Abstract
We isolated a novel rad52 mutation, rad52-L89F, which specifically impairs recombination in rad51Δ cells. rad52-L89F displays phenotypes similar to rad59Δ and encodes a mutant protein impaired in its ability to interact with Rad59. These results support the idea that Rad59 acts in homologous recombination via physical interaction with Rad52.
RAD52 is the only gene required for virtually all homologous recombination events in Saccharomyces cerevisiae. Null mutations in this gene display the most severe phenotype in many different recombination assays (Paques and Haber 1999). The Rad52 protein shows DNA-binding and strand-annealing activities in vitro (Mortensen et al. 1996). In addition, Rad52 physically interacts with both the RecA ortholog Rad51 and Rad59 (Symington 2002).
In contrast to RAD52, RAD51 is required for allelic recombination but not for recombination between DNA repeats (Prado et al. 2003). One-ended recombination events are RAD51 dependent (Davis and Symington 2004), but also occur in the absence of Rad51 (Malkova et al. 1996). Thus, in the absence of Rad51, recombination may occur by single-strand annealing (SSA), which leads to deletions between direct repeats (Lin et al. 1984; Paques and Haber 1999) or break-induced replication, which could give rise to inversions between inverted repeats if followed by SSA (Bartsch et al. 2000; Malagon and Aguilera 2001).
Rad59 is homologous to the amino-terminal half of Rad52 and shares several in vitro activities with Rad52, such as DNA binding and strand annealing (Petukhova et al. 1999; Davis and Symington 2001). It plays an important role in recombination occurring in the absence of Rad51. Thus, rad59Δ mutants present only a slight decrease in inverted-repeat recombination, whereas rad51Δ rad59Δ double mutants show a strong decrease similar to rad52Δ (Bai and Symington 1996).
Isolation of the new rad52-L89F mutation:
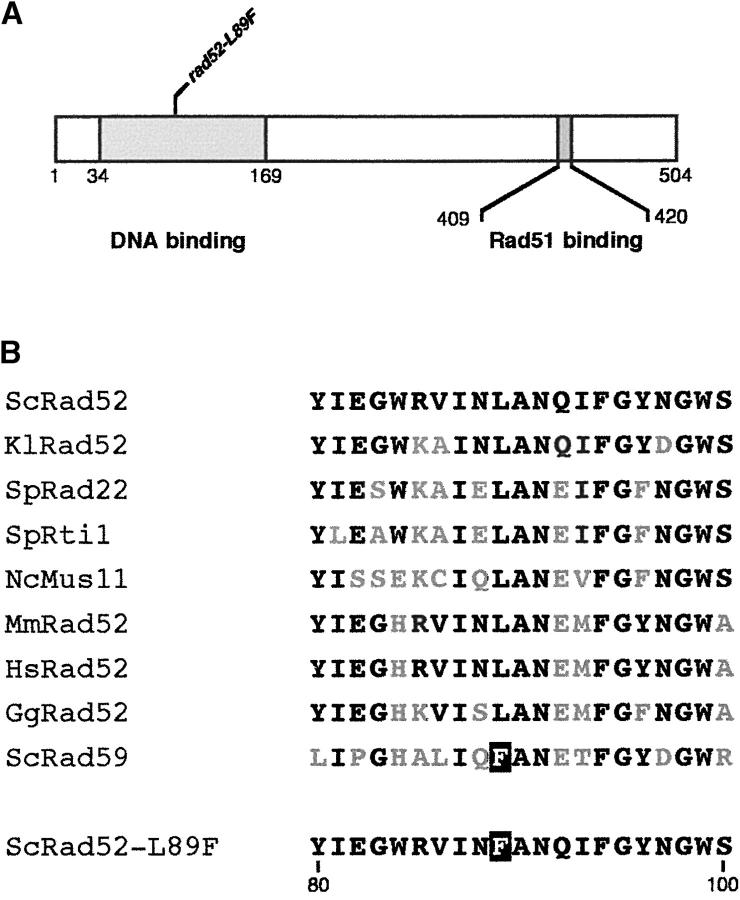
To understand recombination occurring in the absence of Rad51, we searched for mutants with reduced recombination levels in rad51Δ background. To facilitate the search we used a rad51Δ spt6-140 double mutant, which shows high levels of RAD51-independent recombination (Malagon and Aguilera 2001). UV-irradiated cells carrying the chromosomic his3p::INV inverted repeat system (Aguilera and Klein 1988) were screened for low levels of His+ recombinants. This led to the identification of a new rad52 allele. Sequence analysis showed that the mutant allele carried a single T-to-C substitution at position 165, which results in a Leu-to-Phe change in residue 89 (see Figure 1). This residue is located in the amino terminus of Rad52, which is the most conserved part of the protein, in a domain described as being necessary for DNA binding, self-association, and Rad59 interaction (Symington 2002). Interestingly, the rad52-1 mutation, which confers a rad52 null phenotype, is at position 90 (Adzuma et al. 1984). The new mutant allele was named rad52-L89F.
