997 region of the human β3 promoter
We used EMSAs to determine if NFATc1 binds to the potential binding sites in the −1242 to −997 bp, RANKL- and NFATc1-responsive, region of the human β3 promoter. Since there are no established human osteoclast forming cell lines and osteoclasts formed from human primary cells demonstrate significant donor-to-donor variability, we utilized Jurkat T cells as a source of authentic human NFAT proteins. Stimulation of Jurkat T cells with PMA and ionomycin has been shown to induce expression and activation of NFAT family members (Northrop et al., 1994; Noguchi et al., 2004). As shown in Fig. 4A, an oligonucleotide containing a conserved NFAT binding site in addition to the NFAT:AP1 consensus binding site from the β3 promoter bound to factors in nuclear extracts from Jurkat T cells stimulated with PMA and ionomycin. Similar shifts were produced using the commercially available IL-2 NFAT consensus probe as a positive control (Fig. 4B). The identity of the protein moiety was confirmed by supershifting of the retarded band with antibodies to two members of the NFAT family, NFATc1 or NFATc2 (with antibodies 7A6 and G1-D10, respectively). NFAT sites support binding of both NFATc1 and NFATc2 from stimulated Jurkat T cells, since the DNA binding domain and consensus binding sites are conserved between NFAT family members (Rao et al., 1997). However, NFATc1 appears to be the predominant protein binding to the β3 and IL-2 promoter oligoprobes. Competition of the shifted complexes with unlabeled WT β3 oligonucleotides and with the consensus IL-2 NFAT oligonucleotide demonstrates the specificity of the binding reactions. Binding to both the WT β3 promoter probe (Fig. 4A) and the IL-2 consensus probe (Fig. 4B) were competed with both the unlabeled homologous β3 and IL-2 consensus probes. We next introduced mutations into the β3 promoter NFAT consensus sites to identify the sites involved in NFAT binding (refer to Table 1). Previously, NFAT proteins have been shown to bind coordinately with AP-1 (Fos/Jun) proteins on composite DNA elements (Macian et al., 2001; Matsuo et al., 2004). We therefore generated β3 oligonucleotide probes with a mutation in the NFAT component of the NFAT:AP-1 composite site or with a mutation in the adjacent NFAT site (M1 and M2; Table 1). Both the unlabeled M1 and the M2 oligonucleotides competed with the formation of NFATc1 DNA protein complexes. Mutation of both the NFAT binding sites within the β3 oligoprobes (MT1 and 2) resulted in inhibition of competition, indicating that both sites are functional and bind independently of AP-1.
Fig. 4.
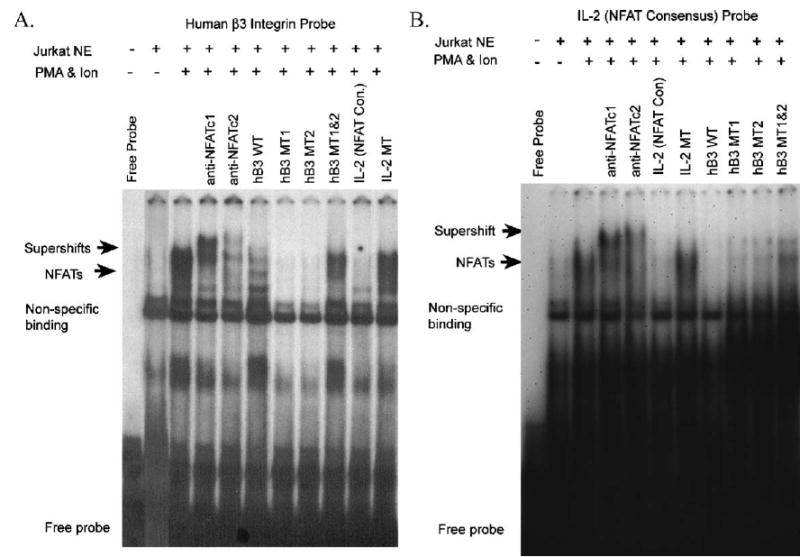
Identification of functional NFATc1 binding sites in the human β3 integrin promoter by EMSA. Competition assays were performed using labeled human β3 oligoprobes containing putative NFATc1 binding sites (A) and an IL-2 oligoprobe with a NFAT consensus site (Santa Cruz). Nuclear extract (NE) proteins were obtained from untreated (PMA−) and PMA and ionomycin-treated (PMA and Ion) Jurkat cells. Extract (−) indicates no extract was added to the binding reaction and migration of the free probe is indicated. Supershift assays were with antibodies to NFATc1 (7A6) (anti-NFATc1) and NFATc2 (G1-D10) (Santa Cruz) (indicated with arrow). Binding reactions were competed with 50× excess of unlabelled oligoprobes including; homologous β3, β3 containing mutations of the NFAT sites, IL-2 consensus, and IL-2 containing a mutated NFAT site (shown in Table 1). Areas of non-specific binding are indicated.
