Abstract
During the first hour after a sublethal dose of ionizing radiation, 72 genes were upregulated threefold or higher in D. radiodurans R1. Thirty-three of these loci were also among a set of 73 genes expressed in R1 cultures recovering from desiccation. The five transcripts most highly induced in response to each stress are the same and encode proteins of unknown function. The genes (ddrA, ddrB, ddrC, ddrD, and pprA) corresponding to these transcripts were deleted, both alone and in all possible two-way combinations. Characterization of the mutant strains defines three epistasis groups that reflect different cellular responses to ionizing radiation-induced damage. The ddrA and ddrB gene products have complementary activities and inactivating both loci generates a strain that is more sensitive to ionizing radiation than strains in which either single gene has been deleted. These proteins appear to mediate efficient RecA-independent processes connected to ionizing radiation resistance. The pprA gene product is not necessary for homologous recombination during natural transformation, but nevertheless may participate in a RecA-dependent process during recovery from radiation damage. These characterizations clearly demonstrate that novel mechanisms significantly contribute to the ionizing radiation resistance in D. radiodurans.
DEINOCOCCUS radiodurans R1 is the type species of a bacterial family distinguished by its ability to tolerate exposure to ionizing radiation (Battista et al. 1999); exponential phase cultures survive doses to 5 kGy without loss of viability. A 5-kGy dose causes massive DNA damage, cleaving the genome of every D. radiodurans cell into multiple, subgenomic fragments (Battista et al. 1999). For most species, this level of DNA damage is lethal, but D. radiodurans has the capacity to reform its genome from these fragments in what appears to be an error-free process. The biochemical details of D. radiodurans's ionizing radiation resistance are poorly understood, but it is clear that proteins needed for cell survival are synthesized in cultures exposed to ionizing radiation. Irradiated cultures cannot recover in the presence of chloramphenicol; this antibiotic prevents restitution of ionizing radiation-induced single-strand (Dean et al. 1969) and double-strand DNA breaks (Kitayama and Matsuyama 1971). We have made identifying the proteins that mediate ionizing radiation resistance a priority in our efforts to better explain D. radiodurans's extreme radioresistance.
In this report we have described the genomic expression profile of D. radiodurans R1 cultures as they recover from a sublethal dose of ionizing radiation and compare that profile with R1 cultures recovering from desiccation (Mattimore and Battista 1996) to define the overlap in the D. radiodurans response to these stresses. Mattimore and Battista (1996) established a link between the desiccation resistance and the radiotolerance of D. radiodurans by demonstrating that a collection of ionizing radiation-sensitive strains were also sensitive to desiccation. The process of desiccation is inherently DNA damaging and dried bacterial cells exhibit a substantial number of DNA double-strand breaks, single-strand breaks, and DNA crosslinks (Dose et al. 1992), DNA damage that is also observed following exposure to ionizing radiation (Ward 1975). D. radiodurans's ability to repair extensive DNA damage renders this organism resistant to both stresses.
Three key results have come from this comparison. First, we find a significant overlap in the genomic expression programs of D. radiodurans cultures recovering from ionizing radiation and desiccation. Second, this organism's remarkable ability to repair ionizing radiation-induced DNA damage does not appear to be related to massive alterations in gene expression or large-magnitude changes in transcript abundance. Third, half of the loci that respond to ionizing radiation and desiccation encode proteins of unknown function, and the five genes most highly induced in response to each stress are the same.
To establish the utility of this comparative study, the genes encoding the five most abundant transcripts were deleted and the resulting strains evaluated for their ability to tolerate exposure to ionizing radiation. None of the genes is essential and all confer enhanced radioresistance on D. radiodurans R1. In addition, all possible pairs of double mutants were generated using the five alleles generated by deletion. Analysis of these strains revealed previously unrecognized RecA-dependent and RecA-independent processes that significantly contribute to the ionizing radiation resistance of this species.
