Abstract
SOL1, the founding member of the S. cerevisiae SOL family, was previously identified as a multi-copy suppressor of the los1 defect in tRNA-mediated nonsense suppression. Here we report that the four-member SOL family is not essential and that individual family members appear to have distinct functions. SOL1–SOL4 are homologous to genes encoding 6-phosphogluconolactonase (6Pgl) involved in the pentose phosphate pathway. Both Sol3p and Sol4p affect this activity. However, Sol4p does not act as a los1 multi-copy suppressor. In contrast, neither Sol1p nor Sol2p, both of which correct the los1 defect in nonsense suppression, possess detectable 6Pgl activity. Rather, Sol1p and Sol2p appear to function in tRNA nuclear export as sol1 and sol2 mutants possess elevated levels of nuclear tRNA. Members of the Sol protein family appear to have different subcellular distributions. Thus, Sol3p and Sol4p likely function in carbohydrate metabolism, while Sol1p and Sol2p appear to have roles in tRNA function and nuclear export, thereby defining an unusual protein family whose individual members are biochemically distinct and spatially dispersed.
EUKARYOTIC precursor-tRNAs (pre-tRNA) differ from their mature counterparts by possession of extra sequences located at the 5′ and 3′ extremities and, for many tRNA families, by the presence of an intron. Pre-tRNAs also lack numerous nucleoside modifications that are present on mature tRNAs and the post-transcriptionally added CCA nucleotides located at the 3′ terminus. Most of the steps of pre-tRNA processing occur in the nucleus, usually in an ordered pathway (Hopper and Phizicky 2003). In yeast, but not in all eukaryotes, removal of introns generally follows end processing (O'Connor and Peebles 1991). Although for Xenopus and humans tRNAs are exported to the cytosol after intron removal (Hopper and Phizicky 2003), there is recent surprising evidence that budding yeast tRNA splicing endonuclease is located on the cytosolic surface of mitochondria (Huh et al. 2003; Yoshihisa et al. 2003). If pre-tRNA splicing indeed occurs in the cytosol rather than in the nucleus in yeast, it could explain why alterations of many, but not all, of the nuclear export machinery components cause defects in pre-tRNA splicing (Hopper et al. 1978, 1980; Kadowaki et al. 1993; Sharma et al. 1996; Simos et al. 1996).
Export of small RNAs from the nucleus to the cytosol often requires a small GTPase, Ran, its regulators, and members of the Ran-binding importin-β family (Görlich and Kutay 1999). Importin-β family members, in addition to binding to Ran, interact with nuclear pore complex components and cargo and shuttle between the nucleus and cytosol. A large body of literature (Arts et al. 1998a,b; Hellmuth et al. 1998; Kutay et al. 1998; Sarkar and Hopper 1998; Lipowsky et al. 1999) documents that the vertebrate importin-β family member, exportin-t, and its yeast homolog, Los1p, serve to export tRNAs from the nucleus to the cytosol. Both human exportin-t and Saccharomyces cerevisiae Los1p have been shown to bind tRNA in a Ran-GTP-dependent mechanism (Arts et al. 1998a; Hellmuth et al. 1998; Kutay et al. 1998) and depletion of either results in nuclear accumulation of tRNAs (Arts et al. 1998b; Sarkar and Hopper 1998). In yeast, deletion of LOS1 also causes accumulation of intron-containing pre-tRNAs (Hopper et al. 1980; Simos et al. 1996). Because LOS1 is an unessential gene (Hurt et al. 1987), tRNA nuclear export in yeast must proceed via at least two pathways. Recent data suggest that parallel tRNA nuclear export pathways also exist in plants and vertebrates (Bohnsack et al. 2002; Calado et al. 2002; Li and Chen 2003).
We previously isolated multi-copy suppressors of S. cerevisiae los1 mutants and identified three members of the S. cerevisiae SOL gene family, SOL1, SOL2, and SOL3. Cells containing individual or any combination of SOL1, SOL2, and SOL3 deletions are viable (Shen et al. 1996). SOL1, the founding member, is an efficient multi-copy suppressor of the loss of nonsense suppression defect of los1 mutants. SOL2 also acts as a los1 suppressor, but to a lesser extent than SOL1. In contrast, SOL3 is a very weak multi-copy suppressor of los1 mutations. Our previous studies of the Sol proteins did not provide a mechanism by which they acted as multi-copy suppressors of los1 (Shen et al. 1996).
The yeast genome contains numerous other examples of reiterated genes, thought to result from genome duplication (Wolfe and Shields 1997; Langkjaer et al. 2003). Although most reiterated genes such as those encoding ribosomal proteins and histones are present in the yeast genome in two copies, reiterations are present in more than two copies (Dolinski et al. 2003; http://www.yeastgenome.org). For example, there are three copies of the genes encoding glyceraldehyde-3-phosphate dehydrogenase (TDH1, TDH2, and TDH3) and four genes encoding alcohol dehydrogenase (ADH1, ADH2, ADH3, and ADH4), tubulin family members (TUB1, TUB2, TUB3, and TUB4), and actin and actin-related proteins (ACT1, ARP1, ARP2, and ARP3). In each of these cases the family members perform related biochemical activities, often in the same subcellular compartment. Here we report that the SOL gene family appears to be an unusual category of reiterated yeast genes.
We revisited the function of the SOL genes because: (1) completion of the S. cerevisiae genome sequence uncovered a fourth family member, SOL4 (Feroli et al. 1997; Tettelin et al. 1997); (2) fluorescence in situ hybridization (FISH) methods to assess intracellular location of tRNAs in yeast have been developed (Sarkar and Hopper 1998); and (3) an enzyme activity in the pentose phosphate pathway for bacterial, protozoan, and human Sol1p (Sol1p/DevB/Pgl) homologs has been uncovered (Collard et al. 1999; Duffieux et al. 2000; Hager et al. 2000; Clarke et al. 2001). Our new studies show that Sol1p and Sol2p function in tRNA nuclear export, which accounts for their effects on tRNA-mediated nonsense suppression. Unlike Sol1p and Sol2p, the other two members of this family, Sol3p and Sol4p, affect 6-phosphogluconolactonase (6Pgl; EC 3.1.1.31) activity. Moreover, the various Sol proteins appear to have different subcellular distributions. Thus, the SOL family may provide a new paradigm of a protein family whose individual members are both spatially dispersed and biochemically distinct.
MATERIALS AND METHODS
Plasmids and constructs:
Plasmids WS37 (referred to as pRSSOL1), WS80 (pRSSOL2), and WS92 (pRSSOL3) with genomic SOL1, SOL2, and SOL3 regions inserted into vector pRS426 were previously described (Shen et al. 1996). YCpSOL1-ET contains SOL1 tagged at the C terminus with a HA epitope (Sol1p-HA) inserted into the low-copy vector YCp50. WS64 (referred to as pRSSOL1-ET) contains the same construct in vector pRS426 (Shen et al. 1996).
A genomic region containing SOL4 was obtained by PCR amplification using primers SOL45 (TGTGTGGGGCCCGTTCAAGAATGG) and SOL43 (CTTCCATGCAGCTGGCCCGAAGC). The template was genomic DNA from strain W303. The PCR product was gel purified, cut with PvuII and ApaI, trimmed with T4 DNA polymerase, and ligated into SmaI-cut pRS426 to generate plasmids pWCS39A (referred to as pRSSOL4) and pWCS39B containing opposite orientations of the SOL4 gene. GST-tagged versions of SOL1 (pGST-SOL1), SOL2 (pGST-SOL2), and SOL4 (pGST-SOL4) in plasmid pYEX 4T-1 were obtained from the arrayed GST-ORF collection generated by Phizicky and colleagues (Martzen et al. 1999).