3.7. Mutation of upstream NFATc1 binding sites in the 1.3 kb human β3 promoter blocks NFATc1 transactivation and RANKL induction
As discussed in 3.6, two specific NFAT binding sites, within 245 bp of the 5′ region of the human β3 promoter, were shown to be functional using EMSAs (Table 1 and Fig. 4). To confirm the role of these NFAT sites in RANKL- and NFAT-dependent human β3 promoter activation, we generated reporter constructs containing mutations of both of the NFAT sites (M1 and M2: Table 1) in the context of the full-length promoter (Fig. 5). This construct, pB3-1.3 M1 and M2 includes mutation of a conserved NFATc1 site, as well as a mutation in the NFAT component of the adjacent NFAT:AP-1 composite site (Table 1). The construct bearing pB3-1.3 M1 and M2 mutations and the wild type construct pB3-1.3, were assessed for RANKL induction and NFATc1 trans-activation by transfection of RAW264.7 cells. As shown in Fig. 5, mutation of these specific conserved NFAT binding sites, in the context of the human β3 promoter construct resulted in a decrease in RANKL induction (Fig. 5A) and a striking reduction in NFATc1 trans-activation (Fig. 5B). NFATc1 trans-activated the pB3-1.3 almost 10 fold, while the mutant construct was induced only 2 fold over untreated constructs. RANKL induction was reduced but not ablated in the double mutant promoter construct, implicating the involvement of transcription factors other than NFAT in the trans-activation of the human β3 promoter in response to RANKL.
Fig. 5.
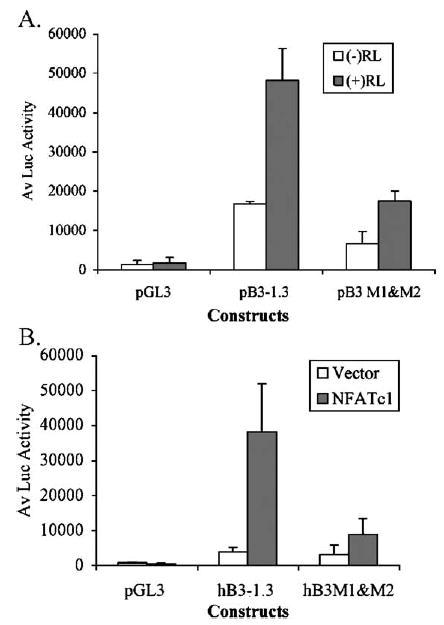
Abrogation of RANKL and NFAT induction of the human β3 integrin promoter by mutation of NFAT sites. RAW264.7 cells were treated with 20 ng/ml RANKL and transfected with the human β3 integrin pB3-1.3 reporter construct (−1242 to +29) or the double mutant pB3 M1 and M2 (as shown in Table 1). The NFATc1 site mutated to M2 is a constituent of the conserved NFAT:AP-1 composite site. Mutations in the pB3-1.3 construct to pB3 M1 and M2 results in reduction in the response to RANKL induction (A) and abrogation of trans-activation by NFATc1 co-transfection (B). Experiments were performed in triplicate and luciferase (Luc) activity was normalized to protein; the average +S.D. is shown and is representative of 2 independent experiments.
