Abstract
Sequence analysis of alphoid repeats from human chromosomes 17, 21 and 13 reveals recurrent diagnostic variant nucleotides. Their combinations define haplotypes, with higher order repeats (HORs) containing identical or closely-related haplotypes tandemly arranged into separate domains. The haplotypes found on homologues can be totally different, while HORs remain 99.8% homogeneous both intrachromosomally and between homologues. These results support the hypothesis, never before demonstrated, that unequal crossovers between sister chromatids accumulate to produce homogenization and amplification into tandem alphoid repeats. I propose that the molecular basis of this involves the diagnostic variant nucleotides, which enable pairing between HORs with identical or closely-related haplotypes. Domains are thus periodically renewed to maintain high intrachromosomal and interhomologue homogeneity. The capacity of a domain to form an active centromere is maintained as long as neither retrotransposons nor significant numbers of mutations affect it. In the presented model, a chromosome with an altered centromere can be transiently rescued by forming a neocentromere, until a restored, fully-competent domain is amplified de novo or rehomogenized through the accumulation of unequal crossovers.
INTRODUCTION
Alpha satellite DNA covers the centromeric region of all human chromosomes over distances often as large as several megabases. It is composed of a basic 171 bp long unit that is organized into tandemly arranged higher order repeats (HORs); HORs contain variable numbers of basic repeats, from 4 (chromosome 2) to as many as 34 (chromosome Y) (1,2). The HORs of a given chromosome are highly homogeneous, with sequence identity often exceeding 99% along the blocks they constitute. The length of the alphoid blocks also varies substantially. For example, on chromosome 21, it can cover from less than 100 kb up to almost 6 Mb (3,4). On chromosome 5, a 6-fold variation has been estimated for D5Z1 (5). Alphoid blocks have been detected in all primate species in which they have been searched for (6), and are recognized as constituting the site where active centromeres are formed in human chromosomes (7).
As suggested by Alexandrov et al. (8), centromeric regions of lower primates only have ‘old’ alpha satellite families based on type A monomeric units. Subsequently, an ancestor to the great apes acquired a new class of monomers, called type B, which has the ability to bind the CENP-B protein. Because the resulting ‘new’ alpha satellite families, which are based on A-B monomers, spread before the human–chimpanzee–gorilla split, human centromeric regions generally contain one alphoid array made of irregular A-B types of monomers and one or several of the older arrays. The old monomeric arrays, which in humans have lost their capacity to form active centromeres, have ceased to be homogenized and exhibit average pairwise sequence identities of ∼72% (9). Moreover, on both chromosomes X and 17 (10,11), the old monomeric arrays have diverged from their human counterparts in exactly the same way as the adjacent non-satellite DNA sequences (>98% sequence identity in the human/chimpanzee comparison).
In contrast, the new arrays continue to be efficiently homogenized within each species, leading to a higher divergence in the human/chimpanzee comparison (only 92–93% sequence identity). The sequence has therefore substantially changed simultaneously with structural changes both between chromosomes within each species and between orthologues between species: HORs are of different lengths and belong to different types of `Suprachromosomal Families' (SFs) (8).
It has been proposed that molecular drive (MD) (12) could account for the homogenization/amplification of repeated sequences within each species. The molecular mechanisms thought to operate during this process essentially include unequal crossing over and gene conversion (13). Smith (14) had previously proposed that the accumulation of unequal crossovers between sister chromatids during gametogenesis could lead to either homogenization or amplification of satellite DNA arrays which, in the absence of meiotic recombination, would otherwise diverge rapidly. On the other hand, gene conversion is the basic mechanism that Schindelhauer and Schwarz (15) invoked to account for their data, which were obtained from the analysis of a series of HORs from a single male X alphoid array.
I have used a strategy developed by Warburton and Willard (16) to perform a detailed analysis of the sequence variations found on several human chromosomes: 17, 21 and 13. On each of these three chromosomes, HORs exhibit diagnostic variant nucleotides that define a limited number of specific haplotypes. HORs of the same haplotype are tandemly arranged into specific domains along the corresponding alphoid arrays. Nevertheless, the overall DNA sequence of the HORs remains 99.8% homogeneous. An unexpected finding from this study is that on each homologue, identical domains are only partially shared, with extreme cases being represented by homologues, which are totally different in that respect in spite of their 99.8% intrachromosomal and inter-homologue sequence identities. This implies the periodic renewal of domains on each individual homologue lineage, which renewal nevertheless maintains high inter-homologue homogeneity. It is best explained by the accumulation of unequal crossovers (14), with pairing between misaligned repeats being either allowed or forbidden by a molecular mechanism involving the diagnostic variant nucleotides found on each chromosome of this study. As meiotic recombination within the alphoid sequences is highly restricted (5,17,18), inter-homologue homogeneity is therefore also mainly ensured by the same basic molecular mechanism.
Finally, the data described in this paper has prompted me to hypothesize how an alphoid domain can function as a centromere, and how it could be restored when its functionality has been greatly altered or even lost: the transient creation of a neocentromere would rescue the chromosome until the accumulation of unequal crossovers creates a domain de novo or re-homogenizes a pre-existing alphoid domain.
MATERIALS AND METHODS
DNA samples
DNA samples were prepared from several sources. Hybrid cell lines: GM10498, GM10321 and GM12506, each containing a single chromosome 17; GM10323 and GM08854 with a single chromosome 21, and GM11689 and GM11767 with a single chromosome 13. All were from Coriell Cell Repositories, as was the GM08729 cell line, which contains two chromosomes 17. Two other DNA samples were prepared from peripheral blood cells (#TOU and #103). Other cell lines, each containing a single rearranged chromosome 21 with unaffected centromeric sequences, were also used: 6918, 9542, 3;21 (19) as well as the chromosome 21 long alphoid array (>1 Mb) from YAC 831B6 (20).
PCR and cloning
DNA samples were PCR amplified using Promega Taq polymerase and associated buffer. Annealing temperature was 54°C. PCR products were resolved by electrophoresis on 1% agarose gels and the DNA fragments of interest were purified using the QIAGEN QIAquick Gel extraction Kit. Cloning was made in the Promega pGEM-T Easy Vector System. Positive individual clones were recovered and grown in 96 well plates. Primers 17-1A and 17-2A were from (21), 17-α1 and 17-α2 were their invcomps. Others were: 17-3A: (5′-TTATGGTCACATAAAAACTG-3′) and 17-4A: (5′-ATCTACTTGCAGTTTCTACAG-3′).
