Abstract
MicroRNA167 (miR167) was shown to cleave auxin responsive factor 8 (ARF8) mRNA in cultured rice cells. MiR167 level was found to be controlled by the presence of auxin in the growth medium. When cells grew in auxin-free medium, miR167 level decreased, resulting in an increase in the level of ARF8 mRNA. Cells growing in the normal growth medium containing auxin showed a reversed trend. It was also shown that expression of OsGH3-2, an rice IAA-conjugating enzyme, was positively regulated by ARF8. Delivery of synthesized miR167 into cells led to decrease of both ARF8 mRNA and OsGH3-2 mRNA. This study provides an evidence in which the exogeneous auxin signal is transduced to OsGH3-2 through miR167 and ARF8 in sequence. This proposed auxin signal transduction pathway, auxin-miR167-ARF8-OsGH3-2, could be, in conjunction with the other microRNA-mediated auxin signals, an important one for responding to exogeneous auxin and for determining the cellular free auxin level which guides appropriate auxin responses.
INTRODUCTION
Phytohormone auxin plays diverse and pivotal roles in many aspects of the growth and development of plants. Its participation in different tissues and/or developmental stages, often in combination with the other hormones, suggests that auxin-mediated plant responses involve complex signal interplay (1–3). Although most of the signaling complexity is not yet understood, a critical step toward understanding of auxin signaling was made recently, when a gene family of auxin responsive factors (ARFs) was recognized in Arabidopsis (4,5). ARFs are the transcription factors that bind to the auxin-responsive cis-acting element in early auxin response genes including Aux/IAA, ACS, SAUR, GH2/4 and GH3 (6–9). There are also several transcription repressors Aux/IAA that dimerize with ARFs to repress transcription of these early auxin response genes. ARFs and Aux/IAA share their respective C-terminal domains for heterodimerization (10,11). Twenty-three ARFs have been found in Arabidopsis (6), implying that these ARFs participates in many different auxin-mediated plant responses. It is known that ARFs 1–4 and 9 repress their respective target genes, while ARFs 5–8 activate them (9,12). In the presence of inducible level of auxin, ARFs homodimerize, while Aux/IAA proteins are proteolyzed via the ubiquitin-pathway (13,14). It is also known that posttranscriptional cleavage of ARF mRNAs by microRNA plays an important role in auxin signaling (15–17).
MicroRNAs are single-stranded RNA molecules of ∼21 nt and are known to regulate expression of many developmentally important transcription factor genes either by cleavage of mRNAs or translational repression (18–21). In plants, cleavage of the target mRNA appears to be a prevalent way of posttranscriptional regulation (15,18,22). A number of pairs of microRNAs and their respective mRNA targets were predicted by database analyses (23), and a few of them were experimentally confirmed. Among these, miR160 was shown to target ARF10, ARF16 and ARF17 (16). Plants transformed with a miR160-resistant form of the ARF17 sequence showed dramatic phenotypic abnormalities due to the inhibition of posttranscriptional regulation of ARF17 by miR160 (16). It was also noted that miR160-directed ARF17 mRNA cleavage was accompanied by the down-regulation of several of the GH3 gene families. GH3 proteins are known to catalyze the conjugation of IAA to the different compounds to control the cellular concentration of free IAA (24,25). In Arabidopsis, the GH3 gene families are composed of the 19 members and they are divided into the three groups according to the similarities of their sequences (9,25). In the rice genome, there appeared to be 12 members of auxin-inducible GH3 genes (26). Participation of microRNA167 (miR167)-mediated cleavage of ARF8 mRNA in the down-regulation of GH3 gene expression is investigated in this study.
Recently, by studying the ARF8 T-DNA insertion mutant (arf8-1) and an overexpression line (ARF8-OX) in Arabidopsis, Tian et al. (27) demonstrated that ARF8 controlled the auxin level in a negative feedback manner by regulating the transcription of the GH3 family of genes. The arf8-1 and the ARF8-OX lines promoted and inhibited lateral root formation, respectively, as well as other developmental defects. It was also demonstrated that expression of the GH3 genes was reduced in the arf8-1 and was increased in the ARF8-OX, indicating that these GH3 genes were positively regulated by ARF8. In the ARF8-OX, the level of free IAA was reduced by 30% in light-grown hypocotyls, most likely due to the repressed GH3-mediated IAA-conjugation activity. It was also shown that two of the three GH3 genes were shown to be auxin-inducible, indicating that an adequate level of free IAA was controlled by ARF8 through the modulation of these GH3 genes. These results collectively suggested that microRNA-mediated cleavage of ARF8 and ARF17 plays a role in controlling the respective downstream GH3 family subsets to fine-tune the level of free IAA.