MATERIALS AND METHODS
Strains, growth conditions, and treatment:
All strains and plasmids used in this study are listed in Table 1. D. radiodurans R1 ATCC13939 and all derivatives were grown at 30° in TGY (0.5% tryptone, 0.3% yeast extract, 0.1% glucose) broth (Mattimore et al. 1995; Earl et al. 2002) or on TGY agar (1.5% agar). Escherichia coli strains were grown at 37° in Luria-Bertani broth. Only cultures in exponential growth (OD600 = 0.08–0.15, 5 × 106 − 1 × 107 cfu/ml) were evaluated for their ability to survive ionizing radiation and desiccation. All cultures were treated at 25°. γ-Irradiation was conducted using a Model 484R 60Co irradiator (J. L. Shepherd & Associates, San Fernando, CA) at a rate of 30 Gy/min. Cultures were desiccated and held at 5% relative humidity for 2 weeks as previously described (Mattimore and Battista 1996). Survival was determined by plating serial dilutions of irradiated cultures in triplicate on TGY plates and incubating at 30°. Most strains were scored for survivors 3 days after plating. Strains TNK103, TNK106, TNK107, TNK108, TNK109, TNK110, and TNK111 form small or slow-growing colonies. These strains were scored 4 days after plating. TNK113 was not scored until 7 days after plating.
TABLE 1.
Strains and plasmids used in this study
| Genes | Description | Reference |
|---|---|---|
| DR0003 | ddrC (DNA damage response C) | This study |
| DR0070 | ddrB | This study |
| DR0326 | ddrD | This study |
| DR0423 | ddrA | Harris et al. (2004) |
| DRA0346 | pprA | Accession no. O32504 |
| Strains | ||
| Deinococcus radiodurans R1 | ATCC13939 | |
| TNK101 | As R1 but ΔddrC::pkat-aadA | This study |
| TNK102 | As R1 but ΔddrB::pkat-cat | This study |
| TNK103 | As R1 but ΔddrD::pkat-kan | This study |
| TNK104 | As R1 but ΔddrA::pkat-hyg | Harris et al. (2004) |
| TNK105 | As R1 but ΔpprA::pkat-aadA | This study |
| TNK106 | As R1 but ΔrecA::pkat-cat | Harris et al. (2004) |
| TNK107 | ΔddrC::pkat-aadA, ΔrecA::pkat-cat | This study |
| TNK108 | ΔddrB::pkat-aadA, ΔrecA::pkat-cat | This study |
| TNK109 | ΔddrD::pkat-kan, ΔrecA::pkat-cat | This study |
| TNK110 | ΔddrA::pkat-hyg, ΔrecA::pkat-cat | Harris et al. (2004) |
| TNK111 | ΔpprA::pkat-aadA, ΔrecA::pkat-cat | This study |
| TNK112 | ΔddrB::pkat-cat, ΔddrC::pkat-aadA, | This study |
| TNK113 | ΔddrC::pkat-aadA, ΔddrD::pkat-kan | This study |
| TNK114 | ΔddrA::pkat-hyg, ΔddrC::pkat-aadA | This study |
| TNK115 | ΔddrC::pkat-aadA, ΔpprA::pkat-hyg | This study |
| TNK116 | ΔddrB::pkat-cat, ΔddrD::pkat-kan | This study |
| TNK117 | ΔddrA::pkat-hyg, ΔddrB::pkat-cat | This study |
| TNK118 | ΔddrB::pkat-cat, ΔpprA::pkat-hyg | This study |
| TNK119 | ΔddrA::pkat-hyg, ΔddrD::pkat-kan | This study |
| TNK120 | ΔddrD::pkat-kan, ΔpprA::pkat-aadA | This study |
| TNK121 | ΔddrA::pkat-hyg, ΔpprA::pkat-aadA | This study |
| TNK122 | As R1 but ΔddrB::pkat-aadA | This study |
| TNK123 | As R1 but ΔpprA::pkat-hyg | This study |
| DH5α-MCR | F mcrA Δ(mrr-hsdRMS-mcrBC) Φ80 lacZΔ15 ΔlacX74 endA1 recA1 deoR Δ(ara-leu) 7697 araD139 galU galK nupG rpsL |
Invitrogen (Grand Island, NY) |
| Plasmids | ||
| pGEM-T | TA cloning vector | Promega (Madison, WI) |
| pTNK101 | pGEM-T::pkatA-cat | This study |
| pTNK102 | pGEM-T::pkatA-kan | This study |
| pTNK103 | pGEM-T::pkatA-aadA | This study |
| pTNK104 | pGEM-T::pkatA-hyg | This study |
| pTNK201 | pGEM-T::ΔddrC::pkatA-aadA | This study |
| pTNK202 | pGEM-T::ΔddrB::pkatA-cat | This study |
| pTNK203 | pGEM-T::ΔddrB::pkatA-aadA | This study |
| pTNK204 | pGEM-T::ΔddrD::pkatA-kan | This study |
| pTNK205 | pGEM-T::ΔddrA::pkatA-hyg | Harris et al. (2004) |
| pTNK207 | pGEM-T::ΔpprA::pkatA-aadA | This study |
| pTNK208 | pGEM-T::ΔpprA::pkatA-hyg | This study |
| pTNK210 | pGEM-T::ΔrecA::pkatA-cat | Harris et al. (2004) |
Gene descriptions:
All genes are identified as described in the published genome sequence (http://www.tigr.org/tigr-scripts/CMR2/GenomePage3.spl?database=gdr). Genes encoding proteins of unknown function that are induced in response to ionizing radiation and desiccation were assigned the designation ddr (DNA damage response), individual genes being differentiated by the letters A–P as indicated in Table 2.