Figure 1.—
Primary structure of the Rad52-L89F protein. (A) Functional domains of Rad52 and localization of the rad52-L89F mutation. (B) Comparative alignment of Rad52 and Rad59 orthologs (Sc, S. cerevisiae; Kl, Kluyveromyces lactis; Sp, Schizosaccharomyces pombe; Nc, Neurospora crassa; Gg, chicken; Mm, mouse; Hs, human) and Rad52-L89F. Residues identical to those of ScRad52 are shown in black and the single substitution of Rad52-L89F is indicated as a solid box. The rad52-L89F mutant was obtained from the hyperrecombinant strain M137-11Ar51k (MATα ade2 can1-100 his3p::INV leu2 lys2-128α rad51Δ::KanMX4 spt6-140 trp1 ura3) by UV mutagenesis. Cells (50 μl) grown in YEPD to an OD660nm of 0.9 were diluted in 6 ml of water and poured into a glass petri dish for irradiation with 45 J/m2 of UV light (λ = 254 nm). After 5 hr of recovery in YEPD in the dark, cells were plated with the appropriate dilutions. Cell survival was 30% and 10,600 clones were screened for low His+ recombination.
Homologous recombination in rad52-L89F:
To understand the types of homologous recombination impaired by rad52-L89F, we determined the effect of rad52-L89F on the frequency of recombination of the his3p::INV system in different rad backgrounds (Figure 2A). Recombination frequencies were reduced only 10- and 2-fold below wild-type levels in rad51Δ and rad59Δ cells, respectively, but 40-fold in the double rad51Δ rad59Δ, consistent with previous reports (Bai and Symington 1996; Shinohara et al. 1998; Malagon and Aguilera 2001). Nevertheless, whereas in rad52Δ cells the reduction was >100-fold, rad52-L89F shows, as do rad51Δ and rad59Δ, only a slight decrease (<5-fold). Interestingly, rad52-L89F rad51Δ mutants showed a synergistic decrease and had the same recombination levels as rad52Δ. In contrast, deletion of RAD59 had no effect in rad52-L89F.
Figure 2.—

Recombination frequencies of the chromosomal inverted repeat system his3p::INV in different rad mutants. (A) Recombination in wild-type, rad51Δ, rad59Δ, rad52Δ, rad52-L89F, rad51Δ rad59Δ, rad52-L89F rad51Δ, and rad52-L89F rad59Δ strains. (B) Recombination in rad52-L89F, rad52-L89F rad51Δ, and rad52-L89F rad59Δ mutants carrying either pRS424 (pRS, negative control) or pRS424-rad52L89F (p52L89F, overexpressing rad52-L89F) plasmids. Recombination frequencies were determined as the median frequency of six independent colonies isolated from YEPD at 30° (Prado et al. 1997). Frequencies are the average and standard deviation (shown in bars) of two to four median values. The rad52-L89F mutation was transferred to other genetic backgrounds by genetic crosses. All radΔ strains used for the recombination assays were derived from M137-11A (MATα ade2 can1-100 his3p::INV leu2 lys2-128α rad51Δ::KanMX4 trp1 ura3) and were constructed by transformation with the corresponding deletion cassettes or by genetic crosses (Malagon and Aguilera 2001).
