Abstract
The function of putative regulatory sequences identified by comparative genomics can be elucidated only through experimentation. Here the effectiveness of using heterologous gene constructs and transgene coplacement to characterize regulatory sequence function is demonstrated. This method shows that a sequence in the Adh 3′-untranslated region negatively regulates expression, independent of gene or chromosomal context.
A major goal of comparative genomics is the identification of gene regulatory sequences. Putative regulatory elements are identified as noncoding sequences that are conserved among species, suggesting that they are under selective constraint for function. A specific regulatory function, however, can be demonstrated only through experimentation. In Drosophila this is often done using transgenic experiments to compare the effects of wild-type and/or mutant forms of a regulatory sequence on expression of either the native or a reporter gene. Typically, expression constructs are cloned in transposable element-based vectors and introduced into the genome by germline transformation (Rubin and Spradling 1982; Spradling and Rubin 1982). A drawback of this method is that transposable elements insert randomly into the genome, where local chromosomal context influences transgene expression. Such position-effect variation (PEV) makes it difficult to compare expression between constructs differing in regulatory elements.
Traditionally PEV is overcome by generating many independent insertions for each construct and then comparing average expression over all transformed lines (e.g., Laurie-Ahlberg and Stam 1987). However, even with large sample sizes PEV may obscure subtle expression differences between two variants. PEV can be reduced by including chromosomal insulator sequences within the vector (Patton et al. 1992), although it still may be significant (Parsch et al. 1997). Gene replacement by homologous recombination is possible in Drosophila melanogaster (Rong and Golic 2000; Gong and Golic 2003), but this method is best applied when a particular gene or chromosomal region is targeted. When putative regulatory elements are being tested with exogenous reporter genes, it is often desirable to compare expression of two variants over multiple chromosomal locations. This is particularly true for elements suspected to impart post-transcriptional regulation, which should function independently from linked transcriptional regulatory sequences.
Siegal and Hartl (1996) described a method, transgene coplacement, to create transformed lines with two variant constructs inserted at the same chromosomal location. This method uses two site-specific recombination systems, the Cre/loxP system of bacteriophage P1 and the FLP/FRT system of Saccharomyces cerevisiae. By placing the two experimental gene constructs in a single transposable element vector, in which each gene is flanked by a different site-specific recombinase target sequence, it is possible to insert both constructs into the same genomic location and later remove one or the other by introducing the appropriate recombinase. The advantage of this method is that expression of constructs at the same chromosomal location can be compared, thereby controlling PEV. This feature of transgene coplacement has been taken advantage of to study transvection (Chen et al. 2002) and enhancer-promoter specificity (Butler and Kadonaga 2001). While these studies were largely qualitative, a major advantage of transgene coplacement is in the quantitative comparison of gene expression. For this, multiple transformed lines are generated for each construct pair and expression differences between variants are tested with a paired t-test (or nested ANOVA). As long as expression between coplaced genes is correlated, statistical power to detect differences is increased over traditional, nonpaired approaches (Siegal and Hartl 1998). Previously the effectiveness of transgene coplacement in controlling PEV was quantitatively demonstrated using the Drosophila Adh gene flanked by its native regulatory sequences (Siegal and Hartl 1998). Here I show that this method also controls PEV in experiments using an exogenous reporter gene with a minimal promoter sequence, demonstrating the power of this approach for investigating the function of regulatory sequences independent of their local genomic environment.
Comparative sequence analysis identified an 8-bp sequence in the Adh 3′-untranslated region (3′-UTR) that is conserved throughout the Drosophila genus (Parsch et al. 1997). Transgenic experiments using the D. melanogaster Adh gene demonstrated that this sequence is a negative regulatory element: Deletion of the 8-bp motif leads to a twofold increase in Adh expression (Parsch et al. 1999). The location of this motif and its similarity to other known regulatory elements suggest that it functions post-transcriptionally (Parsch et al. 2000). To test if this sequence imparts negative regulation on a heterologous reporter gene, constructs containing the human cytomegalovirus (CMV) promoter, the Escherichia coli lacZ coding sequence, and the Adh 3′-UTR were introduced into the D. melanogaster genome using the pP[wFl] vector (Siegal and Hartl 1996; Figure 1). Two different forms of the Adh 3′-UTR were used: one wild type and one with a precise deletion of the conserved 8-bp motif (Figure 1). To ensure that all transformed lines maintained identical genetic backgrounds, only third chromosome insert lines were used for site-specific recombination by crossing to a transgenic fly stock containing FLP- and CRE-expressing transgenes on balanced third chromosomes (Siegal and Hartl 1998). Following FLP or CRE excision, genotypes were confirmed by PCR amplification of the 3′ end of the transgene and digestion with HinfI, which distinguishes wild-type and mutant Adh 3′-UTRs. β-Galactosidase activity of 10 pairs of independently coplaced genes was compared over several developmental stages (Table 1). A highly significant correlation in expression between coplaced genes was observed at all stages (Table 1; Figure 2). The observed correlation coefficients are similar to those reported for coplaced Adh genes and imply a threefold increase in the power to detect statistically significant differences between constructs over traditional, unpaired methods (Siegal and Hartl 1998).