Yeast strains:
We used the following published yeast strains: X2316-3C (MATα LOS1 SUP4 ade2-1 can1-100 lys1-1 his5-2 trp5-48 ura3-1; Hopper et al. 1980); 201-1-5 (MATα los1-1 SUP4 ade2-1 can1-100 lys1-1 his5-2 trp5-48 ura3-1; Hopper et al. 1980); SS700 (MATα los1::kanr SUP4 ade2-1 can1-100 lys1-1 his5-2 trp5-48 ura3-1; Feng and Hopper 2002); YD05055 (MATa los1::kanMX4 his3Δ1 leu2Δ0 met15Δ0 ura3Δ0; Winzeler et al. 1999); WCS2 (MATα LOS1 SUP4 ade2-1 can1-100 lys1-1 his5-2 trp5-48 ura3-1 sol1::URA3; Shen et al. 1996); WCS7 (MATα LOS1 SUP4 ade2-1 can1-100 lys1-1 his5-2 trp5-48 ura3-1 sol2::URA3; Shen et al. 1996); WCS9 (MATα LOS1 SUP4 ade2-1 can1-100 lys1-1 his5-2 trp5-48 ura3-1 sol1::ura3 sol2::URA3; Shen et al. 1996); WCS18 (MATα LOS1 SUP4 ade2-1 can1-100 lys1-1 his5-2 trp5-48 ura3-1 sol1::ura3 sol2::ura3 sol3::URA3; Shen et al. 1996); W303 (MATα ade2-1 ura3-1 his3-1,115 trp1-1 leu2-3,112).
Strains MLW104 (MATα LOS1 SUP4 ade2-1 can1-100 lys1-1 his5-2 trp5-48 ura3-1 sol4::hphr) and MLW114 (MATα LOS1 SUP4 ade2-1 can1-100 lys1-1 his5-2 trp5-48 ura3-1 sol1::ura3 sol2::ura3 sol3::URA3 sol4::hphr) were constructed by replacing SOL4 with a sol4::hphr cassette by one-step gene disruption (Guldener et. al. 1996) in X2316-3C and WCS18, respectively. The plasmid pAG32 (Goldstein and McCusker 1999) containing the hphr cassette was used as a template, and the sol4::hphr cassette was created by PCR amplification using oligonucleotide primers WHIT15 (CGTATACTGATCGCTTGCCTTCGCAAGGGATGGAAATCCCAGGTCGACGGATCCCCGG) and WHIT16 (CTGGGTTTATGCTTCGGGAGTAAGCTCCAACATACGCTGGTGGATCTGATATCATCGA). The resulting strains were confirmed by Southern blot analysis and PCR using primers WHIT19 (GATCGCTTGCCTTCGCAAGGGATGG) and WHIT20 (CTGGGTTTATGCTTCGGGAGTAAGC). MLW115 (MATα LOS1 SUP4 ade2-1 can1-100 lys1-1 his5-2 trp5-48 ura3-1 sol1::ura3 sol2::ura3 sol3::ura3 sol4::hphr) was derived from MLW114 by selecting for a ura3 variant by growth on 5-fluoroortic acid.
Growth and suppression assays:
To assess growth, yeast strains were grown to saturation in liquid media that selects for the plasmid of interest and then 10 μl of serial dilutions of cells were spotted onto solid media and incubated for 3–4 days at the indicated temperatures. To assess nonsense suppression of ade2-1, cells were either replica plated or spotted to solid media containing reduced levels of adenine and incubated 2–4 days at the indicated temperatures. Cells with wild-type levels of nonsense suppression gave rise to white colonies, whereas cells defective in nonsense suppression gave rise to pink or red colonies.
Measurement of 6-phosphogluconolactonase activity:
6Pgl activity was assayed as previously described (Collard et al. 1999). Protein extracts were prepared at 4° in 50 mm Tris-HCl (pH 7.5), 5 mm EDTA, 1 mm DTT containing protease inhibitors (5 μg/ml leupeptin, pepstatin, and aprotinin and 0.5 mm PMSF) utilizing glass beads. The activity assay follows the production of NADPH by the dehydrogenases that flank the phosphogluconolactonase in a coupled reaction. Glucose-6-phosphate dehydrogenase (EC 1.1.1.49) converts glucose-6-phosphate and NADP+ to NADPH and 6-phosphoglucono-1,5-lactone, the substrate for 6Pgl, which converts this molecule to 6-phosphogluconate. 6-Phosphogluconate dehydrogenase (EC 1.1.1.44) then converts 6-phosphogluconate to ribulose-5-phosphate with production of NADPH observed spectrophotometrically. Assays were started by the addition of 2.5 units glucose-6-phosphate dehydrogenase (yeast, Sigma, St. Louis) to 1 ml 25 mm HEPES, pH 7.1, 2 mm MgCl2 50 μm glucose-6-phosphate (Sigma) 200 μm NADP+ (Sigma). Absorbance at 340 nm was monitored for 8 min at room temperature at which time 1.5 units 6-phosphogluconate dehydrogenase and protein extract (50 μg) were added and the A340 was measured for an additional 10 min.
Immunofluorescence and fluorescence in situ hybridization:
Indirect immunofluorescence was carried out as previously described (Shen et al. 1993). Primary mouse monoclonal antibody 12CA5 against the HA epitope (Babco, lot L588b) was used at 1:500 dilution to detect Sol1p-HA. The cellular location of both constitutive and induced levels of GST-tagged Sol2p and Sol4p were detected employing anti-GST [Santa Cruz, GST(12), lot A126] at a 1:500 dilution. Cells were induced for 1 hr by addition of Cu2SO4 to a final concentration of 0.5 mm. Cy3-conjugated goat anti-mouse IgG (Jackson ImmunoResearch Labs, West Grove, PA) was used at a 1:200 dilution to locate the primary antibody.
FISH was performed according to a modification of a published procedure (Feng and Hopper 2002) using the previously described oligonucleotide probes to detect tRNAMet, tRNATyr, and poly(A)-containing RNAs (Sarkar and Hopper 1998). The following modifications were incorporated: cells were grown on defined media; all cultures were incubated at 37° for 2 hr prior to fixing; the slides were not placed in a desiccator; prehybridization was conducted at 39°; hybridization proceeded at 43°; washes with 2× SSC were conducted at 50°; and blocking and antibody incubation steps proceeded for 2.5 hr.
Fluorescence images were observed using a Nikon Microphot-FX microscope and were captured with a SenSys charge-coupled device camera (Photometrics, Tucson, AZ) using QED software (QED Imaging, Pittsburgh). Images were assembled using Adobe Photoshop 5.0.
Bioinformatics:
Sequence analysis was performed as previously described (Azad et al. 2001). BLAST searches were conducted at the National Center for Biotechnology Information BLAST server (Altschul et al. 1997) and CLUSTAL X (Thompson et al. 1997) was utilized to generate alignments. Amino acid residues were color coded on the basis of the BLOSUM 62 scoring matrix (Henikoff and Henikoff 1992) and background shading reflects the percentage of identity/similarity within a column of the alignment. The dendrogram in Figure 1 was produced with the Draw N-J Tree and NJplot options of CLUSTAL X. Schematics were assembled on the basis of alignment and structural information. Structures were generated and viewed with Swiss-Pdb Viewer (Guex and Peitsch 1997).