Sequence analysis
Individual clones were sequenced either with an ABI 377 sequencer or a MegaBACE (Amersham). In both cases, sequence reactions were performed with the DYEnamic ET Terminator kit of Amersham. In the former case, templates were PCR products of individual clones, using either SP6 or T7 as primer. In the second case, the plasmids of individual clones were amplified with the TempliPhi kit from Amersham prior to the sequence reaction.
Allele specific oligonucleotide (ASO) analysis
The four diagnostic variant nucleotides which were detected in a preliminary analysis of a few clones from the PCR product obtained with the two primers 17α1/17α2 were further analysed using the ASO approach. The oligonucleotide pairs were as follows: TCAAATCCCAGAGTTGAAC and TCAAATCCCCGAGTTGAAC, CAGAAGCATTCTCGAAGC and CAGAAGCATCCTCGAAGC, TAAAAACTATACAGATGCA and TAAAAACTACACAGATGCA and TGAAACACTATTTTTCTAG and TGAAACACTGTTTTTCTAG. Hybridization conditions were at 5°C under the average calculated Tm for each pair. Clones were dotted in double on to Hybond plus from Amersham and hybridized with labelled oligonucleotides.
Preparative pulsed field gel electrophoresis
DNA from hybrid cell line GM 10 498 was prepared in agarose plugs as described in (3). Agarose plugs were treated by various restriction endonucleases, with SacI providing a good resolution of five DNA fragments (F1–F5). They were recovered from a preparative agarose gel with the gel extraction kit QIAquick (QIAGEN). In order to localize the unlabelled fragments on the gel, the two external lanes were Southern blotted and hybridized overnight at 65°C in 5× SSPE, 1% SDS, 0.1 mg/ml non-fat powdered milk with a chromosome 17 specific alphoid probe, and washed in stringent conditions (0.2× SSPE, 0.1% SDS at 65°C).
RESULTS
The alphoid array of chromosome 17 homologues exhibits unshared domains
In contrast with the centromeric regions of most other human chromosomes, that of chromosome 17 only contains a single alphoid array. By pulsed field gel electrophoresis, the alphoid array size was shown to vary from 2.8 to 3.7 Mb between three hybrid cell lines that each retained a single chromosome 17 (22). It has been characterized as being composed of tandemly repeated 16mer (2712 nt) HORs with as much as 99% sequence homogeneity (16,23). Polymorphic HORs, namely 15, 14, 13, 12, 11 and 9mers, are often detected. They result from unequal crossing over events between two 16mer elements which have subsequently been amplified. A high proportion of the chromosome 17 homologues exhibit a large array with the 13mer version (16).
Hybrid cell lines GM 10498 and GM 10321 (Corriel Cell Repository) both retain a single human chromosome 17, which was estimated by pulsed field gel electrophoresis to cover more than 2 and 3 Mb, respectively (data not shown). Their DNA was PCR amplified with primers 17-1A/17-2A (Figure 1A). The presence of the 16mer variant on both hybrids was confirmed by PCR with primers 17-3A/17-4A. In spite of the deletion of three copies, the 13mer HOR variant was still recognized by both primers and was therefore also detectable (16).
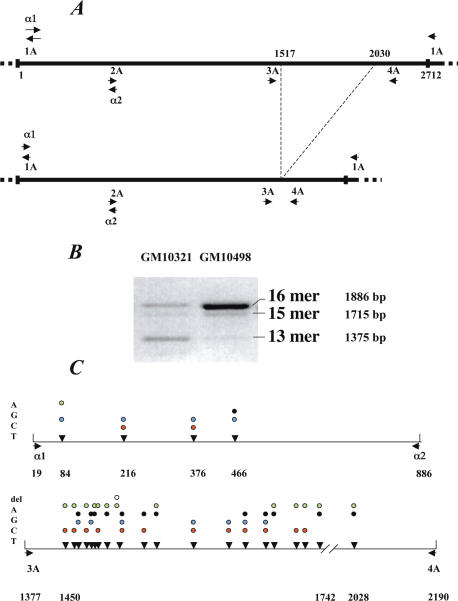
Figure 1.
(A) The chromosome 17 satellite alpha HOR is 2712 bp long. The primers chosen for PCR are indicated in the Materials and Methods section. On PCR amplification with 17-1A/17-2A, a DNA fragment of 1886 bp is generated when the HOR is a 16mer, or diminished by one, two or three… integral basic repeat units (171 bp). This figure represents the deletion by three basic repeat units leading to the 13mer HOR. (B) PCR DNA fragments obtained with 17-1A/17-2A with the two hybrid cell lines GM10498 and GM10321 analysed in this study. (C) The PCR products with 17-α1/17-α2 and 17-1A/17-2A are shown together with the positions () of the diagnostic variant nucleotides and the alternative nucleotides they exhibit.
To detect polymorphic nucleotides along the 16mer repeats, I used the strategy of Warburton and Willard (16), which consists of PCR amplifying the 16mer HORs of the homogeneous core (>99% sequence identity), thus yielding a subset that is representative of the hundreds to thousands of copies present along the alphoid array. Most of the divergent copies, which are localized on both edges of the array, are excluded from targeting and therefore from amplification. The PCR product is then cloned and a number of clones are sequenced.
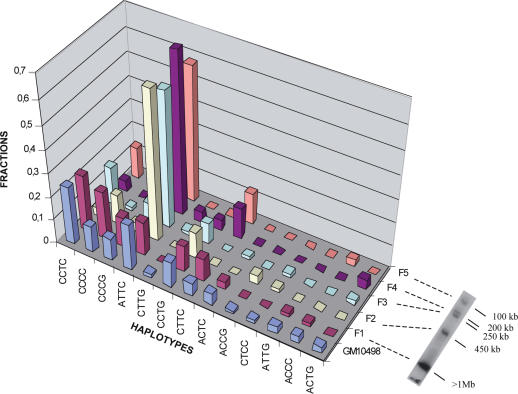
I first used primers 17-α1/17-α2, which generate 857 bp long PCR DNA fragments (Figure 1A). Sequencing of a few of the resulting clones revealed the existence of four diagnostic variant nucleotides. These, as well as those described later in this paper, correspond to positions which in most cases exhibit two alternative nucleotides (16), as is the case for most single nucleotide polymorphisms (SNPs). They do not include the extremely limited proportion (see later) of sporadic variations, which were not detected in a recurrent manner and which therefore only appeared in isolated cloned repeats. I then used these diagnostic variant nucleotides to establish whether or not the different haplotypes they define are clustered. For this purpose, genomic DNA from hybrid cell line GM10 498 was resolved on an agarose gel by pulsed field gel electrophoresis into five DNA fragments ranging from about 100 kb to more than 1 Mb (Figure 2). Each DNA fragment was analysed using the ASO method to determine its haplotype content. The largest fragment (F1) clearly exhibited a haplotype distribution close to that of the entire GM10 498 alphoid DNA array, while the smaller fragments (fragments F2–F5) had specific distributions. I therefore concluded that similar haplotypes are indeed clustered overall, with their corresponding repeats being thus mainly organized in different alphoid domains along the array.