TABLE 2.
The overlap in loci that respond as exponential phase populations ofD. radiodurans R1 recover from ionizing radiation and desiccation
| Accurate mass tag | Identifier | Gene name/annotation |
|---|---|---|
| DNA metabolism | ||
| − | DR0596 | ruvB/Holliday junction DNA helicase |
| + | DR2340 | recA/recombinase |
| + | DR1771 | uvrA/exinuclease ABC, subunit A |
| + | DR2275 | uvrB/exinuclease ABC, subunit B |
| + | DR1913 | gyrA/DNA gyrase, subunit A |
| + | DR0906 | gyrB/DNA gyrase, subunit B |
| Adaptation to oxidative stress | ||
| + | DR2220 | terB/tellurium resistance protein |
| − | DR2224 | terZ/tellurium resistance protein |
| Putative regulatory proteins | ||
| + | DR2338 | cinA/competence-inducible protein |
| RNA metabolism | ||
| − | DR2339 | ligT/2′-5′ RNA ligase, putative |
| − | DR1262 | rsr/ribonucleotide Ro/SS-A-related protein |
| Protein fate | ||
| − | DR1114 | Heat-shock protein, HSP20 family |
| Transport | ||
| + | DR1709 | NRAMP protein |
| Unknown function | ||
| − | DR0003 | ddrC/hypothetical |
| + | DR0070 | ddrB/hypothetical |
| + | DR0194 | ddrE/conserved hypothetical |
| + | DR0219 | ddrF/hypothetical |
| + | DR0227 | ddrG/hypothetical |
| + | DR0326 | ddrD/hypothetical |
| + | DR0423 | ddrA/hypothetical |
| − | DR0438 | ddrH/hypothetical |
| − | DR0659 | frnE/predicted dithiol-disulfide isomerase |
| − | DR0997 | ddrI/predicted cyclic nucleotide-binding domain/CRP family |
| + | DR1263 | ddrJ/conserved hypothetical |
| − | DR1264 | ddrK/hypothetical |
| + | DR1439 | ddrL/hypothetical |
| − | DR1440 | ddrM/hypothetical |
| − | DR2441 | ddrN/hypothetical |
| − | DR2574 | ddrO/predicted helix-turn-helix XRE-family |
| + | DRA0346 | pprA/DNA damage repair protein |
| + | DRB0100 | ddrP/hypothetical |
| − | DRB0141 | hicB/uncharacterized |
RNA isolation:
Total RNA was extracted from 1-liter cultures of irradiated and unirradiated D. radiodurans cultures using TRI Reagent (Molecular Research Center, Cincinnati), following the manufacturer's instructions after disrupting cells with glass beads. Total RNA derived from each sample condition was treated with 10 units DNase I (Ambion, Austin, TX) and purified using RNeasy Minikit columns (QIAGEN, Valencia, CA). RNA quality and quantity were evaluated by determining UV absorbance at 260 and 280 nm.
Microarray design and construction:
PCR primers were designed to amplify each open reading frame present in the fully sequenced R1 genome (White et al. 1999). PCR products represent internal portions of annotated sequences with a size range between 100 and 800 bp. Primer pairs were designed for 3180 ORFs. PCR products were generated by combining 20 ng of genomic DNA from strain R1 with oligonucleotide primer pairs (0.2 μmol each, average Tm = 55°) and 0.1 units Taq DNA polymerase (Perkin-Elmer, Wellesley, MA) in a total volume of 100 μl. The other reaction components were as specified by the manufacturer except that 0.3 m betaine was included in the reaction to aid in denaturing D. radiodurans genomic DNA. PCR amplification successes were scored (single band, correct size, >50 ng/ul). Failed reactions were repeated with an additional 2% success, for an overall efficiency of 93%. PCR products were spotted onto Superamine glass slides (Telechem, Sunnyvale, CA) using an MD Gen III Array Spotter (Molecular Dynamics, Sunnyvale, CA) at a redundancy of either 1.5 or 3.0. PCR products were immobilized to the slide surface using a Stratalinker UV crosslinker (Stratagene, La Jolla, CA). All slides were stored in a desiccator at room temperature.