Figure 1.—
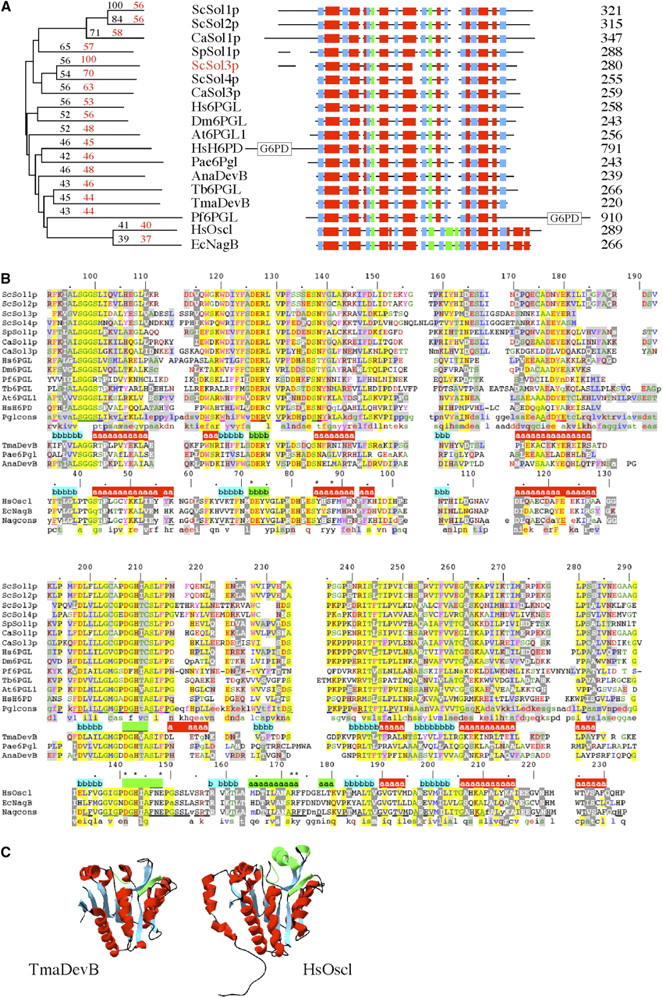
Comparison of Sol and Nag families. (A) Dendrogram showing similarities between selected members of the Sol family and two members of the Nag family. Percentage of similarities to ScSol1p (shown in black) and ScSol3p (shown in red) are overlayed on the CLUSTAL X generated dendrogram (see materials and methods). ScSol1p, ScSol2p, ScSol3p, and ScSol4p, S. cerevisiae Sol family members; CaSol1p and CaSol3p, Candida albicans; SpSol1p, Schizosaccharomyces pombe; Hs6PGL, H. sapiens; Dm6PGL, Drosophila melanogaster; At6PGL1, Arabidopsis thaliana; Tb6PGL, Trypanosoma brucei; Pae6Pgl, Pseudomonas aeruginosa; AnaDevB, Anabaena sp.; and TmaDevB, T. maritima. HsH6PD, H. sapiens, and Pf6PGL, Plasmodium falciparum, are two members of this family that contain both 6Pgl and G6PD domains. HsOscl, human oscillin (GNP1, glucosamine-6-phosphate deaminase), and EcNagB, E. coli, are two members of the Nag family. A schematic is also presented showing the relationships between these two families. Helices are shown in red boxes and sheets are shown in blue boxes. Green boxes represent regions of special importance to the catalytic activity of the Nag family (Rudino-Pinera et al. 2002; Arreola et al. 2003). Protein lengths are indicated to the right. (B) Alignment of Sol and Nag proteins. CLUSTAL X alignment of selected family members, beginning with residue 91 of ScSol1p. Other members are the same as in A. Color coding of amino acids is turned on when at least 10% of the residues in a column are identical or similar on the basis of the BLOSUM 62 matrix. LIVM, blue; YFW, magenta; STAGC, green; DE, red; KRQHN, brown; P, dark blue. Background shading reflects the percentage of identity/similarity within a column: 15–49%, light gray background; 50–74%, yellow background; >74%, yellow background with the consensus line underlined. Similar residues that have higher conservation in the Nag family than in the Sol family are indicated by white type on a dark gray background. Structural information is coded as above, helices are indicated on a red background, and sheets are indicated on a blue background, while a green background represents regions of importance to catalytic activity. Catalytic residues in the Nag family are indicated by asterisks. (C) Structural comparison of the Sol and Nag families. Shown are views of the same face of TmaDevB (1PBT; Kim et al. 2003) and HsOscl (Arreola et al. 2003). Color coding is the same as in A and B. Structures were generated from the crystallographic coordinates with Swiss-Pdb Viewer.
RESULTS
The S. cerevisiae SOL family is unessential:
Previously we identified SOL1 as a multi-copy suppressor of the defect in tRNA-mediated nonsense suppression of los1 mutations (Shen et al. 1996). At that time we also identified, isolated, and tested two additional members of the SOL gene family. Sol2p has the highest similarity to Sol1p (84%; Figure 1A), whereas Sol3p has 56% similarity to Sol1p. The level of multi-copy suppression of los1 mutants correlated with the similarity to Sol1p (i.e., SOL1>SOL2>>>SOL3; Shen et al. 1996). Since then the genome sequence of S. cerevisiae has been completed and one additional member of the SOL gene family was discovered (SOL4). SOL4 is located on the right arm of chromosome VII (YGR248w; Feroli et al. 1997; Tettelin et al. 1997) between CPD1 and MGA1 and encodes a protein with 36% identity and 54% similarity to Sol1p, but Sol4p is more similar to Sol3p, sharing 49% identity and 70% similarity (Figure 1A).
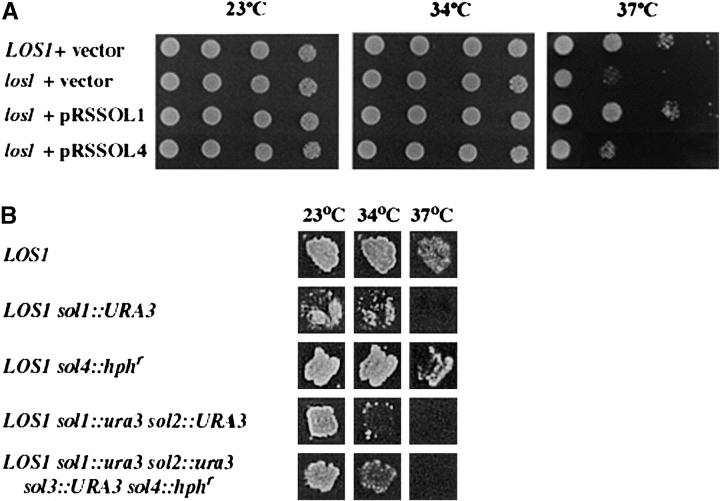
SOL4 was cloned by PCR from strain W303 and the sequence proved to be identical to that from strain S288C (Tettelin et al. 1997; Saccharomyces Genome Database, http://www.yeastgenome.org/). When provided on multi-copy plasmids pWCS39A or pWCS39B to strain SS700 (relevant genotype los1::kanr), SOL4 did not confer nonsense suppression activity (not shown), nor did it correct the slow growth at 37° phenotype of los1 mutant cells (Feng and Hopper 2002; Figure 2A; Table 1). SOL4 was disrupted/deleted with a hph drug resistance cassette in strain X2316-3C (relevant genotype: LOS1), yielding strain MLW104 (relevant genotype sol4::hphr). Whereas deletion of SOL1 in a LOS1 background causes a reduction in tRNA-mediated nonsense suppression, deletion of SOL4 in strain X2316-3C results in no observable effect upon nonsense suppression (Figure 2B; Table 1). SOL4, like SOL3, appears to have little effect upon tRNA function unlike SOL1 and SOL2. The data indicate that individual members of the SOL family may have different functions.
