Abstract
Traits under relaxed selection are expected to become reduced or disappear completely, a process called vestigialization. In parthenogenetic populations, traits historically involved in sexual reproduction are no longer under selection and potentially subject to such reduction. In Leptopilina clavipes, thelytokous (parthenogenetic) populations are infected by Wolbachia bacteria. Arrhenotokous populations do not harbor Wolbachia. When antibiotics are applied to infected females, they are cured from their infection and males arise. Such males are capable of producing offspring with uninfected females, but with lower fertilization success than sexual males. This can be attributed to the lack of selection on male fertility in thelytokous lines. In this study we used this variation in L. clavipes male fertility to determine the genetic basis of this trait. Males from cured thelytokous populations were crossed to females from uninfected populations. Using AFLP markers, a genetic linkage map was generated, consisting of five linkage groups and spanning a total distance of 219.9 cM. A single QTL of large effect (explaining 46.5% of the phenotypic variance) was identified for male fertility, which we call male fertility factor (mff). We discuss possible mechanisms underlying the effect of mff, as well as mechanisms involved in vestigialization of traits involved in sexual reproduction.
VESTIGIALIZATION of traits has been reported in a wide variety of organisms, such as the eye reduction found in cave-dwelling fish (Astyanax fasciatus; Wilkens 1988) and in mole rats (Spalx ehrenbergi; Nevo 1991) or the reduction of wings in some birds found on islands where predators are absent and food is abundant (McNab 1994). Two distinct classes of mechanisms are proposed to explain the reduction in traits (that at some point renders them nonfunctional). Most explanations consider trait loss to be an adaptive process that involves indirect selection. This occurs as a consequence of antagonistic pleiotropy; the redundant trait is reduced through selection on a correlated trait, for instance, via resource allocation (Fong et al. 1995). The other mechanism that has been proposed does not require adaptive evolution and involves the random accumulation of mutations in genes coding for traits that are no longer actively maintained by selection (Muller 1949).
Characters under relaxed selection are expected to become reduced or even disappear completely (Fong et al. 1995). In parthenogenetically reproducing populations, traits involved in sexual reproduction are no longer under selection and thus potentially subject to vestigialization. Decay of female courtship traits was first reported in parthenogenetic strains of Drosophila mercatorum (Carson et al. 1982). Flies of the parthenogenetic strains showed a reduced mating propensity when mated to conspecific males, compared to females of sexual strains. Although the accumulation of mutations in genes coding for female sexual behavior was proposed as the explanation in this example (Carson et al. 1982), indirect selection as a consequence of energy economy or antagonistic pleiotropy cannot be ruled out.
A special case of parthenogenesis occurs when it is induced by intracellular bacteria of the genus Wolbachia (α-proteobacteria). Parthenogenesis-inducing (PI) Wolbachia are restricted to species with haplodiploid sex determination (Huigens and Stouthamer 2003), as occurs in the arthropod groups of Hymenoptera, Thysanoptera, Coleoptera, and Acari (Wrensch and Ebbert 1993). The most common mode of haplodiploid reproduction is arrhenotoky where males develop from unfertilized haploid eggs and females from fertilized diploid eggs. In thelytoky or parthenogenesis, females develop from unfertilized eggs and there are no males (White 1973; Luck et al. 1993). Wolbachia is involved in many cases of haplodiploid parthenogenesis (Stouthamer 1997), but not in all (Beukeboom and Pijnacker 2000).
Females infected by PI Wolbachia can be cured from their infection by high temperature or antibiotic treatment (Stouthamer et al. 1990). This results in the production of males from unfertilized eggs. In most cases, however, these males are sexually not fully functional (e.g., Zchori-Fein et al. 1992, 1995; Pijls et al. 1996; Gottlieb and Zchori-Fein 2001; but see Stouthamer and Kazmer 1994). As males are nonexistent in parthenogenetic strains, genes coding for male sexual traits are not actively maintained by selection and random mutations in those genes are thus not removed by selection. Mutations can accumulate or, by means of antagonistic pleiotropy, even be actively selected for if they improve the parthenogenetic performance of females (Pijls et al. 1996; Werren 1998).
Leptopilina clavipes (Hartig; Hymenoptera: Figitidae) is a parasitoid wasp of fungi-breeding Drosophila larvae that occurs in Western Europe (Nordlander 1980). In northwestern Europe, populations of L. clavipes are thelytokous (Driessen et al. 1990), which is Wolbachia induced (Werren et al. 1995; Schidlo et al. 2002). Recently, uninfected arrhenotokous individuals were found in Spain (Pannebakker et al. 2004b). Previous experiments have shown that antibiotic curing of infected females is possible (Schidlo et al. 2002). The males from these experimental crosses are capable of producing viable offspring with uninfected females from sexual populations, but with a lower fertilization success than sexual males (B. A. Pannebakker, unpublished results). Fertilization success can be easily measured in haplodiploids through the sex ratio of the offspring because only successful fertilizations result in female offspring. Although successful matings are still possible, selection has been relaxed for sufficient time to allow for genetic divergence in traits involved in male reproduction, thereby creating variation in fertilization capacity between the wild type and cured males. Infection with Wolbachia followed by vestigialization has thus provided variation between populations, which in turn can be used for linkage mapping of male fertilization capacity. This, together with the possibility of crossing the low-fertilizing males to wild-type sexual females, makes L. clavipes an ideal system in which to study the genetic basis of fertilization capacity. The haplodiploid genetics further facilitate mapping of male traits because the haploid F2 mapping population renders dominant markers such as amplified fragment length polymorphism (AFLPs) always informative (Antolin et al. 1996).
The objectives of the present study were to generate an AFLP linkage map of L. clavipes and to map quantitative trait loci (QTL) involved in male fertilization capacity. Crosses were made between cured males and wild-type sexual females. The F1 hybrid females were set up as virgins to create haploid recombinant F2 males. These were then used to construct a linkage map based on the segregation of AFLP markers and to identify QTL linked to male fertilization capacity. The results are discussed in the light of the two proposed mechanisms for vestigialization.