Increasing amounts of evidence have been accumulated to conclude that proper microRNA-mediated auxin signaling is essential for guiding normal auxin-mediated plant responses. As a principal component of the RNA-induced silencing complex, AGO1 protein has a pleiotropic effect on the processing of microRNA cleavage of the target mRNAs. It was reported that ARF17 was over-accumulated and the expression of several auxin-inducible GH3 genes were down-regulated in hypocotyls of ago1 mutant Arabidopsis (28). It was also shown that miR164 cleaved the NAC1 transcription factor, which controls auxin-mediated emergence of lateral roots (29,30). These lines of evidence indicate that various microRNAs control the functions of multiple components in the transduction of auxin signals. In this study, an evidence is provided to demonstrate miR167-directed posttranscriptional down-regulation of ARF8 and OsGH3-2 in cultured rice cells. By demonstrating the control of miR167 level by exogeneous auxin, we propose an auxin signaling pathway from exogeneous auxin to cellular free auxin via the stepwise signal relay miR167-ARF8-OsGH3-2.
MATERIALS AND METHODS
Plant materials and growth conditions
Rice suspension cells were derived from the ovules (germinal vesicles) of Oryza sativa Japonica cv. Dongjin. The culture was maintained in the CHU (N6) medium (31) containing 3% sucrose, 2 mg/l 2,4-D, 0.2 mg/l kinetin and vitamins (pH 5.8). The culture was routinely transferred to fresh 60 ml CHU(N6) medium every week and was grown in the dark with gentle shaking (0.1128 g) at 25°C. For the acetate culture, sucrose was replaced with 10 mM potassium acetate.
Preparation of double-stranded RNA (dsRNA) (ARF8) and miR167 and their delivery into cells
To prepare dsRNA (ARF8), the DNA templates were amplified by PCR, which introduced T7 RNA polymerase-binding sequences at the 5′ end of the both strands. The primer sequences for ARF8 are 5′-TAATACGACTCACTATAGGGAGACCCACAAGCAAAGATGGT-3′ (forward) and 5′-TAATACGACTCACTATAGGGAGAATGCACATCCTCAGGTGA-3′ (reverse). The underlined parts represent the T7 promoter region. The 0.48 kb internal region of the ARF8 coding sequence was amplified. The products were transcribed in the both directions by T7 RNA polymerase using the Megascript T7 Transcription Kit (Ambion, USA) to generate both sense and antisense RNAs. After the DNA templates were removed by DNaseI, RNA was purified by phenol-chloroform extraction and was precipitated using ethanol. The pellet was resuspended in 0.1% DEPC-treated water and annealed into dsRNA at 65°C. The dsRNA was quantified using GeneQuant (Pharmacia Biotech, USA) and stored at −20°C. Two complementary strands of miR167 were commercially synthesized (Samchully Pharm, Korea). The sense strand (5′-UGAAGCUGCCAGCAUGAUCTT-3′) and antisense strand (5′-GAUCAUGCUGGCAGCUUCATT-3′) were annealed into dsRNAs at 30°C before being delivered into cells.
To deliver dsRNA(ARF8), cationic oligopeptide of 12 arginine polymer (POA; Peptron Inc., Korea) was used to form a dsRNA/POA complex by electrostatic interaction, as described in a previous paper from our laboratory (32). CHU (N6) medium was removed from 1 ml of 3 day old rice suspension cells (∼1 g fresh cell) and the cells were rinsed with phosphate-buffered saline (PBS) buffer. Cells were mixed with 30 μg of POA-complexed dsRNA or with 100 nM of synthesized microRNA alone without POA in a final volume of 500 μl. To allow penetration into cells, these mixtures were shaken (0.1128 g) for 1 h at 25°C. Fresh CHU (N6) medium (5 ml) was added and incubated for the designated periods.
RT–PCR analysis
Total RNA was prepared from the samples using TRI reagent (Molecular Probes Diagnostics, USA) according to the manufacturer's instructions. Approximately 1 μg of total RNA was used to prepare cDNA using a RevertAid™ First Strand cDNA Synthesis Kit (Fermentas, Lithuania). Oligo(dT)18 primer was used for PCR amplification in a reaction volume of 20 μl. PCRs were run for 30 cycles at the annealing temperature of 58°C (for ARF8) or 55°C (for GH3). The following gene-specific primers were used for PCR amplification, ARF8, 5′-AGCAAGGAAATGATCCACGGT-3′ (forward) and 5′-ACTGACATAACCGGACAATCG-3′ (reverse); OsGH3-2, 5′-GCAGTACATCCCCACGCTCAA-3′ (forward) and 5′-GCTCGTGTACTCCACGATGCT-3′ (reverse). The rice actin gene (forward; 5′-TCCATCTTGGCATCTCTCAG-3′; and reverse; 5′-GTACCCGCATCAGGCATCTG-3′) was used as an internal control.