3.8. TAT-transduced dominant-negative NFATc1 and cell-permeable 11R-VIVIT peptide attenuate RANKL induction of the human β3 promoter
Results from our transfection experiments indicate that the calcineurin/NFAT signaling pathway mediates RANKL-induced β3 integrin expression through direct binding of NFATc1 to specific upstream sites in the human β3 promoter. To determine whether the pharmacological blockade of calcineurin/NFAT signaling affects β3 integrin promoter activity, we employed two independent methods of blockade of calcineurin/NFAT signaling: a TAT-transducible dominant-negative (dn) NFATc1 protein (TAT-dn NFATc1), and a cell-permeable calcineurin inhibitory peptide, 11R-VIVIT. Expression of the dnNFATc1 has been shown to effectively block NFAT mediated gene expression in T cells (Northrop et al., 1994). The dnNFATc1 has the DNA binding domain deleted (Northrop et al., 1994), the protein thereby acts by binding to calcineurin so on translocation to the nucleus it is unable to bind to the DNA binding domain in the promoter (Northrop et al., 1994). RAW264.7 cells were transfected as described above and treated with or without RANKL. Cells were treated with 200 nM TAT-dnNFATc1 or hemagglutinin (HA) tagged TAT–(TAT–HA) as a control (Fig. 6A). This concentration is within the functionally effective range and has been shown to be effective in osteoclast differentiation assays (Abu-Amer et al., 2001; Clohisy et al., 2004). To confirm our findings with the TAT-dnNFAT proteins, we used 11R-VIVIT, a cell-permeable NFAT inhibitory peptide (Fig. 6B). 11R-VIVIT interferes selectively with calcineurin–NFAT interaction without affecting calcineurin phosphatase activity, thereby blocking dephosphorylation of NFAT and preventing translocation to the nucleus. This peptide potently inhibits NFAT activation without affecting the expression of other genes that require calcineurin (Aramburu et al., 1999). Retroviral transfection with VIVIT-GFP has been shown to inhibit osteoclast differentiation induced by RANKL (Hirotani et al., 2004). Cells were treated with 2 μM 11R-VIVIT or the vehicle, DMSO, as a control. This concentration is within the range shown to inhibit IL2 mRNA transcription in Jurkat T cells (Noguchi et al., 2004). As shown in Fig. 6A, treatment with TAT-dnNFATc1 attenuated the response of the transfected human β3 promoter to RANKL treatment as compared to the TAT protein control. Treatment with 11R-VIVIT also reduced the RANKL-induction of human β3 promoter compared to vehicle alone (DMSO), as shown in Fig. 6B.
Fig. 6.