To determine whether the effect of rad52-L89F on recombination was due to a leaky activity of Rad52-L89F, we wondered if its overexpression could reestablish wild-type recombination. As can be seen in Figure 2B, multicopy rad52-L89F partially suppressed the recombination defect of rad52-L89F up to levels of rad59Δ, but had no effect in rad59Δ or rad51Δ backgrounds (Figure 2B). Therefore, rad52-L89F causes the same recombination phenotype as rad59Δ, regardless of a putative leakiness of the Rad52-L89F activity.
The recombination phenotypes of rad52-L89F are indeed similar to those of the previously characterized rad52-R70K allele in RAD and rad51Δ backgrounds, although they differ when Rad59 is not present (Bai et al. 1999). This suggests that a Rad52 amino-terminal domain covering at least the residues from 70 to 89 is essential for recombination in the absence of Rad51. Interestingly, both residues 89 and 70 are conserved in all known Rad52 orthologs and the L89F and R70K changes make the terminal domain of the mutant Rad52 proteins more similar to Rad59 (Figure 1B).
Repair of MMS damage in rad52-L89F:
To further characterize genetic interaction between rad52-L89F and RAD59, methyl methanesulfonate (MMS) sensitivity was determined by cell survival after different time exposures to 0.5% MMS. The rad52-L89F mutant showed a weaker MMS sensitivity than rad52Δ, but much stronger than rad59Δ (Figure 3A), in contrast to its low recombination defect. Nevertheless, as in recombination, rad52-L89F sensitivity was not affected in rad59Δ background. Therefore, these data support the idea that rad52-L89F behaves like rad59Δ. In any case, Rad52-L89F was leaky, as suggested from the observation that overexpression of Rad52-L89F enhanced MMS resistance of rad52-L89F strains (Figure 3B).
Figure 3.—
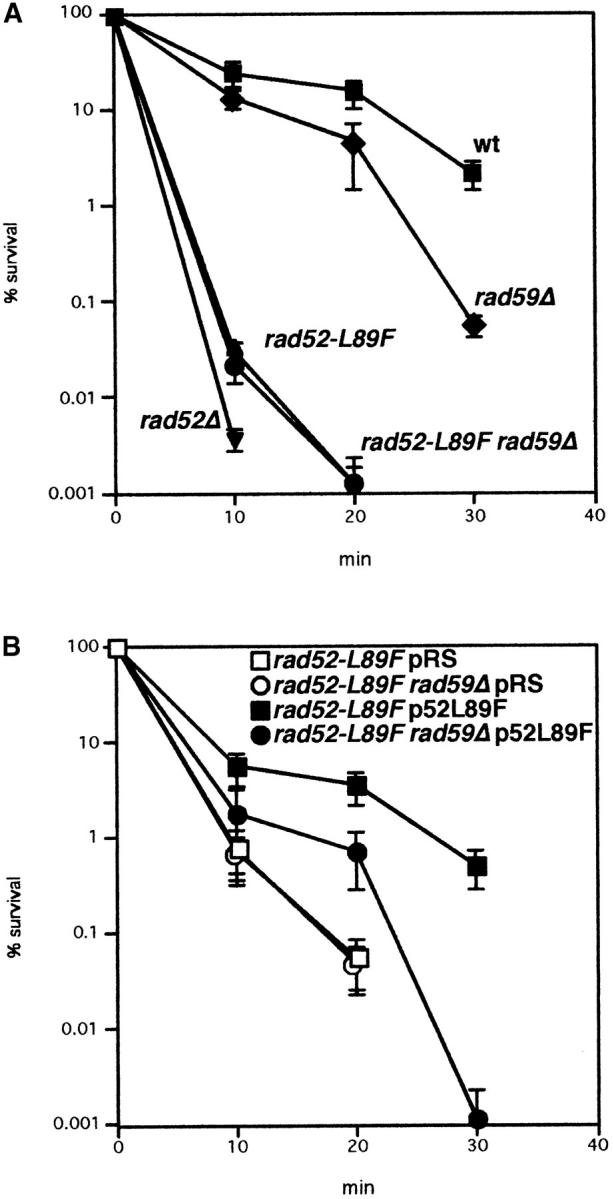
MMS sensitivity of different rad strains. (A) Sensitivity of wild-type, rad59Δ, rad52Δ, rad52-L89F, and rad52-L89F rad59Δ mutants. (B) Sensitivity of rad52-L89F and rad52-L89F rad59Δ mutants carrying either pRS424 (pRS, negative control) or pRS424-rad52L89F (p52L89F, overexpressing rad52-L89F) plasmids. Cells were exposed to 0.5% MMS for 0, 10, 20, and 30 min before plating onto YEPD. For each strain, the percentage of survival is referred to the value of cells not exposed to MMS (0 min), taken as 100%. Data are the average and standard deviation (shown in bars) of two to four independent experiments.