RNA gel blot analysis
MiR167 was detected as described by Lau et al. (33), with the following modifications. Low molecular weight (LMW) RNAs containing miR167 were precipitated with the 0.5% PEG8000/0.5 M NaCl, 10 μg of yeast tRNA and 3 vol of ethanol for 2 h at −70°C. The precipitates were dissolved in 0.1% DEPC-treated water. LMW RNAs were separated in a 15% denaturing polyacrylamide gel and were subjected to blot-hybridization analysis. A small RNA (miR167a, 5′-TAGATCATGCTGGCAGCTTCA-3′) probe was labeled with [γ32P]ATP by T4 kinase (TaKaRa, Japan). The filter was pre-hybridized in 5× SSC, 20 mM Na2HPO4 (pH 7.2), 7% SDS, 2× Denhardt's Solution and 1 mg of denatured salmon sperm DNA. Hybridization was carried out under the same conditions with the addition of a [γ-32P]ATP-end-labeled oligonucleotide probe. After hybridization, the filter was washed four times in non-stringent solution [3× SSC, 25 mM Na2HPO4 (pH 7.5), 5% SDS and 10× Denhardt's Solution] and once in stringent solution (1× SSC and 1% SDS) at 50°C. The dried filter was exposed to X-ray film at −70°C.
For mRNA detection, 10 μg of total RNA was fractionated by electrophoresis on a 1.2% (w/v) agarose gel containing formaldehyde. It was transferred to a nylon membrane (NEN GeneScreen™, USA) overnight, and probed with PCR-amplified fragments of the target RNA (OsGH3-2) labeled with [α32P]dCTP by random priming. Hybridization was performed at 50°C by using Rapid-hyb Buffer Solution (Amersharm, USA) according to the manufacturer's instructions.
Primer extension and 5′-RACE
Primer extension analysis was carried out as described previously by Kim and co-workers (34), with some modifications. Total RNA (10 μg) was used for annealing to the 5′ end-labeled primer (5′-ATGCACATCCTCAGGTGAAAG-3′) at 35°C for 16 h and for subsequent extension using Super Script™II (invitrogen, CA) at 42°C for 2 h. Samples were separated by 6% denaturing polyacrylamide gel and autoradiographed.
To determine the internal cleavage site in ARF8 mRNA, RNA ligase-mediated rapid amplification of cDNA ends (5′-RACE) was performed with the GeneRacer Kit (Invitrogen, USA). Total RNA was isolated from 3 day old suspension cells. The experimental details were essentially followed by the manufacturer's instructions. A unique gene-specific DNA fragment (0.54 kb) was amplified. Gel-purified PCR products were cloned into pGEM-T Easy vector (Promega, USA) for DNA sequencing.
RESULTS AND DISCUSSION
MiR167-directed cleavage of ARF8 mRNA
MiR167 is a microRNA of 21 nt in length and it is present in several isoforms. These isoforms present in Arabidopsis thaliana, O.sativa, Sorghum bicolor and Zea mays (Figure 1A), suggesting that the miR167 sequences are highly conserved among these evolutionary distant species. MiR167 was shown to be expressed stably in cultured rice cells grown either on sucrose or acetate as the sole carbon sources (Figure 1B). Considering that cells growing on sucrose multiply much faster than cells on acetate, it was assumed that the cellular miR167 level was independent of growth rate and/or carbon source. In contrast, miR167 level was found to be sensitive to the presence of auxin in the growth medium, as will be shown later (Figure 5A), justifying the studies of the miR167-dependent auxin signaling in these cells.
Figure 1.
Conserved miR167 sequences among different plants and its expression in sucrose and acetate cultures. (A) The miR167 isogene sequences of O.sativa, A.thaliana, S.bicolor and Z.mays. These miR167 isogenes are differed in their 3′ end sequences. (B) The levels of miR167 in cells grown either in sucrose (3%) or acetate (10 mM). RNA blot was performed to detect miR167 in the total RNAs harvested from cells grown either in sucrose (lane suc) or acetate (lane ace) as a single carbon sources. tRNA and 5S rRNA were used as the internal RNA controls (ethidium bromide-stained bands at bottom).