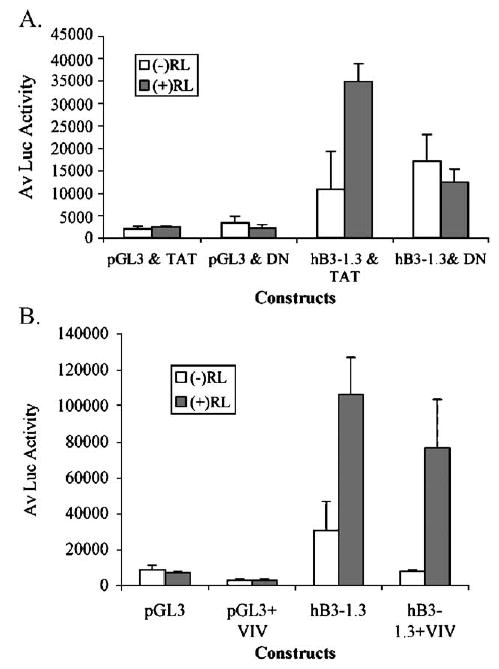
Inhibition of NFAT/calcineurin signaling attenuates RANKL induction of the human β3 promoter. RAW264.7 cells were induced to differentiate into osteoclasts with 20 ng/ml RANKL. On day 1, cells were transfected with the human β3 integrin promoter reporter construct pB3-1.3 or pGL3 basic. Cells were transduced with 200 TAT-or TAT-dnNFATc1 fusion proteins (A) or 11R-VIVIT (B) from the day of seeding. Cells were grown 72 h post-transfection and then assayed for luciferase (Luc) activity. Experiments were performed in triplicate and Luc activity was normalized to protein; the average of the replicates+S.D. is shown and is representative of 2 independent experiments.
4. Discussion
RANKL has been shown to act downstream of its receptor, RANK, by signaling through TRAF6 and c-Fos pathways (Takayanagi et al., 2002). RANKL selectively induces NFATc1 expression via these two pathways and triggers a sustained NFATc1-dependent transcriptional program during osteoclast differentiation (Takayanagi et al., 2002). In these studies we have shown that 1.3 kb of genomic DNA upstream of the human β3 gene, with > 80% homology to the mouse gene, is sufficient to act as a promoter in RAW264.7 cells and, importantly, this fragment is responsive to RANKL-induced signaling. Consistent with a direct effect of RANKL-induced transcription factors on the β3 promoter, treatment with greater than 2.5 ng/ml RANKL induced expression of the β3 reporter construct over 72 h post-transfection. We identified multiple putative NFAT transcription factor binding sites in the β3 promoter and therefore sought to ascertain if NFATc1 mediated RANKL-induction of the promoter.
NFATc1 has been identified as having a major role in regulating the promoters of multiple osteoclast genes, including cath K, TRAP, CTR and OSCAR (Anusaksathien et al., 2001; Matsumoto et al., 2004; Matsuo et al., 2004; Kim et al., 2005b), further implicating its essential role in terminal differentiation of functional osteoclasts (Ishida et al., 2002; Takayanagi et al., 2002; Hirotani et al., 2004). Ectopic expression of NFATc1 causes precursor cells to undergo efficient differentiation in the absence of RANKL signaling (Takayanagi et al., 2002), while blockade of NFATc1 signaling by transduction with a NFAT-calciunerin pathway inhibitory peptide motif, VIVIT-RT (Hirotani et al., 2004), cyclosporin A (Ishida et al., 2002), or FK506 attenuates osteoclast spreading and resorptive activity.
A recent study has shown that retroviral expression of a constitutively active NFATc1 can induce osteoclast-like cell formation in RAW264.7 cells. Induction of the expression of the β3 integrin accompanied osteoclast-like cell formation (Hirotani et al., 2004), however, a direct effect on the β3 promoter was not established. Through homology analysis and transcription factor binding site screening we identified multiple putative NFAT transcription factor binding sites in the 1.3 kb human β3 integrin promoter. Of the putative NFAT binding sites, 4 were conserved between mouse and human, 3 within the 5′ 245 bp of the 1.3 kb promoter, and 1 which comprises an NFAT;AP1 composite consensus site. We therefore sought to ascertain if the RANKL-induction of the human β3 promoter was mediated by binding to the region containing three of the conserved NFAT sites. We found that upon deletion of 245 bp from the 5′ end of the 1.3 kb human β3 promoter, the response to RANKL was significantly attenuated. This suggests that the majority of RANKL induction of the β3 promoter is mediated by sequences in the first 245 bp of the 1.3 kb promoter, however, other promoter sites may contribute to the regulation of the β3 gene by RANKL.
