Abstract
Parallels between vertebrate and Drosophila hematopoiesis add to the value of flies as a model organism to gain insights into blood development. The Drosophila hematopoietic system is composed of at least three classes of terminally differentiated blood cells: plasmatocytes, crystal cells, and lamellocytes. Recent studies have identified transcriptional and signaling pathways in Drosophila involving proteins similar to those seen in human blood development. To identify additional genes involved in Drosophila hematopoiesis, we have conducted a P-element-based genetic screen to isolate mutations that affect embryonic crystal cell development. Using a marker of terminally differentiated crystal cells, we screened 1040 P-element-lethal lines located on the second and third chromosomes and identified 44 individual lines that affect crystal cell development. Identifying novel genes and pathways involved in Drosophila hematopoiesis is likely to provide further insights into mammalian hematopoietic development and disorders.
DESPITE the obvious differences in cell type and functionality between vertebrate and Drosophila blood systems, parallels in their developmental mechanisms are remarkable (reviewed by Evans et al. 2003). Such shared developmental and functional mechanisms have prompted the use of Drosophila as a model organism to further investigate the genetic control of hematopoietic cell differentiation.
Both vertebrate and Drosophila hematopoiesis involve distinct, terminally differentiated lineages derived from common progenitor cells. Mammalian hematopoietic cells differentiate into two main branches: the lymphoid and myeloid lineages (reviewed by Dzierzak and Medvinsky 1995). Differentiation, function, and lineage hierarchy of Drosophila blood cells, or hemocytes, are most similar to those of the vertebrate myeloid lineage (reviewed by Orkin 2000). The Drosophila hematopoietic system is composed of at least three classes of terminally differentiated hemocytes: plasmatocytes, crystal cells, and lamellocytes, which participate in development and immune response (reviewed by Evans et al. 2003; Meister and Lageaux 2003).
Plasmatocytes are the most abundant hemocyte type in Drosophila and are commonly referred to as macrophages. Accordingly, they function to engulf apoptotic cells and debris as well as play a role in immune response by eliminating pathogens (Tepass et al. 1994; Lanot et al. 2001). Crystal cells, which compose ∼5% of the hemocyte population, participate in immune responses and wound healing through melanization. The paracrystaline inclusions within the cells are thought to contain Pro-Phenoloxidase A1 (ProPO A1; Rizki et al. 1980), an enzyme that is similar to tyrosinase and is important in the biosynthesis of melanin (Rizki et al. 1985). Unlike plasmatocytes and crystal cells, which are found in all developmental stages, lamellocytes have been observed only in Drosophila larvae and increase in number during immune challenge (Lanot et al. 2001; Sorrentino et al. 2002).
Drosophila hemocytes have dual sites of origin. Early hemocytes arising from the mesoderm of the embryonic head region are detected throughout development and into adulthood (Holz et al. 2003). A split of the hemocyte population into plasmatocytes and crystal cells occurs at an early stage. Crystal cells form a small, cohesive cell group that remains clustered around the embryonic proventriculus (Lebestky et al. 2000), whereas plasmatocytes migrate throughout the entire embryo (Tepass et al. 1994). A second population of hemocytes that differentiate in the late larva and during metamorphosis to populate the pupa and adult are derived from a second blood-forming tissue, the lymph gland, which is situated next to the dorsal blood vessel (aorta/heart) of the larva.
Over 20 genes have been identified in mammalian blood cell differentiation, including genes that encode transcription factors, recombinases, signaling molecules, transmembrane receptors, and secreted factors (reviewed by Orkin 1996) that can act positively and/or antagonistically in the regulation of hematopoiesis. Many molecules play a role in both vertebrate and Drosophila hematopoiesis, including transcriptional regulators such as GATA, friend of GATA (FOG), and acute myeloid leukemia-1 (AML-1), as well as the signaling transduction molecules Notch, Janus kinase/signal transducer and activator of transcription (JAK/STAT), and NFκB of the Toll/Cactus pathway (reviewed by Evans et al. 2003).
Serpent (srp) is one of five GATA factors in Drosophila and is required for both embryonic and larval blood development (reviewed by Rehorn et al. 1996; Lebestky et al. 2000; Fossett and Schulz 2001). A second GATA factor, Pannier (Pnr), is required for development of the heart (Rehorn et al. 1996; Gajewski et al. 1999) and larval blood (Mandal et al. 2004). Srp is the earliest known factor expressed in hemocyte precursors as its expression is first detected in the procephalic mesoderm (Tepass et al. 1994; Rehorn et al. 1996). Expression of srp is necessary for the differentiation of plasmatocytes and crystal cells in the embryo.
FOG is a zinc finger protein that functions as a transcriptional coregulator. Mammalian FOG1 binds directly to GATA-1 and has a similar loss-of-function phenotype as GATA-1 (reviewed by Cantor and Orkin 2001 and Fossett and Schulz 2001; Chang et al. 2002). The corepressor C-terminal binding protein (CtBP) and FOG together regulate hematopoietic lineage commitment in mammals. The Drosophila FOG ortholog, U-shaped (Ush), is expressed in all hemocyte precursors throughout embryonic and larval hematopoiesis (Fossett and Schulz 2001). Physical and genetic interaction between Ush and Srp has been demonstrated to repress crystal cell fate in prohemocytes (Fossett et al. 2003; Waltzer et al. 2003). Accordingly, ush is downregulated in crystal cell precursors and ush mutants exhibit an increase in crystal cell number.
AML-1, or Runx1, was first isolated as a fusion partner in a chromosomal translocation associated with AML (reviewed by Lutterbach and Hiebert 2000; Rabbitts 1994) and is necessary for definitive hematopoiesis in mammals (Okuda et al. 1996; Wang et al. 1996). Drosophila Lozenge (Lz), a transcription factor that has 71% identity to the Runt domain of the human protein AML-1, is necessary for crystal cell development during embryonic and larval hematopoiesis (Daga et al. 1996; Lebestky et al. 2000). lz has been shown to function downstream of srp; however, additional interactions between these transcription factors initiate hemocyte commitment to the crystal cell lineage (Fossett et al. 2003; Waltzer et al. 2003) by stage 11 of Drosophila embryonic development. An interesting parallel between mammals and Drosophila is the relationship between AML-1 and FOG. In mammals, AML-1 is a positive regulator of myeloid differentiation, while FOG1 is antagonistic. Similarly in Drosophila, Lz is required for proper crystal cell development (Lebestky et al. 2000), while Ush acts as a negative regulator of crystal cell differentiation (Fossett et al. 2003; Waltzer et al. 2003).
srp-expressing cells that differentiate into plasmatocyte precursors also express the transcription factor Glial cells missing (Gcm) (Bernardoni et al. 1997), which is the primary regulator of glial cell differentiation in the nervous system (Hosoya et al. 1995; Jones et al. 1995). There are two gcm genes in Drosophila, and Gcm homologs have also been identified in vertebrates (gcm-1 and gcm-2); however, no hematopoietic function has been associated with these (Altshuller et al. 1996; Kim et al. 1998; Gunther et al. 2000; Schreiber et al. 2000). Drosophila gcm-1 and gcm-2 double mutants show only a 40% reduction in presumptive plasmatocytes, and only a fraction of mature plasmatocyte markers are detected in the remaining cells (Akiyama et al. 1996; Alfonso and Jones 2002), suggesting gcm is unlikely to be the only determinant of plasmatocyte differentiation.
Signal transduction pathways common to both Drosophila and mammalian hematopoiesis include Notch, JAK/STAT, NFκB, and receptor tyrosine kinases (RTKs). The Notch pathway is important in many Drosophila developmental processes, including cell fate decisions and cell proliferation in the nervous system, mesoderm, and imaginal discs (Artavanis-Tsakonas et al. 1999). Notch signaling has also been shown to be required for the development of embryonic and larval crystal cells and in the proliferation of hemocytes (Duvic et al. 2002; Lebestky et al. 2003). A specific and distinct role for individual Notch receptors has been identified in mammalian hematopoiesis as well (Walker et al. 2001; Kumano et al. 2003; Saito et al. 2003). While it has been well established that both the JAK/STAT and Toll/Cactus pathways are involved in blood cell production during immune response, it also appears that each participates in normal blood development as well (Mathey-Prevot and Perrimon 1998; Qiu et al. 1998). JAK/STAT signaling seems to play a specific role in blood cell differentiation and NFκB plays a role in blood cell proliferation (Luo et al. 1997). The RTK, vascular and endothelial growth factor receptor (VEGFR), mediates blood cell migration in mammals (reviewed by Traver and Zon 2002), while another RTK, c-Kit, is important for the proliferation and maintenance of myeloid progenitors (Kelly and Gilliland 2002). Similarly, the Drosophila platelet-derived growth factor and VEGF-receptor-related (PVR) protein is expressed in hemocytes and is required for the proper migration (Heino et al. 2001; Cho et al. 2002) and survival (Brückner et al. 2004) of plasmatocytes.