Figure 2.—
Growth and suppression assays. (A) Ability of multi-copy SOL1 and multi-copy SOL4 to suppresses the los1 growth defect. LOS1 and los1::kanr cells containing pRS426, pRSSOL1, or pRSSOL4 were grown to saturation, serially diluted, and aliquots were spotted onto solid media lacking uracil and incubated at 23°, 34°, or 37° for 3 days. (B) Effect of deletion of SOL genes upon tRNA-mediated ade2-1 nonsense suppression at various temperatures. Strains were patched onto YEPD and then replicate plated onto defined media lacking adenine and incubated at the indicated temperatures for 3 days.
TABLE 1.
Summary of the functions and location ofSOL gene family members
| SOL1 | SOL2 | SOL3 | SOL4 | |
|---|---|---|---|---|
| Multi-copy los1 suppressor | ++ | + | − | − |
| Deletion causes defects in tRNA-mediated nonsense suppression | ++ | + | −/+ | − |
| 6Pgl activity in yeast extracts | − | − | ++ | ++ |
| Subcellular location of encoded protein | Nucleus and cytosol | Cytosol | ND | Cytosol |
| Deletion causes nuclear tRNA accumulation | + | + | ND | − |
++, strong; +, intermediate; −/+, weak; −, undetectable; ND, not determined.
SOL4 was also deleted from strain WCS18 (relevant genotype: LOS1 sol1::ura3 sol2::ura3 sol3::URA3), which lacks SOL1, SOL2, and SOL3. Deletion of the entire SOL family (MLW114 or its ura3 derivative, MLW115) resulted in viable cells showing that this family is unessential. Cells containing the quadruple sol1-sol4 deletion do not appear to be significantly less efficient in nonsense suppression than cells with the sol1 sol2 double deletion (Figure 2B).
Sol3p and Sol4p possess 6Pgl activity, but Sol1p and Sol2p do not:
S. cerevisiae Sol proteins are part of the large conserved Sol/DevB/Pgl protein family. The Sol/DevB/Pgl family is related to another protein family, Nag1p/oscillin/glucose 6-phosphate deaminase/NagB, at the sequence and structural level (Figure 1). Members of the Nag family, NagB from Escherichia coli and oscillin/GNP1 from Homo sapiens, have been studied in detail and catalytic and allosteric site residues have been identified (Rudino-Pinera et al. 2002; Arreola et al. 2003). A crystal structure of one member of the Sol family, TmaDevB (Figure 1), from the bacteria Thermotoga maritima has been determined (1PBT; Kim et al. 2003). Although the overall structures of the Sol and Nag proteins are very similar (Figure 1C) and several of the catalytic residues are conserved between these two families, there are also significant differences in other catalytic residues and a region of the Nag proteins known as the active site lid is completely different in the Sol proteins (Figure 1B; compare HsOscl residues 162–182 and ScSol1p residues 225–238; Rudino-Pinera et al. 2002; Arreola et al. 2003).
Sol family members are present in most eukaryotes (>150 in the full alignment) and a large number of eubacteria (Figure 1). Nag proteins are present in a majority of eukaryotes, but appear to be absent from plants and Saccharomycetes (except Candida), the very organisms that possess multiple genes for the Sol proteins. Eubacteria lacking SOL (devB/pgl) genes usually have a nagB gene, whereas other eubacteria contain genes for both. Vertebrates as well as a few single-cell eukaryotes have an additional form of the Sol/DevB/Pgl family, a fusion protein containing a Sol/Pgl domain as well as a glucose 6-phosphate dehydrogenase (G6PD) domain (Figure 1A, HsH6PD and Pf6PGL; Shen et al. 1996; Collard et al. 1999; Clarke et al. 2001). A few eubacteria also have additional homologs of this family. E. coli has genes for agaI (putative galactosamine 6-phosphate isomerase), and yieK, a member of this family with no known function. No members of the Sol/DevB/Pgl family are found in Archaea.
Some of the SOL/devB/pgl homologs encode 6-phosphogluconolactonase activity (Collard et al. 1999; Duffieux et al. 2000; Hager et al. 2000; Clarke et al. 2001). 6Pgl (EC 3.1.1.31) catalyzes the second step of the pentose phosphate pathway, the conversion of 6-phosphogluconolactone to 6-phosphogluconate. We tested the S. cerevisiae SOL family for 6Pgl activity, employing a previously described assay that monitors production of NADPH by the successive activity of the enzymes glucose 6-phosphate dehydrogenase, 6Pgl, and 6-phosphogluconate dehydrogenase (Collard et al. 1999). Given the anticipated redundancy of members of the Sol protein family, our strategy was to assay cells possessing only a single member of this family. Therefore we employed strain MLW115 lacking all four members and supplied this strain with each SOL gene individually via a multi-copy vector.
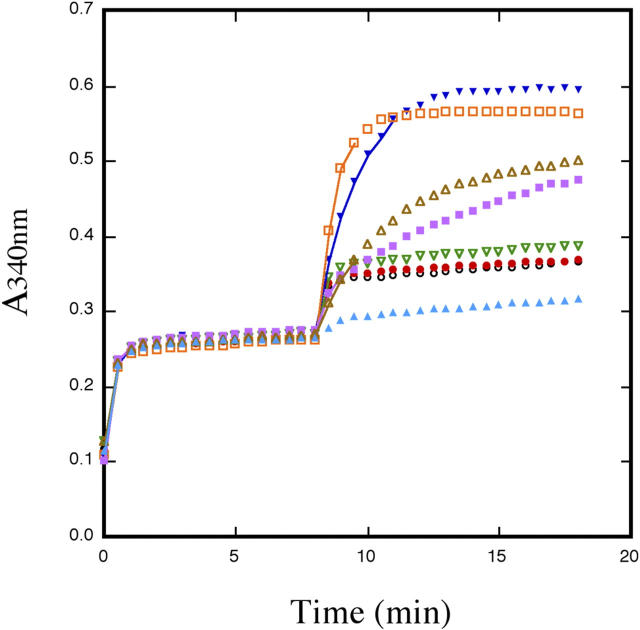
Compared to the wild-type strain (X2316-3C) containing the vector pRS426, the strain with the entire SOL family deleted (MLW115) containing pRS426 had very low, if any, 6Pgl activity. Cells containing multi-copy plasmids with functional SOL1 or SOL2 genes do not possess any greater 6Pgl activity than cells containing the vector only (Figure 3). In contrast, cells containing multi-copy plasmids with SOL3 or SOL4 provided activity beyond wild-type levels (Figure 3; Table 1). Thus, as expected, members of the Sol protein family appear to encode 6Pgl activity; however, unexpectedly Sol1p and Sol2p do not provide cells with 6Pgl activity.