Figure 2.
Haplotypes obtained with 17-α1/17-α2 in the five DNA fragments (F1–F5) generated upon SacI digestion of GM10498 DNA prepared in agarose plugs. Pulsed field gel electrophoresis (data not shown) allowed an estimation of the DNA fragment lengths as about 100, 200, 250, 450 kb, and more than 1 Mb, for F1 to F5, respectively.
PCR with the 17-α1/17-α2 primers could not, however, distinguish between the different HORs (16, 15, 14, 13mers…) present on the chromosome 17 alphoid array. Moreover, the number of diagnostic variant nucleotides was relatively small. I therefore analysed the defined region using primers 17-3A/17-4A, which could distinguish between the 13 and 16mer HORs (Figure 1A).
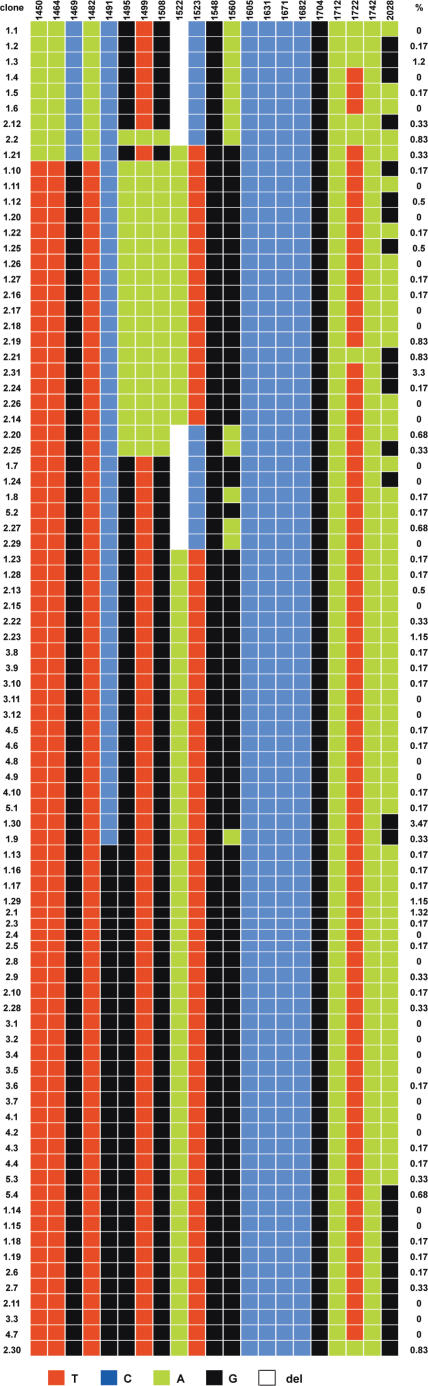
This greatly increased the number of diagnostic variant nucleotides, which could clearly be attributed to either the 16 or 13mer HOR, depending upon the PCR DNA fragment cloned. A number of the polymorphic nucleotides were similar to those detected by Warburton and Willard (16). Figure 3 shows the rather small number of haplotypes detected with GM10498, as well as the percentage of sporadic mutations detected in each of the 87 sequenced repeats. The haplotypes could be grouped into several subsets, presumably representing alphoid domains made of identical or closely-related haplotypes.
Figure 3.
The haplotypes of the 87 sequenced clones of GM10498 DNA are represented with the alternative nucleotides found at each diagnostic position of the 17-3A / 17-4A portion of the 16mer HOR (Figure 1). The percentage of sporadic mutations of each clone is also indicated, averaging 0.18 ± 0.22%. Clones 1.3, 2.31, 2.23, 1.30, 1.29 and 2.1 have been arbitrarily excluded from the calculation because they exhibited more than 1% mutation.
When other alphoid arrays from chromosome 17 homologues were analysed in a similar manner, a striking and unexpected feature appeared, exemplified by the comparison between the two chromosomes 17 16mer alphoid arrays of the two hybrid cell lines GM10498 and GM10321: they did not exhibit a single common haplotype. In spite of this, any two 16mer repeats compared to their respective sequences showed 99.7% identity (Figure 4), with the sporadic nucleotide variation being on average 0.2%. Six of the 87 clones analysed, however, exhibited more than 1% sporadic nucleotide variation.
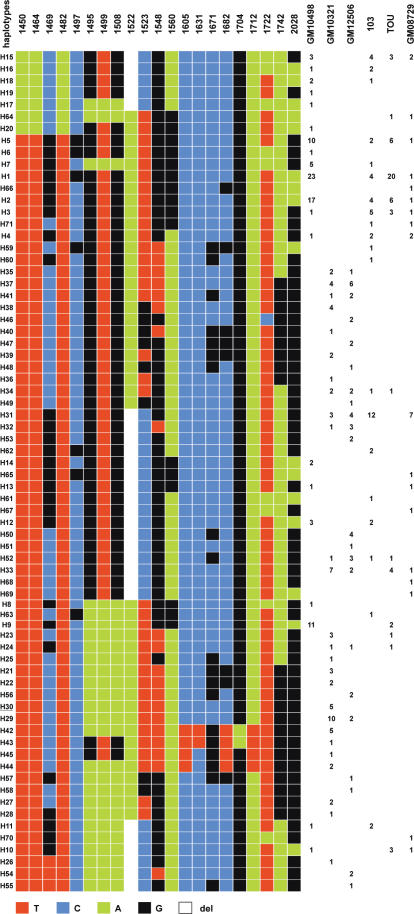
Figure 4.