The parallels between vertebrate and Drosophila hematopoiesis have proven flies to be a useful model system in which to dissect the role of the many signaling pathways involved in blood development. Drosophila genetics allows for comprehensive screens for mutations that disrupt a particular biological process. To identify additional genes involved in Drosophila hematopoiesis, we have conducted a P-element screen to isolate mutations that affect crystal cell development in embryos. Crystal cells, composing only 5% of the total hemocyte population (amounting to ∼36 cells in a wild-type embryo), are ideal for a screen of this nature because the total number can be accurately determined. In the current screen we utilized the expression pattern of ProPO A1, a highly specific and sensitive marker for crystal cells. Out of 1040 lines screened, we identified 44 with an abnormal number and/or distribution of crystal cells. These lines were subjected to further phenotypic characterization to establish if other developmental defects exist in these mutants. Using antibodies against the Engrailed and Twist proteins we established that none of the 44 mutations had generalized defects in germ-band patterning and gastrulation, respectively. Collagen IV in situ hybridization allowed visualization of plasmatocytes and revealed the subset of mutations with defects in both crystal cells and plasmatocytes.
MATERIALS AND METHODS
Fly stocks:
All stocks and crosses used were maintained at 25°, unless mentioned otherwise. A collection of lethal lines with P-element insertions on the second and third chromosomes from the Bloomington P-lethal collection was screened. The lines are referred to by their P-line numbers throughout the study. All available genotypes are listed in Table 1. P lines were balanced over either CyO (Pw+ Kr-Gal4, UAS-GFP) or TM3 (Pw+ Kr-Gal4, UAS-GFP) balancer chromosomes (Casso et al. 1999). The GFP balancers allowed the identification of homozygous mutant embryos, which were analyzed in all subsequent experiments except where indicated. The following stocks were obtained from the Bloomington Stock Center: Oregon-R, Canton-S, ft4, stg4, lab2, twr1, put135, osp29, neur1, cbx05704, Catsup1, sytN6, Gug03928, dppH46, scw11, tkv7, sax4, witB11, Med5, shn1, twi-Gal4, and UAS-nuclacZ. Temperature-sensitive alleles for twr1 and put135 were collected at the nonpermissive temperature of 30°. Sec61α [EP(2)2567] was obtained from the Szeged Stock Center. Mutant alleles obtained from other laboratories include: srpneo45 and srp-Gal4 from R. Reuter, pap53 from J. Botas, U2af38ΔE18 from Donald Rio, corto420 from F. Peronnet, smt3l(2)k01211 from J. Schnorr, FRT(3R)CtBPRev19 from S. Parkhurst, Sin3A from G. Rubin, hrg1 from T. Murata, Mad10 and Mad12 from R. Padgett, brkM68 and UAS-brk from C. Rushlow, ebiE4 from S. L. Zipursky, and UAS-Rab5S43N from M. A. Gonzalez-Gaitan.
TABLE 1.
Summary of the mutants identified in the crystal cell screen
| P line | Map position | Gene | Candidate gene | Description | Avg. no. of cc |
|---|---|---|---|---|---|
| P10198 | 78 A2–5 | papL7062 | Component of transcription mediator complex (TRAP240) |
9.9 | |
| P10199 | 78 A5–6 | l(3)L5541L5541 | CG10581 | DUF265, unknown function | 12.9 |
| CG32434 | sec7, PH domain | ||||
| P10382 | 24 A1–2 | fork04703 | Protein ser/thr kinase | 13.8 | |
| P10412 | 98 C | No CG in area | 13.4 | ||
| P10419 | 27 C7 | smt3k06307 | Ubiquitin like, nuclear protein tag (SUMO-1) |
7.6 | |
| P10448 | 21 B7–8 | U2af38k14504 | Component of splicesome | 12.5 | |
| P10492 | 29 C1–2 | snRNA:U6atac:29Bk01105 | Nuclear mRNA splicing via U12-type spliceosome |
10.8 | |
| P10527 | 26 D6–9 | l(2)k03201k03201 | Sec61α | Protein transport, cell death | 10.7 |
| P10542 | 46 F1–2 | l(2)k04308k04308 | CG30011 | pnt/SAM transcription factor | 12.3 |
| P10555 | 55 C9–10 | l(2)08770k04808 | CG30118 | Mth_Ecto domain | 21.2 |
| P10572 | 37 B8–12 | Catsupk05424 | Regulator of catecholamine metabolism |
17.5 | |
| P10586 | 23 B1–2 | sytk05909 | Ca2+ phospholipid binding, synaptic vesicle fusion |
10.8 | |
| P10596 | 21 B4–6 | l(2)k06019k06019 | kis | myb-binding domain, DEAD/DEAH box helicase |
9 |
| P10604 | 47 F1–2 | Fppsk06103 | Sterol biosynthesis, cell division and enlargement |
10.5 | |
| P10664 | 46 C1–2 | l(2)k07237k07237 | cbx | Ubiquitin-conjugating enzyme | 11.8 |
| CG12744 | Zinc finger transcription factor | ||||
| P10675 | 38 B4 | l(2)k07614k07614 | fok | 9.9 | |
| Klp38B | Chromokinesin, bipoplar spindle assembly |
||||
| P10676 | 43 E15–16 | l(2)07619k07619 | hrg | Adenlyltransferase | 11.4 |
| P10692 | 24 D7–E1 | fatk07918 | Contains 34 cadherin repeats | 16 | |
| P10756 | 51 D3–5 | l(2)k08015k08015 | CG10228 | Regulation of nuclear pre-mRNA | 10.3 |
| P10770 | 39 F1–3 | l(2)k08110k08110 | CG11628 | Guanyl-nt exchange factor, intracellular signaling |
17.1 |
| P10786 | 22 E1–2 | Rab5k08232 | Small monomeric GTPase, Dpp trafficking |
11.4 | |
| P10791 | 46 C6–8 | l(2)k08601k08601 | PKa-R2 | cAMP-dep protein kinase R2 | 14.1 |
| P10848 | 30 E1–3 | FKBP59k09010 | Peptidyl isomerase, protein folding | 17.3 | |
| P11066 | 53 D11–13 | l(2)k12701k12701 | CG6301 | 15.6 | |
| P11121 | 26 F3–5 | l(2)k14206k14206 | CG11098 | Putative SMC chromosome segregation ATPase |
10.3 |
| CG13769 | Putative leucine-rich ribonuclease inhibitor type domain |
||||
| P11123 | 28 F1–2 | l(2)k14308k14308 | CG8451 | Sodium-dependent multivitamin transporter |
14.2 |
| CG8419 | B-box zinc, C3HC4 ring finger transcription factor |
||||
| P11166 | 21 C2–3 | ebik16213 | WD40 repeats, F-box, binds E3 ubiquitin ligases |
13.6 | |
| P11174 | 36 A4–5 | l(2)k16215k16215 | CG5953 | MADF domain, transcription factor | 13.2 |
| P11342 | 43 D1–2 | ALDH-III03610 | Oxidizes fatty and aromatic aldehydes | 20.5 | |
| P11525 | 99 A5–7 | stg01235 | Tyrosine phosphatase, controls G2/M transition (Cdc 25) |
7.9 | |
| P11527 | 84 A1–2 | lab01241 | Homebox transcription factor | 9.8 | |
| P11538 | 89 B1–3 | srp01549 | GATA-type zinc finger transcription factor |
1 | |
| P11590 | 87 D9–11 | CtBP03463 | Transcription corepressor | 13.3 | |
| P11622 | 66 C13/73 D3 | l(3)04069b04069b | CG6983 | 35.9 | |
| Gug | Transcription corepressor | ||||
| P11663 | 84 A1–2 | twr05614 | Affect eye and macrochaetae | 14.1 | |
| P11716 | 82 E5–7 | corto07128b | RNA transcription factor, component of the centrosome |
14.5 | |
| P11745 | 88 C9–10 | put10460 | ser/thr protein kinase, type II TGFβ receptor |
13.5 | |
| P12045 | 34 A1–2 | l(2)rK639rK639 | No CG in area | 16.9 | |
| P12046 | 35 B1–4 | osprJ571 | Putative ligand carrier, component of the cytoskeleton |
11.8 | |
| P12059 | 57 A3–4 | l(2)s4831s4831 | CG13434 | 10.9 | |
| CG30153 | |||||
| P12124 | 85 C5–9 | neurj6B12 | E3 type ubiquitin ligase | 8.8 | |
| P12196 | 59 E1–2 | l(2)s4830s4830 | Or59a | Olfactory receptor | 14.1 |
| CG5357 | Putative component of ribosome | ||||
| P12346 | 55 D1–2 | Prp1907838 | Pre-mRNA splicing factor, component of splicesome |
12.8 | |
| P12350 | 49 B3–6 | Sin3A08269 | Transcription corepressor | 14.1 |
P lines have been listed in order of their line numbers and are referred to as such throughout this study. Corresponding alleles of each line are listed under Gene. Unknown lines with more than one candidate gene listed have a P-element insertion mapped between the loci of two genes. Avg no. of cc, the average number of crystal cells found in homozygous P-line embryos at stage 13–14.
Phenotypic analysis:
Embryo collections and in situ hybridizations were performed in 30-well collection boxes as previously described (Hummel et al. 1997). Antisense ProPO A1, GFP, and Collagen IV (CIV) digoxygenin-labeled RNA probes were made from 2.3 kb ProPO A1, 0.74 kb GFP, and 1.6 kb CIV cDNAs as previously described (Daga et al. 1996). Rabbit antibodies to β-Gal, GFP, Phospho-Histone H3, and Twist were used. Mouse antibody to Engrailed was used. The Engrailed antibody developed by C. Goodman was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development. The GFP and the Phospho-Histone H3 antibody were obtained from Sigma (St. Louis) and Upstate Innovative Signaling Solutions, respectively. In situ hybridization and immunohistochemistry protocols were as previously described (Tepass et al. 1994).