Figure 3.—
6-Phosphogluconolactonase assay. Yeast glucose-6-phosphate dehydrogenase was incubated with glucose-6-phosphate and the production of NADPH was monitored at 340 nm for 8 min. Cell extract (50 μg protein) and 1.5 units 6-phosphogluconate dehydrogenase were added to the reaction mix and the absorbance at 340 nm was monitored for an additional 10 min. Black open circles, strain MLW115 with vector pRS426; red solid circles, MLW115 + pRSSOL1; green open triangles, MLW115 + WS80 (pRSSOL2); dark-blue solid triangles, MLW115 + WS92 (pRSSOL3); orange open squares, MLW115 + pRSSOL4; gold open triangles, MLW115 + pGST-SOL4; purple solid squares, X2316-3C + pRS426 vector; light-blue solid triangles, no extract. Note that even though GST-SOL4 transcription is regulated by copper, cells grown under noninducing conditions possess significant activity.
The Sol family members have different subcellular distributions:
As phosphogluconolactonase catalysis should occur in the cytosol, we anticipated that Sol3p and Sol4p would be cytoplasmic proteins. If suppression of the los1::kanr phenotype by multi-copy Sol1p and Sol2p occurs by altering tRNA biogenesis, then one might anticipate that these two proteins would be located in the nucleus. We determined the cellular location of functional tagged versions of these proteins employing indirect immunofluorescence and cell fractionation.
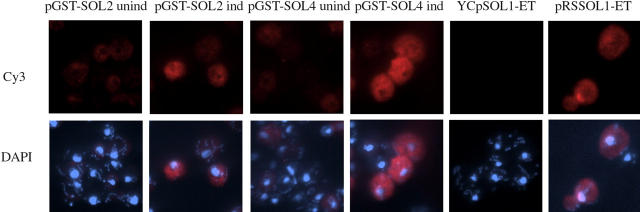
Amino-terminal GST-tagged versions of Sol2p and Sol4p obtained from the arrayed collection of GST-tagged yeast genes (Martzen et al. 1999) were active as assessed by the multi-copy los1 suppression assay for Sol2p (not shown) or the coupled 6Pgl assay for Sol4p (Figure 3) and each gave rise to only full-length protein as detected by Western analysis using anti-GST antibody (not shown). A GST-tagged version of Sol3p appeared to be proteolytically cleaved as assessed by Western analysis (not shown) and therefore its location was not studied. Gene expression of the GST-tagged constructs is inducible by copper. In the absence of copper, cells containing pGST-SOL2 and pGST-SOL4 express low levels of a cytosolic, apparently nuclear-excluded, signal recognized by anti-GST (Figure 4). The cytosolic signals are significantly increased in cell cultures induced by copper addition (Figure 4). Thus, as anticipated, Sol4p appears to be a cytosolic protein; surprisingly, Sol2p also appears to be cytosolic.
Figure 4.—
Indirect immunofluorescence location of Sol proteins. Strain SS700 containing pGST-SOL2 or pGST-SOL4, either uninduced (unind) or induced for 1 hr by addition of Cu2SO4 to a final concentration of 0.5 mm (ind), YCpSOL1-ET, or pRSSOL1-ET. (Top) Cells were incubated with mouse anti-GST at a dilution of 1:500 or with mouse anti-HA at a dilution of 1:500; Cy3 anti-mouse antibody was used as secondary antibody. (Bottom) The overlay of DNA staining via DAPI and Cy3 detection of the mouse antibodies.
The amino-terminal GST-tagged Sol1p was not functional. Therefore, we employed a previously constructed functional version of Sol1p (Shen et al. 1996) tagged at the carboxyl terminus with the HA epitope that generates full-length protein to study localization. Wild-type cells containing SOL1-HA on a centromere-containing vector (YCpSOL1-ET) had no detectable signal using anti-HA (Figure 4). In contrast, cells containing a multi-copy vector harboring the SOL1-HA construct (pRSSOL1-ET) produced a protein with a predominant, but not exclusive, nuclear location (Figure 4). Employing cell fractionation procedures followed by Western analysis, we were able to assess the distribution of Sol1p-HA encoded by YCpSOL1-ET. In agreement with the immunofluorescence results, Sol1p-HA was detected primarily in the nuclear fraction (not shown). Sol1p, Sol2p, and Sol4p had the same subcellular distributions in los1 mutant and wild-type cells (not shown).
A recent genome-wide project reported Sol1-GFP, Sol3-GFP, and Sol4-GFP to be located in the nucleus and the cytosol, whereas Sol2-GFP was reported to be nuclear excluded (Huh et al. 2003). One possible reason that the data for Sol4p are not in complete agreement could be construct functionality as it is unknown whether the GFP-tagged SOL constructs are active. Alternatively, protein expression levels might account for the differences as Huh et al. (2003) studied integrated genes with endogenous regulation whereas our studies utilized multi-copy plasmids.
In sum, our data show that there is a significant nuclear pool of Sol1p under the conditions in which it acts as a los1 suppressor. In contrast, the other members of the family that are poorer suppressors (i.e., Sol2p) or that lack detectable suppressor activity (i.e., Sol4p) appear to reside primarily in the cytosol (Table 1).
Sol1p, but not Sol4p, functions in tRNA nuclear export:
Previously we employed in vivo labeling procedures to assess whether Sol1p suppresses los1 defects by affecting tRNA biogenesis. No effects by Sol1p overexpression or by sol1 and sol2 deletion upon tRNA processing or accumulation were detected (Shen et al. 1996). To readdress how the Sol1 and Sol2 proteins affect tRNA-mediated nonsense suppression, we assessed whether these proteins affect the distribution of tRNAs between the nucleus and the cytosol employing FISH.
Cells defective in Los1p have increased nuclear pools of tRNAMet and tRNATyr, but not of poly(A) RNA (Sarkar and Hopper 1998). If Sol1p and/or Sol2p corrected the los1 phenotype via action of an alternative tRNA nuclear export pathway, one might expect that los1 cells containing multi-copy SOL1 would evidence reduced tRNA nuclear pools compared to los1 cells with vector alone. Indeed, los1 cells possessing multi-copy LOS1 have reduced tRNA nuclear pools (Feng and Hopper 2002). Despite the prediction, no apparent changes in nuclear levels of tRNAMet and tRNATyr in los1 cells possessing multi-copy SOL1 were observed (data not shown). However, because FISH is primarily a qualitative technique, we cannot rule out the possibility that overexpression of SOL1 causes a reduction of tRNA nuclear pools.