Hirotani et al. demonstrated the role of the calcineurin/NFAT signaling pathway in the regulation of osteoclast differentiation, defining calcineurin as an essential downstream effector of the RANKL-induced signal transduction pathway for osteoclast differentiation (Hirotani et al., 2004). By inhibiting calcineurin with either the immunosuppressant drugs cyclosporin A, FK506, or by retroviral-mediated expression of the specific calcineurin inhibitory peptide VIVIT, RANKL-induced differentiation of RAW264.7 cells into mature multinucleated osteoclasts was inhibited. These studies provided evidence that activation of the NFATc1 transcription factor is sufficient to initiate a genetic program that results in the specification of the mature functional osteoclast cell phenotype (Hirotani et al., 2004).
Since NFATc1 plays an important role in osteoclast differentiation (Ishida et al., 2002; Takayanagi et al., 2002; Hirotani et al., 2004) and deletion of 3 conserved NFAT sites, including an NFAT:AP1 site, decreased RANKL induction of the β3 promoter, we sought to investigate the role of NFATc1 in the regulation of the human β3 integrin gene. Co-transfection of the full-length human β3 promoter with a human NFATc1 expression construct resulted in a dose-dependent trans-activation of the promoter. Significantly, this induction was abrogated with the 5′ 245 bp deletion in the β3 integrin promoter, which was also the predominant region responsible for RANKL induction of the promoter. While these studies identified the human β3 promoter region responsive to RANKL and NFAT induction, and strongly implicated that the conserved NFAT sites 5′ in the promoter mediate these effects, they do not show direct NFAT-promoter interaction and allow the possibility for indirect or secondary transcriptional effects. We used EMSA to test for direct NFAT interactions and sequence-specific binding to the putative NFAT sites of the β3 promoter.
We confirmed by EMSA that human NFATc1 binds specifically to two conserved NFAT sites (M1 and M2), including the NFAT:AP1 composite consensus site (M2), in the upstream region of the human β3 integrin promoter. We used nuclear extracts prepared from Jurkat T cells for EMSA analysis since they are a human cell line that has been well characterized with regard to NFAT expression (Northrop et al., 1994; Noguchi et al., 2004). These results were verified by binding to an NFAT consensus oligonucleotide from the IL-2 promoter. Specificity was demonstrated by competition of the labeled IL-2 (NFAT consensus) and human β3 oligonucleotide probes with homologous unlabeled human β3 or NFAT consensus oligonucleotide probes. Oligonucleotides containing mutations of the IL-2 consensus NFAT site failed to compete with NFAT:DNA complexes. Mutation of only one of the two adjacent NFAT sites within the 5′ region of the β3 integrin promoter still enabled partial competition with binding of the wild type sequence. Mutation of both adjacent NFAT sites (M1 and M2) resulted in failure to compete with DNA protein complex formation, suggesting that both adjacent sites effectively bind NFAT proteins. The identity of the protein moiety was confirmed by super-shift of the retarded bands with antibodies to both NFATc1 and NFATc2. Therefore, while the promoter contains numerous potential NFAT binding sites, our deletion and EMSA data suggested that the upstream NFAT sites (−997 relative to TSS), including the NFAT:AP1 site in the β3 promoter, are the principal regulators of the β3 integrin gene in response to RANKL. Furthermore, subsequent mutation of these sites attenuated RANKL induction of the human β3 promoter and blocked trans-activation by NFATc1. These data indicate that NFATc1 can directly bind to the β3 promoter and identify the predominant sites responsible for RANKL-induced NFAT regulation. Recent work has demonstrated that NFAT and AP-1 cooperatively bind to the TRAP promoter (Matsuo et al., 2004). Therefore, mechanisms in addition to NFATc1 signaling may be involved in regulating the human β3 gene downstream of the RANKL signaling cascade in osteoclast differentiation, as additional transcription factors were identified within this region, including GATA and Ets factors.