Counting methods:
Crystal cells from 10 embryos for each line were counted at 40× magnification on a Zeiss compound microscope. The average number of cells hybridized to ProPO A1 per embryo was calculated and graphed using Excel (Microsoft) and Delta Graph (SPSS, Chicago). The number of cells staining for α-Phospho-H3 was determined in the head region of stage 14 embryos (n = 3). The head region for this purpose was defined as the tissue demarcated by the anterior edge of the amnioserosa and the anterior end of the embryo.
Molecular analysis:
The sequence of genes interrupted by P-element inserts was obtained from the Berkeley Drosophila Genome Project (BDGP; http://www.fruitfly.org/p_disrupt/). The locations of the inserts of four P lines not sequenced by BDGP, P10412, P10596, P10675, and P11066, were identified using inverse PCR and sequencing. Blast searches were performed using the standard nucleotide-nucleotide BLAST [blastn] program provided by the NCBI database (http://www.ncbi.nlm.nih.gov:80/BLAST/).
RESULTS AND DISCUSSION
Screen for crystal cell mutants:
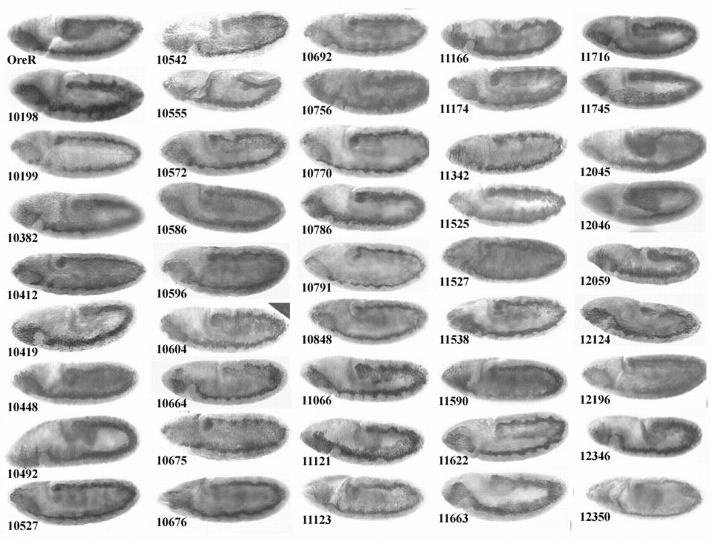
In Drosophila, most components of the hematopoietic developmental hierarchy are still unknown. We have conducted a genetic screen to identify genes involved in this process. This screen utilizes in situ hybridization with ProPO A1, a marker specific for terminally differentiated crystal cells. A collection of 1040 P-element lethal lines generated and mapped by the Drosophila Genome Project, covering the second and third chromosomes, was screened. To distinguish the homozygous mutant embryos from those of other genotypes, the P-element lines were rebalanced with balancer chromosomes containing Kruppel (Kr)-Gal4 driving UAS-GFP (Casso et al. 1999). The screen identified 44 mutants that affect crystal cell development (Figure 1). Forty-two demonstrate a reduction while crystal cell mislocalization is apparent in one line (P11622), and there is one line (P10555) with both a reduction and mislocalization of crystal cells.
Figure 1.—
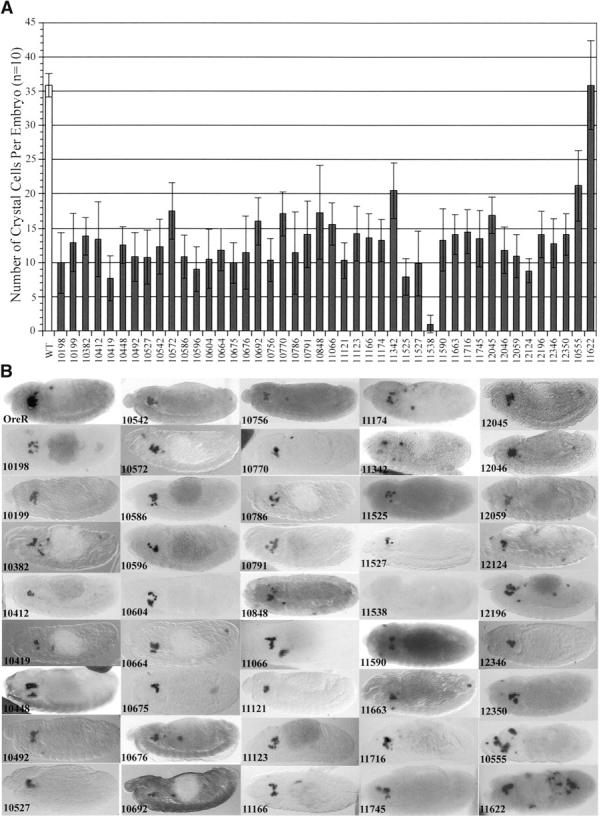
Mutations affecting crystal cell development. (A) Quantitation of the effect of P-line mutants on crystal cell development. The chart shows the average number of crystal cells per embryo (n = 10) as marked by ProPO A1 riboprobe. The wild-type average of 36 is the combined averages of Ore-R, Canton-S, heterozygous nonmutant P10579/Cyo, Kr-GFP, and homozygous nonmutant P10579 embryos. (B) Stage 13–14 embryos of wild-type (Ore-R) and mutant P lines hybridized with ProPO A1 riboprobe marking terminally differentiated crystal cells. Wild-type mature crystal cells can be detected by ProPO A1 hybridization as early as stage 12 and are grouped in two clusters of 18 each in the head mesoderm. Lines P10198 to P12350 show a reduction in the number of crystal cells present. P10555 and P11622 show a mislocalization of the mature crystal cells.
The number of crystal cells present in wild-type flies was determined by counting the number of ProPO A1-positive cells in Ore-R, Canton-S, heterozygous nonmutant 10579/CyO, Kr-GFP, and homozygous nonmutant P10579 embryos. P10579, used as a control, contains a P-element insertion that does not affect crystal cells. The combined average of the control fly lines established that wild-type flies have a relatively invariant number (36 ± 2.2) of crystal cells located in two bilateral clusters in the head region of stage 14 embryos. The number of crystal cells in the P-line mutants ranges from 3 to 57% of wild type, with the median number of crystal cells in a mutant embryo being 12.5 (Figure 1A). P11622 was different from the rest in that it showed wild-type numbers of crystal cells that were grossly mislocalized over the entire embryo.
Of the 44 mutants found, 24 of the lines have P-element insertions that are within previously characterized genes (Table 1). To verify the location of each P-element insertion and its effect on crystal cell development, we tested available loss-of-function mutant alleles of the known genes identified in our screen. Of the 24 known genes, 18 had available loss-of-function alleles, all of which also demonstrate a reduction in crystal cell number, consistent with results from the P lines (Figure 2).
Figure 2.—
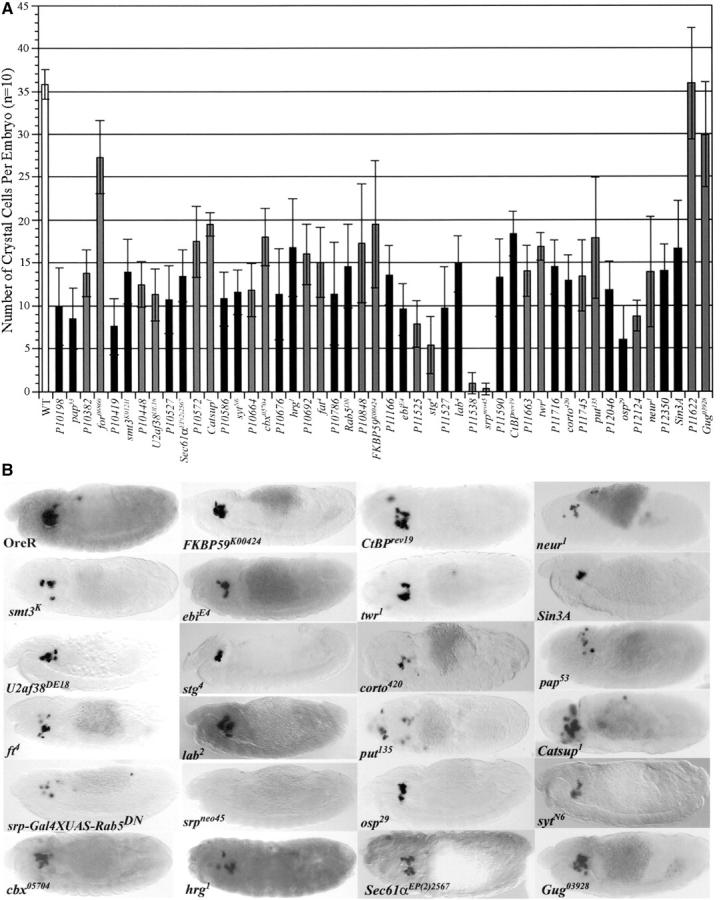
Loss-of-function alleles of known and candidate genes of P-line mutants. (A) Quantitation of the effect of loss-of-function alleles of known and candidate genes on crystal cell development. The chart shows the average number of crystal cells per embryo as marked by ProPO A1 riboprobe (n = 10). The wild-type average of 36 is the combined averages of Ore-R, Canton-S, heterozygous nonmutant P10579/Cyo, Kr-GFP, and homozygous nonmutant P10579 embryos. The complete Rab5DN genotype is srp-Gal4/UAS-Rab5DN. for06860 is the strongest allele available; however, is not a null allele, thus accounting for the reduced crystal cell effect as compared to line 10382. (B) Stage 13–14 mutant embryos were hybridized with ProPO A1 riboprobe. All recapitulate the crystal cell reduction of their corresponding P line except for Gug, which is unlikely to be the insertion site of P11622, as it does not recapitulate the mislocalized phenotype.