MATERIALS AND METHODS
L. clavipes stocks:
Two L. clavipes lines were used in the experiment. The infected thelytokous line KBH-NL00 originated from Klein Beekermark, s' Heerenbergh, The Netherlands (N 51° 55.38′, E 06° 12.95′). The uninfected arrhenotokous line DC-E00 originated from Duna Continental, El Montgrí, Spain (N 42° 04.84′, E 03° 08.66′). Both founder populations were collected in the spring of 2000 (Pannebakker et al. 2004b). The cultures were maintained in the laboratory on Drosophila phalerata larvae at 20°, light to dark (L:D) of 16:8 and 65% relative humidity. D. phalerata were reared on a medium containing mushroom (Agaricus bisporus), water, dry yeast (Saccharomyces cerevisiae Hansen), and agar. The fungicides nipagin and propionic acid were added to combat mold in the medium.
Curing experiments and crosses were done using Drosophila subobscura as a host. The fly larvae were reared on a patch of live baker's yeast (S. cerevisiae) suspension on a medium of water, dry yeast (S. cerevisiae), and agar at 20°, L:D of 16:8, and 65% relative humidity.
Antibiotic treatment:
To induce male offspring from the thelytokous line, infected females were cured from their Wolbachia infection using antibiotics applied in honey (0.5% rifampicin) and in the host medium (0.2% rifampicin) as described by Schidlo et al. (2002). Curing was done using D. subobscura as a host on a live baker's yeast patch on agar. The curing treatment was done at 25° (L:D of 16:8 and 65% relative humidity), because a higher temperature increases curing efficiency (Schidlo et al. 2002).
Crosses:
Six males from the cured KBH-NL00 line were successfully crossed to highly inbred females from the uninfected DC-E00 line. As a control, eight males from the uninfected DC-E00 line were crossed to females from the same line. These and all other matings were observed at 20°. After mating, females were allowed to oviposit on a patch of fly larvae. Ovipositions, except where indicated, were done on a standard patch of 125 second instar D. subobscura larvae for 24 hr at 25°. The larvae were further incubated at 25°, L:D of 16:8 and 65% relative humidity. After 2 weeks, pupae were washed out of the medium and isolated in gel capsules (Lamepro, Raamsdonkveer, The Netherlands). The sex of the emerging F1 wasps was scored (Table 2) and the offspring of the mating pair producing the highest sex ratio (i.e., proportion of males) was used for further analysis.
TABLE 2.
Phenotypic data for male fertilization capacity measured as sex ratio (proportion males) of the offspring when each is mated to a standard DC-E00 female
| Cured (KBH-NL00) | Uninfected (DC-E00) | Recombinant F2 | |
|---|---|---|---|
| Sex ratio | 0.564 ± 0.196 (6) | 0.171 ± 0.090 (8) | 0.349 ± 0.245 (68) |
Means, standard deviations, and sample sizes (in parentheses) are given.
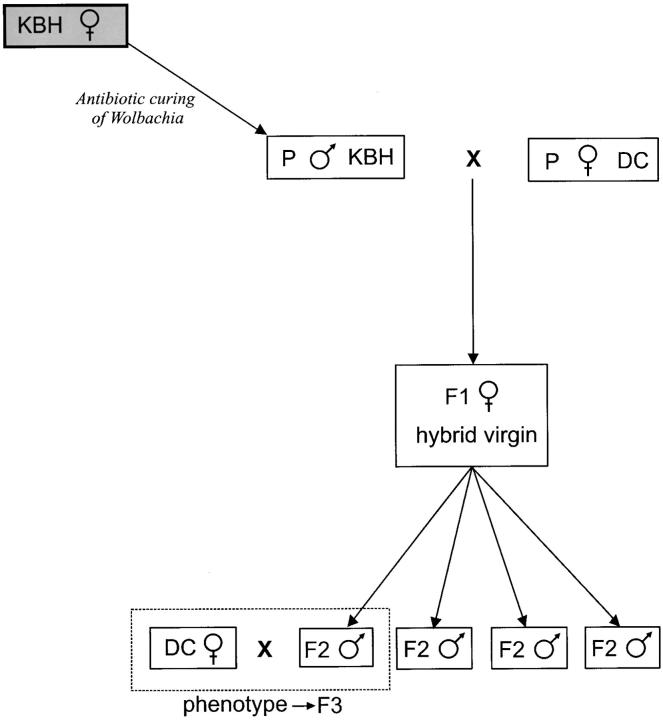
Female F1 offspring (n = 6) of this cross were allowed to oviposit as virgins to produce the F2 males for mapping. To determine the F2 male phenotype, 72 F2 males were successfully crossed to virgin uninfected DC-E00 females. After mating, females were allowed to oviposit on a standard patch. The phenotype of the F2 males was determined by scoring the sex of the emerging F3 offspring (Figure 1).
Figure 1.—
Crossing scheme used in this study. An antibiotic-induced male from the thelytokous line KBH-NL00 is crossed to a female from the arrhenotokous line DC-E00. Six virgin hybrid F1 females produced the recombinant F2 males (n = 72), which were phenotyped by crossing each male with a virgin arrhenotokous female from DC-E00.
Phenotypic analysis:
Sex-ratio data are usually binomially distributed and, therefore, are best analyzed using generalized linear models (GLMs; Wilson and Hardy 2002). Phenotypic data were analyzed by fitting GLMs with a binomial error structure and a logit link function using R statistical software (version 1.7.1; Ihaka and Gentleman 1996). Significance of line effect was assessed by comparing the Aikake Information Criterion (AIC) of the initial model with the AIC after including the line effects in the model. An F-test was used to determine the significance of the line effect and hence of the difference between the parental lines.