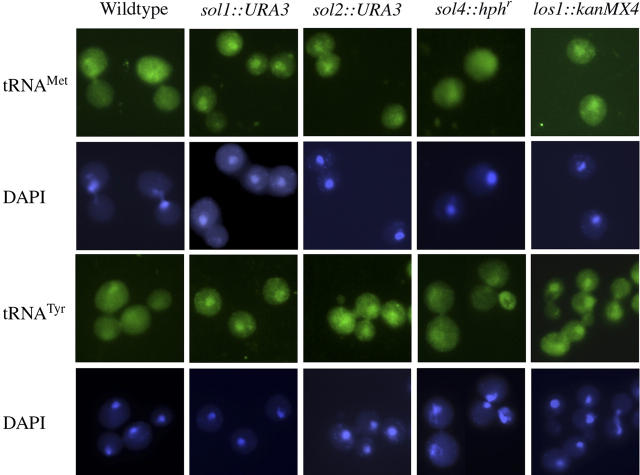
To address a possible role for the Sol proteins in tRNA nuclear export in a different way, we performed FISH analyses for LOS1 cells possessing deletions of individual and combinations of the SOL genes (Figure 5 and data not shown). Wild-type and sol4::hphr cells possess very low levels of nuclear tRNAMet, whereas sol1::URA3 and sol2::URA3 (Figure 5, top two rows) and sol1::ura3 sol2::ura3 sol3::ura3 sol4::hphr (not shown) possess significant tRNAMet nuclear pools, comparable to los1::kanMX4 cells (Figure 5, top two rows). Similarly, wild-type and sol4::hphr cells possess low, but detectable, levels of nuclear tRNATyr, and the levels are increased in sol1::URA3 and sol2::URA3 cells (Figure 5, bottom two rows) as well as in cells missing all four SOL genes (not shown). The data show that Sol1p and Sol2p affect tRNA nuclear export whereas Sol4p has no detectable role in this process (Table 1).
Figure 5.—
Sol1p and Sol2p apparently function in the distribution of tRNA between the nucleus and the cytosol. Fluorescence in situ hybridization analysis of tRNA location in wild-type (X2316-3C), sol1::URA3 (WCS2), sol2::URA3 (WCS7), sol4::hphr (MLW104), and los1::kanMX4 (YD05055) mutant cells as indicated. (First row) Hybridization employing probe for tRNAMet. (Second row) The same cells counterstained with DAPI to detect DNA. (Third row) Hybridization employing probe for tRNATyr. (Fourth row) The same cells as in third row, counterstained with DAPI.
DISCUSSION
Sol1p–Sol4p—an unusual example of a protein family whose individual members are biochemically distinct and spatially dispersed:
Here we show that the Sol protein family is apparently involved in both tRNA nuclear export and carbon metabolism. There is precedent for individual proteins functioning in multiple biochemical pathways. For example, some mitochondrial aminoacyl tRNA synthetases function in group I pre-mRNA splicing, in addition to bringing amino acids to the ribosome during translation (see Myers et al. 2002 for a recent summary). Dual function of the aminoacyl synthetases apparently involves the ability to fold/stabilize a tRNA-like structure of the group I introns prior to RNA-mediated catalysis. There are also numerous examples of dinucleotide-dependent enzymes able to catalyze various metabolic reactions (e.g., aconitase, glyceraldehyde-3-phosphate dehydrogenase, and lactate dehydrogenase) that also possess the ability to bind nucleic acids and to catalyze reactions involving nucleic acids (Hentze 1994; Pioli et al. 2002). It has been proposed that dinucleotide binding domains may also function in RNA binding, allowing for dual activities of this class of proteins (Hentze 1994).
In contrast to the above examples, it appears that individual Sol proteins do not possess multiple activities. Rather there appears to be a division of labor among family members. There is also precedent for individual members of gene families to function in distinct, but related, processes. The S. cerevisiae cytosolic Gal1 and Gal3 proteins provide such an example. Gal1p is a galactokinase affecting galactose metabolism. Gal3p tethers the negative transcription regulator, Gal80p, in the cytosol, allowing the positive regulator, Gal4p, to recruit the transcription complex to DNA in the nucleus (Peng and Hopper 2002). Both Gal1p and Gal3p bind galactose; however, Gal3p does not possess kinase activity (Platt et al. 2000 and references within). There are other examples of protein families in which the individual members act in related processes in separate subcellular compartments. For example, two different highly related human oxidative demethylases act to repair alkylation damage to DNA in the nucleus and RNA in the cytosol (Aas et al. 2003) and thus present a protein family with one member able to bind DNA in the nucleus and the other member able to bind RNA in the cytosol.
The Sol family appears to differ from other protein families with multiple functions in that Sol members appear to be spatially dispersed and affect the biochemically distinct processes, tRNA nuclear export and carbon metabolism. Sol1p, the best los1 suppressor, has a predominant nuclear pool, whereas Sol3p and Sol4p, which have little or no detectable effect upon los1 suppression or tRNA biogenesis, are located predominantly in the cytosol (Huh et al. 2003; Figure 4). However, we did not detect a nuclear pool for Sol2p even though it acts as a multi-copy suppressor of los1 (albeit less than Sol1p) and its deletion affects nonsense suppression (Shen et al. 1996) and the distribution of tRNA between the nucleus and the cytosol.
Although we do not know if Sol3p and Sol4p would affect tRNA biogenesis if directed to the nucleus, it does not appear that Sol1p or Sol2p affect carbon metabolism since cells containing either Sol1p or Sol2p on multi-copy vectors do not possess detectable 6Pgl activity in cellular extracts. Whereas Sol1p and Sol2p are clearly homologous with Sol3p and Sol4p, they are only ∼50% similar in amino acid sequence, rendering it difficult to assign regions of Sol1p and Sol2p that could function in tRNA nuclear export. We are unaware of another example of a protein family whose individual members are spatially dispersed and function in distinct biochemical pathways; thus, the Sol proteins may define a new type of protein family.
Role of Sol1p and Sol2p in tRNA subcellular distribution:
Deletion of SOL1 and/or SOL2 causes defects in tRNA-mediated nonsense suppression and accumulation of tRNAs in the nucleus. The data indicate that Sol1p and Sol2p participate directly or indirectly in tRNA nuclear export. Overexpressed Sol1p or Sol2p suppress, to varying extents, los1 defects in tRNA-mediated nonsense suppression, but have no detectable effect upon pre-tRNA splicing. That suppression is as effective in cells with los1 null alleles as in cells with the los1-1 point mutation likely means that Sol1p and Sol2p function in a pathway that operates in parallel to Los1p. If Sol1p and Sol2p serve a direct role in Los1p-independent tRNA nuclear export, then the fact that cells missing both the Los1p export pathway and the Sol1p/Sol2p genes (i.e., los1 sol1 sol2) are viable (Shen et al. 1996) could indicate that there are greater than two pathways for exporting tRNAs from the nucleus in S. cerevisiae.
To date, several other gene products have been implicated in Los1p-independent tRNA nuclear export. These include Cca1p that adds the C, C, and A nucleotides to the tRNA 3′ termini (Feng and Hopper 2002), aminoacyl-tRNA synthetases that catalyze addition of amino acids to mature tRNA 3′ termini (Lund and Dahlberg 1998; Sarkar et al. 1999; Grosshans et al. 2000; Azad et al. 2001), the translation factor eEF1-A (Grosshans et al. 2001), and tRNA modification proteins such as Pus1p (Simos et al. 1996). We do not know whether Sol1p and Sol2p function in the same pathways as any of these other gene products or, indeed, whether they function directly in the nuclear export process.