To identify genes disrupted in the remaining 20 P lines, the P-element flanking region sequences were obtained from the Drosophila Genome Project when available or were determined in this study. These sequences were used in BLAST searches to identify candidate genes for each of the 20 unknown P lines. One to two candidate genes were identified for 18 of the 20 unknown P lines (Table 1). As with the known genes, the three candidate genes with available loss-of-function alleles were tested for recapitulation of the reduced crystal cell phenotype observed in the P lines. These three are P10527, P10664, and P10676, which mapped to the Sec61α, crossbronx (cbx), and hiiragi (hrg) loci, respectively. Mutant alleles of these genes all show a reduction in crystal cells consistent with the corresponding P line mutations (Figure 2).
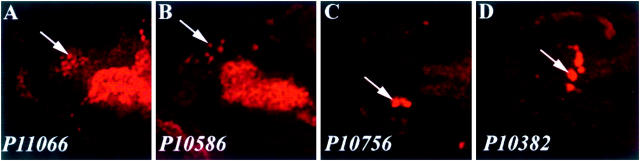
Eleven of the 44 P lines behave as enhancer traps and demonstrate expression in hemocytes and/or crystal cells (Figure 3). Heterozygous and homozygous embryos were examined in this experiment. P10604, P10664, P10848, P11121, P10675, P11066, P10586, P10676, P12059, and P10756 contain a lacZ within the P element that is expressed in embryonic hemocytes in the head region of stage 11 embryos. As examples, P11066 and P10586 are shown (Figure 3, A and B). LacZ expression in P10756 and P10382 is detected in crystal cells of stage 14 embryos (Figure 3, C and D). These enhancer trap lines nicely illustrate an expression pattern consistent with the likely role of these genes in one or more stages of hematopoietic development.
Figure 3.—
Enhancer trap expression patterns of P lines. Four examples of P lines contain P-lacZ inserts expressing lacZ in hemocytes. Embryos of all stages were stained with α-β-gal antibody. Arrows indicate a representative hemocyte in each embryo. (A and B) Stage 11 embryos in which the enhancer trap lines show expression in hemocytes. (C and D) Stage 14 embryos in which enhancer trap expression is evident in mature crystal cells.
Characterization of isolated mutants:
Crystal cells constitute a class of hemocytes derived from the head mesoderm. Genes that, when mutated, cause a change in the number of crystal cells could act directly on the specification of this cell type; alternatively, they could function at an earlier step during the formation of hemocytes or of mesoderm in general. For example, a double mutation of twist and snail that has no mesoderm also lacks hemocytes, including crystal cells (V. Hartenstein, unpublished data); likewise, in bicaudal, where early events in the specification of the anterior body axis fail to take place, head structures, including head mesoderm and all hemocytes, are missing (Tepass et al. 1994). Finally, changes in the number of crystal cells could be due to a generalized effect on cell division. To distinguish between these different steps at which the genes uncovered in this screen might act, we used the markers Collagen IV (plasmatocytes; Yasothornsrikul et al. 1997), Twist (mesoderm; Furlong et al. 2001), Engrailed (metamerically reiterated expression along antero-posterior axis; Bejsovec and Wieschaus 1993), and Phosphohistone-H3 (marker for cells entering mitosis; Hendzel et al. 1997).
All mutations showed grossly normal staining with anti-Twist and anti-Engrailed antibodies, indicating that changes in crystal cell number were not caused by a global defect in axis formation, gastrulation, or segmentation (Figures 4 and 5). To assess if the mutations contribute to general cell proliferation defects, we employed the α-Phosphohistone-H3 (Phospho-H3) antibody, which identifies cells in late G2 through anaphase when chromatin is condensed (Hendzel et al. 1997). To quantitate the number of cells entering mitosis we chose the head region of stage 14 embryos in which only neuroblasts continue dividing. Choosing this later stage maximizes our chance of detecting general proliferation defects in mutants with significant maternal components, and the relatively small number of cells dividing at this time allows accurate quantitation. Six mutant lines were identified with α-Phospho-H3 as having cell-cycle defects (Figure 6): P10198, P11166, P11525, P11663, P12045, and P12196. These were all shown to have a significant increase in the number of α-Phospho-H3-positive cells, indicating that cells were arrested in G2 or mitosis at a higher than normal rate. Alternatively, the elevated number of α-Phospho-H3-positive cells may elicit an escalated apoptotic response leading to a decrease in the number of differentiated crystal cells. Included in this group are two previously known cell cycle regulators, ebi and stg. Ebi functions in normal G1 arrest of cells in the peripheral nervous system and central nervous system. Boulton et al. (2000) discovered an increase in BrdU incorporation of cells in ebi mutants. Failure to arrest in G1 results in an increased number of cells that are Phospho-H3 positive in this mutant. stg mutants have been shown to arrest cells in G2 phase, thus accounting for the increase in α-Phospho-H3 staining (Neufeld et al. 1998).
Figure 4.—
Characterization of the effect of crystal cell mutants on mesoderm development. Stage 8 embryos of wild-type (Ore-R) and mutant P lines stained with α-Twi antibody are shown. All 44 P-line mutants demonstrate proper Twist expression and mesoderm patterning.
Figure 5.—

Characterization of the effect of crystal cell mutants on anterior/posterior segmentation. Stage 10 embryos of wild-type (Ore-R) and mutant P lines stained with α-En antibody are shown. All 44 P-line mutants demonstrate wild-type expression of Engrailed.
Figure 6.—
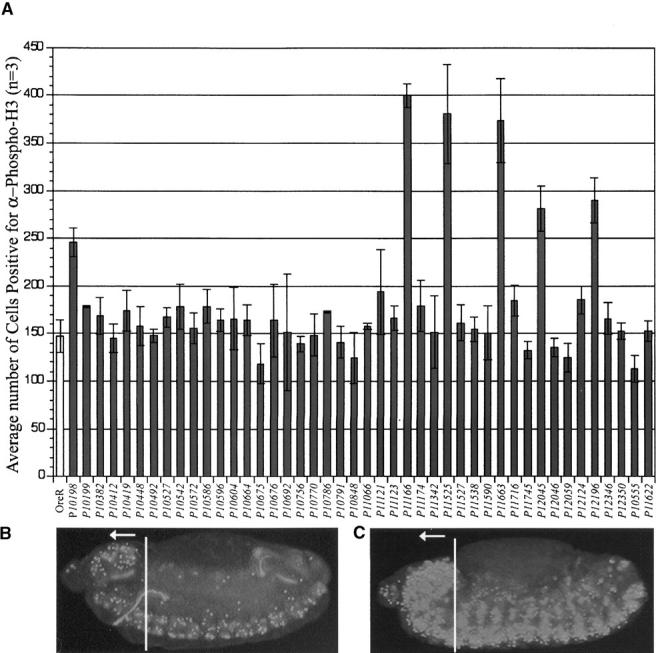
Quantitation of the effect of crystal cell mutants on the cell cycle. (A) The average number of α-Phospho-H3-positive cells in the head region per embryo (n = 3). (B) α-Phospho-H3 staining in a stage 14 wild-type (Ore-R) embryo. (C) Stage 14 stg01235 embryo shown here as an example of increased α-Phospho-H3-positive cells. In B and C, the line indicates the defined head region for this analysis as the anterior-most edge of the amnioserosa to the anterior end of the embryo.
We found that six crystal cell-reduction mutant lines also demonstrate a significant reduction in the number of plasmatocytes (Figure 7). These are P10848, P11525, P11538, P11716, P11745, and P10676. Plasmatocytes were found to be mislocalized in P10555, where instead of an even distribution throughout the entire embryo, plasmatocytes were clustered in the anterior one-third of this mutant. Similarly, P11622 also exhibits a mislocalization of plasmatocytes into large clumps throughout the embryo (Figure 7).