Male reproductive system:
Reproductive tracts from 15 males of the KBH-NL00 line and 15 males of the DC-E00 line were visually inspected for abnormalities and viable sperm presence in the testes and seminal vesicles. The reproductive tracts were dissected in Insect Ringers and observed using a Nikon TS100-F phase-contrast microscope (Melville, NY). Photographs of representative reproductive tracts were taken using a Nikon TS100-F phase contrast microscope and a Nikon CoolPix 4500 digital camera.
DNA isolation:
DNA from the parental male and female and from 72 F2 males was extracted using a Nucleon BACC2 kit (Amersham Pharmacia Biotech, Buckinghamshire, UK) according to the manufacturer's protocol. DNA was precipitated in absolute ethanol and washed with 70% ethanol after which the DNA was dissolved in 25 μl sterile water.
AFLP analysis:
Markers were generated using the AFLP technique (Vos et al. 1995). In addition to the A/T cutting EcoRI/MseI restriction enzyme combination from the original protocol (Vos et al. 1995), AFLP reactions were also performed with the combination HindIII/HhaI that cuts at G/C restriction sites to acquire a more even marker distribution. For each restriction enzyme combination, 7 μl of DNA of each individual was restricted and ligated with T4 DNA ligase (New England Biolabs, Beverly, MA) to enzyme-specific adaptors (Table 1) at 37° for 2 hr after which the ligase was inactivated by heating for 10 min at 65°. The restriction-ligation mixture was used as template in the preselective PCR, using the core primers with one selective nucleotide at the 3′-end (Table 1) and AFLP core mix (Applied Biosystems, Foster City, CA) in a total reaction volume of 15 μl. Preselective PCR was done under the following conditions: 72° for 2 min, 94° for 2 min followed by 20 cycles of 94° for 5 sec, 56° for 30 sec, and 72° for 2 min. Prior to the selective PCR, the preselective mixture was diluted 1:10 in Tris-EDTA (TE) buffer. The selective PCR was performed with primers similar to those used in the preselective reaction, but with extra selective bases at the 3′-end (Table 1). On the basis of their level of polymorphism, three primer combinations for the EcoRI/MseI enzymes and one for the HindIII/HhaI enzymes were used in the selective PCR amplification (Table 1). The EcoRI and HindIII primers were fluorescently labeled for subsequent gel analysis. For selective PCR amplification, a touchdown reaction was used with 94° for 5 sec, 65° for 30 sec, and 72° for 2 min, followed by 8 cycles in which the annealing temperature was lowered to 56° in 1° steps. This was followed by 24 cycles at an annealing temperature of 56° and an extension at 72° for 35 min.
TABLE 1.
Sequences of adapters, primers, and primer combinations used in the AFLP analysis
| Adapter | EcoRI | 5′-CTCGTAGACTGCGTACC-3′ |
|---|---|---|
| 3′-CATCTGACGCATGGTTAA-5′ | ||
| MseI | 5′-GACGATGAGTCCTGAG-3′ | |
| 3′-TACTCAGGACTCAT-5′ | ||
| HindIII | 5′-CTCGTAGACTGCGTACC-3′ | |
| 3′-CTGACGCATGGTCGA-5′ | ||
| HhaI | 5′-GACGATGAGTCCTGATCG-3′ | |
| 3′-GCTACTCAGGACTA-5′ | ||
| Primers | EcoRI core | 5′-GACTGCGTACCAATTC-3′ |
| MseI core | 5′-GATGAGTCCTGAGTAA-3′ | |
| HindIII core | 5′-GACTGCGTACCAGCTT-3′ | |
| HhaI core | 5′-GATGAGTCCTGATCGC-3′ | |
| Preamplification | EcoRI | Core+A (E01) |
| MseI | Core+C (M02) | |
| HindIII | Core+A (Hi01) | |
| HhaI | Core+C (Hh02) | |
| Selective amplification | EcoRI | Core+AAC (E32) |
| Core+ACA (E35) | ||
| Core+AGG (E41) | ||
| MseI | Core+CA (M15) | |
| HindIII | Core+ACA (Hi35) | |
| HhaI | Core+CAT (Hh50) | |
| Primer combinations used | E32M15, E35M15, E41M15, Hi35Hh50 |
PCR products were visualized on 5% Long Ranger polyacrylamide gels (BMA Bio Products, Vallensbaek Strand, Denmark) running on an ABI 377 DNA Sequencer (Applied Biosystems). Data were processed using GENESCAN 3.1 (Applied Biosystems) software. Samples were visually checked for correct aligning of the size standard and corrected if necessary. The fluorescent profiles were imported into GENOGRAPHER 1.60 software (J. J. Benham; http://hordeum.oscs.montana.edu/genographer) and presence or absence of fragments was scored between 50 and 500 bp.
Linkage analysis:
The analysis was based on 72 recombinant F2 males originating from eight F1 females. JoinMap (version 3.0; Van Ooijen and Voorrips 2001) software was used to analyze the data and to generate a linkage map. Mendelian segregation (i.e., 1:1 ratio for a haploid mapping population) of the markers in the F2 population was tested using the χ2 test implemented in JoinMap. Markers showing highly significant segregation distortion (P < 0.001) were excluded from linkage analysis.
Grouping of the markers was done using a minimum logarithm of odds (LOD) score of 2 and a maximum recombination fraction of 0.400 as thresholds. Kosambi's mapping function (Kosambi 1944) was used to translate recombination fractions into map distances (in centimorgans). Linkage groups were visualized using MapChart (version 2.1; Voorrips 2002).