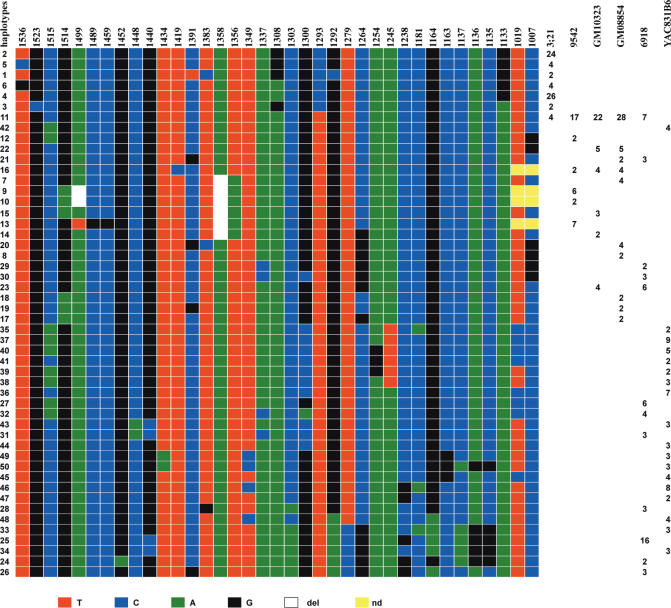
Representation of the 71 haplotypes detected in the nine chromosomes 17 of this study (one for the hybrid cell lines GM10498, GM10321 and GM12506; two for individuals 103 and TOU, and hybrid cell line GM08729). They have been ordered by the multiple sequence alignment program clustalW (47). The number of clones exhibiting a given haplotype is indicated for each sample.
In order to exclude the possibility that this originated from modifications which could have arisen during the establishment or propagation of the hybrid cell lines, I also analysed DNA samples extracted from peripheral blood cells of several individuals (Figure 4). Although chromosome 17 homologues could only be analysed in pairs, they exhibited the same domain organization characteristics. In addition, for a total of nine chromosome 17 homologues, no new diagnostic variant nucleotide was observed when compared to those found in GM10498 and GM10321. This indicates that regardless of the amplification/homogenization mechanisms that may be acting, it can be excluded that the sporadic mutations which are introduced (0.2% on average) are spread to the repeats of the different domains of the array.
I also examined the 13mer HOR of several chromosomes 17. They behave in a similar way to the 16mer repeats (data not shown).
Chromosome 21 and 13 alphoid arrays also exhibit domains which vary largely from homologue to homologue
At this point, it was necessary to establish whether the observations made for chromosome 17 could be extended to the whole human karyotype. I examined chromosomes 21 (D21Z1) and 13 (D13Z1), because they share 99.7% sequence homology between their respective centromeric alphoid HORs (24). This approach allowed me to determine whether or not they also shared diagnostic variant nucleotides. Hybrid cell lines bearing a single, different chromosome 21 or 13 were examined (six chromosomes 21 and two chromosomes 13).
Similar observations were made with both chromosomes. The existence of a large set of haplotypes was also a characteristic of their alphoid centromeric arrays. However, the average dispersion was larger (see Discussion), as judged by the total number of haplotypes shared by the six chromosome 21 homologues. In Figure 5, those differing by one diagnostic variant nucleotide are grouped so that only 50 are shown out of the 78 originally detected. This larger dispersion is not true, however, for all homologues: 3;21, for instance, is highly homogeneous in that respect. Haplotypes were again only partially shared between the different homologues. One was found to be common to most chromosome 21 arrays. The number of diagnostic variant nucleotides was also larger, although the frequency of sporadic mutations among the repeats was the same as for chromosome 17, again with slight variation between homologues. The tandem organization of close haplotypes was not checked, but presumably also holds true for both chromosomes (see Discussion).
Figure 5.
Representation of the haplotypes detected in the six chromosomes 21 of this study. For clarity, different haplotypes that are identical except for one diagnostic variant nucleotide have been grouped so that their number is reduced from the original 78 to 50 ordered by the multiple sequence alignment program clustalW.
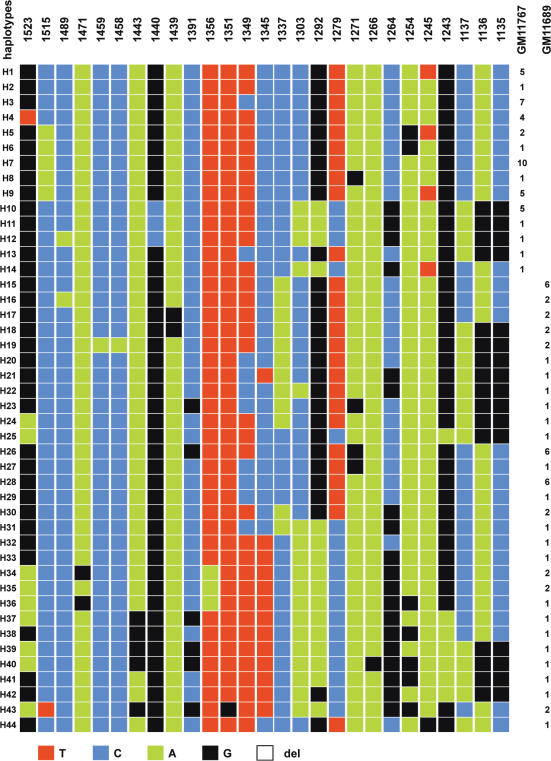
I was interested to determine whether or not the chromosome 13 homologues share the same haplotypes as those found on chromosome 21. In fact, they are different, as only a minority of the diagnostic variant nucleotides are shared between the two chromosomes. This, however, must be tempered by the fact that on one chromosome (namely, chromosome 21), the nucleotide found on the other (chromosome 13) is identical to one of the two alternative nucleotides, and vice versa, thanks to the high sequence homology between the two HORs. Though limited to two homologues, the same observation could be made as for chromosomes 17 and 21, as no overlap between the haplotypes of the two homologues could be found (Figure 6). This suggests a complete renewal in the lineage ancestral to one of the two. Most probably, the renewal occurred more recently in an ancestor of GM11767, as the dispersion of the haplotypes in this cell line is much less pronounced than in GM11689.
Figure 6.
Representation of the haplotypes detected in the two chromosomes 13 of the two hybrid cell lines GM11767 and GM11689.
DISCUSSION AND CONCLUSIONS
Alpha satellite DNA constitutes the most rapidly-evolving fraction of the primate genomes. It has been proposed by Dover (12) that its evolution is driven by a process called MD. For satellite DNAs of several related Drosophila species (the `500' and `360' families), the process of variant spreading has been rapid enough relative to the mutation rate to make the transition stages detectable (25). Chromosome-specific sequences of chimpanzee alpha satellites show in contrast high sequence divergence from those of their human orthologues. However, high homogeneity is observable between repeats belonging to the same array, and they exhibit the same type of organization into subsets (26,27). Human and chimpanzee alpha satellite DNAs have therefore been homogenized within their respective lineages by concerted evolution. One prediction of MD is not, however, fulfilled, as each individual chromosome exhibits its own sequence and structural specificities.