We were able to demonstrate a reduction in the RANKL response of the human β3 promoter by blockade of calcineurin/NFAT signaling. To do so we generated a TAT-dnNFATc1 fusion protein in which the DNA binding domain was deleted. The protein is able to interact with calcineurin, but on translocation to the nucleus it is unable to interact with DNA (Northrop et al., 1994). These findings were confirmed with the calcineurin-pathway inhibitor 11R-VIVIT (Aramburu et al., 1999). The specific inhibitory action of the 11R-VIVIT peptide has recently been demonstrated in Jurkat cells (Noguchi et al., 2004). In our studies, the effects of 11R-VIVIT on regulation of the β3 integrin promoter verified the effects of transducing cells with the TAT-dnNFATc1 fusion protein. While these studies do not address regulation of the endogenous β3 integrin gene, Hirotani has demonstrated induction of the β3 integrin gene in RAW264.7 by constituitive expression of NFATc1 (Hirotani et al., 2004).
Together our data supports a significant role for NFATc1 in the mediation of RANKL induction of the human β3 gene. We conclude that specific NFAT binding sites (M1 and M2) located within the 1.3 kb human β3 gene promoter are the predominant sites required for NFAT trans-activation and the preponderance of RANKL-inducible activity. Transcription factors other than NFATc1 may also contribute to modulation of the RANKL induction of β3 gene expression. Together these results confirm the role of NFATc1 signaling pathway in osteoclast differentiation and establish the promoter region of the β3 gene as a direct target of NFATc1 in RANKL-dependent osteoclast formation.
Acknowledgments
The author of this paper holds a National Health and Medical Research Council (Aust) CJ Martin Fellowship (I.D. 200078). This work was supported by National Institute of Health Grants NIAMS R01 AR45472 (to SRG), NIAMS R01 AR47229 (to KPM) and aided by a grant from the Orthopaedic Research and Education Foundation (# 00-020, to KPM). We thank Dr P. Bray for the human β3 construct and Dr G.R. Crabtree for the NFATc1 expression plasmid.
References
- Abu-Amer Y, Dowdy SF, Ross FP, Clohisy JC, Teitelbaum SL. TAT fusion proteins containing tyrosine 42-deleted IkappaBalpha arrest osteoclastogenesis. J Biol Chem. 2001;276:30499–30503. doi: 10.1074/jbc.M104725200. [DOI] [PubMed] [Google Scholar]
- Anusaksathien O, et al. Tissue-specific and ubiquitous promoters direct the expression of alternatively spliced transcripts from the calcitonin receptor gene. J Biol Chem. 2001;276:22663–22674. doi: 10.1074/jbc.M007104200. [DOI] [PubMed] [Google Scholar]
- Aramburu J, Yaffe MB, Lopez-Rodriguez C, Cantley LC, Hogan PG, Rao A. Affinity-driven peptide selection of an NFAT inhibitor more selective than cyclosporin A. Science. 1999;285:2129–2133. doi: 10.1126/science.285.5436.2129. [DOI] [PubMed] [Google Scholar]
- Baron R. Molecular mechanisms of bone resorption by the osteoclast. Anat Rec. 1989;224:317–324. doi: 10.1002/ar.1092240220. [DOI] [PubMed] [Google Scholar]
- Becker-Hapak M, McAllister SS, Dowdy SF. TAT-mediated protein transduction into mammalian cells. Methods. 2001;24:247–256. doi: 10.1006/meth.2001.1186. [DOI] [PubMed] [Google Scholar]
- Clohisy JC, Hirayama T, Frazier E, Han SK, Abu-Amer Y. NF-kB signaling blockade abolishes implant particle-induced osteoclastogenesis. J Orthop Res. 2004;22:13–20. doi: 10.1016/S0736-0266(03)00156-6. [DOI] [PubMed] [Google Scholar]