Alternatively, Sol1p and Sol2p may serve a regulatory role in tRNA nuclear export; for example, Sol1p and Sol2p could communicate cytosolic nutrient levels to the tRNA biogenesis/nuclear export machinery, assuring tRNA export only when growth conditions are appropriate. There is precedent for such communication involving amino acid availability. For example, Stp1p, which was identified by its role in tRNA biogenesis (Wang and Hopper 1988), actually functions in the pathway that signals external amino acid availability to the nucleus (Andréasson and Ljungdahl 2002). It is also known that amino acid deprivation causes tRNA nuclear accumulation, presumably via decreases in nuclear levels of charged tRNAs (Sarkar et al. 1999; Grosshans et al. 2000). Although a priori there was no expectation for a connection between the pentose phosphate pathway and tRNA biogenesis/nuclear export, it is feasible Sol1p and Sol2p interact with substrates or products of this pathway and communicate carbon availability to the tRNA biogenesis and/or nuclear export machinery, thereby functioning analogously to amino acid sensing. One could perhaps distinguish between direct vs. regulatory roles if Sol1p and Sol2p interactors were known. Unfortunately, to date, neither the genome-wide two hybrid searches (Uetz et al. 2000; http://portal.curagen.com/) nor the genome-wide copurification studies (Gavin et al. 2002; Ho et al. 2002; http://www.mdsp.com/yeast/) have identified Sol1p or Sol2p interactors.
Defects of genes in the Los1p-dependent pathway, LOS1, RNA1 (the Ran GAP), and PRP20 (the Ran GEF), as well as defects of some of the nucleoporins cause accumulation of intron-containing pre-tRNAs (Hopper et al. 1978, 1980; Kadowaki et al. 1993; Sharma et al. 1996; Simos et al. 1996), consistent with the recent reports that pre-tRNA splicing may occur in the cytosol (Huh et al. 2003; Yoshihisa et al. 2003). Curiously, none of the known components of the Los1p-independent pathway(s) appears to affect pre-tRNA splicing. It is difficult to rectify nuclear accumulation of mature spliced tRNAs with the idea that splicing occurs in the cytosol. One feasible, but unorthodox, idea is that in cells with defective Cca1p, tRNA charging enzymes, Sol1p or Sol2p, etc., there is retrograde movement of tRNA from the cytosol to the nucleus.
Acknowledgments
We thank J. McCusker for reagents and information to generate hygromycin B resistant knockout cassettes. We also thank K. Stauffer and H. Shaheen for numerous scientific interactions and help in manuscript preparation. This work was supported by an award from the American Heart Association (predoctoral fellowship to R.L.H.) and a grant from the National Institutes of Health to A.K.H.
References
- Aas, P. A., M. Otteriel, P. O. Falnes, C. B. Våggbø, G. Skorpen et al., 2003. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature 421: 859–863. [DOI] [PubMed] [Google Scholar]
- Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang et al., 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andréasson, C., and P. O. Ljungdahl, 2002. Receptor-mediated endoproteolytic activation of two transcription factors in yeast. Genes Dev. 16: 3158–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arreola, R., B. Valderrama, M. L. Morante and E. Horjales, 2003. Two mammalian glucosamine-6-phosphate deaminases: a structural and genetic study. FEBS Lett. 551: 63–70. [DOI] [PubMed] [Google Scholar]
- Arts, G. J., M. Fornerod and I. W. Mattaj, 1998. a Identification of a nuclear export receptor for tRNA. Curr. Biol. 8: 305–314. [DOI] [PubMed] [Google Scholar]
- Arts, G. J., S. Kuersten, P. Romby, B. Ehresmann and I. W. Mattaj, 1998. b The role of exportin-t in selective nuclear export of mature tRNAs. EMBO J. 17: 7430–7441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad, A. K., D. R. Stanford, S. Sarkar and A. K. Hopper, 2001. Role of nuclear pools of aminoacyl-tRNA synthetases in tRNA nuclear export. Mol. Biol. Cell 12: 1381–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack, M. T., K. Regener, B. Schwappach, R. Saffrich, E. Paraskeva et al., 2002. Exp5 exports eEF1A via tRNA from nuclei and synergizes with other transport pathways to confine translation to the cytoplasm. EMBO J. 21: 6205–6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calado, A., N. Treichel, E.-C. Müller, A. Otto and U. Kutay, 2002. Exportin-5-mediated nuclear export of eukaryotic elongation factor 1A and tRNA. EMBO J. 21: 6216–6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, J. L., D. A. Scopes, O. Sodeinde and P. J. Mason, 2001. Glucose-6-phosphate dehydrogenase-6-phosphogluconolactonase: a novel bifunctional enzyme in malaria parasites. Eur. J. Biochem. 268: 2013–2019. [DOI] [PubMed] [Google Scholar]
- Collard, F., J. F. Collet, I. Gerin, M. Veiga-da-Cunha and E. Van Schaftingen, 1999. Identification of the cDNA encoding human 6-phosphogluconolactonase, the enzyme catalyzing the second step of the pentose phosphate pathway. FEBS Lett. 459: 223–226. [DOI] [PubMed] [Google Scholar]
- Dolinski, K., R. Balakrishnan, K. R. Christie, M. C. Costanzo, S. S. Dwight et al., 2003 Saccharomyces genome database (http://www.yeastgenome.org/).
- Duffieux, F., J. Van Roy, P. A. M. Michels and F. R. Opperdoes, 2000. Molecular characterization of the first two enzymes of the pentose-phosphate pathway of Trypanosoma brucei. Glucose-6-phosphate dehydrogenase and 6-phosphogluconolactonase. J. Biol. Chem. 275: 27559–27565. [DOI] [PubMed] [Google Scholar]
- Feng, W., and A. K. Hopper, 2002. A Los1p-independent pathway for nuclear export of intronless tRNAs in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 99: 5412–5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feroli, F., G. Carignani, A. Pavanello, P. Guerreiro, D. Azevedo et al., 1997. Analysis of a 17.9 kb region from Saccharomyces cerevisiae chromosome VII reveals the presence of eight open reading frames, including BRF1 (TFIIIB70) and GCN5 genes. Yeast 13: 373–377. [DOI] [PubMed] [Google Scholar]
- Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch et al., 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147. [DOI] [PubMed] [Google Scholar]
- Goldstein, A. L., and H. H. McCusker, 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15: 1541–1553. [DOI] [PubMed] [Google Scholar]
- Görlich, D., and U. Kutay, 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15: 607–660. [DOI] [PubMed] [Google Scholar]
- Grosshans, H., E. Hurt and G. Simos, 2000. An aminoacylation-dependent nuclear tRNA export pathway in yeast. Genes Dev. 14: 830–840. [PMC free article] [PubMed] [Google Scholar]
- Grosshans, H., F. Lecointe, H. Grosjean, E. Hurt and G. Simos, 2001. Pus1p-dependent tRNA pseudouridinylation becomes essential when tRNA biogenesis is compromised in yeast. J. Biol. Chem. 276: 46333–46339. [DOI] [PubMed] [Google Scholar]
- Guex, N., and M. C. Peitsch, 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18: 2714–2723. [DOI] [PubMed] [Google Scholar]
- Guldener, U., S. Heck, T. Fielder, J. Beinhauer and J. H. Hegemann, 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24: 2519–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager, P. W., M. W. Calfee and P. V. Phibbs, 2000. The Pseudomonas aeruginosa devB/SOL homolog, pgl, is a member of the hex regulon and encodes 6-phosphogluconolactonase. J. Bacteriol. 182: 3934–3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmuth, K., D. M. Lau, F. R. Bischoff, M. Kunzler, E. Hurt et al., 1998. Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol. Cell. Biol. 18: 6374–6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S., and J. G. Henikoff, 1992. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 89: 10915–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze, M. W., 1994. Enzymes as RNA-binding proteins: A role for (di)nucleotide-binding domains? Trends Biochem. Sci. 19: 99–142. [DOI] [PubMed] [Google Scholar]
- Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore et al., 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180–183. [DOI] [PubMed] [Google Scholar]
- Hopper, A. K., and E. M. Phizicky, 2003. tRNA transfers to the limelight. Genes Dev. 17: 162–180. [DOI] [PubMed] [Google Scholar]
- Hopper, A. K., F. Banks and V. Evangelides, 1978. A yeast mutant which accumulates precursor tRNAs. Cell 14: 211–219. [DOI] [PubMed] [Google Scholar]
- Hopper, A. K., L. D. Schultz and R. A. Shapiro, 1980. Processing of intervening sequences: a new yeast mutant which fails to excise intervening sequences from precursor tRNAs. Cell 19: 741–751. [DOI] [PubMed] [Google Scholar]
- Huh, W.-I., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson et al., 2003. Global analysis of protein localization in budding yeast. Nature 425: 686–691. [DOI] [PubMed] [Google Scholar]
- Hurt, D. J., S. S. Wang, Y. H. Lin and A. K. Hopper, 1987. Cloning and characterization of LOS1, a Saccharomyces cerevisiae gene that affects tRNA splicing. Mol. Cell. Biol. 7: 1208–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki, T., D. Goldfarb, L. M. Spitz, A. M. Tartakoff and M. Ohno, 1993. Regulation of RNA processing and transport by a nuclear guanine nucleotide release protein and members of the Ras superfamily. EMBO J. 12: 2929–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y., A. Joachimiak, A. Edwards, T. Skarina and A. Savchenko, 2003 The crystal structure analysis of Tm1154, oxidoreductase from Thermotoga maritima. Protein Data Bank (http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1PBT).