Still, it remains that the large differences in haplotypes between homologues observed in this study need to be explained. This is a difficult task because of our ignorance of when the diagnostic variant nucleotides were introduced, of their mutation rates, and of the relative contributions of several potential molecular mechanisms which presumably operate at the same time, each with its own rate, bias….
The alphoid arrays of the whole human karyotype are made of several haplotypic domains which vary substantially between homologues
As first shown with the 16mer HOR of the chromosome 17 alphoid array, a limited number of diagnostic variant nucleotides (21 over 813 bp) were detected in each cloned and sequenced repeat. They are found in various combinations, thus forming haplotypes. The chromosome 17 alphoid array is, therefore, composed of repeats which differ at fixed positions along the HOR, while the number of sporadic mutations is extremely low, 0.2% on average.
When several chromosome 17 homologues were examined, the number of haplotypes increased but remained limited compared to the extremely large number of potential ones (71 for a total of nine homologues). Unexpectedly, haplotypes were only partially shared among the nine homologues of this study. Conversion is the simplest and most probable mechanism that could account for the establishment of this haplotype diversity, as it can operate between homologues at different stages of the cell cycle and because it can involve only a portion of the repeats. Though recombination in the centromeric regions is highly restricted but not totally excluded (5,17,18), rare meiotic crossing over events could also have contributed to their spread to other homologues.
Strikingly, the two homologues from GM10498 and GM10321 do not share a single haplotype, indicating that during the evolution of these alphoid array lineages exchanges between them were extremely limited if not totally absent. They nevertheless maintained 99.8% sequence homology. Interestingly, no new diagnostic variant nucleotides were detected in any of the additional homologues analysed. A plausible explanation would be that most sporadic mutations have been introduced too recently to have spread to a detectable level. Increasing the number of analysed homologues could therefore reveal new haplotypes with new diagnostic variant nucleotides.
Analyses of chromosome 21 and 13 homologues also evidenced the existence of haplotypes defined by diagnostic variant nucleotides specific for each chromosome (37 and 27, respectively, over 622 bp). It is interesting to note that although the chromosome 21 and 13 alphoid arrays share 99.7% homology (24), the haplotypes they exhibit are totally exclusive of each other. As with chromosome 17, the degree of dispersion of the haplotypes was variable from homologue to homologue. In one chromosome 21 (YAC831B6), a large dispersion is observed, while it is highly reduced on 3;21. The same holds true for chromosome 13, where GM11689 is largely dispersed, in contrast to GM11 767.
Clustering of the HORs bearing the same or closely-related haplotypes was shown by analysing those detected within the portion of the chromosome 17 HOR between 17α1 and 17α2 (Figure 1A) after resolution of the whole array into several DNA fragments by pulsed field gel electrophoresis (Figure 2). Obviously, this is true for the entire length of the chromosome 17 HOR (GM10 498), including the portion between 17-1A and 17-2A (Figure 1A). Concerning chromosome 21, the results obtained with (3;21) also strongly support this conclusion: the haplotypes are extremely reduced in number (Figure 5). They were therefore recently generated and must be mostly tandemly arranged into separate domains. Examination of the few pure, long-enough alphoid contigs available in databases either from chromosome 17 or from chromosome 21 shows that their clustered HORs indeed bear either identical or very closely-related haplotypes (data not shown).
The variant diagnostic nucleotides serve as a molecular basis for homogenization/amplification and diversification into variable domains within the human centromeric alpha satellite arrays
The results obtained in this study are best explained by the process proposed by Smith (14). As with sister chromatid exchanges, which occur between strictly identical sequences, unequal crossing over would only be allowed between alpha satellite HORs that exhibit the same haplotype. This is in agreement with the observation that the presence of mismatches between recombining DNA molecules strongly inhibits homologous recombination (28,29). Homogenization / amplification through the accumulation of crossovers would thus create a new domain or re-homogenize an old one in a process which could be either continuous or saltatory. I therefore propose that each haplotype serves as a basis in this process for the molecular recognition between repeats. Closely-related haplotypes do not seem, however, to be totally excluded from pairing and subsequent crossing-over. This implies that during pairing non-identical diagnostic variant nucleotides can occasionally be accepted.
As an important consequence of this suggested mechanism, and without necessitating any other, HORs would remain highly homogeneous even though they are periodically renewed. On each homologue, the homogenization / amplification process would occur according to the availability of particular haplotypes. This explains why it leads to variable domains on different homologues. Therefore, the putative basis of molecular recognition in pairing explains per se why homogenization is maintained not only intrachromosomally but also inter-homologues.
Overall, chromosomes 21 and 13 exhibit different haplotypes. They do share, however, a number of positions at which the SNPs are identical. This indicates that the two chromosomes had their alphoid arrays in frequent contact until the ‘crosstalking’ between them dramatically diminished, after which a number of new SNPs appeared on each of them at different positions. One cannot estimate, however, when this occurred during the evolution of these alphoid arrays.
If, as supposed above, new haplotypes are also periodically introduced, this would imply continuous sequence changes over time and their spread to homologues and, eventually, but at a lower rate, to non-homologues. To explain the paradox of fast-evolving centromeric sequences in higher eukaryotes, a centromere drive model has been suggested (30,31) in which the expansion of satellite DNA, and presumably the homogenization of particular satellite domains, would provide an advantage for the unique remaining cell that is available for fertilization during female meiosis II. Talbert et al. (32) stressed that the tendency of centromere satellite DNAs to rapidly diverge, combined with the ability of CENP-C to adaptively evolve, are involved in this meiotic drive, as in the molecular coevolution process of Dover and Flavell (33). To reach the fertilization stage of the remaining unique cell, the maternal chromosomes of the primary germ cells (PGCs) undergo several tens of mitoses during which amplification or homogenization can occur. Presumably selection is acting during this process (34). If this is the case, it could positively select for the centromeres with newly amplified or homogenized alphoid sequences.
What status for gene conversion in the evolution of alpha satellites?
Even if unequal crossing over can account for most observations made on tandemly-arranged repeated DNA sequences like alpha satellites, one cannot exclude that other molecular mechanisms also participate in their formation. Large-scale amplification cannot be excluded a priori, although it would only explain large variations in array lengths and intrachromosomal homogenization. Even if this process might occur occasionally (35), it cannot account for inter-homolog homogenization.