Figure 7.—
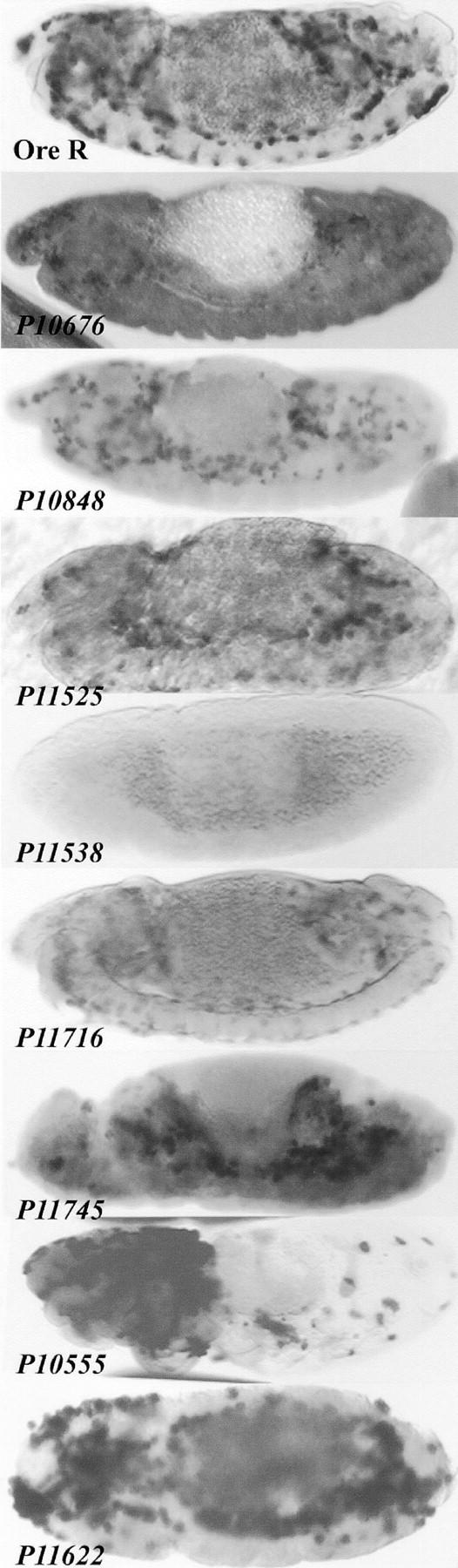
Characterization of the effect of crystal cell mutants on plasmatocytes. Stage 13–14 embryos of wild-type and mutant P lines hybridized with CIV riboprobe are shown. P10676, P10848, P11525, P11538, P11716, and P11745 show a reduction in the number of plasmatocytes. Crystal cell mislocalization mutants P10555 and P11622 also show a mislocalization of the plasmatocytes.
Classes of genes that reduce crystal cell development:
With the exception of the P line interrupting srp, we did not observe a complete elimination of crystal cells in any of our mutants although they show significant reductions. There can be a number of reasons for this partial loss. First, approximately half of the P lines that interrupt previously characterized genes have a maternal complement of RNA. Future analysis of germline clones for each of the P-line mutants will determine if maternal contribution is responsible for the observed partial development of crystal cells. As a test case, we found that ebi germline clones have a significantly more severe defect in crystal cells than that seen with the zygotic loss-of-function P line (data not shown). Second, P-line insertions tend to cause hypomorphic rather than complete null mutations. However, we tested all available null alleles corresponding to our mutants and these still eliminate only a fraction of the crystal cell population. Finally, the incompleteness in the number of crystals cells lost could reflect the flexible nature of the blood development system, allowing multiple developmental signal response pathways that lead to cell differentiation.
By excluding general defects in the patterning of the embryo, we were able to show that the P-line mutants likely have a direct role in crystal cell development. Of course, this does not preclude the function of these genes in other tissues. In fact, many genes identified in the screen are pleiotropic. Our results do suggest that the observed defects in hematopoiesis are not secondary consequences of gross patterning defects. The expression pattern of several of the enhancer trap lines in embryonic hemocytes and the fact that eight of the mutants identified affect both crystal cells and plasmatocytes also indicate that these P-element insertions interrupt genes involved in blood development. Known and candidate genes isolated in this screen can be placed into four groups: transcriptional regulators, signaling molecules, cell proliferation regulators, and other miscellaneous factors.
Transcriptional regulators:
srp, a GATA factor homolog required for all hematopoiesis in Drosophila, was found in the screen to ablate crystal cells. This is reasonable as Srp is necessary for the expression of Gcm and Lz, the transcription factors that define the two main branches of Drosophila hematopoiesis (Lebestky et al. 2000; Fossett et al. 2003; Waltzer et al. 2003). Mutations in genes encoding transcriptional corepressors, CtBP and Sin3A, cause a significant reduction in the number of crystal cells. CtBP plays several roles as a corepressor that binds short-range repressors such as Knirps, Snail, Hairless, and Kruppel (Nibu et al. 1998). In Xenopus, CtBP interacts with FOG to suppress GATA factors and thereby block erythroid development. U-shaped has also been shown to repress crystal cell development; however, loss of CtBP does not cause an increase in crystal cell numbers as with Ush. CtBP also interferes with both Dpp and Notch signaling pathways (reviewed by Morel et al. 2001; Turner and Crossley 2001). Sin3A forms a complex with histone deacytelases, RpAp46/48, histone-binding proteins, and with many other proteins to act as a transcriptional repressor (reviewed by Ahringer 2000). Thus, Sin3A could play a role in repressing a signaling factor, allowing for the preferential development of the crystal cell lineage over plasmatocytes.
Signaling pathways:
One factor with a relationship to the Toll pathway (Smt3) and two with roles in Notch signaling (Neur and Hrg) were detected in the screen. smt3k06307 and neuralized (neurj6B12) were found to cause a particularly strong reduction of crystal cells. Smt3 is the Drosophila homolog of SUMO-1, an ubiquitin-like protein, which may conjugate with other proteins and alter their function. One such role of Smt3 is to regulate the nuclear localization and transcriptional activation of Dorsal, a well-characterized component of the Toll signaling pathway, which is involved in both hematopoiesis and immunity (Qiu et al. 1998; Wu and Anderson 1998; Bhaskar et al. 2000, 2002). The loss of smt3 may lead to the loss of Dorsal import and interfere with normal NFκB-like signaling, which, one may speculate, is needed for proper crystal cell development. The E3 type ubiquitin ligase, Neur, was discovered as a neurogenic gene required to direct neuroectodermal cells from a neural to an epidermal fate (Boulianne et al. 1991; Lai and Rubin 2001). Neur positively regulates Notch signaling in a non-cell-autonomous manner by targeting Delta for internalization and subsequent degradation (Lai et al. 2001). Loss of Notch signaling, which regulates Lz expression in crystal cell precursors, is one possible explanation for the decrease in crystal cells caused by neurj6B12. hiiragi (hrg), a candidate gene for 10676, encodes an adenyltransferase that is important in the regulation of the Notch pathway (Bachmann and Knust 1998). Hrg downregulates the transcription of Serrate during larval wing development (Murata et al. 1996). Therefore, Hrg may be important in the temporal regulation of Notch signaling during crystal cell development. Note that previous work has established that the Notch pathway is essential for crystal cell development (Lebestky et al. 2003).
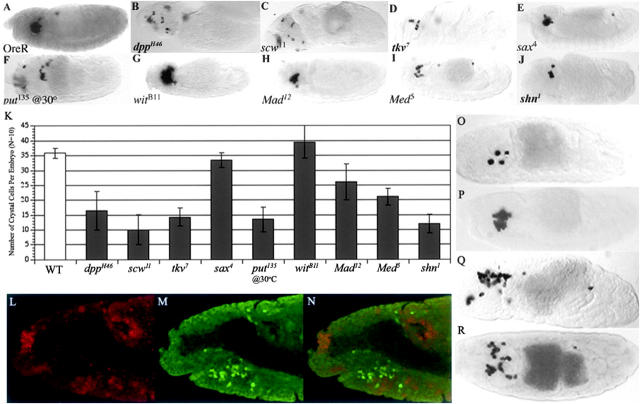
Four mutants could be linked to the Decapentaplegic (Dpp) pathway: punt (put), Rab-protein 5 (Rab5), labial (lab), and poils aux pattes (pap). Drosophila Dpp is a TGF-β family member, and the TGF-β pathway has been known to play a pleiotropic role in all stages of mammalian hematopoiesis (reviewed by Ruscetti and Bartelmez 2001); however, its role in Drosophila hematopoiesis has not been previously explored. Loss-of-function mutants in the Dpp pathway, dppH46, scw11, tkv7, put135, Mad12, Med5, and shn1, all cause significant reductions in the number of crystal cells while mutants in the second type I Dpp receptor, sax, and the second type II Dpp receptor, wit, do not (Figure 8, A–K).
Figure 8.—
Characterization of the effect of Dpp pathway mutants on crystal cells. (A–J) Stage 13–14 mutant embryos were hybridized with ProPO A1 riboprobe. The graph in K demonstrates the number of crystal cells found in each mutant (n = 10). (L–N) Double staining of stage 11 srp-Gal4 × UAS-βGal embryos with (L) α-pMAD (red) and (M) α-βGal (green) antibodies. (N) The merged image of α-pMAD and α-βGal staining shows α-pMAD-positive cells are adjacent to the α-βGal-positive cells. (O–R) Stage 14 embryos hybridized with ProPO A1 riboprobe. (O) Ectopic expression of brinker driven by twi-Gal4. (P) Ectopic expression of brinker driven by srp-Gal4. (Q) Ectopic expression of dpp driven by twi-Gal4. (R) brkM68.
Dpp acts in an indirect and non-cell-autonomous manner during Drosophila blood development. Active Dpp signaling, marked by pMad staining, is detected in cells adjacent to, but not colocalizing with, Srp-positive cells in the head mesoderm (Figure 8, L–N). brk, a negative regulator of Dpp target genes, is able to block crystal cell development when ectopically expressed throughout the mesoderm using the twi-Gal4 driver, but not when expressed in prohemocytes using a srp-Gal4 driver (PFigure 8, O and P). These studies support an indirect role of Dpp in crystal cell development and a requirement of Dpp signaling in the head mesoderm adjacent to Srp-positive prohemocytes.