QTL analysis:
Complete data (i.e., sex-ratio and molecular data) were available for 60 F2 males. Together with the linkage map based on the complete set of 72 F2 males, these data were analyzed using Windows QTL Cartographer (version 2.0; Wang et al. 2001–2003). Two types of interval mapping analysis were done: interval mapping (IM; Lander and Botstein 1989) and composite interval mapping (CIM; Zeng 1993, 1994). IM analysis calculates the likelihood ratio of no QTL present vs. QTL present at any position in the interval between two markers or at the markers themselves (Lander and Botstein 1989; Doerge 2002). The likelihood ratio is expressed in the LOD score. The threshold for QTL presence was determined by performing 1000 permutations prior to the analysis corresponding to a genome-wide significance level of 5% (Churchill and Doerge 1994; Doerge and Churchill 1996) and was set to a LOD of 2.00.
In CIM analysis, the interval containing the QTL affecting the trait is localized and combined with multiple regression on marker loci outside of the interval under consideration (cofactors) to reduce variation associated with segregating QTL elsewhere in the genome (Zeng 1994; Doerge 2002). In this way the likelihood of detecting QTL is improved, as well as the precision of the estimated map position (Zeng 1994). The cofactors were chosen by forward selection-backward elimination stepwise regression, with critical P-values set at 0.01 and a window size of 10 cM to exclude potential cofactors tightly linked to the test interval. The significance level was set to 5% and determined by performing 1000 permutations prior to the analysis (Churchill and Doerge 1994; Doerge and Churchill 1996), resulting in a genome-wide LOD threshold of 2.27.
RESULTS
Phenotypic differences:
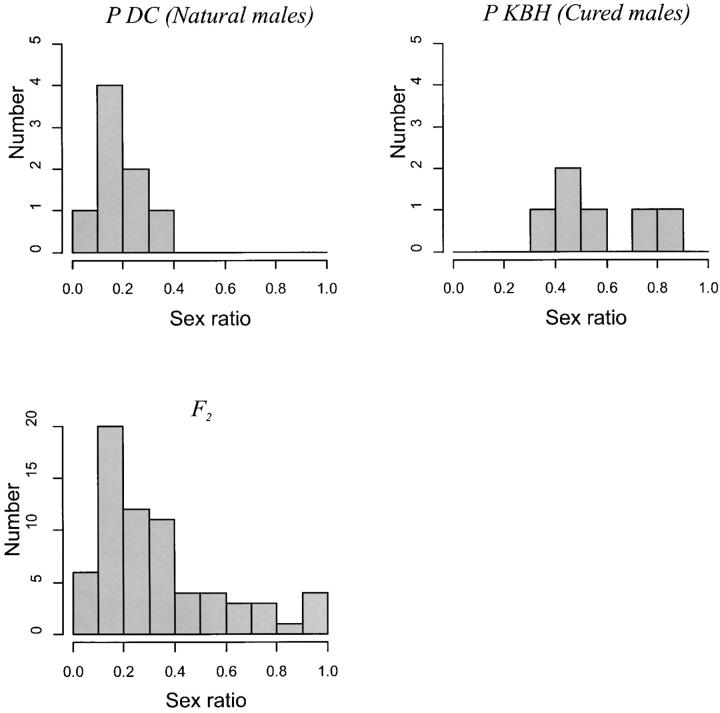
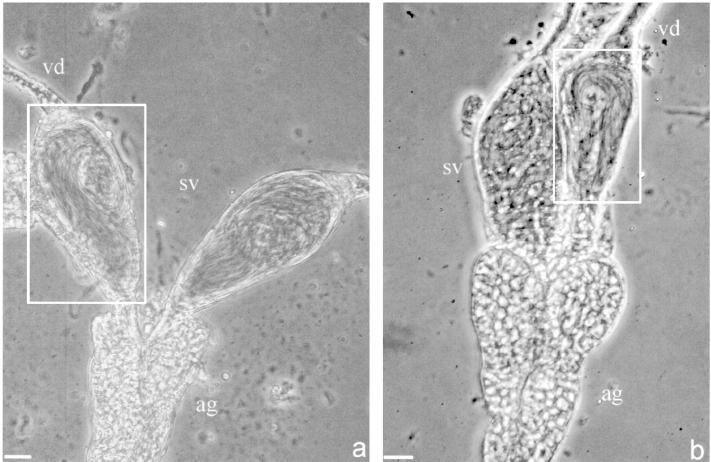
The parental lines differed significantly in sex ratio (F2,12 = 139.29, P < 0.0001; Table 2, Figure 2), whereas the recombinant F2 males showed an intermediate sex ratio. The parental distributions for sex ratio were represented within the F2 distribution (Figure 2). This is consistent with the segregation of one or few genes of large effect on male fertilization capacity and contrasts with the results from crosses to F2 in other systems (e.g., Fishman et al. 2002). No differences were observed between the male reproductive tracts from the parental lines. Viable sperm was found in the seminal vesicles and testes of both parental lines (Figure 3).
Figure 2.—
Phenotypic distributions of sex ratio (proportion of male offspring) in parental and F2 hybrid L. clavipes populations.
Figure 3.—
Representative male reproductive tracts of L. clavipes males from the uninfected line DC-E00 (a) and from the infected line KBH-NL00 (b). Boxes indicate seminal vesicles containing mature sperm. vd, vas deferens; sv, seminal vesicles filled with mature sperm; ag, assesory gland. Bars, 10 μm.