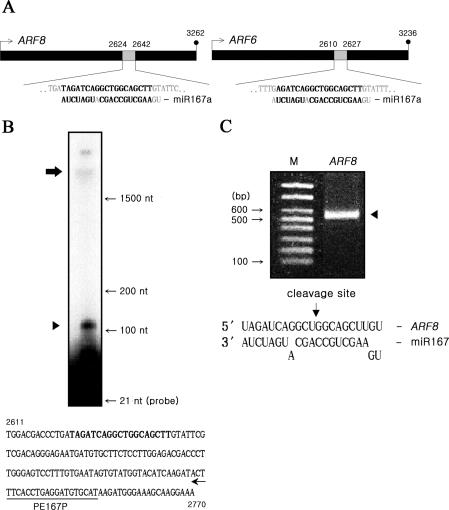
Database analysis predicted that miR167 targets both ARF8 and ARF6 in Arabidopsis and rice (23,35). The miR167 sequence was near perfectly complementary to the internal sequence of these two respective target mRNAs in rice (Figure 2A). Bona fide cleavage of ARF8 mRNA by miR167 was demonstrated by primer extension analyses in which a single primer of the 3′-proximal region of the ARF8 gene generated the expected 117 bp 3′-cleavage products (Figure 2B). The specific cleavage site of the ARF8 mRNA was determined by 5′-RACE (rapid amplification of cDNA ends), which takes advantage of the monophosphate present at the 5′ terminus of the 3′ cleavage fragment (15,36). When RNA isolated from 3 day old rice cells was subjected to 5′-RACE, a distinct PCR band was observed for the ARF8 target (Figure 2C). Cloning and DNA sequencing of this amplified product mapped the 5′ end of the cleavage products to the designated position at the middle of the region complementary to miR167 (Figure 2C). These results clearly indicated that miR167 cleaved the specific site within the ARF8 mRNA in rice, confirming that miR167-ARF8 auxin signal transduction operates also in rice as in Arabidopsis.
Figure 2.
MiR167 cleavage of ARF8 mRNA. (A) The regions within the ARF8(left)/ARF6(right) coding sequences complementary to the miR167a (expanded diagrams). Nucleotide mismatches are shown in light–dark color. (B) Primer extension analysis to confirm the cleavage of the target ARF8 mRNA. The upper arrow represents the full-size mRNA, and the bottom arrowhead the expected 117 bp 3′ end cleavage products ARF8 mRNA. The underlined portion represents the primer-binding region. (C) 5′-RACE to determine the cleavage site. Nested PCR product (lane ARF8), spanning between the primer and cleavage sites, was used for cloning into a vector for DNA sequencing. Lane M represents 100 bp DNA ladders. The ARF8 mRNA cleavage site is designated at the 11th base from the 5′ end, between U and G indicated by the arrow (bottom).
Confirmation of miR167-ARF8-OsGH3-2 regulatory pathway
Prior to investigating whether GH3 expression is regulated by ARF8 also in rice cells, Rice GH3 homologs were searched. In Arabidopsis, mRNA levels of these GH3 genes, DFL1, AtGH3a and At1g28130 were shown to be decreased by ∼25–50% in mutant arf8-1, and increased by ∼50% in the ARF8 overexpression line (27), suggesting that these three members of the GH3 family are the targets of the ARF8. According to the sequence homology to its Arabidopsis counterpart, the GH3 genes were shown to be present in rice (26). Among the rice GH3 homologs (OsGH3) to these Arabidopsis genes, the published OsGH3-2 sequence (26) exhibited the highest homology to the Arabidopsis DFL1 counterpart (73% homology at the 390 bp span in blast search) and OsGH3-2 was used to monitor ARF8 control of GH3 gene expression.
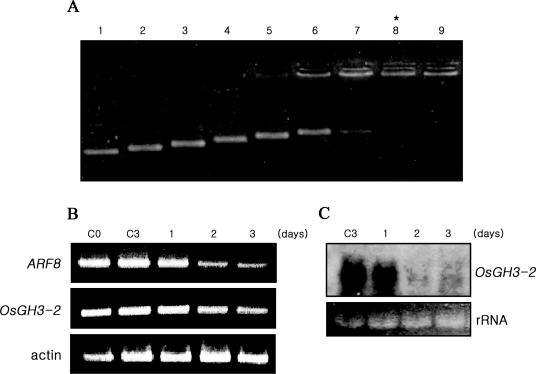
To investigate ARF8 control of OsGH3-2 expression, we attempted to inhibit ARF8 expression by posttranscriptional gene silencing (PTGS). To perform PTGS, 0.48 kb dsRNA, which corresponds to the coding sequence of ARF8, was synthesized in vitro and was delivered exogeneously into cells, using cationic oligopeptide polyarginine-12mer (POA) as a carrier (32) (Figure 3A). Both RT–PCR and northern analyses demonstrated that the levels of ARF8 and OsGH3-2 mRNAs dropped upon delivery of ARF8 dsRNA (Figure 3B and C). These results demonstrated that expression of OsGH3-2 was positively regulated by ARF8, indicating that ARF8 regulation of OsGH3-2 expression is also conserved in rice as in Arabidopsis.