- Kutay, U., G. Lipowsky, E. Izaurralde, F. R. Bischoff, P. Schwarzmaier et al., 1998. Identification of a tRNA-specific nuclear export receptor. Mol. Cell 1: 359–369. [DOI] [PubMed] [Google Scholar]
- Langkjaer, R. B., P. F. Cliften, M. Johnston and J. Piskur, 2003. Yeast genome duplication was followed by asynchronous differentiation of duplicated genes. Nature 421: 848–852. [DOI] [PubMed] [Google Scholar]
- Li, J., and X. Chen, 2003. PAUSED, a putative exportin-t, acts pleiotropically in Arabidopsis development but is dispensable for viability. Plant Physiol. 132: 1913–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipowsky, G., F. R. Bischoff, E. Izaurralde, U. Kutay, S. Schafer et al., 1999. Coordination of tRNA nuclear export with processing of tRNA. RNA 5: 539–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund, E., and J. E. Dahlberg, 1998. Proofreading and aminoacylation of tRNAs before export from the nucleus. Science 282: 2082–2085. [DOI] [PubMed] [Google Scholar]
- Martzen, M. R., S. M. McCraith, S. L. Spinelli, F. M. Torres, S. Fields et al., 1999. A biochemical genomics approach for identifying genes by the activity of their products. Science 286: 1153–1155. [DOI] [PubMed] [Google Scholar]
- Myers, C. A., B. Kuhla, S. Cusack and A. M. Lambowitz, 2002. tRNA-like recognition of group I introns by a tyrosyl-tRNA synthetase. Proc. Natl. Acad. Sci. USA 99: 2630–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor, J. P., and C. L. Peebles, 1991. In vivo pre-tRNA processing in Saccharomyces cerevisiae. Mol. Cell. Biol. 11: 425–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, G., and J. E. Hopper, 2002. Gene activation by interaction of an inhibitor with a cytoplasmic signaling protein. Proc. Natl. Acad. Sci. USA 99: 8548–8553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pioli, P. A., B. J. Hamilton, J. C. Connolly, G. Brewer and W. F. C. Rigby, 2002. Lactate dehydrogenase is an AU-rich element-binding protein that directly interacts with AUF1. J. Biol. Chem. 277: 35738–35745. [DOI] [PubMed] [Google Scholar]
- Platt, A., H. C. Ross, S. Hankin and H. J. Reece, 2000. The insertion of two amino acids into a transcriptional inducer converts it into a galactokinase. Proc. Natl. Acad. Sci. USA 97: 3154–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudino-Pinera, E., S. Morales-Arrieta, S. P. Rojas-Trejo and E. Horjales, 2002. Structural flexibility, an essential component of the allosteric activation in Escherichia coli glucosamine-6-phosphate deaminase. Acta Crystallogr. 58: 10–20. [DOI] [PubMed] [Google Scholar]
- Sarkar, S., and A. K. Hopper, 1998. tRNA nuclear export in Saccharomyces cerevisiae: in situ hybridization analysis. Mol. Biol. Cell 9: 3041–3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar, S., A. K. Azad and A. K. Hopper, 1999. Nuclear tRNA aminoacylation and its role in nuclear export of endogenous tRNAs in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 96: 14366–14371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, K., E. Fabre, H. Tekotte, E. C. Hurt and D. Tollervey, 1996. Yeast nucleoporin mutants are defective in pre-tRNA splicing. Mol. Cell. Biol. 16: 294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, W. C., D. Selvakumar, D. R. Stanford and A. K. Hopper, 1993. The Saccharomyces cerevisiae LOS1 gene involved in pre-tRNA splicing encodes a nuclear protein that behaves as a component of the nuclear matrix. J. Biol. Chem. 268: 19436–19444. [PubMed] [Google Scholar]
- Shen, W.-C., D. R. Stanford and A. K. Hopper, 1996. Los1p, involved in yeast pre-tRNA splicing, positively regulates members of the SOL gene family. Genetics 143: 699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simos, G., H. Tekotte, H. Grosjean, A. Segref, K. Sharma et al., 1996. Nuclear pore proteins are involved in the biogenesis of functional tRNA. EMBO J. 15: 2270–2284. [PMC free article] [PubMed] [Google Scholar]
- Tettelin, H., M. L. Agostoni Carbone, K. Albermann, M. Albers, J. Arroyo et al., 1997. The nucleotide sequence of Saccharomyces cerevisiae chromosome VII. Nature 387(6632 Suppl.): 81–84. [PubMed] [Google Scholar]
- Thompson, J. D., T. J. Plewniak, F. Jeanmougin and D. G. Higgins, 1997. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson et al., 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403: 623–627. [DOI] [PubMed] [Google Scholar]
- Wang, S. S., and A. K. Hopper, 1988. Isolation of a yeast gene involved in species-specific pre-tRNA processing. Mol. Cell. Biol. 8: 5140–5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson et al., 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906. [DOI] [PubMed] [Google Scholar]
- Wolfe, K. H., and D. C. Shields, 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 422: 708–713. [DOI] [PubMed] [Google Scholar]
- Yoshihisa, T., K. Yunoki-Esaki, C. Ohshima, N. Tanaka and T. Endo, 2003. Possibility of cytoplasmic pre-tRNA splicing: the yeast tRNA splicing endonuclease mainly localizes on the mitochondria. Mol. Biol. Cell 14: 3266–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]