Linkage analysis:
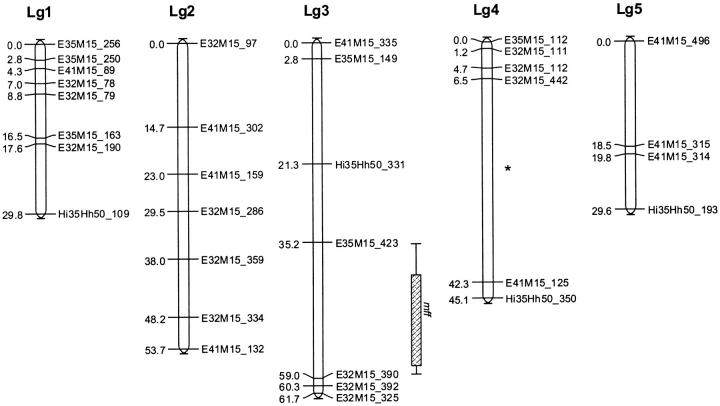
Four AFLP primer combinations resulted in 55 fragments that were polymorphic between the two parental lines and were used in the linkage analysis. Seven fragments were excluded prior to the analysis because of significant segregation distortion (χ2 ≥ 10.82, d.f. = 1, P < 0.01). Another 9 fragments were identical to other markers and excluded from analysis because they map at identical positions and thus provide no extra information. The final map included 32 (82.1%) of the remaining 39 markers, which, at a LOD of 2.0, grouped into five linkage groups that spanned over 219.9 cM (Figure 4). The five linkage groups most likely represent the five chromosomes of L. clavipes (Pannebakker et al. 2004a). The average distance between markers was 6.9 cM. Four of the five linkage groups did not split further until LOD 5.0; linkage group 4, however, did split into two groups at LOD 3.0 (Figure 4). In the final map, 7 markers (17.9%) remained unlinked.
Figure 4.—
Linkage map of L. clavipes showing the position of the mff as located with interval mapping and composite interval mapping. Box shows the 1-LOD support interval and whiskers the 2-LOD support interval for mff from composite interval mapping. (*) The point where linkage group 4 splits at LOD > 2.0.
QTL analyses:
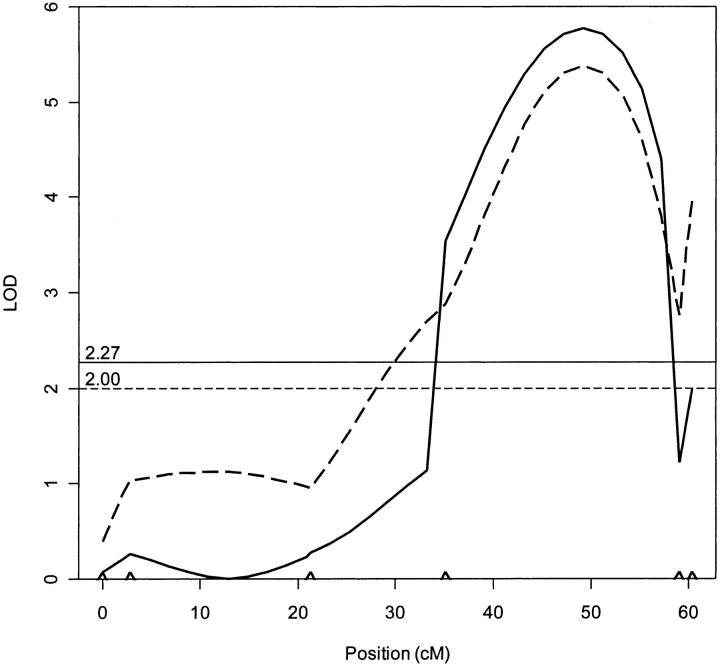
The IM analysis for male fertility (sex ratio) revealed a single genomic region of large effect (Table 3, Figures 4 and 5), explaining 46.5% of the observed phenotypic variance. We call this putative QTL male fertility factor (mff). The LOD score for this QTL was well above the genome-wide LOD threshold of 2.00 as determined prior to the analyses by permutation. The same QTL was detected by CIM analysis and no additional QTL were found to be segregating for male fertility (Table 3, Figure 5).
TABLE 3.
QTL for themale fertility factor as estimated by IM and CIM
| Analysis | Mapping (linkage group, position) |
Flanking markers | LOD score | Additive effect | % explained phenotypic variance |
|---|---|---|---|---|---|
| IM | 3, 49.2 | 5.38 | 0.1707 | 46.49 | |
| E35M15_423 | 2.88 | 19.52 | |||
| E32M15_390 | 2.76 | 19.20 | |||
| CIM | 3, 49.2 | 5.77 | 0.1611 | 41.45 | |
| E35M15_423 | 3.00 | 18.02 | |||
| E32M15_390 | 4.45 | 25.57 |
Shown are the position, flanking markers, LOD scores, additive effects, and percentage of explained phenotypic variance.
Figure 5.—
Likelihood map for linkage group 3 indicating the presence of a single QTL under IM (dotted line) and CIM (solid line). LOD thresholds based on 1000 permutations are given for IM (horizontal dotted line) and CIM (horizontal solid line). Carets indicate the markers on linkage group 3.
Sex-ratio data are usually binomially distributed (Wilson and Hardy 2002), whereas QTL analysis requires the phenotypic trait to be normally distributed (Lander and Botstein 1989). Therefore, the analysis was repeated after log transformation of the data, which resulted in an increase of the LOD scores, but left the position of the QTL unchanged as well as our conclusion that it explained about half of the phenotypic variance (data not shown). Because the results are not altered by log transformation, only the results of the untransformed data are shown, which allows for a better biological interpretation.
DISCUSSION
Linkage map:
Using the advantage of male haploidy in Hymenoptera, we were able to quickly generate a genetic linkage map for L. clavipes. The map consisted of five linkage groups that probably represent the five chromosomes of L. clavipes (Pannebakker et al. 2004a). The total map distance spanned 219.9 cM, which is small compared to those published for other Hymenoptera (Hunt and Page 1995; Antolin et al. 1996; Laurent et al. 1998; Gadau et al. 1999). Although considerable variation in map sizes occurs in the Hymenoptera (Gadau et al. 2000), the high percentage (17.9%) of unlinked loci indicates that the linkage map of L. clavipes is not saturated. Increasing the number of markers or repeating the experiment with a larger F2 mapping would probably increase the size of the linkage map (Liu 1998; Lynch and Walsh 1998).