- Dolgilevich S, Zaidi N, Song J, Abe E, Moonga BS, Sun L. Transduction of TAT fusion proteins into osteoclasts and osteoblasts. Biochem Biophys Res Commun. 2002;299:505–509. doi: 10.1016/s0006-291x(02)02664-5. [DOI] [PubMed] [Google Scholar]
- Engleman VW, et al. A peptidomimetic antagonist of the alpha(v)beta3 integrin inhibits bone resorption in vitro and prevents osteoporosis in vivo. J Clin Invest. 1997;99:2284–2292. doi: 10.1172/JCI119404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grano M, et al. Adhesion properties and integrin expression of cultured human osteoclast-like cells. Exp Cell Res. 1994;212:209–218. doi: 10.1006/excr.1994.1136. [DOI] [PubMed] [Google Scholar]
- Hirotani H, Tuohy NA, Woo JT, Stern PH, Clipstone NA. The calcineurin/nuclear factor of activated T cells signaling pathway regulates osteoclastogenesis in RAW264.7 cells. J Biol Chem. 2004;279:13984–13992. doi: 10.1074/jbc.M213067200. [DOI] [PubMed] [Google Scholar]
- Horton MA, Taylor ML, Arnett TR, Helfrich MH. Arg-Gly-Asp (RGD) peptides and the anti-vitronectin receptor antibody 23C6 inhibit dentine resorption and cell spreading by osteoclasts. Exp Cell Res. 1991;195:368–375. doi: 10.1016/0014-4827(91)90386-9. [DOI] [PubMed] [Google Scholar]
- Ishida N, et al. Large scale gene expression analysis of osteoclastogenesis in vitro and elucidation of NFAT2 as a key regulator. J Biol Chem. 2002;277:41147–41156. doi: 10.1074/jbc.M205063200. [DOI] [PubMed] [Google Scholar]
- Kim, K., et al., 2005a. NFATc1 induces OSCAR gene expression during TRANCE-mediated osteoclastogenesis. J. Biol. Chem.
- Kim, Y., Sato, K., Asagiri, M., Morita, I., Soma, K., Takayanagi, H., 2005b. Contribution of NFATc1 to the transcriptional control of immunoreceptor OSCAR but not TREM-2 during osteoclastogenesis. J. Biol. Chem. [DOI] [PubMed]
- Kitazawa S, Ross FP, McHugh K, Teitelbaum SL. Interleukin-4 induces expression of the integrin alpha v beta 3 via transactivation of the beta 3 gene. J Biol Chem. 1995;270:4115–4120. doi: 10.1074/jbc.270.8.4115. [DOI] [PubMed] [Google Scholar]
- Klingenhoff A, Frech K, Quandt K, Werner T. Functional promoter modules can be detected by formal models independent of overall nucleotide sequence similarity. Bioinformatics. 1999;15:180–186. doi: 10.1093/bioinformatics/15.3.180. [DOI] [PubMed] [Google Scholar]
- Kodama H, et al. Congenital osteoclast deficiency in osteopetrotic (op/op) mice is cured by injections of macrophage colony-stimulating factor. J Exp Med. 1991;173:269–272. doi: 10.1084/jem.173.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong YY, et al. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature. 1999;397:315–323. doi: 10.1038/16852. [DOI] [PubMed] [Google Scholar]
- Li CF, Ross FP, Cao X, Teitelbaum SL. Estrogen enhances alpha v beta 3 integrin expression by avian osteoclast precursors via stabilization of beta 3 integrin mRNA. Mol Endocrinol. 1995;9:805–813. doi: 10.1210/mend.9.7.7476964. [DOI] [PubMed] [Google Scholar]
- Macian F, Lopez-Rodriguez C, Rao A. Partners in transcription: NFAT and AP-1. Oncogene. 2001;20:2476–2489. doi: 10.1038/sj.onc.1204386. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, et al. Essential role of p38 mitogen-activated protein kinase in cathepsin K gene expression during osteoclastogenesis through association of NFATc1 and PU. 1 J Biol Chem. 2004;279:45969–45979. doi: 10.1074/jbc.M408795200. [DOI] [PubMed] [Google Scholar]