To investigate the effect of ectopic dpp throughout the mesoderm, we expressed UAS-dpp with a twi-Gal4 driver. Ectopic Dpp signaling in the mesoderm causes mislocalization of crystal cells to further posterior within the embryo (Figure 8Q). A null allele of brk, which has been shown to cause an upregulation of Dpp target genes (Jazwinska et al. 1999a,b; reviewed by Nakayama et al. 2000), phenocopied this phenotype (Figure 8R), thereby indicating a role for Dpp target genes in the patterning of the mesoderm to establish a background in which normal crystal cell development may occur. Additionally, put10460 also causes a reduction in the number of plasmatocytes in the embryo.
Cell proliferation regulators:
Two crystal cell mutants with known roles in cell proliferation are fat and ebi. A member of the Cadherin superfamily, Fat is important in cell-cell adhesion and plays an autonomous role in the regulation of cell proliferation (Mahoney et al. 1991). Loss of fat function may cause a defect in the proliferation of crystal cells due to a misregulation of proliferation signals. ebi, which encodes a GTP-binding molecule containing WD40 repeats and an F-box domain, causes a significant reduction in crystal cells. WD40 repeats can bind E3 type ubiquitin ligases and bring them in contact with F-box-bound proteins that are to be degraded. Ebi regulates cell proliferation by limiting the entry of cells into the S-phase of the cell cycle during neuronal development (Boulton et al. 2000) and we show, with α-Phospho-H3 staining, that ebi also affects cell cycle regulation during embryogenesis. However, in the regulation of Suppressor of Hairless during photoreceptor development, ebi functions downstream of EGF signaling in cell fate specification (Tsuda et al. 2002). Given that ebik16213 does not affect plasmatocytes, it is possible that the role of ebi in crystal cell development is in patterning the precursor and not in the regulation of cell proliferation.
Other bona fide cell cycle factors identified by the screen include String (Stg), a Cdc25 homolog, Centrosomal and chromosomal factor (Ccf or Corto), and Twisted bristle roughened eye (Twr). Stg is a nuclear tyrosine phosphatase that controls the G2/M transition of the cell cycle by dephosphorylating Cdc2, a mitotic kinase (reviewed by Edgar 1994; Edgar et al. 1994a,b). stg01235 causes a strong reduction in the number of crystal cells and also reduces the number of plasmatocytes. Little is known about twr except that hypomorphic alleles affect the eye and macrochaetae (Lewis et al. 1980). Corto is necessary for proper condensation of mitotic chromosomes and the maintenance of chromosome structure during mitosis and interphase (Kodjabachian et al. 1998). Corto is also a putative regulator of Hox genes (Lopez et al. 2001). papL7062 and two unknown mutant lines, P12045 and P12196, were also identified by this study to affect the cell cycle; however, their role in this process remains to be investigated.
Other factors:
Eleven mutants were identified in the screen that correspond to genes that have interesting functions, but do not fit into any one category: U2 small nuclear riboprotein auxiliary factor 38 (U2af38), Prp19, foraging (for), Farnesyl pyrophosphate synthase (Fpps), FK506 binding protein 59 (FKBP59), Aldehyde dehydrogenase type III (Aldh-III), outspread (osp), synaptotagmin (syt), Sec61α, Catecholamines up (Catsup), and crossbronx (cbx). RNA splicesome components U2af38 and Prp19, both necessary for RNA splicing (Tarn et al. 1993; Rudner et al. 1996), were found to affect crystal cell development. Loss of RNA splicing can lead to the improper processing of many important developmental proteins. However, it is unclear why the development of hematopoietic cells should be particularly sensitive to the function of these proteins. for encodes a serine/threonine kinase involved in larval feeding behavior (Renger et al. 1999) and is expressed in both the head mesoderm and mature crystal cells, consistent with a potential role in crystal cell development. Fpps encodes a protein required for sterol biosynthesis, which in plants is important in membrane stability, cell growth, proliferation, and respiration (Gaffe et al. 2000). FKBP59 encodes a peptidylprolyl isomerase, which has been indicated to function as a molecular chaperone during protein folding. Consistent with a role in crystal cell development, FKBP59 has been shown to have a unique expression pattern in embryonic lymph glands (Zaffran 2000). Interestingly, FKBP59 is also expressed in a cell type- and developmental stage-specific pattern during mouse male germ cell differentiation (Sananes et al. 1998). Aldh-III, which causes a weak reduction of crystal cells, is known as the tumor-associated Aldh due to its upregulation in several human tumor types (Park et al. 2002). Aldh's are considered to be general detoxifying enzymes that oxidize toxic biogenic and xenobiotic aldehydes (reviewed by Yoshida et al. 1998). osp encodes a putative transmembrane receptor involved in developmental processes (Ashburner et al. 1999). Syt is a calcium phospholipid-binding protein that aids in synaptic vesicle fusion by facilitating the formation of the SNARE complex (reviewed by Tucker and Chapman 2002). Sec61α encodes a protein thought to be involved in protein transport and signal recognition particle-dependent membrane targeting and translocation. Catsup is a negative regulator of tyrosine hydroxylase, the limiting factor in catecholamine metabolism (Stathakis et al. 1999). Cbx encodes an ubiquitin-conjugating enzyme that is involved in spermatogenesis (Castrillon et al. 1993).
These studies have documented several signaling pathways, transcriptional regulators, and other factors that were not previously known to be involved in Drosophila hematopoiesis. We have used the terminal marker (ProPO A1) for crystal cells in our screen, which in principle allows us to identify mutations encompassing the entire span of crystal cell differentiation from the earliest precursor specification to late expression of ProPO A1. This larger collection of mutants can now be further classified according to their developmental defects using earlier markers such as Lozenge. In future work we hope to determine the individual role of each of these genes in this important developmental process. Given the conservation of genes known to be involved in mammalian and Drosophila hematopoiesis, it is not unreasonable to expect that a subset of the genes identified in this study will be determined to have functions in hematopoiesis across various species.
Acknowledgments
We thank Josh Deigan for his work with Engrailed. We thank members of the Banerjee lab for helpful discussions. We thank J. Botas, G. Campbell, L. Fessler, K. Fujimoto, M.A. Gonzalez-Gaitan, D.K. Hoshizaki, M. Krasnow, T. Murata, S. Parkhurst, F. Peronnet, D. Rio, C. Rushlow, S. Roth, J. Schnorr, K.K. Yokoyama, and the Bloomington Stock Center for fly stocks and reagents. We thank T. Hummel for the design of the collection boxes. This work was supported by National Institutes of Health grant RO1 HD9948-27 to U.B., by U.S. Public Health Service National Research Service Award GM07104 to A.B.M. and A.L.H, and by a University of California, Los Angeles, Dissertation Year Fellowship to A.B.M.
References
- Ahringer, J., 2000. NuRD and SIN3 histone deacetylase complexes in development. Trends Genet. 16: 351–356. [DOI] [PubMed] [Google Scholar]
- Akiyama, Y., T. Hosoya, A. M. Poole and Y. Hotta, 1996. The gcm-motif: a novel DNA-binding motif conserved in Drosophila and mammals. Proc. Natl. Acad. Sci. USA 93: 14912–14916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfonso, T. B., and B. W. Jones, 2002. gcm2 promotes glial cell differentiation and is required with glial cells missing for macrophage development in Drosophila. Dev. Biol. 248: 369–383. [DOI] [PubMed] [Google Scholar]
- Altshuller, Y., N. G. Copeland, D. J. Gilbert, N. A. Jenkins and M. A. Frohman, 1996. Gcm1, a mammalian homolog of Drosophila glial cells missing. FEBS Lett. 393: 201–204. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas, S., M. D. Rand and R. J. Lake, 1999. Notch signaling: cell fate control and signal integration in development. Science 284: 770–776. [DOI] [PubMed] [Google Scholar]
- Ashburner, M., S. Misra, J. Roote, S. E. Lewis, R. Blazej et al., 1999. An exploration of the sequence of a 2.9-Mb region of the genome of Drosophila melanogaster: the Adh region. Genetics 153: 179–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann, A., and E. Knust, 1998. Positive and negative control of Serrate expression during early development of the Drosophila wing. Mech. Dev. 76: 67–78. [DOI] [PubMed] [Google Scholar]