QTL analysis:
Using the genetic variation generated under relaxed selection, we were able to determine the presence of a single QTL with large effect on male fertilization capacity in L. clavipes. This QTL (mff) explained 41.5% (CIM)—46.5% (IM) of the phenotypic variance for male fertilization capacity, which is in the upper range of detected QTL effects reported (Tanksley 1993; Kearsey and Farquhar 1998). However, when using sample sizes comparable to the present study (N < 100), (a) the effects of statistically significant QTL are overestimated, and (b) closely linked QTL with effects in the same direction can give the appearance of a single QTL of larger effect (Tanksley 1993; Beavis 1994; Kearsey and Farquhar 1998; Barton and Keightley 2002). Thus, although the actual effect of mff is likely to be smaller than observed, reduced male fertilization capacity in L. clavipes can still be assigned to a single genomic region on linkage group 3. It is unclear whether a single gene accounts for the effect of mff, although several studies have shown that single genes can be responsible for QTL of large effect (e.g., Frary et al. 2000). Furthermore, mutation research in Drosophila shows no evidence for clustering of genes involved in male fertility, which are, in contrast, distributed throughout the genome (Castrillon et al. 1993; Hackstein et al. 2000).
The phenotype of the F2 males was determined by scoring the sex ratio of the offspring resulting from crosses with virgin DC-E00 females. Although the virgin females were genetically similar and the experiment was done under controlled conditions, we cannot exclude an effect of small environmental fluctuations. As fertilization behavior and consequently offspring sex ratio in Hymenoptera is under female control (Godfray 1994), environmental fluctuations may have influenced the observed phenotypes. Eliminating these fluctuations will probably reduce variation and thus the phenotypic effect attributable to mff.
Different mechanisms could account for the phenotypic effect of mff. When under relaxed selection in parthenogenetic lines, every male component of the fertilization process is prone to mutation—from spermatogenesis, sperm transfer, surviving the storage in the female spermatheca, and sperm-egg interactions at fertilization to zygotic death after fertilization. Comparison of the reproductive tracts of males induced from parthenogenetic lines with those from uninfected lines revealed no obvious differences in morphology or sperm quantity. Hence, the mechanism underlying the reduced male fertilization capacity is likely to act later in the fertilization process. Examples of genes that may be involved in reduced male fertility can be found in studies of male sterility in Drosophila. In an extensive screen of induced male sterility mutants in D. melanogaster, Castrillon et al. (1993) classified the phenotype of 14 of 83 mutations as semisterile, i.e., capable of producing a limited number of progeny compared to the wild type (comparable to the effect of mff). Of these semisterile mutants only pointless (ptl) results in a normal reproductive tract and wild-type levels of motile sperm, but ptl males transfer only few or no sperm to the female spermatheca, yielding low offspring numbers. However, the total number of mutations known to cause male sterile phenotypes in Drosophila is estimated as up to 1500 (Hackstein et al. 2000), and hence ptl provides only a single candidate gene from what is likely to be a substantially larger group.
The mechanism underlying the vestigialization of traits involved in sexual reproduction can be examined by comparing the QTL involved in loss of sexual function in males obtained by curing females from other infected L. clavipes populations. When loss of sexual function is due to antagonistic pleiotropy, males induced from other infected populations are expected to show a similarly reduced fertilization capacity that is induced by a QTL at the same genomic location. This is because under antagonistic pleiotropy the loss of sexual function is due to selection on traits that are adaptive in the parthenogenetic strain. When the selection favoring these traits is similar in the different populations, the same sexual trait is expected to decline in each population. However, when mutation accumulation is the underlying mechanism, different sexual functions should be corrupted in isolated populations due to the stochastic nature of the process (Cooper and Lenski 2000). In a controlled laboratory experiment with Escherichia coli bacteria, Cooper and Lenski (2000) used this approach to show that antagonistic pleiotropy was the mechanism underlying loss of function during ecological specialization.
Studies reporting on the sexual function of males from other PI Wolbachia-infected species have not revealed comparable cases of reduced male fertilization capacity. Males induced from infected lines successfully inseminate uninfected females and produce similar numbers of diploid offspring as males from uninfected lines do (Pijls et al. 1996; Arakaki et al. 2000), produce sperm but fail to inseminate uninfected females (Zchori-Fein et al. 1992, 1995), or fail to produce any fertile sperm (Gottlieb and Zchori-Fein 2001). This suggests either variation in the time since infection among species in combination with mutation accumulation as the primary mechanism for loss of male sexual function or variation in the mode of selection among infected species in which antagonistic pleiotropy is involved in disruption of male function.
In conclusion, we have determined the genetic architecture of reduced male fertilization capacity using the variation generated by vestigialization of sexual function in parthenogenetic L. clavipes infected by Wolbachia. Using a biometric approach, Wilkens (1988) used a similar approach in cave-dwelling fish to determine the genetic basis of several traits, including eyeball size, pigmentation, feeding behavior, and taste receptors. In our genetic analysis we found a single factor, mff, responsible for nearly 50% of the phenotypic variation in male fertilization capacity in L. clavipes. Since genetic regulation of developmental processes has been well conserved during evolution (Rubin et al. 2000; Venter et al. 2001) and the phenotypes of male sterility are strikingly similar among Drosophila, mice, and humans (Hackstein et al. 2000), further insights from L. clavipes could be relevant to understanding the genetic basis of human male infertility. However, before homologs can be sought in the genetic model systems, high-resolution mapping and positional cloning are needed to identify the mff gene in L. clavipes.
Acknowledgments
We thank Natasha Schidlo for help in crosses between the parental lines and Tom van Dooren for statistical advice. B.J.Z. was supported by the Netherlands Organization for Scientific Research, Postdoc Universitaire Loopbaan Stimuleringsprogramma grant no. 805-48-002.