In contrast, gene conversion could be an alternative to unequal crossing over. This is indeed the basic mechanism that Schindelhauer and Schwarz (15) invoked to explain their data obtained from the analysis of a series of HORs from a single male X alphoid array. While they don't exclude unequal crossing over as one of the mechanisms contributing to the homogenization, they favour gene conversion as an explanation for both intrachromosomal and inter-homologue homogenization. Their basic argument rests on the apparent lack of higher-order haplotypic repeats, as they found 40 different haplotypes within 45 independent sequences originating from five randomly located PACs. When the different haplotypes are compared to each other, however, one can observe closer haplotyping within each PAC and see that the haplotypes differ by very few diagnostic variant nucleotides. An analysis of the limited number of published sequences that are available from X homologues (15,36) covering the same part of the 2 kb X HORs makes it possible, however, to conclude that the X alpha satellite DNA array behaves similarly to those of chromosomes 17, 21 and 13: variant haplotypes are only partially shared between the different X homologues.
Therefore, and in contrast to their conclusion, I suggest that conversion is actually acting mainly to introduce divergence between alpha satellite DNA copies that were initially homogenized in tandem, thus leading to the diversification observed in this study. This does not exclude, however, that gene conversion could also contribute to some extent to the homogenization between copies belonging to different domains.
What makes an alphoid locus competent for centromere activity, and how could a chromosome with an altered centromere be rescued
We don't know if amplification/homogenization of alphoid domains occurs because sister chromatid exchanges are particularly frequent within centromeric sequences due to their structure, or if this phenomenon has been positively selected for because of functional reasons.
Csink and Henikoff (37) have proposed a model in which most of the satellite DNA array serves as a spacer to protect the centromere locus from sequences which would otherwise have a detrimental effect on centromere activity. In particular, they emphasized the role of retrotransposons, which could destroy the ability of the array to form an active centromere. We have extended this concept by proposing, based on observations, that the alpha satellite domain, within which DNA sequences are recruited to form an active centromere, must be uninterrupted by L1s or any other retrotransposons and simultaneously be highly homogeneous in sequence (38). High homogeneity in sequence of alphoid arrays increases their efficiency to create human artificial chromosomes (HACs), notably when the assays are performed with synthetic alpha satellite arrays (39,40). In contrast, when the array is interrupted by a retrotransposon (41) or is irregular in sequence -BAC 5 from chromosome 22 in Kouprina et al. (42)-, HACs are not formed.
The data provided here are in good agreement with a centromere formed within an alphoid domain that fulfills the two above conditions. It will, however, be exposed to the introduction of L1 retrotransposons and/or mutations, which could alter its centromere-forming capacity for reasons which are not yet clear. A single retrotransposon might be sufficient to eliminate the capacity of an alphoid domain to become or remain a centromere-forming locus. From Figure 3, it appears that few copies exhibit >1% divergence. It might very well be that those copies within an otherwise highly homogeneous domain could destroy, by disrupting its homogeneity, its capacity to form a centromere, although we have no precise idea of the acceptable upper limit of divergence.
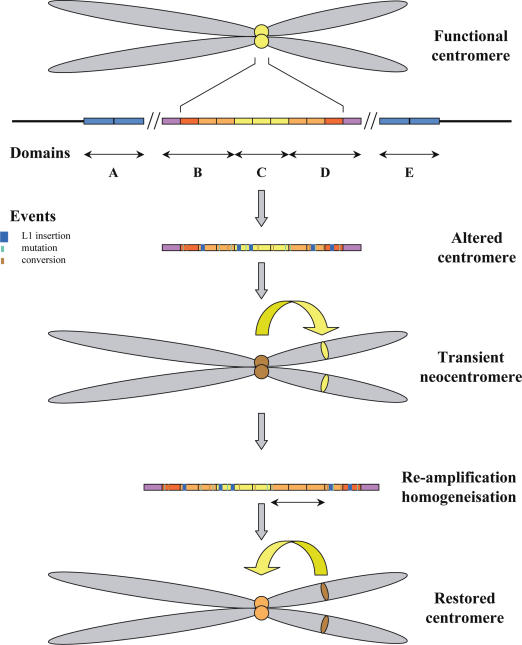
When a centromere locus is altered or destroyed with respect to centromere function, the chromosome, and hence the potential individual, could be lost unless the homogenization/amplification process described above creates a new locus that is competent for the function (Figure 7). One could imagine therefore that a transiently-formed neocentromere could temporarily rescue the affected chromosome until the activity of the old endogenous centromere is restored. Among the 70 neocentric chromosomes so far described (43), six have been observed to be meiotically transmitted. Of these six, four are neodicentric: one each on chromosomes 4 (44), 3 (45), 8 (43) and the Y chromosome (46). In these four cases, the original endogenous centromere is still present but is inactivated. These cases all fit with my suggestion of chromosome rescue by the transient formation of a neocentromere. It is particularly interesting to note that the Y alphoid array has undergone a deletion which could have affected its capacity to form a centromere, hence its inactive state (46).
Figure 7.
A fully functional centromere is normally formed within an alphoid array when a domain that is both highly homogeneous and devoid of retrotransposons is present. Given the large number of alphoid sequences, several domains can fulfill these conditions. Here, domain C is supposed to be the centromere locus. With time, a number of undermining events can occur, such as L1 retrotransposition and accumulation of nucleotide changes, so that the centromere can become altered in its functionality. The chromosome could then be either lost or rescued by the formation of a neocentromere, which would transiently substitute for the altered centromere until a fully functional centromere is again formed through the accumulation of unequal crossovers.
Amor et al. (44) proposed that pseudodicentric–neocentric chromosomes in healthy individuals, and the ability of neocentric activity to form in euchromatic sites in preference to pre-existing alphoid domains, provide direct evidence for an inherent mechanism of human centromere repositioning and karyotype evolution `in progress'. This is supported by the work of Ventura et al. (45), who also suggest, based on an evolutionary comparison between a human neocentromere mapping to 3q26 and the positions of centromeres in Old and New World monkeys, that the formation of neocentromeres in humans and the emergence of new centromeres during the course of evolution share a common mechanism.
These two suggestions are not exclusive of each other. If one considers the transient formation of neodicentrics as a system that is able to rescue chromosomes bearing an altered centromere, it is interesting to note that the four cases described so far were detected by chance, as they were not associated with an abnormal phenotype, which is the case for most of the other `normal' neocentromeres. This implies that such cases might be relatively common, in accordance with a presumed relatively high rate of repair by amplification/homogenization. Whether or not this is an evolutionarily-selected system remains pure speculation.