Probe preparation, microarray hybridization, and data acquisition:
Hybridization probes were made as described in Peterson et al. (2000). The cDNAs were purified with QIAGEN QIAquick PCR purification columns (QIAGEN, Valencia, CA). NHS-Cy5 and NHS-Cy3 were coupled to amino-allyl containing cDNA (Amersham Biosciences, Piscataway, NJ) as described (http://www.derisilab.ucsf.edu).
Between one and four DNA microarray slides were prehybridized by incubation in 50 ml of 5× SSC, 0.1% SDS, and 1.0% bovine serum albumin for 1 hr at 42°. Slides were washed four times with vigorous agitation in distilled water followed by slide washing and dried with compressed air.
All slides were scanned at both 532 and 635 nm visible light using a Genepix 4000 imager (Axon, Union City, CA). TIGR Spotfinder (http://www.tigr.org/software/tm4/spotfinder.html) was used to quantify hybridization signals that were three times above the local background in one of the two channels (Cy3 or Cy5). The data were normalized using total intensity normalization. Due to pervasive low-magnitude gene induction values associated with D. radiodurans's response to IR and since independent biological replicates varied by as much as threefold, we set conservative criteria for categorizing gene induction. Only genes with an average expression ratio greater than threefold in at least two independent experiments were considered induced. Averages were based on at least four hybridization experiments and included at least two independent biological samples. Aberrant values were removed from consideration in calculating averages. In most cases this occurred due to weak signal intensity in one channel resulting in ratios with high variability.
Quantitative real-time PCR:
Two micrograms of each DNase I-treated, purified RNA sample were converted to cDNA using SUPERSCRIPT II RNase H-Reverse Transcriptase (Invitrogen, Carlsbad, CA) combined with 25 pmol of random hexamers to initiate synthesis. Conditions for this reaction followed the manufacturer's instructions.
Approximately 100 bp of unique sequence from the genes of interest was amplified using the following primer sets: DR2340up and DR2340dwn, DR1343up and DR1343dwn, DR0003up and DR0003dwn, DR0070up and DR0070dwn, DR0326up and DR0326dwn, DR0423up and DR0423dwn, and DRA0346up and DRA0346dwn (supplemental Table 1 at http://www.genetics.org/supplemental/). The PCR reaction (50 μl) for amplifying these genes contained the appropriate primers at a final concentration of 0.2 μm, 1 μl of the cDNA template, and SYBR Green PCR core reagents (Applied Biosystems, Foster City, CA). Amplifications were carried out by incubating reactions at 95° for 3 min prior to 40 cycles of 30 sec at 95° followed by 30 sec at 65° and 30 sec at 72°. Data were collected and analyzed at each 72° interval. Amplification was followed by melting-curve analysis consisting of 80 cycles of 55° at 10-sec intervals with 0.5° increments per cycle. Reactions were then held at 23° until analysis.
Each 96-well plate consisted of standard curves for each primer set run in duplicate. Standard curves were constructed using cDNA obtained from the unirradiated wild-type organism. A dilution series (1 − 1 × 10−4) of each experimental sample was generated and run in duplicate. Negative controls without cDNA template were run on every plate analyzed.
All assays were performed using the iCycler iQTM real-time detection system (Bio-Rad, Hercules, CA). All data were PCR baseline subtracted before threshold cycle values were designated and standard curves were constructed. Mean concentrations of each transcript in each sample were calculated from the standard curves generated using each primer set. Induction levels were determined by dividing the calculated concentration of the irradiated sample by the concentration of the unirradiated sample for each strain. The mean concentration of the glyceraldehyde 3-phosphate dehydrogenase (gap) transcript, a housekeeping gene whose expression is unaffected by ionizing radiation, was also determined before and after irradiation for each strain.