Rad52-L89F is affected in its ability to interact with Rad59:
The similarity of phenotypes between rad59Δ and rad52-L89F could be explained if in the rad52-L89F mutant the levels of Rad59 protein were reduced, as reported for rad52Δ mutants (Davis and Symington 2001). Nevertheless, this was not the case because the levels of Rad59 protein in rad52-L89F were similar to those of wild-type cells (Figure 4A).
Figure 4.—
Rad52-L89F-Rad59 interaction. (A) SDS-PAGE analysis of Rad59 protein in wild-type, rad52Δ, and rad52-L89F strains. Total protein extract (5 μg) was loaded for each strain. Coomassie staining (top) and Western blot using αRad59 polyclonal antibody (bottom) are shown. The position in the gel at which Rad59 migrates is indicated. (B) Purification of Rad59-GST fusion protein in wild-type, rad52Δ, and rad52-L89F strains. Purification of GST in the wild type was included as a negative control. Protein expression was under the control of the CUP1 promoter and was induced by addition of CuSO4 to a final concentration of 0.5 mm. Coomassie staining (top) and Western blot using αRad52 polyclonal antibody (bottom) of total cell extracts (left) and purified fractions (right) are shown. The positions in the gel at which Rad59-GST, Rad52, and GST migrate are indicated. For the preparation of extracts, cells were grown in 500 ml of SC-Leu to an OD660nm of 0.8. Cells were harvested, washed with water, and resuspended in one pellet volume of 50 mm Tris-HCl, pH 7.5, 1 mm EDTA, 4 mm MgCl2, 5 mm DTT, 10% glycerol, 1 m NaCl. Leupeptin and pepstatin A were added to a final concentration of 2 μg/ml and 1 μg/ml, respectively. Extracts were made with glass beads (McCraith and Phizicky 1990), followed by supplementation with 1 mm PMSF and 30 min centrifugation at 14,000 rpm in a JA-20 Beckman rotor. For Rad59::GST protein purification, supernatant was incubated with 1/100 volume of glutathione-Sepharose 4B (A. P. Biotech) previously equilibrated in 50 mm potassium phosphate pH 7.2, 1 mm DTT, 0.5 mm EDTA, 10% glycerol, 1% Triton X-100, 1 m NaCl. Samples were washed twice with 500 bead volume of 50 mm potassium phosphate pH 7.2, 1 mm DTT, 0.5 mm EDTA, 10% glycerol, 1% Triton X-100, 0.5 m NaCl. Proteins were eluted by boiling in 1× loading buffer for 5 min.