- Bejsovec, A., and E. Wieschaus, 1993. Segment polarity gene interactions modulate epidermal patterning in Drosophila embryos. Development 119: 501–517. [DOI] [PubMed] [Google Scholar]
- Bernardoni, R., V. Vivancos and A. Giangrande, 1997. glide/gcm is expressed and required in the scavenger cell lineage. Dev. Biol. 191: 118–130. [DOI] [PubMed] [Google Scholar]
- Bhaskar, V., S. A. Valentine and A. J. Courey, 2000. A functional interaction between dorsal and components of the Smt3 conjugation machinery. J. Biol. Chem. 275: 4033–4040. [DOI] [PubMed] [Google Scholar]
- Bhaskar, V., M. Smith and A. J. Courey, 2002. Conjugation of Smt3 to dorsal may potentiate the Drosophila immune response. Mol. Cell. Biol. 22: 492–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulianne, G. L., A. de la Concha, J. A. Campos-Ortega, L. Y. Jan and Y. N. Jan, 1991. The Drosophila neurogenic gene neuralized encodes a novel protein and is expressed in precursors of larval and adult neurons. EMBO J. 10: 2975–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton, S. J., A. Brook, K. Staehling-Hampton, P. Heitzler and N. Dyson, 2000. A role for Ebi in neuronal cell cycle control. EMBO J. 19: 5376–5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brückner, K. L., L. Kockel, P. Ducheck, C. M. Luque, P. Rorth et al., 2004. The PDGF/VEGF receptor controls blood cell survival in Drosophila. Dev. Cell 7: 73–84. [DOI] [PubMed] [Google Scholar]
- Cantor, A. B., and S. H. Orkin, 2001. Hematopoietic development: a balancing act. Curr. Opin. Genet. Dev. 11: 513–519. [DOI] [PubMed] [Google Scholar]
- Casso, D., F. A. Ramirez-Weber and T. B. Kornberg, 1999. GFP-tagged balancer chromosomes for Drosophila melanogaster. Mech. Dev. 88: 229–232. [DOI] [PubMed] [Google Scholar]
- Castrillon, D. H., P. Gonczy, S. Alexander, R. Rawson, C. G. Eberhart et al., 1993. Toward a molecular genetic analysis of spermatogenesis in Drosophila melanogaster: characterization of male-sterile mutants generated by single P element mutagenesis. Genetics 135: 489–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, A. N., A. B. Cantor, Y. Fujiwara, M. B. Lodish, S. Droho et al., 2002. GATA-factor dependence of the multitype zinc-finger protein FOG-1 for its essential role in megakaryopoiesis. Proc. Natl. Acad. Sci. USA 99: 9237–9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, N. K., L. Keyes, E. Johnson, J. Heller, L. Ryner et al., 2002. Developmental control of blood cell migration by the Drosophila VEGF pathway. Cell 108: 865–876. [DOI] [PubMed] [Google Scholar]
- Daga, A., C. A. Karlovich, K. Dumstrei and U. Banerjee, 1996. Patterning of cells in the Drosophila eye by Lozenge, which shares homologous domains with AML1. Genes Dev. 10: 1194–1205. [DOI] [PubMed] [Google Scholar]
- Duvic, B., J. A. Hoffmann, M. Meister and J. Royet, 2002. Notch signaling controls lineage specification during Drosophila larval hematopoiesis. Curr. Biol. 12: 1923–1927. [DOI] [PubMed] [Google Scholar]
- Dzierzak, E., and A. Medvinsky, 1995. Mouse embryonic hematopoiesis. Trends Genet. 11: 359–366. [DOI] [PubMed] [Google Scholar]
- Edgar, B. A., 1994. Cell cycle. Cell-cycle control in a developmental context. Curr. Biol. 4: 522–524. [DOI] [PubMed] [Google Scholar]
- Edgar, B. A., D. A. Lehman and P. H. O'Farrell, 1994. a Transcriptional regulation of string (cdc25): a link between developmental programming and the cell cycle. Development 120: 3131–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, B. A., F. Sprenger, R. J. Duronio, P. Leopold and P. H. O'Farrell, 1994. b Distinct molecular mechanisms regulate cell cycle timing at successive stages of Drosophila embryogenesis. Genes Dev. 8: 440–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, C. J., V. Hartenstein and U. Banerjee, 2003. Thicker than blood: conserved mechanisms in Drosophila and vertebrate hematopoiesis. Dev. Cell 5: 673–690. [DOI] [PubMed] [Google Scholar]
- Fossett, N., and R. A. Schulz, 2001. Functional conservation of hematopoietic factors in Drosophila and vertebrates. Differentiation 69: 83–90. [DOI] [PubMed] [Google Scholar]
- Fossett, N., K. Hyman, K. Gajewski, S. H. Orkin and R. A. Schulz, 2003. Combinatorial interactions of serpent, lozenge, and U-shaped regulate crystal cell lineage commitment during Drosophila hematopoiesis. Proc. Natl. Acad. Sci. USA 100: 11451–11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong, E. E., E. C. Andersen, B. Null, K. P. White and M. P. Scott, 2001. Patterns of gene expression during Drosophila mesoderm development. Science 293: 1629–1633. [DOI] [PubMed] [Google Scholar]
- Gaffe, J., J. P. Bru, M. Causse, A. Vidal, L. Stamitti-Bert et al., 2000. LEFPS1, a tomato farnesyl pyrophosphate gene highly expressed during early fruit development. Plant Physiol. 123: 1351–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajewski, K., N. Fossett, J. D. Molkentin and R. A. Schulz, 1999. The zinc finger proteins Pannier and GATA4 function as cardiogenic factors in Drosophila. Development 126: 5679–5688. [DOI] [PubMed] [Google Scholar]
- Gunther, T., Z. F. Chen, J. Kim, M. Priemel, J. M. Rueger et al., 2000. Genetic ablation of parathyroid glands reveals another source of parathyroid hormone. Nature 406: 199–203. [DOI] [PubMed] [Google Scholar]
- Heino, T. I., T. Karpanen, G. Wahlstrom, M. Pulkkinen, U. Eriksson et al., 2001. The Drosophila VEGF receptor homolog is expressed in hemocytes. Mech. Dev. 109: 69–77. [DOI] [PubMed] [Google Scholar]
- Hendzel, M. J., Y. Wei, M. A. Mancini, A. Van Hooser, T. Ranalli et al., 1997. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 106: 348–360. [DOI] [PubMed] [Google Scholar]
- Holz, A., B. Bossinger, T. Strasser, W. Janning and R. Klapper, 2003. The two origins of hemocytes in Drosophila. Development 130: 4955–4962. [DOI] [PubMed] [Google Scholar]
- Hosoya, T., K. Takizawa, K. Nitta and Y. Hotta, 1995. glial cells missing: a binary switch between neuronal and glial determination in Drosophila. Cell 82: 1025–1036. [DOI] [PubMed] [Google Scholar]
- Hummel, T., K. Schimmelpfeng and C. Klambt, 1997. Fast and efficient egg collection and antibody staining from large numbers of Drosophila strains. Dev. Genes Evol. 207: 131–135. [DOI] [PubMed] [Google Scholar]
- Jazwinska, A., N. Kirov, E. Wieschaus, S. Roth and C. Rushlow, 1999. a The Drosophila gene brinker reveals a novel mechanism of Dpp target gene regulation. Cell 96: 563–573. [DOI] [PubMed] [Google Scholar]
- Jazwinska, A., C. Rushlow and S. Roth, 1999. b The role of brinker in mediating the graded response to Dpp in early Drosophila embryos. Development 126: 3323–3334. [DOI] [PubMed] [Google Scholar]
- Jones, B. W., R. D. Fetter, G. Tear and C. S. Goodman, 1995. glial cells missing: a genetic switch that controls glial versus neuronal fate. Cell 82: 1013–1023. [DOI] [PubMed] [Google Scholar]
- Kelly, L. M., and D. G. Gilliland, 2002. Genetics of myeloid leukemias. Annu. Rev. Genomics Hum. Genet. 3: 179–198. [DOI] [PubMed] [Google Scholar]
- Kim, J., B. W. Jones, C. Zock, Z. Chen, H. Wang et al., 1998. Isolation and characterization of mammalian homologs of the Drosophila gene glial cells missing. Proc. Natl. Acad. Sci. USA 95: 12364–12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodjabachian, L., M. Delaage, C. Maurel, R. Miassod, B. Jacq et al., 1998. Mutations in ccf, a novel Drosophila gene encoding a chromosomal factor, affect progression through mitosis and interact with Pc-G mutations. EMBO J. 17: 1063–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumano, K., S. Chiba, A. Kunisato, M. Sata, T. Saito et al., 2003. Notch1 but not Notch2 is essential for generating hematopoietic stem cells from endothelial cells. Immunity 18: 699–711. [DOI] [PubMed] [Google Scholar]
- Lai, E. C., and G. M. Rubin, 2001. Neuralized is essential for a subset of Notch pathway-dependent cell fate decisions during Drosophila eye development. Proc. Natl. Acad. Sci. USA 98: 5637–5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, E. C., G. A. Deblandre, C. Kintner and G. M. Rubin, 2001. Drosophila neuralized is a ubiquitin ligase that promotes the internalization and degradation of delta. Dev. Cell 1: 783–794. [DOI] [PubMed] [Google Scholar]
- Lanot, R., D. Zachary, F. Holder and M. Meister, 2001. Postembryonic hematopoiesis in Drosophila. Dev. Biol. 230: 243–257. [DOI] [PubMed] [Google Scholar]
- Lebestky, T., T. Chang, V. Hartenstein and U. Banerjee, 2000. Specification of Drosophila hematopoietic lineage by conserved transcription factors. Science 288: 146–149. [DOI] [PubMed] [Google Scholar]