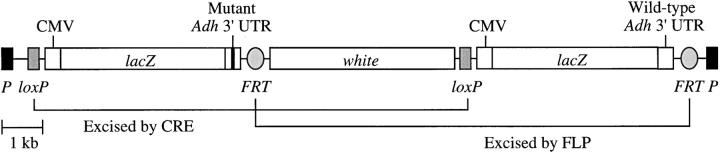
Figure 1.—
Vector used for transgene coplacement. A fragment containing the entire Adh 3′-UTR (bases 1674–2049 from Kreitman 1983) was PCR amplified from wild-type and mutant (bases 1762–1769 deleted) Adh constructs (Parsch et al. 1999) and cloned into the pCR2.1-TOPO vector (Invitrogen, Carlsbad, CA). It was then introduced as either a BamHI-XhoI (wild-type) or an SpeI (mutant) fragment into the BamHI/XhoI or XbaI sites of pCMV-SPORT-βgal (Invitrogen), which contains the human CMV promoter and the E. coli lacZ coding sequence. After introduction of an XhoI (wild-type) or BamHI linker (mutant) into the vector's SapI site, the entire expression construct was cloned as either an XhoI or a BamHI fragment into the XhoI or BamHI sites of pP[wFl] (Siegal and Hartl 1996). Proper orientation was confirmed by restriction analysis. In the final vector, the two constructs are flanked by different site-specific recombinase target sequences (loxP and FRT). Introduction of either CRE or FLP recombinase allows for precise removal of one or the other construct and results in two variant transgenes inserted at the same chromosomal location. P represents the boundaries of the transposable element inserted into the genome. The white gene (eye color) is used as a selectable marker.
TABLE 1.
Comparison of coplaced gene expression
| Age (days) | Ra | Pb | Ratioc |
|---|---|---|---|
| Larvae | 0.90 | 0.0004 | 1.09 |
| 2–4 | 0.93 | 0.0001 | 1.07 |
| 4–6 | 0.89 | 0.0005 | 1.21 |
| 6–8 | 0.75 | 0.0130 | 1.18 |
| 8–10 | 0.78 | 0.0070 | 1.16 |
| All | 0.94 | <0.0001 | 1.16 |
Assays were performed on third instar larvae and adults using the procedures and lines given in Figure 2.
Correlation coefficient of activity of coplaced genes.
P-value of correlation.
Average ratio of mutant to wild-type transgene activity.
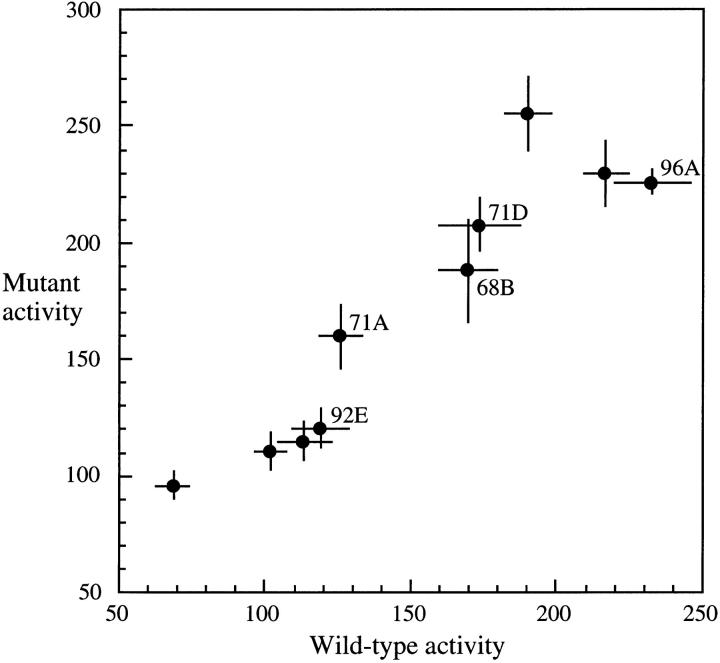
Figure 2.—
β-Galactosidase activity of 10 pairs of coplaced genes (see Figure 1) inserted at different locations of the D. melanogaster third chromosome. For five of the lines, chromosomal location of unique insertions was determined by in situ hybridization of a lacZ probe to polytene chromosomes and is indicated on the graph. Insertions of the remaining five lines were mapped to the third chromosome by genetic crosses with marked chromosome stocks. Activity is given as nanomoles of o-nitrophenyl-β-d-galactopyranoside hydrolyzed per minute at pH 7.5 and 37° per milligram of total protein. Average activity over all adult stages (2–10 days) is shown. At each stage, four separate assays were performed, each using pooled soluble protein extracts from five male flies. Bars indicate SEM.