Figure 3.
dsRNA(ARF8) delivery for PTGS of ARF8 expression. (A) Formation of the dsRNA/POA complex. To evaluate an optimum complex formation, various concentrations of POA were mixed with a fixed concentration of dsRNA (ARF8) (lane 1, 100 ng of dsRNA only; lanes 2–9, 12.5, 25, 37.5, 50, 62.5, 75, 87.5 and 100 ng of POA mixed with 100 ng of dsRNA, respectively). As the POA concentrations increased, the mobilities of dsRNA gradually were gradually retarded. The POA concentration at the lane 8 was chosen to be the most effective delivery-complex, as dsRNA appeared to be protected from ethidium bromide staining by the presumed POA shell (31). (B) RT–PCR analysis of PTGS of the ARF8 and OsGH3-2 expressions in dsRNA (30 μg)-delivered cells. Unlike the control cells harvested at the day 0 (lane C0) and day 3 (lane C3) following the treatment with PBS buffer only, dsRNA-treated cells exhibited gradual and apparent decreases in the RT–PCR products of both ARF8 and OsGH3-2 in cells grown for 1–3 days (lanes 1–3) following the dsRNA treatments. Rice actin was used as an internal control. (C) RNA blot analysis of OsGH3-2 mRNA. To further confirm the PTGS of OsGH3-2 transcripts, the same total RNAs, used for the RT–PCR analyses (B), were subjected to northern hybridization. While the OsGH3-2 transcripts remained in the untreated controls (C3), they decreased in dsRNA-treated cells (lanes 1–3).
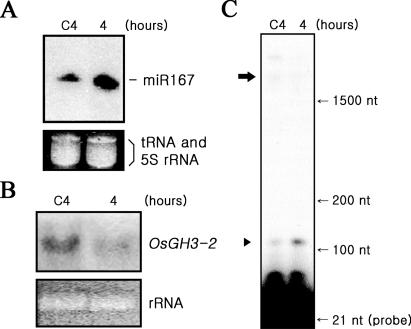
To determine whether ARF8 regulation of OsGH3-2 is controlled by miR167, artificially synthesized miR167 was delivered into rice cells. In cells harvested 4 h after the delivery of miR167, the cellular miR167 level increased while the OsGH3-2 mRNA level decreased (Figure 4A and B). It was also shown that, compared to untreated control cells, cleavage of ARF8 mRNA increased evidently in these miR167-treated cells (Figure 4C), indicating that miR167-mediated cleavage of ARF8 mRNA led to down-regulation of OsGH3-2. These lines of experiments clearly demonstrated that the stepwise signal transduction miR167-ARF8- OsGH3-2 operates in rice cells.
Figure 4.
MiR167 control of ARF8-regulation of OsGH3-2. (A) Increased level of cellular miR167 upon delivery of synthesized miR167. To cells growing in auxin-containing medium, 100 nM of artificially synthesized miR167 was delivered and its cellular level was measured at 4 h after the delivery (lane 4). Lane C4 represents the PBS-treated control. (B) RNA blot assay of OsGH3-2 mRNA. Northern hybridization was performed using the same total RNAs used in A using OsGH3-2 coding region as the probe. (C) Increased cleavage of ARF8 mRNA. The same cells, used in (A and B), were examined for the cleavage of ARF8 mRNA by primer extension. The 117 bp 3′-cleavage products of ARF8 mRNA increased in miR167-treated cells (lane 4), compared with the untreated control (lane C4). The upper arrow and the bottom arrowhead represent the full-size mRNA and the 3′ end cleavage product, respectively.
Control of miR167 level by auxin
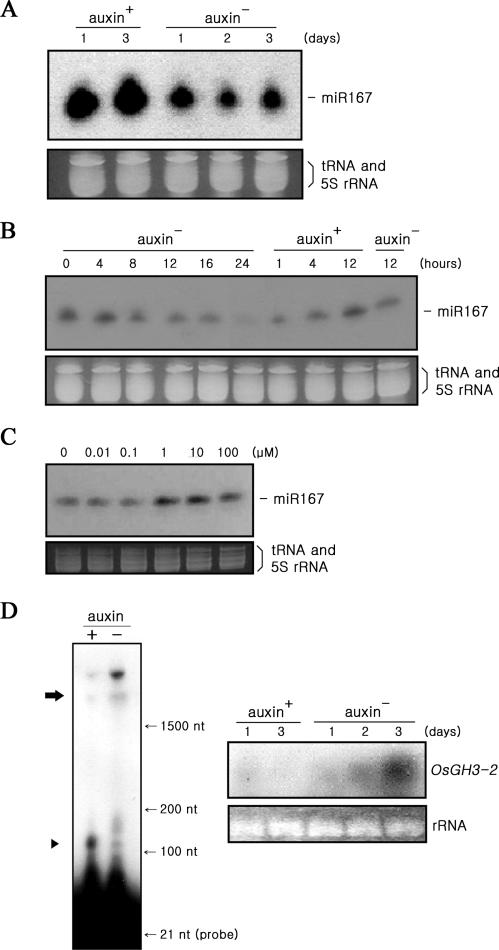
To investigate whether the level of miR167 and the miR167-ARF8-OsGH3-2 signal transduction are controlled by exogeneous auxin, they were analyzed in the presence and absence of auxin in the medium. When cells, growing on normal CHU(N6) medium containing 9 μM of synthetic auxin 2,4-dichlorophenoxyacetic acid (2,4-D), were transferred to auxin-free medium, miR167 level dropped significantly (Figure 5A), indicating that miR167 level responds to the presence of auxin in the medium. It appeared that it responded insignificantly to the presence of the other phytohormones such as cytokinin, ABA and gibberellin (data not shown). It was shown that miR167 level was gradually decreased when cells grown on auxin-containing medium transferred to auxin-free medium (left six auxin− columns, Figure 5B). MiR167 accumulated during growth on auxin medium (0 h) was disappeared after 24 h following the transfer. Alternating increase and decrease of miR167 level were evident when cells growing on auxin-free medium were transferred to auxin-containing medium (three auxin+ columns) and to auxin-free medium (right auxin− column). Following the transfers, cellular levels of auxin should change