Acknowledgments
The author wish to thank Anne-Marie Laurent, Sophie Vidal and Conchita Ferraz for sequence determination; Jacques Demaille for his interest and access to sequencing facilities; Sylvain Hubac for participating as a student at the first steps of this study; Jacques Puechberty for numerous discussions and help. The author warmly thanks Gabby Dover for thorough reading of the manuscript and numerous helpful suggestions. The author wish to thank Peter Folette for the editing work. The author also acknowledge financial support by Centre National de la Recherche Scientifique, the European Union within the Network contract no. HPRN-CT-2000-00089 and Fondation Jerome Lejeune (Paris). Funding to pay the Open Access publication charges for this article was provided by Fondation Jerome Lejeune.
Conflict of interest statement. None declared.
REFERENCES
- 1.Haaf T., Willard H.F. Organization polymorphism and molecular cytogenetics of chromosome-specific alpha-satellite DNA from the centromere of chromosome 2. Genomics. 1992;13:122–128. doi: 10.1016/0888-7543(92)90211-a. [DOI] [PubMed] [Google Scholar]
- 2.Tyler-Smith C., Brown W.R. Structure of the major block of alphoid satellite DNA on the human Y chromosome. J. Mol. Biol. 1987;195:457–470. doi: 10.1016/0022-2836(87)90175-6. [DOI] [PubMed] [Google Scholar]
- 3.Marçais B., Bellis M., Gerard A., Pages M., Boublik Y., Roizès G. Structural organization and polymorphism of the alpha satellite DNA sequences of chromosomes 13 and 21 as revealed by pulsed field gel electrophoresis. Hum. Genet. 1991;86:311–316. doi: 10.1007/BF00202418. [DOI] [PubMed] [Google Scholar]
- 4.Lo A.W.I., Liao G.C.C., Rocchi M., Choo K.H.A. Extreme reduction of chromosome-specific alpha-satellite array is unusually common in human chromosome 21. Genome Res. 1999;9:895–908. doi: 10.1101/gr.9.10.895. [DOI] [PubMed] [Google Scholar]
- 5.Puechberty J., Laurent A.M., Gimenez S., Billault A., Brun-Laurent M.E., Calenda A., Marçais B., Prades C., Ioannou P., Yurov Y., et al. Genetic and physical analyses of the centromeric and pericentromeric regions of human chromosome 5: recombination across 5cen. Genomics. 1999;56:274–287. doi: 10.1006/geno.1999.5742. [DOI] [PubMed] [Google Scholar]
- 6.Willard H.F. Evolution of alpha satellite. Curr. Opin. Genet. Dev. 1991;1:509–514. doi: 10.1016/s0959-437x(05)80200-x. [DOI] [PubMed] [Google Scholar]
- 7.Schueler M.G., Higgins A.W., Rudd M.K., Gustashaw K., Willard H.F. Genomic and genetic definition of a functional human centromere. Science. 2001;294:109–115. doi: 10.1126/science.1065042. [DOI] [PubMed] [Google Scholar]
- 8.Alexandrov I., Kazakov A., Tumeneva I., Shepelev V., Yurov Y. Alpha-satellite DNA of primates: old and new families. Chromosoma. 2001;110:253–266. doi: 10.1007/s004120100146. [DOI] [PubMed] [Google Scholar]
- 9.Rudd M.K., Willard H.F. Analysis of the centromeric regions of human genome assembly. Trends Genet. 2004;20:529–533. doi: 10.1016/j.tig.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 10.Schueler M.G., Dunn J.M., Bird C.P., Ross M.T., Viggiano L., Rocchi M., Willard H.F., Green E.D. Progressive proximal expansion of the primate X chromosome centromere. Proc. Natl Acad. Sci. USA. 2005;102:10563–10568. doi: 10.1073/pnas.0503346102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rudd M.K., Wray G.A., Willard H.F. The evolutionary dynamics of α-satellite. Genome Res. 2006;16:88–96. doi: 10.1101/gr.3810906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dover G.A. Molecular drive: a cohesive mode of species evolution. Nature. 1982;299:111–117. doi: 10.1038/299111a0. [DOI] [PubMed] [Google Scholar]
- 13.Charlesworth B., Sniegowski P., Stephan W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994;371:215–220. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- 14.Smith G.P. Evolution of repeated DNA sequences by unequal crossing over. Science. 1976;191:198–212. doi: 10.1126/science.1251186. [DOI] [PubMed] [Google Scholar]
- 15.Schindelhauer D., Schwarz T. Evidence for a fast intrachromosomal conversion mechanism from mapping of nucleotide variants within a homogeneous α-satellite DNA array. Genome Res. 2002;12:1815–1826. doi: 10.1101/gr.451502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warburton P.E., Willard H.F. Interhomologue sequence variation of alpha satellite DNA from human chromosome 17: evidence for concerted evolution along haplotypic lineages. J. Mol. Evol. 1995;41:1006–1015. doi: 10.1007/BF00173182. [DOI] [PubMed] [Google Scholar]
- 17.Mahtani M.M., Willard H.F. Physical and genetic mapping of the human X chromosome centromere: repression of recombination. Genome Res. 1998;8:100–110. doi: 10.1101/gr.8.2.100. [DOI] [PubMed] [Google Scholar]
- 18.Laurent A.M., Meizhang,Li., Sherman S., Roizès G., Buard J. Recombination across the centromere of disjoined and non-disjoined chromosome 21. Hum. Mol. Genet. 2003;12:2229–2239. doi: 10.1093/hmg/ddg220. [DOI] [PubMed] [Google Scholar]
- 19.Bosch A., Nunes V., Patterson D., Estivill X. Isolation and characterization of 14 CA-repeat microsatellites from human chromosome 21. Genomics. 1993;18:151–155. doi: 10.1006/geno.1993.1442. [DOI] [PubMed] [Google Scholar]
- 20.De Sario A., Roizès G., Allegre N., Bernardi G. A compositional map of the cen-q21 region of human chromosome 21. Gene. 1997;194:107–113. doi: 10.1016/s0378-1119(97)00177-7. [DOI] [PubMed] [Google Scholar]
- 21.Warburton P.E., Greig G.M., Haaf T., Willard H.F. PCR amplification of chromosome-specific alpha satellite DNA: definition of centromeric STS markers and polymorphic analysis. Genomics. 1991;11:324–333. doi: 10.1016/0888-7543(91)90139-6. [DOI] [PubMed] [Google Scholar]
- 22.Warburton P.E., Willard H.F. Genomic analysis of sequence variations in tandemly repeated DNA evidence for localized homogeneous sequence domains within arrays of α-satellite DNA. J. Mol. Biol. 1990;216:3–16. doi: 10.1016/s0022-2836(05)80056-7. [DOI] [PubMed] [Google Scholar]