- Matsuo K, et al. Nuclear factor of activated T-cells (NFAT) rescues osteoclastogenesis in precursors lacking c-Fos. J Biol Chem. 2004;279:26475–26480. doi: 10.1074/jbc.M313973200. [DOI] [PubMed] [Google Scholar]
- McHugh KP, et al. Mice lacking beta3 integrins are osteosclerotic because of dysfunctional osteoclasts. J Clin Invest. 2000;105:433–440. doi: 10.1172/JCI8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh KP, Kitazawa S, Teitelbaum SL, Ross FP. Cloning and characterization of the murine beta(3) integrin gene promoter: identification of an interleukin-4 responsive element and regulation by STAT-6. J Cell Biochem. 2001;81:320–332. doi: 10.1002/1097-4644(20010501)81:2<320::aid-jcb1047>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Nagahara H, et al. Transduction of full-length TAT fusion proteins into mammalian cells: TAT-p27Kip1 induces cell migration. Nat Med. 1998;4:1449–1452. doi: 10.1038/4042. [DOI] [PubMed] [Google Scholar]
- Nakamura I, et al. Role of alpha(v)beta(3) integrin in osteoclast migration and formation of the sealing zone. J Cell Sci. 1999;112 (Pt 22):3985–3993. doi: 10.1242/jcs.112.22.3985. [DOI] [PubMed] [Google Scholar]
- Noguchi H, et al. A new cell-permeable peptide allows successful allogeneic islet transplantation in mice. Nat Med. 2004;10:305–309. doi: 10.1038/nm994. [DOI] [PubMed] [Google Scholar]
- Northrop JP, et al. NF-AT components define a family of transcription factors targeted in T-cell activation. Nature. 1994;369:497–502. doi: 10.1038/369497a0. [DOI] [PubMed] [Google Scholar]
- Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- Shinar DM, Schmidt A, Halperin D, Rodan GA, Weinreb M. Expression of alpha v and beta 3 integrin subunits in rat osteoclasts in situ. J Bone Miner Res. 1993;8:403–414. doi: 10.1002/jbmr.5650080404. [DOI] [PubMed] [Google Scholar]
- Takayanagi H, et al. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev Cell Biol. 2002;3:889–901. doi: 10.1016/s1534-5807(02)00369-6. [DOI] [PubMed] [Google Scholar]
- Tanaka S, et al. Macrophage colony-stimulating factor is indispensable for both proliferation and differentiation of osteoclast progenitors. J Clin Invest. 1993;91:257–263. doi: 10.1172/JCI116179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa-Garcia M, Li L, Riely G, Bray PF. Isolation and characterization of a TATA-less promoter for the human beta 3 integrin gene. Blood. 1994;83:668–676. [PubMed] [Google Scholar]
- Wilhide CC, et al. The human integrin beta3 gene is 63 kb and contains a 5′-UTR sequence regulating expression. Blood. 1997;90:3951–3961. [PubMed] [Google Scholar]
- Wingender E, et al. The TRANSFAC system on gene expression regulation. Nucleic Acids Res. 2001;29:281–283. doi: 10.1093/nar/29.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, et al. The murine mutation osteopetrosis is in the coding region of the macrophage colony stimulating factor gene. Nature. 1990;345:442–444. doi: 10.1038/345442a0. [DOI] [PubMed] [Google Scholar]
- Zhu HJ, Ross FP, Cao X, Teitelbaum SL. Phorbol myristate acetate transactivates the avian beta 3 integrin gene and induces alpha v beta 3 integrin expression. J Cell Biochem. 1996;61:420–429. doi: 10.1002/(SICI)1097-4644(19960601)61:3%3C420::AID-JCB9%3E3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]