We tested the possibility that Rad52-L89F was impaired in its ability to interact with Rad59. For this purpose, we purified Rad59 fused to the glutathione S-transferase (Rad59::GST) from wild-type, rad52Δ, and rad52-L89F strains overexpressing the GST-fusion protein. Rad59::GST is functional, as it rescues the MMS sensitivity of rad59Δ. As negative control we purified GST from a wild-type strain overexpressing GST. As can be seen in Figure 4B, Rad52-L89F protein was present in cell extracts at levels lower than those of Rad52. Indeed, other missense mutations in the amino terminus reduce the levels of Rad52 (Asleson and Livingston 2003). As expected, wild-type Rad52 copurified with Rad59::GST, but not with GST. However, no Rad52-L89F protein was detected in the purified Rad59::GST fraction. Since Rad52-L89F protein levels are reduced, we cannot rule out the possibility of a nondetected weak interaction with Rad59 even though our results suggest that Rad52-L89F is affected in its ability to interact with Rad59.
Biological significance of the in vivo Rad52-Rad59 interaction:
Both human and yeast Rad52 proteins form multimeric ring structures (Shinohara et al. 1998; Stasiak et al. 2000; Ranatunga et al. 2001), and Rad59 has also been reported to self-associate (Davis and Symington 2003). It would be interesting to know whether Rad52 and Rad59 could form heteromeric ring structures (Symington 2002). This is supported by the fact that the Rad52 regions necessary and sufficient for self-interaction and Rad59 binding coincide (Davis and Symington 2003).
Our study confirms that the amino terminus of Rad52 is important for its interaction with Rad59. The reduced ability of Rad52-L89F to interact with Rad59 could at least partially explain the rad59Δ-like recombination phenotype of the rad52-L89F mutant. It could cause a reduction of the presence of Rad59 at recombination centers, leading to a rad59 phenocopy. As Rad59 is essential in recombination occurring in the absence of Rad51-dependent strand exchange, this would explain why the recombination phenotype of rad52-L89F is specifically observed in a rad51 background.
The MMS sensitivity of rad52-L89F is much more severe than its recombination defect. Other mutations in RAD52 and other RAD genes have been reported to separate recombinational and DNA repair phenotypes (Mortensen et al. 2002; Symington 2002). In our case, the lower amount of stable Rad52-L89F protein present in the cell (Figure 4B) is sufficient for spontaneous recombination, but not for the repair of MMS-induced damage. Consistently, overexpression of Rad52-L89F significantly suppresses the MMS sensitivity phenotype (Figure 3B).
Concluding remarks:
A novel rad52 mutation (rad52-L89F), identified by its specific effect in recombination occurring in the absence of Rad51, encodes a mutant Rad52 protein impaired in its ability to interact with Rad59. This, together with the strong similarity of recombination and repair phenotypes of rad52-L89F and rad59Δ, suggests that Rad59-Rad52 interaction is essential for the role of Rad59 in recombination in the absence of Rad51-mediated strand exchange.
Acknowledgments
We are grateful to L. Symington for yeast strain gifts, P. Sung and W. Heyer for antibodies, F. Prado for critical reading of the manuscript, and D. Haun for style supervision. This work was supported by grants from the Spanish Ministry of Science and Technology (BMC2000-0439 and SAF2003-00204) and the Regional Government of Andalusia (CVI0102). F.C.-L. was the recipient of predoctoral training grants from the Spanish Ministry of Education and Ministry of Health.