Antibiotic resistance cassettes:
Four antibiotic resistance cassettes were constructed by separately fusing (i) the aadA gene (accession no. AF458479), (ii) the cat gene of pBC (Stratagene Cloning Systems, La Jolla, CA), (iii) the kan gene of pGPS3 (New England Biolabs, Beverly, MA), and (iv) the hyg gene of Streptomyces hygroscopicus (accession no. X03615) to the 120 bp of DNA sequence immediately upstream of the katA (DR1998) gene of D. radiodurans R1. Genes spliced downstream of this sequence, which is designated pkatA, are constitutively expressed in R1 (Funayama et al. 1999). Each antibiotic resistance gene was spliced to pkatA by overlap extension in vitro during a polymerase chain reaction (Horton et al. 1989). The primers used in these reactions are listed in supplemental Table 2 at http://www.genetics.org/supplemental/. The fused fragments were amplified and cloned into pGEM-T (Promega, Madison WI) by T-A cloning. The sequence of pGEM-T can be found at http://wheat.pw.usda.gov/~lazo/methods/pro/tb150.html#ix. The resulting plasmids (pTNK101, pTNK102, pTNK103, and pTNK104) were propagated in the E. coli strain DH5α-MCR.
Strain construction:
Genes were deleted using techniques described previously (Funayama et al. 1999; Ruan et al. 2004). Fragments corresponding to ∼1000 bp of DNA sequence immediately upstream and downstream of each gene to be deleted were amplified and spliced to the 5′and 3′ ends of an antibiotic resistance cassette by overlap extension during a polymerase chain reaction. The resulting fragment was transformed into exponential phase cells, and recombinants were selected on TGY plates containing an appropriate antibiotic. Since D. radiodurans is multigenomic, individual antibiotic resistant colonies were screened to establish whether they were homozygous for the deletion. Pairs of primers that anneal outside each gene's coding sequence (supplemental Table 2) were used to amplify diagnostic PCR fragments, establishing whether the strain was homozygous for the marker that replaced the gene of interest. The details of each construction are given below.
TNK101 ΔddrC (ΔDR0003) was created by splicing the pkatAaadA (StrR) cassette to the 1.1 kbp of genomic DNA sequence immediately upstream and the 1.1 kbp immediately downstream of DR0003. This ΔddrC::pkatA-aadA hybrid fragment was cloned into pGEM-T (Promega), creating pTNK201. pTNK201 was propagated in E. coli DH5α-MCR. The hybrid fragment was PCR amplified using primers tnk0003FW2 and tnk0003RV3 and used to transform an R1 culture by standard methods (Ruan et al. 2004). Recombinants were selected on TGY plates containing 8 μg/ml streptomycin. The deletion of ddrA was confirmed by PCR using the primers that anneal to sequences flanking DR0003 (supplemental Table 2). These primers generate a 0.9-kbp fragment when R1 genomic DNA is used as a template in the reaction and a 1.2-kbp fragment if ddrC is replaced by the pkatA-aadA cassette. Amplification of genomic DNA isolated from the strain designated TNK101 produced only the 1.2-kbp fragment. The ΔddrB (ΔDR0070) strains TNK102 and TNK122, the ΔddrD (ΔDR0326) strain TNK103, the ΔddrA (ΔDR0423) strain TNK104, the ΔpprA (ΔDRA0346) strains TNK105 and TNK123, and the ΔrecA (ΔDR2340) strain TNK106 were constructed in the same manner as TNK101.
To permit the generation of all possible pairs of double mutants with these alleles, it was necessary to create two ΔddrB strains. In TNK102 ddrB is replaced with the pkatA-cat cassette and in TNK122 ddrB is replaced with the pkatA-aadA cassette. In each construction the cassette was spliced to PCR fragments derived from the sequence 0.8 kbp upstream and 0.8 kbp downstream of ddrB. The hybrid fragment ΔddrB::pkatA-cat or ΔddrB::pkatA-aadA was cloned into pGEM-T to generate pTNK202 or pTNK203, respectively. The fragments used to create the deletions were amplified by PCR using tnk0070FW2 and tnk0070RV3 and transformed into exponential phase cultures of R1. Recombinants were obtained on TGY plates containing 3 μg/ml chloramphenicol for ΔddrB::pkatA-cat candidates or TGY plates with 8 μg/ml streptomycin for ΔddrB::pkatA-aadA candidates. Genomic DNA isolated from putative recombinants was PCR screened to establish that the disruption was homozygous, using primers listed in supplemental Table 2.