- 23.Waye J.S., Willard H.F. Structure organization and sequence of alpha satellite DNA from human chromosome 17: evidence for evolution by unequal crossing-over and an ancestral pentamer repeat shared with the human chromosome X. Mol. Cell. Biol. 1986;6:3156–3165. doi: 10.1128/mcb.6.9.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jorgensen A.L., Bostock C.J., Bak A.L. Homologous subfamilies of human alphoid repetitive DNA on different nucleolus organizing chromosomes. Proc. Natl Acad. Sci. USA. 1987;84:1075–1079. doi: 10.1073/pnas.84.4.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strachan T., Webb D., Dover G.A. Transition stages of molecular drive in multiple-copy DNA families in Drosophila. EMBO J. 1985;4:1701–1708. doi: 10.1002/j.1460-2075.1985.tb03839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jorgensen A.L., Jones C., Bostock C.J., Bak A.L. Different subfamilies of alphoid repetitive DNA are present on the human and chimpanzee homologous chromosomes 21 and 22. EMBO J. 1987;3:1691–1696. doi: 10.1002/j.1460-2075.1987.tb02419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jorgensen A.L., Laursen H.B., Jones C., Bak A.L. Evolutionary different alphoid repeat DNA on homologous chromosomes in human and in chimpanzee. Proc. Natl Acad. Sci. USA. 1992;89:3310–3314. doi: 10.1073/pnas.89.8.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng C., Capecchi M.R. Reexamination of gene targeting frequency as a function of the extent of homology between the targeting vector and the target locus. Mol. Cell. Biol. 1992;12:3365–3371. doi: 10.1128/mcb.12.8.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Te Riele H., Mandaag E.R., Berns A. Highly efficient gene targeting in embryonic stem cells through homologous recombination with isogeneic DNA constructs. Proc. Natl Acad. Sci. USA. 1992;89:5128–5132. doi: 10.1073/pnas.89.11.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henikoff S., Ahmad K., Malik H.S. The centromere paradox: stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- 31.Malik H.S., Henikoff S. Adaptive evolution of Cid a centromere-specific histone in Drosophila. Genetics. 2001;157:1293–1298. doi: 10.1093/genetics/157.3.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Talbert P.B., Bryson T.D., Henikoff S. Adaptive evolution of centromere proteins in plants and animals. J. Biol. 2004;3:18. doi: 10.1186/jbiol11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dover G.A., Flavell R.B. Molecular coevolution: DNA divergence and the maintenance of function. Cell. 1984;38:622–623. doi: 10.1016/0092-8674(84)90255-1. [DOI] [PubMed] [Google Scholar]
- 34.Motta P.M., Nottola S.A., Makabe S. Natural history of the female germ cell from its origin to full maturation through prenatal ovarian development. Eur. J. Obstet. Gynecol. Reprod. Biol. 1997;75:5–10. doi: 10.1016/s0301-2115(97)00216-9. [DOI] [PubMed] [Google Scholar]
- 35.Marçais B., Charlieu J.P., Allain B., Brun E., Bellis M., Roizès G. On the mode of evolution of alpha satellite DNA in human populations. J. Mol. Evol. 1991;33:42–48. doi: 10.1007/BF02100194. [DOI] [PubMed] [Google Scholar]
- 36.Durfy S.J., Willard H.F. Patterns of intra- and interarray sequence variation in alpha satellite from the X chromosome: evidence for short-range homogenization of tandemly repeated DNA sequences. Genomics. 1989;5:810–821. doi: 10.1016/0888-7543(89)90123-7. [DOI] [PubMed] [Google Scholar]
- 37.Csink A.K., Henikoff S. Something from nothing: the evolution and utility of satellite repeats. Trends Genet. 1998;14:200–204. doi: 10.1016/s0168-9525(98)01444-9. [DOI] [PubMed] [Google Scholar]
- 38.Laurent A.M., Puechberty J., Roizès G. Hypothesis: for the worst and for the best L1Hs retrotransposons actively participate in the evolution of the human centromeric alphoid sequences. Chromosome Res. 1999;7:305–317. doi: 10.1023/a:1009283015738. [DOI] [PubMed] [Google Scholar]
- 39.Basu J., Stromberg G., Compitello G., Willard H.F., Van Bokkelen G. Rapid creation of BAC-based human artificial chromosome vectors by transposition with synthetic alpha-satellite arrays. Nucleic Acids Res. 2005;33:587–596. doi: 10.1093/nar/gki207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ebersole T., Okamoto Y., Noskov V.N., Kouprina N., Kim J-H., Leem S.H., Barrett J.C., Masumoto H., Larionov V. Rapid generation of long synthetic tandem repeats and its application for analysis in human artificial chromosome formation. Nucleic Acids Res. 2005;33:e130. doi: 10.1093/nar/gni129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Masumoto H., Ikeno M., Nakano M., Okazaki T., Grimes B., Cooke H., Suzuki N. Assay of centromere function using a human artificial chromosome. Chromosoma. 1998;107:406–416. doi: 10.1007/s004120050324. [DOI] [PubMed] [Google Scholar]
- 42.Kouprina N., Ebersole T., Koriabine M., Pak E., Rogozin I.B., Katoh M., Oshimura M., Ogi K., Peredelchuk M., Solomon G., et al. Cloning of human centromeres by transformation-associated recombination in yeast and generation of functional human artificial chromosomes. Nucleic Acids Res. 2003;31:922–934. doi: 10.1093/nar/gkg182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amor D.J., Bentley K., Ryan J., Perry J., Wong L., Slater H., Choo K.H. Human centromere repositioning `in progress'. Proc. Natl Acad. Sci. USA. 2004;101:6542–6547. doi: 10.1073/pnas.0308637101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ventura M., Weigl S., Carbone L., Cordon M.F., Misceo D., Teti M., D'addabbo,P., Wandall A., Bjorck E., de Jong P.J., et al. Recurrent sites for new centromere seeding. Genome Res. 2004;14:1696–1703. doi: 10.1101/gr.2608804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Warburton P.E. Chromosomal dynamics of human neocentromere formation. Chromosome Res. 2004;12:617–626. doi: 10.1023/B:CHRO.0000036585.44138.4b. [DOI] [PubMed] [Google Scholar]
- 46.Tyler-Smith C., Gimelli G., Giglio S., Floridia G., Pandaya A., Terzoli G., Warburton T.E., Earnshaw W.C., Zuffardi O. Transmission of a fully functional human neocentromere through three generations. Am. J. Hum. Genet. 1999;64:1440–1444. doi: 10.1086/302380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighing position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]