The ratio of mutant to wild-type β-galactosidase activity of coplaced transgenes averaged 1.16 over all developmental stages (Table 1), indicating that the 8-bp Adh 3′-UTR motif imparts significant negative regulation on the reporter gene (paired t-test, t = 3.28, P = 0.01). If the paired information of coplaced genes is not considered, then the difference between mutant and wild-type activity is not significant (t-test, t = 0.77, P = 0.45). This difference in statistical power is due to the relatively large PEV and illustrates the advantage of transgene coplacement over the traditional method. Given the sample size and PEV observed here, an average activity difference of 34% would be required to detect a significant difference by a standard t-test. For a paired t-test, a difference of only 9% would be necessary.
Deletion of the 8-bp 3′-UTR element in the reporter gene results in a 16% increase in expression, while deletion of this same element in the native Adh gene results in a twofold increase in expression (Parsch et al. 1999). The difference in magnitude may be attributable to epistatic interactions between the 3′-UTR and other regions of the Adh gene. For example, a long-range RNA secondary structure pairing between Adh exon 2 and the 3′-UTR has been shown to play a role in Adh expression (Parsch et al. 1997; Baines et al. 2004). Such interactions cannot occur when the Adh 3′-UTR is fused to the E. coli lacZ coding sequence. Adh expression also may be influenced by intronic sequences (Laurie and Stam 1994; Chen and Stephan 2003) and synonymous codon usage (Carlini and Stephan 2003). The example presented here demonstrates the utility of using heterologous reporter gene constructs and transgene coplacement to determine the functional role of conserved noncoding sequences independent of their gene or chromosomal context.
Acknowledgments
I thank M. Siegal and D. Hartl for providing the pP[wFl] vector and FLP/CRE-expressing fly stocks, H. Gebhart for technical assistance in the laboratory, and J. Baines, T. Hambuch, and L. Rose for critical reading of the manuscript. This research was supported by funds from the University of Munich (LMU).
References
- Baines, J. F., J. Parsch and W. Stephan, 2004. Pleiotropic effect of disrupting a conserved sequence involved in a long-range compensatory interaction in the Drosophila Adh gene. Genetics 166: 237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, J. E. F., and J. T. Kadonaga, 2001. Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 15: 2515–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlini, D. B., and W. Stephan, 2003. In vivo introduction of unpreferred synonymous codons into the Drosophila Adh gene results in reduced levels of ADH protein. Genetics 163: 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J. L., K. L. Huisinga, M. M. Viering, S. A. Ou, C. T. Wu et al., 2002. Enhancer action in trans is permitted throughout the Drosophila genome. Proc. Natl. Acad. Sci. USA 99: 3723–3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., and W. Stephan, 2003. Compensatory evolution of a precursor messenger RNA secondary structure in the Drosophila melanogaster Adh gene. Proc. Natl. Acad. Sci. USA 100: 11499–11504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, W. J., and K. G. Golic, 2003. Ends-out, or replacement, gene targeting in Drosophila. Proc. Natl. Acad. Sci. USA 100: 2556–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitman, M., 1983. Nucleotide polymorphism at the alcohol dehydrogenase locus of Drosophila melanogaster. Nature 304: 412–417. [DOI] [PubMed] [Google Scholar]
- Laurie, C. C., and L. F. Stam, 1994. The effect of an intronic polymorphism on alcohol dehydrogenase expression in Drosophila melanogaster. Genetics 138: 379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie-Ahlberg, C. C., and L. F. Stam, 1987. Use of P-element-mediated transformation to identify the molecular basis of naturally occurring variants affecting Adh expression in Drosophila melanogaster. Genetics 115: 129–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsch, J., S. Tanda and W. Stephan, 1997. Site-directed mutations reveal long-range compensatory interactions in the Adh gene of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 94: 928–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsch, J., W. Stephan and S. Tanda, 1999. A highly conserved sequence in the 3′-untranslated region of the Drosophila Adh gene plays a functional role in Adh expression. Genetics 151: 667–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsch, J., J. A. Russell, I. Beerman, D. L. Hartl and W. Stephan, 2000. Deletion of a conserved regulatory element in the Drosophila Adh gene leads to increased alcohol dehydrogenase activity but also delays development. Genetics 156: 219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton, J. S., X. V. Gomes and P. K. Geyer, 1992. Position-independent germline transformation in Drosophila using a cuticle pigmentation gene as a selectable marker. Nucleic Acids Res. 20: 5859–5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong, Y. S., and K. G. Golic, 2000. Gene targeting by homologous recombination in Drosophila. Science 288: 2013–2018. [DOI] [PubMed] [Google Scholar]
- Rubin, G. M., and A. C. Spradling, 1982. Genetic transformation of Drosophila with transposable element vectors. Science 218: 348–353. [DOI] [PubMed] [Google Scholar]
- Siegal, M. L., and D. L. Hartl, 1996. Transgene coplacement and high efficiency site-specific recombination with the Cre/loxP system in Drosophila. Genetics 144: 715–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegal, M. L., and D. L. Hartl, 1998. An experimental test for lineage-specific position effects on alcohol dehydrogenase (Adh) genes in Drosophila. Proc. Natl. Acad. Sci. USA 95: 15513–15518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling, A. C., and G. M. Rubin, 1982. Transposition of cloned P-elements into Drosophila germ line chromosomes. Science 218: 341–347. [DOI] [PubMed] [Google Scholar]