References
- Adzuma, K., T. Ogawa and H. Ogawa, 1984. Primary structure of the RAD52 gene in Saccharomyces cerevisiae. Mol. Cell. Biol. 4: 2735–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera, A., and H. L. Klein, 1988. Genetic control of intrachromosomal recombination in Saccharomyces cerevisiae. Isolation and genetic characterization of hyper-recombination mutations. Genetics 119: 779–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asleson, E. N., and D. M. Livingston, 2003. Investigation of the stability of yeast rad52 mutant proteins uncovers post-translational and transcriptional regulation of Rad52p. Genetics 163: 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, Y., and L. S. Symington, 1996. A Rad52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev. 10: 2025–2037. [DOI] [PubMed] [Google Scholar]
- Bai, Y., A. P. Davis and L. S. Symington, 1999. A novel allele of RAD52 that causes severe DNA repair and recombination deficiencies only in the absence of RAD51 or RAD59. Genetics 153: 1117–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch, S., L. E. Kang and L. S. Symington, 2000. RAD51 is required for the repair of plasmid double-stranded DNA gaps from either plasmid or chromosomal templates. Mol. Cell. Biol. 20: 1194–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, A. P., and L. S. Symington, 2001. The yeast recombinational repair protein Rad59 interacts with Rad52 and stimulates single-strand annealing. Genetics 159: 515–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, A. P., and L. S. Symington, 2003. The Rad52-Rad59 complex interacts with Rad51 and replication protein A. DNA Repair 2: 1127–1134. [DOI] [PubMed] [Google Scholar]
- Davis, A. P., and L. S. Symington, 2004. RAD51-dependent break-induced replication in yeast. Mol. Cell. Biol. 24: 2344–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, F. L., K. Sperle and N. Sternberg, 1984. Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol. Cell. Biol. 4: 1020–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malagon, F., and A. Aguilera, 2001. Yeast spt6-140 mutation, affecting chromatin and transcription, preferentially increases recombination in which Rad51p-mediated strand exchange is dispensable. Genetics 158: 597–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova, A., E. L. Ivanov and J. E. Haber, 1996. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc. Natl. Acad. Sci. USA 93: 7131–7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCraith, S. M., and E. M. Phizicky, 1990. A highly specific phosphatase from Saccharomyces cerevisiae implicated in tRNA splicing. Mol. Cell. Biol. 10: 1049–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen, U. H., C. Bendixen, I. Sunjevaric and R. Rothstein, 1996. DNA strand annealing is promoted by the yeast Rad52 protein. Proc. Natl. Acad. Sci. USA 93: 10729–10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen, U. H., N. Erdeniz, Q. Feng and R. Rothstein, 2002. A molecular genetic dissection of the evolutionarily conserved N terminus of yeast Rad52. Genetics 161: 549–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques, F., and J. E. Haber, 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63: 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petukhova, G., S. A. Stratton and P. Sung, 1999. Single strand DNA binding and annealing activities in the yeast recombination factor Rad59. J. Biol. Chem. 274: 33839–33842. [DOI] [PubMed] [Google Scholar]
- Prado, F., J. I. Piruat and A. Aguilera, 1997. Recombination between DNA repeats in yeast hpr1Δ cells is linked to transcription elongation. EMBO J. 16: 2826–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado, F., F. Cortes-Ledesma, P. Huertas and A. Aguilera, 2003. Mitotic recombination in Saccharomyces cerevisiae. Curr. Genet. 42: 185–198. [DOI] [PubMed] [Google Scholar]
- Ranatunga, W., D. Jackson, J. A. Lloyd, A. L. Forget, K. L. Knight et al., 2001. Human RAD52 exhibits two modes of self-association. J. Biol. Chem. 276: 15876–15880. [DOI] [PubMed] [Google Scholar]
- Shinohara, A., M. Shinohara, T. Ohta, S. Matsuda and T. Ogawa, 1998. Rad52 forms ring structures and co-operates with RPA in single-strand DNA annealing. Genes Cells 3: 145–156. [DOI] [PubMed] [Google Scholar]
- Stasiak, A. Z., E. Larquet, A. Stasiak, S. Muller, A. Engel et al., 2000. The human Rad52 protein exists as a heptameric ring. Curr. Biol. 10: 337–340. [DOI] [PubMed] [Google Scholar]
- Symington, L. S., 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66: 630–670. [DOI] [PMC free article] [PubMed] [Google Scholar]