gradually as time progressed. It was noted that miR167 level was significantly increased within 1 h following the transfer to auxin-containing medium from auxin-free medium (Figure 5B), suggesting that auxin effect on miR167 is rapid. It was also shown that the level of miR167 appeared to be saturated at the concentrations of higher than 1 μM 2.4-D, while the level was decreased significantly at the concentrations lower than 1 μM (Figure 5C). Lowered level of miR167 in cell grown in auxin-free medium was shown to be accompanied by decreased cleavage of ARF8 mRNA (increased ARF8 full-length mRNA) and increased OsGH3-2 transcripts (Figure 5D). Delivery of synthesized miR167 into cells grown on auxin-free medium for 3 d (Figure 6A) led to an increase of the 3′-cleavage products of ARF8 mRNA (Figure 6B), resulting in decrease of OsGH3-2 transcripts (Figure 6C), substantiating the participation of miR167 in this auxin signal pathway. These results clearly demonstrated that the presence of auxin in the medium controlled the level of miR167 and in turn miR167-mediated regulation of ARF8 and OsGH3-2.
Figure 5.
Auxin control of the miR167 level. (A) Decrease of the miR167 level in cells growing in the auxin-free medium. The levels of miR167 in cells harvested at the first and second days following the growth in medium containing 9 μM of 2,4-D (left two lanes) and in cells harvested 1–3 days following the transfer of cells from the auxin-containing medium to the auxin-free medium (right three lanes). (B) Changes of the miR167 level following transfer between auxin+ and auxin− media. Cells growing on auxin+ medium were transferred to auxin− medium, and measured miR167 at the designated times following the transfer (left six auxin− columns). These cells grown on auxin- medium for 24 h were transferred to auxin+ medium to measure miR167 at the designated times (the three auxin+ columns). These cells grown on auxin+ medium for 12 h were transferred back to aux− medium and measured miR167 at 12 h after transfer (right auxin- column). (C) Auxin-concentration effects on the miR167 level. The miR167 levels were measured in cells growing for 24 h on the medium containing the designated concentrations of 2,4-D. (D) Auxin control of cleavage of ARF8 mRNA and of OsGH3-2 expression. (Left column) While ARF8 mRNA remained intact in cells grown in auxin-free medium (right lane), they were efficiently cleaved in cells grown in auxin-containing medium (left lane). (Right column) Northern blot analyses of the expression of the OsGH3-2 gene in cells growing in auxin-free medium (right three lanes) and in auxin-containing medium (left two lanes). The strongest OsGH3-2 signal is shown in cells grown for three days in auxin-free medium.
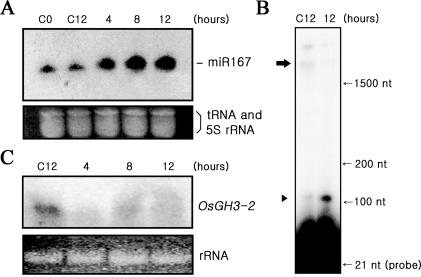
Figure 6.
Delivery of miR167 into cells grown in auxin-free medium. (A) Delivery of miR167 (100 nM) into cells growing in the auxin-free medium. The levels of miR167 were measured in cells, which have grown in auxin-free medium for three days, harvested at 4, 8 and 12 h following the delivery of miR167 (right three lanes) and in untreated control cells (left two lanes). Lane C12 represents cells harvested 12 h following the treatment of PBS buffer only in the absence of miR167. (B) Increased cleavage of ARF8 mRNA in miR167-delivered cells. Cleavage products of ARF8 mRNA are shown in cells harvested 12 h following the delivery of miR167 (lane 12), but not in the control (lane C12). (C) RNA blot assay of OsGH3-2 expression in miR167-delivered cells. While OsGH3-2 mRNA remained in the untreated control (lane C12), it appeared down-regulated in miR167-delivered cells (right three lanes).
These experiments performed in cultured rice cells provided an direct evidence that miR167 is a signal mediator which controls OsGH3-2 expression via the cleavage of ARF8 mRNA. Although use of cells harboring miR167-resistant form of ARF8 could have confirmed further (we could not obtain these cells), applications of PTGS by delivering dsRNA (ARF8) and monitoring of cleavage of ARF8 mRNA and the level of OsGH3-2 transcripts following the delivering of the synthesized miR167 allowed us to carefully examine the connections of the auxin signaling pathway.