References
- Antolin, M. F., C. F. Bosio, J. Cotton, W. Sweeney, M. R. Strand et al., 1996. Intensive linkage mapping in a wasp (Bracon hebetor) and a mosquito (Aedes aegypti) with single-strand conformation polymorphism analysis of random amplified polymorphic DNA markers. Genetics 143: 1727–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakaki, N., H. Noda and K. Yamagishi, 2000. Wolbachia-induced parthenogenesis in the egg parasitoid Telenomus nawai. Entomol. Exp. Appl. 96: 177–184. [Google Scholar]
- Barton, N. H., and P. D. Keightley, 2002. Understanding quantitative genetic variation. Nat. Rev. Genet. 3: 11–21. [DOI] [PubMed] [Google Scholar]
- Beavis, W. D., 1994 The power and deceit of QTL experiments: lessons from comparative QTL studies, pp. 252–268 in Proceedings of the 49th Annual Corn and Sorghum Industry Research Conference. American Seed Trade Association, Washington, DC.
- Beukeboom, L. W., and L. P. Pijnacker, 2000. Automictic parthenogenesis in the parasitoid Venturia canescens (Hymenoptera: Ichneumonidae) revisited. Genome 43: 939–944. [DOI] [PubMed] [Google Scholar]
- Carson, H. L., L. S. Chang and T. W. Lyttle, 1982. Decay of female sexual behavior under parthenogenesis. Science 218: 68–70. [DOI] [PubMed] [Google Scholar]
- Castrillon, D. H., P. Gonczy, S. Alexander, R. Rawson, C. G. Eberhart et al., 1993. Toward a molecular-genetic analysis of spermatogenesis in Drosophila melanogaster—characterization of male-sterile mutants generated by single P element mutagenesis. Genetics 135: 489–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill, G. A., and R. W. Doerge, 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, V. S., and R. E. Lenski, 2000. The population genetics of ecological specialization in evolving Escherichia coli populations. Nature 407: 736–739. [DOI] [PubMed] [Google Scholar]
- Doerge, R. W., 2002. Mapping and analysis of quantitative trait loci in experimental populations. Nat. Rev. Genet. 3: 43–52. [DOI] [PubMed] [Google Scholar]
- Doerge, R. W., and G. A. Churchill, 1996. Permutation tests for multiple loci affecting a quantitative character. Genetics 142: 285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driessen, G., L. Hemerik and J. J. M. van Alphen, 1990. Drosophila species, breeding in the stinkhorn (Phallus impudicus Pers.) and their larval parasitoids. Netherlands J. Zool. 40: 409–427. [Google Scholar]
- Fishman, L., A. J. Kelly and J. H. Willis, 2002. Minor quantitative trait loci underlie floral traits associated with mating system divergence in Mimulus. Evolution 56: 2138–2155. [DOI] [PubMed] [Google Scholar]
- Fong, D. W., T. C. Kane and D. C. Culver, 1995. Vestigialization and loss of nonfunctional characters. Annu. Rev. Ecol. Syst. 26: 249–268. [Google Scholar]
- Frary, A., T. C. Nesbitt, S. Grandillo, E. van der Knaap, B. Cong et al., 2000. fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science 289: 85–88. [DOI] [PubMed] [Google Scholar]
- Gadau, J., R. E. Page and J. H. Werren, 1999. Mapping of hybrid incompatibility loci in Nasonia. Genetics 153: 1731–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadau, J., R. E. Page, J. H. Werren and P. Schmid-Hempel, 2000. Genome organization and social evolution in Hymenoptera. Naturwissenschaften 87: 87–89. [DOI] [PubMed] [Google Scholar]
- Godfray, H. C. J., 1994 Parasitoids: Behavioral and Evolutionary Ecology. Princeton University Press, Princeton, NJ.
- Gottlieb, Y., and E. Zchori-Fein, 2001. Irreversible thelytokous reproduction in Muscidifurax uniraptor. Entomol. Exp. Appl. 100: 271–278. [Google Scholar]
- Hackstein, J. H. P., R. Hochstenbach and P. L. Pearson, 2000. Towards an understanding of the genetics of human male infertility: lessons from flies. Trends Genet. 16: 565–572. [DOI] [PubMed] [Google Scholar]
- Huigens, M. E., and R. Stouthamer, 2003 Parthenogenesis associated with Wolbachia, pp. 247–266 in Insect Symbiosis, edited by K. Bourtzis and T. A. Miller. CRC Press, Boca Raton, FL.
- Hunt, G. J., and R. E. Page, 1995. Linkage map of the honey-bee, Apis mellifera, based on RAPD markers. Genetics 139: 1371–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka, R., and R. Gentleman, 1996. R: a language for data analysis and graphics. J. Comput. Graph. Stat. 5: 299–314. [Google Scholar]
- Kearsey, M. J., and A. G. L. Farquhar, 1998. QTL analysis in plants: Where are we now? Heredity 80: 137–142. [DOI] [PubMed] [Google Scholar]
- Kosambi, D. D., 1944. The estimation of map distances from recombination values. Ann. Eugen. 12: 172–175. [Google Scholar]
- Lander, E. S., and D. Botstein, 1989. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent, V., E. Wajnberg, B. Mangin, T. Schiex, C. Gaspin et al., 1998. A composite genetic map of the parasitoid wasp Trichogramma brassicae based on RAPD markers. Genetics 150: 275–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, B. H., 1998 Statistical Genomics: Linkage, Mapping, and QTL Analysis. CRC Press, Boca Raton, FL.
- Luck, R. F., R. Stouthamer and L. Nunney, 1993 Sex determination and sex ratio patterns in parasitic Hymenoptera, pp. 442–476 in Evolution and Diversity of Sex Ratio in Haplodiploid Insects and Mites, edited by D. L. Wrensch and M. A. Ebbert. Chapman & Hall, New York.
- Lynch, M., and B. Walsh, 1998 Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA.