- Lebestky, T., S. H. Jung and U. Banerjee, 2003. A Serrate-expressing signaling center controls Drosophila hematopoiesis. Genes Dev. 17: 348–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, R. A., B. T. Wakimoto, R. E. Denell and T. C. Kaufman, 1980. Genetic analysis of the Antennapedia gene complex (ANT-C) and adjacent chromosomal regions of Drosophila melanogaster. Genetics 95: 383–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez, A., D. Higuet, R. Rosset, J. Deutsch and F. Peronnet, 2001. corto genetically interacts with Pc-G and trx-G genes and maintains the anterior boundary of Ultrabithorax expression in Drosophila larvae. Mol. Genet. Genomics 266: 572–583. [DOI] [PubMed] [Google Scholar]
- Luo, J. S., R. Kammerer, H. Schultze and S. von Kleist, 1997. Modulations of the effector function and cytokine production of human lymphocytes by secreted factors derived from colorectal-carcinoma cells. Int. J. Cancer 72: 142–148. [DOI] [PubMed] [Google Scholar]
- Lutterbach, B., and S. W. Hiebert, 2000. Role of the transcription factor AML-1 in acute leukemia and hematopoietic differentiation. Gene 245: 223–235. [DOI] [PubMed] [Google Scholar]
- Mahoney, P. A., U. Weber, P. Onofrechuk, H. Biessmann, P. J. Bryant et al., 1991. The fat tumor suppressor gene in Drosophila encodes a novel member of the cadherin gene superfamily. Cell 67: 853–868. [DOI] [PubMed] [Google Scholar]
- Mandal, L., U. Banerjee and V. Hartenstein, 2004. Evidence for a fruitfly hemangioblast and similarities between lymph-gland hematopoiesis in fruitfly and mammal aorta-gonadal-mesonephros mesoderm. Nat. Genet. 38: (in press). [DOI] [PubMed]
- Mathey-Prevot, B., and N. Perrimon, 1998. Mammalian and Drosophila blood: JAK of all trades? Cell 92: 697–700. [DOI] [PubMed] [Google Scholar]
- Meister, M., and M. Lageaux, 2003. Drosophila blood cells. Cell Microbiol. 9: 573–580. [DOI] [PubMed] [Google Scholar]
- Morel, V., M. Lecourtois, O. Massiani, D. Maier, A. Preiss et al., 2001. Transcriptional repression by suppressor of hairless involves the binding of a hairless-dCtBP complex in Drosophila. Curr. Biol. 11: 789–792. [DOI] [PubMed] [Google Scholar]
- Murata, T., K. Ogura, R. Murakami, H. Okano and K. K. Yokoyama, 1996. hiiragi, a gene essential for wing development in Drosophila melanogaster, affects the Notch cascade. Genes Genet. Syst. 71: 247–254. [DOI] [PubMed] [Google Scholar]
- Nakayama, T., Y. Cui and J. L. Christian, 2000. Regulation of BMP/Dpp signaling during embryonic development. Cell. Mol. Life Sci. 57: 943–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld, T. P., A. F. de la Cruz, L. A. Johnston and B. A. Edgar, 1998. Coordination of growth and cell division in the Drosophila wing. Cell 93: 1183–1193. [DOI] [PubMed] [Google Scholar]
- Nibu, Y., H. Zhang, E. Bajor, S. Barolo, S. Small et al., 1998. dCtBP mediates transcriptional repression by Knirps, Kruppel and Snail in the Drosophila embryo. EMBO J. 17: 7009–7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda, T., J. van Deursen, S. W. Hiebert, G. Grosveld and J. R. Downing, 1996. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84: 321–330. [DOI] [PubMed] [Google Scholar]
- Orkin, S. H., 1996. Development of the hematopoietic system. Curr. Opin. Genet. Dev. 6: 597–602. [DOI] [PubMed] [Google Scholar]
- Orkin, S. H., 2000. Diversification of haematopoietic stem cells to specific lineages. Nat. Rev. Genet. 1: 57–64. [DOI] [PubMed] [Google Scholar]
- Park, K. S., S. Y. Cho, H. Kim and Y. K. Paik, 2002. Proteomic alterations of the variants of human aldehyde dehydrogenase isozymes correlate with hepatocellular carcinoma. Int. J. Cancer 97: 261–265. [DOI] [PubMed] [Google Scholar]
- Qiu, P., P. C. Pan and S. Govind, 1998. A role for the Drosophila Toll/Cactus pathway in larval hematopoiesis. Development 125: 1909–1920. [DOI] [PubMed] [Google Scholar]
- Rabbitts, T. H., 1994. Chromosomal translocations in human cancer. Nature 372: 143–149. [DOI] [PubMed] [Google Scholar]
- Rehorn, K. P., H. Thelen, A. M. Michelson and R. Reuter, 1996. A molecular aspect of hematopoiesis and endoderm development common to vertebrates and Drosophila. Development 122: 4023–4031. [DOI] [PubMed] [Google Scholar]
- Renger, J. J., W. D. Yao, M. B. Sokolowski and C. F. Wu, 1999. Neuronal polymorphism among natural alleles of a cGMP-dependent kinase gene, foraging, in Drosophila. J. Neurosci. 19: RC28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizki, T. M., R. M. Rizki and E. H. Grell, 1980. A mutant affecting the crystal cells in Drosophila melanogaster. Roux's Arch. Dev. Biol. 188: 91–99. [DOI] [PubMed] [Google Scholar]
- Rizki, T. M., R. M. Rizki and R. A. Bellotti, 1985. Genetics of a Drosophila phenoloxidase. Mol. Gen. Genet. 201: 7–13. [DOI] [PubMed] [Google Scholar]
- Rudner, D. Z., R. Kanaar, K. S. Breger and D. C. Rio, 1996. Mutations in the small subunit of the Drosophila U2AF splicing factor cause lethality and developmental defects. Proc. Natl. Acad. Sci. USA 93: 10333–10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruscetti, F. W., and S. H. Bartelmez, 2001. Transforming growth factor beta, pleiotropic regulator of hematopoietic stem cells: potential physiological and clinical relevance. Int. J. Hematol. 74: 18–25. [DOI] [PubMed] [Google Scholar]
- Saito, T., S. Chiba, M. Ichikawa, A. Kunisato, T. Asai et al., 2003. Notch2 is preferentially expressed in mature B cells and indispensable for mariginal zone B lineage development. Immunity 18: 675–685. [DOI] [PubMed] [Google Scholar]
- Sananes, N., E. E. Baulieu and C. Le Goascogne, 1998. Stage-specific expression of the immunophilin FKBP59 messenger ribonucleic acid and protein during differentiation of male germ cells in rabbits and rats. Biol. Reprod. 58: 353–360. [DOI] [PubMed] [Google Scholar]
- Schreiber, J., E. Riethmacher-Sonnenberg, D. Riethmacher, E. E. Tuerk, J. Enderich et al., 2000. Placental failure in mice lacking the mammalian homolog of glial cells missing, GCMa. Mol. Cell. Biol. 20: 2466–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrentino, R. P., Y. Carton and S. Govind, 2002. Cellular immune response to parasite infection in the Drosophila lymph gland is developmentally regulated. Dev. Biol. 243: 65–80. [DOI] [PubMed] [Google Scholar]
- Stathakis, D. G., D. Y. Burton, W. E. McIvor, S. Krishnakumar, T. R. Wright et al., 1999. The catecholamines up (Catsup) protein of Drosophila melanogaster functions as a negative regulator of tyrosine hydroxylase activity. Genetics 153: 361–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarn, W. Y., K. R. Lee and S. C. Cheng, 1993. The yeast PRP19 protein is not tightly associated with small nuclear RNAs, but appears to associate with the spliceosome after binding of U2 to the pre-mRNA and prior to formation of the functional spliceosome. Mol. Cell. Biol. 13: 1883–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepass, U., L. I. Fessler, A. Aziz and V. Hartenstein, 1994. Embryonic origin of hemocytes and their relationship to cell death in Drosophila. Development 120: 1829–1837. [DOI] [PubMed] [Google Scholar]
- Traver, D., and L. I. Zon, 2002. Walking the walk: migration and other common themes in blood and vascular development. Cell 108: 731–734. [DOI] [PubMed] [Google Scholar]
- Tsuda, L., R. Nagaraj, S. L. Zipursky and U. Banerjee, 2002. An EGFR/Ebi/Sno pathway promotes delta expression by inactivating Su(H)/SMRTER repression during inductive notch signaling. Cell 110: 625–637. [DOI] [PubMed] [Google Scholar]
- Tucker, W. C., and E. R. Chapman, 2002. Role of synaptotagmin in Ca2+-triggered exocytosis. Biochem. J. 366: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, J., and M. Crossley, 2001. The CtBP family: enigmatic and enzymatic transcriptional co-repressors. BioEssays 23: 683–690. [DOI] [PubMed] [Google Scholar]
- Walker, L., A. Carlson, H. T. Tan-Pertel, G. Weinmaster and J. Gasson, 2001. The Notch receptor and its ligands are selectively expressed during hematopoietic development in the mouse. Stem Cells 19: 543–552. [DOI] [PubMed] [Google Scholar]
- Waltzer, L., G. Ferjoux, L. Bataille and M. Haenlin, 2003. Cooperation between the GATA and RUNX factors Serpent and Lozenge during Drosophila hematopoiesis. EMBO J. 22: 6516–6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q., T. Stacy, J. D. Miller, A. F. Lewis, T. L. Gu et al., 1996. The CBFbeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell 87: 697–708. [DOI] [PubMed] [Google Scholar]
- Wu, L. P., and K. V. Anderson, 1998. Regulated nuclear import of Rel proteins in the Drosophila immune response. Nature 392: 93–97. [DOI] [PubMed] [Google Scholar]
- Yasothornsrikul, S., W. J. Davis, G. Cramer, D. A. Kimbrell and C. R. Dearolf, 1997. viking: identification and characterization of a second type IV collagen in Drosophila. Gene 198: 17–25. [DOI] [PubMed] [Google Scholar]
- Yoshida, A., A. Rzhetsky, L. C. Hsu and C. Chang, 1998. Human aldehyde dehydrogenase gene family. Eur. J. Biochem. 251: 549–557. [DOI] [PubMed] [Google Scholar]
- Zaffran, S., 2000. Molecular cloning and embryonic expression of dFKBP59, a novel Drosophila FK506-binding protein. Gene 246: 103–109. [DOI] [PubMed] [Google Scholar]