It was also shown that the level of miR167 was controlled by exogeneous auxin, although we do not know whether auxin effects on the miR167 level is direct. According to the experiments carried out in Arabidopsis seedling, there were little changes in the levels of miR160, miR164 and miR167, when it was treated with 10 μM of IAA (16). In a similar experimental condition, there was a modest but consistent increase of the miR164 level upon treatment with 10 μM of synthetic auxin NAA (29). It is speculative that, depending on the auxin concentrations and target tissues, a microRNA is chosen to operate a respective auxin signal pathway.
Being established that GH3 participates in the control of cellular free IAA levels by its IAA-conjugating activity, the proposed pathway may represent a scenario in which exogeneous auxin determines its own cellular level in a positive feedback manner. Auxin repression of OsGH3-2 is expected to increase cellular free IAA level. Knowing that some GH3 genes are auxin-inducible, negative feedback control can also be adopted in determining cellular free IAA level (27). This notion strongly suggests that isozymes of ARFs and GH3s form a network in addition to microRNAs. It is likely that selected isozymes in particular developmental stages and tissues would adopt positive or negative loops in determining the free IAA levels. To make things more complicated, one microRNA can target more than one ARF, as the case of miR167 whch is predicted to target both ARF8 and ARF6. In a preliminary study, ARF6 mRNA was also cleaved by miR167 in rice cells (data not shown). It is speculative that these two ARF mRNAs are differentially chosen depending on the concentration of auxin and/or some other factors.
Transcriptional control of auxin responses involves another branch in addition to the microRNA/ARF-mediated one. Aux/IAA repressor is known to be proteolyzed in the presence of auxin (13,14). Recently, it was reported that this protein degradation is mediated by the Transport Inhibitor Response1 (TIR1) which is found to be the auxin receptor (37,38). Auxin is also possible to control directly ARF8 and/or GH3 without involving microRNA. It should be a very intriguing question how auxin controls these multiple targets in a concerted way to determine the final cellular free IAA concentration to perform the auxin responses. By exploring more detailed studies of the other ARFs and their respective downstream GH3 targets, a broad overview of microRNA-mediated auxin signaling circuits will likely be delineated. Confirmation of this auxin-miR167-ARF8-OsGH3-2 pathway proposed in this study could be an important step toward understanding of the complex auxin signal interplay.
Acknowledgments
We thank Drs M.S. Yang and W.S. Jung for providing rice cells. This work was supported by a grant (20050301034405) from BioGreen 21 Program, Rural Development Administration, Republic of Korea. Funding to pay the Open Access publication charges for this article was provided by the Sungkyunkwan University, Korea.
Conflict of interest statement. None declared.
REFERENCES
- 1.Swarup R., Parry G., Graham N., Allen T., Bennett M. Auxin cross-talk: integration of signaling pathways to control plant development. Plant Mol. Biol. 2002;49:411–426. doi: 10.1007/978-94-010-0377-3_12. [DOI] [PubMed] [Google Scholar]
- 2.Friml J. Auxin transport—shaping the plant. Curr. Opin. Plant Biol. 2003;6:7–12. doi: 10.1016/s1369526602000031. [DOI] [PubMed] [Google Scholar]
- 3.Jurgens G. Growing up green: cellular basis of plant development. Mech. Dev. 2003;120:1395–1406. doi: 10.1016/j.mod.2003.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Guilfoyle T.J., Hagen G., Ulmasov T., Murfett J. How does auxin turn on genes? Plant Physiol. 1998;118:341–347. doi: 10.1104/pp.118.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guilfoyle T.J., Ulmasov T., Hagen G. The ARF family of transcription factors and their role in plant hormone-responsive transcription. Cell Mol. Life Sci. 1998;54:619–627. doi: 10.1007/s000180050190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abel S., Theologis A. Early genes and auxin action. Plant Physiol. 1996;111:9–17. doi: 10.1104/pp.111.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ulmasov T., Hagen G., Guilfoyle T.J. Activation and repression of transcription by auxin-response factors. Proc. Natl Acad. Sci. USA. 1999;96:5844–5849. doi: 10.1073/pnas.96.10.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ulmasov T., Hagen G., Guilfoyle T.J. Dimerization and DNA binding of auxin response factors. Plant J. 1999;19:309–319. doi: 10.1046/j.1365-313x.1999.00538.x. [DOI] [PubMed] [Google Scholar]
- 9.Hagen G., Guilfoyle T. Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol. Biol. 2002;49:373–385. [PubMed] [Google Scholar]
- 10.Rogg L.E., Bartel B. Auxin signaling: derepression through regulated proteolysis. Dev. Cell. 2001;1:595–604. doi: 10.1016/s1534-5807(01)00077-6. [DOI] [PubMed] [Google Scholar]