- McNab, B. K., 1994. Energy-conservation and the evolution of flightlessness in birds. Am. Nat. 144: 628–642. [Google Scholar]
- Muller, H. J., 1949. The Darwinian and modern conceptions of natural selection. Proc. Am. Philos. Soc. 93: 459–470. [PubMed] [Google Scholar]
- Nevo, E., 1991. Evolutionary theory and processes of active speciation and adaptive radiation in subterranean mole rats, Spalax ehrenbergi superspecies, in Israel. Evol. Biol. 25: 1–125. [Google Scholar]
- Nordlander, G., 1980. Revision of the genus Leptopilina Forster, 1869, with notes on the status of some other genera (Hymenoptera, Cynipoidea: Eucoilidae). Entomol. Scand. 11: 428–453. [Google Scholar]
- Pannebakker, B. A., L. P. Pijnacker, B. J. Zwaan and L. W. Beukeboom, 2004. a Cytology of Wolbachia-induced parthenogenesis in Leptopilina clavipes (Hymenoptera: Figitidae). Genome 47: 299–303. [DOI] [PubMed] [Google Scholar]
- Pannebakker, B. A., B. J. Zwaan, L. W. Beukeboom and J. J. M. van Alphen, 2004. b Genetic diversity and Wolbachia infection of the Drosophila parasitoid Leptopilina clavipes in western Europe. Mol. Ecol. 13: 1119–1128. [DOI] [PubMed] [Google Scholar]
- Pijls, J. W. A. M., H. J. van Steenbergen and J. J. M. van Alphen, 1996. Asexuality cured: the relations and differences between sexual and asexual Apoanagyrus diversicornis. Heredity 76: 506–513. [Google Scholar]
- Rubin, G. M., M. D. Yandell, J. R. Wortman, G. L. G. Miklos, C. R. Nelson et al., 2000. Comparative genomics of the eukaryotes. Science 287: 2204–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schidlo, N. S., B. A. Pannebakker, B. J. Zwaan, L. W. Beukeboom and J. J. M. Van Alphen, 2002. Curing thelytoky in the Drosophila parasitoid Leptopilina clavipes (Hymenoptera: Figitidae). Proc. Exp. Appl. Entomol. Netherlands Entomol. Soc. 13: 93–96. [Google Scholar]
- Stouthamer, R., 1997 Wolbachia-induced parthenogenesis, pp. 102–124 in Influential Passengers: Inherited Microorganisms and Arthropod Reproduction, edited by S. L. O'Neill, A. A. Hoffmann and J. H. Werren. Oxford University Press, Oxford.
- Stouthamer, R., and D. J. Kazmer, 1994. Cytogenetics of microbe-associated parthenogenesis and its consequences for gene flow in Trichogramma wasps. Heredity 73: 317–327. [Google Scholar]
- Stouthamer, R., R. F. Luck and W. D. Hamilton, 1990. Antibiotics cause parthenogenetic Trichogramma (Hymenoptera/Trichogrammatidae) to revert to sex. Proc. Natl. Acad. Sci. USA 87: 2424–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley, S. D., 1993. Mapping polygenes. Annu. Rev. Genet. 27: 205–233. [DOI] [PubMed] [Google Scholar]
- Van Ooijen, J. W., and R. E. Voorrips, 2001 JoinMap Version 3.0, Software for the Calculation of Genetic Linkage Maps. Plant Research International, Wageningen, The Netherlands.
- Venter, J. C., M. D. Adams, E. W. Myers, P. W. Li, R. J. Mural et al., 2001. The sequence of the human genome. Science 291: 1304. [DOI] [PubMed] [Google Scholar]
- Voorrips, R. E., 2002. MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93: 77–78. [DOI] [PubMed] [Google Scholar]
- Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. Vandelee et al., 1995. AFLP—a new technique for DNA-fingerprinting. Nucleic Acids Res. 23: 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S., C. J. Basten and Z-B. Zeng, 2001–2003 Windows QTL Cartographer 2.0. Department of Statistics, North Carolina State University, Raleigh, NC.
- Werren, J. H., 1998 Wolbachia and speciation, pp. 245–260 in Endless Forms: Species and Speciation, edited by D. J. Howard and S. H. Berlocher. Oxford University Press, New York.
- Werren, J. H., W. Zhang and L. R. Guo, 1995. Evolution and phylogeny of Wolbachia: reproductive parasites of arthropods. Proc. R. Soc. Lond. B Biol. Sci. 261: 55–63. [DOI] [PubMed] [Google Scholar]
- White, M. J. D., 1973 Animal Cytology and Evolution. Cambridge University Press, Cambridge, UK.
- Wilkens, H., 1988. Evolution and genetics of epigean and cave Astyanax fasciatus (Characidae, Pisces): support of the neutral mutation theory. Evol. Biol. 23: 271–367. [Google Scholar]
- Wilson, K., and I. C. W. Hardy, 2002 Statistical analysis of sex ratios: an introduction, pp. 48–92 in Sex Ratios: Concepts and Research Methods, edited by I. C. W. Hardy. Cambridge University Press, Cambridge, UK.
- Wrensch, D. L., and M. A. Ebbert, 1993 Evolution and Diversity of Sex Ratio in Insects and Mites. Chapman & Hall, New York.
- Zchori-Fein, E., R. T. Roush and M. S. Hunter, 1992. Male production induced by antibiotic treatment in Encarsia formosa (Hymenoptera: Aphelinidae), an asexual species. Experientia 48: 102–105. [Google Scholar]
- Zchori-Fein, E., O. Faktor, M. Zeidan, Y. Gottlieb, H. Czosnek et al., 1995. Parthenogenesis-inducing microorganisms in Aphytis (Hymenoptera, Aphelinidae). Insect Mol. Biol. 4: 173–178. [DOI] [PubMed] [Google Scholar]
- Zeng, Z-B., 1993. Theoretical basis for separation of multiple linked gene effects in mapping quantitative trait loci. Proc. Natl. Acad. Sci. USA 90: 10972–10976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Z-B., 1994. Precision mapping of quantitative trait loci. Genetics 136: 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]