- 11.Liscum E., Reed J.W. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 2002;49:387–400. [PubMed] [Google Scholar]
- 12.Tiwari S.B., Hagen G., Guilfoyle T. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell. 2003;15:533–543. doi: 10.1105/tpc.008417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dharmasiri S., Estelle M. The role of regulated protein degradation in auxin response. Plant Mol. Biol. 2002;49:401–409. [PubMed] [Google Scholar]
- 14.Kepinski S., Leyser O. Ubiquitination and auxin signaling: a degrading story. Plant Cell. 2002;14:S81–S95. doi: 10.1105/tpc.010447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kasschau K.D., Xie Z., Allen E., Llave C., Chapman E.J., Krizan K.A., Carrington J.C. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell. 2003;4:205–217. doi: 10.1016/s1534-5807(03)00025-x. [DOI] [PubMed] [Google Scholar]
- 16.Mallory A.C., Bartel D.P., Bartel B. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell. 2005;17:1360–1375. doi: 10.1105/tpc.105.031716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J.W., Wang L.J., Mao Y.B., Cai W.J., Xue H.W., Chen X.Y. Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell. 2005;17:2204–2216. doi: 10.1105/tpc.105.033076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Llave C., Xie Z., Kasschau K.D., Carrington J.C. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 19.Zeng Y., Yi R., Cullen B.R. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl Acad. Sci. USA. 2003;100:9779–9784. doi: 10.1073/pnas.1630797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartel D. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 21.Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang G., Reinhart B.J., Bartel D.P., Zamore P.D. A biochemical framework for RNA silencing in plants. Genes Dev. 2003;17:49–63. doi: 10.1101/gad.1048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rhoades M., Reinhart B., Lim L., Burge C., Bartel B., Bartel D. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 24.Ljung K., Hull A.K., Kowalczyk M., Marchant A., Celenza J., Cohen J.D., Sandberg G. Biosynthesis, conjugation, catabolism and homeostasis of indole-3-acetic acid in Arabidopsis thaliana. Plant Mol. Biol. 2002;49:249–272. [PubMed] [Google Scholar]
- 25.Staswick P.E., Serban B., Rowe M., Tiryaki I., Maldonado M.T., Maldonado M.C., Suza W. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell. 2005;17:616–627. doi: 10.1105/tpc.104.026690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jain M., Kaur N., Tyagi A.K., Khurana J.P. The auxin-responsive GH3 gene family in rice (Oryza sativa) Funct. Integr. Genomics. 2006;6:36–46. doi: 10.1007/s10142-005-0142-5. [DOI] [PubMed] [Google Scholar]
- 27.Tian C.E., Muto H., Higuchi K., Matamura T., Tatematsu K., Koshiba T., Yamamoto K.T. Disruption and overexpression of auxin response factor 8 gene of Arabidopsis affect hypocotyl elongation and root growth habit, indicating its possible involvement in auxin homeostasis in light condition. Plant J. 2004;40:333–343. doi: 10.1111/j.1365-313X.2004.02220.x. [DOI] [PubMed] [Google Scholar]
- 28.Sorin C., Bussell J.D., Camus I., Ljung K., Kowalczyk M., Geiss G., Mckhann H., Garcion C., Vaucheret H., Sandberg G., et al. Auxin and light control of adventitious rooting in Arabidopsis require ARGONAUTE1. Plant Cell. 2005;17:1343–1359. doi: 10.1105/tpc.105.031625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo H.S., Xie Q., Fei J.F., Chua N.H. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell. 2005;17:1376–1386. doi: 10.1105/tpc.105.030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie Q., Frugis G., Colgan D., Chua N.H. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 2000;14:3024–3036. doi: 10.1101/gad.852200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chu C.C., Wang C.S., Sun C.S., Hus C., Ying K.C., Chu C.Y., Bi F.Y. Establishment of an efficient medium for anther culture of rice through comparative experiments on the nitrogen sources. Sci. Sin. 1975;18:659–668. [Google Scholar]
- 32.Unnamalai N., Kang B.G., Lee W.S. Cationic oligopeptide-mediated delivery of dsRNA for post-transcriptional gene silencing in plant cells. FEBS Lett. 2004;566:307–310. doi: 10.1016/j.febslet.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 33.Lau N.C., Lim L.P., Weinstein E.G., Bartel D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 34.Lee Y., Kim M., Han J., Yeom K.H., Lee S., Baek S.H., Kim V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reinhart B.J., Weinstein E.G., Rhoades M.W., Bartel B., Bartel D.P. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mallory A.C., Reinhart B.J., Jones-Rhoades M.W., Tang G., Zamore P.D., Barton M.K., Bartel D.P. MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. EMBO J. 2004;23:3356–3364. doi: 10.1038/sj.emboj.7600340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dharmasiri N., Dharmasiri S., Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 38.Kepinski S., Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]