TNK103 ΔddrD (ΔDR0326) was deleted using a ΔddrD::pkatA-kan cassette built into pTNK204. The appropriate fragment was PCR amplified using tnk0326FW2 and tnk0326RV3 (supplemental Table 2) and used to transform into exponential phase cultures. This deletion cassette is a hybrid fragment in which the pkatA-kan cassette is spliced to the sequence 0.9 kbp upstream and 1.3 kbp downstream of the ddrD coding region. Recombinants were selected on TGY plates containing 10 mg/ml kanamycin. Primers (supplemental Table 2) flanking ddrD were used to verify the deletion. These primers amplify a 2.8-kbp fragment from R1 and a 3.1-kbp fragment corresponding to pkatA-npt when ddrD is deleted.
Deletion of ddrA (DR0423) was performed as described previously (Harris et al. 2004). The ΔddrA::pkatA-hyg cassette from pTNK205 was used to transform exponential phase cells. The ΔddrA::pkatA-hyg cassette is a hybrid fragment in which pkatA-hyg is joined to the 1.0-kbp upstream region and 0.9-kbp downstream region of ddrA. Recombinants were selected on TGY agar containing 37.5 μg/ml hygromycin. Primers were designed to anneal outside the coding sequence of ddrA (Table 2). These primers amplify a 0.85-kbp fragment from R1 and a 1.3-kbp fragment corresponding to the katA-npt fusion when ddrD is deleted.
The ΔpprA (ΔDRA0346) strains, TNK105 and TNK123, were generated using the ΔpprA::pkatA-aadA cassette from pTNK207 or the ΔpprA::pkatA-hyg cassette from pTNK208, respectively. For each cassette, the sequence 0.85 kbp upstream and 0.85 kbp downstream of pprA was fused to pkatA-aadA or pkatA-hyg. Recombinants were selected on TGY agar containing 8 or 37.5 μg/ml streptomycin. Deletions were confirmed using primers annealing to flanking sequences to pprA (supplemental Table 2). Amplification with tnkA0346FW2 and tnkA0346RV3 generates a 0.9-kbp fragment from R1 genomic DNA and a 1.2-kbp fragment corresponding to the katA-aadA cassette. Amplification with tnkA0346FW5 and tnkA0346RV7 produces a 2.6-kbp product from R1 genomic DNA and a 2.8-kbp fragment if pprA is replaced by the katA-hyg cassette.
Deletion of recA (DR2340) was accomplished using a ΔrecA::pkatA-cat cassette from pTNK210, as described previously (Harris et al. 2004). This hybrid fragment was constructed by joining pkatA-cat to the 1.6 kbp upstream and 1.2 kbp downstream of the recA coding region. Recombinants were selected on a TGY plate containing 3 μg/ml chloramphenicol and the deletion was screened for using primers annealing to sequences flanking to recA (supplemental Table 2). These primers amplified a 1.5-kbp fragment if the recA coding sequence remained and a 1.3-kbp fragment corresponding to the katA-cat cassette that was used to replace recA.
Double mutants were created in the same manner as that of single mutants. TNK101, TNK102, TNK103, TNK104, TNK105, and TNK122 were used as the parental strains and these strains were transformed with another allele carrying a compatible antibiotic resistance marker.
RESULTS
Identification of loci in D. radiodurans R1 induced in response to 3 kGy ionizing radiation:
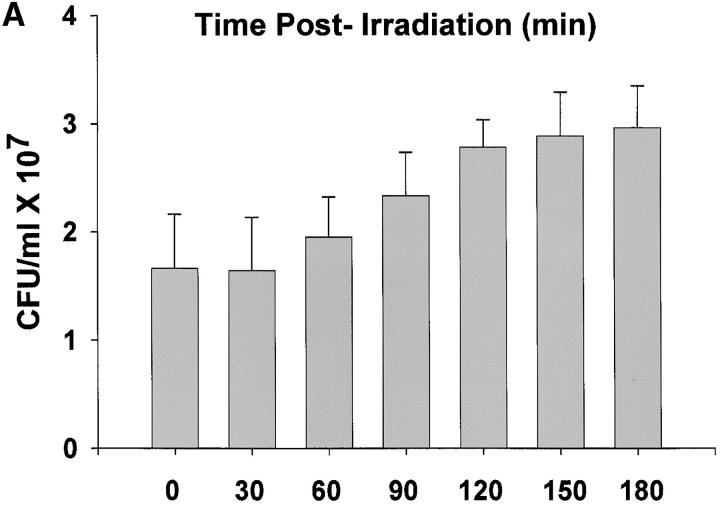
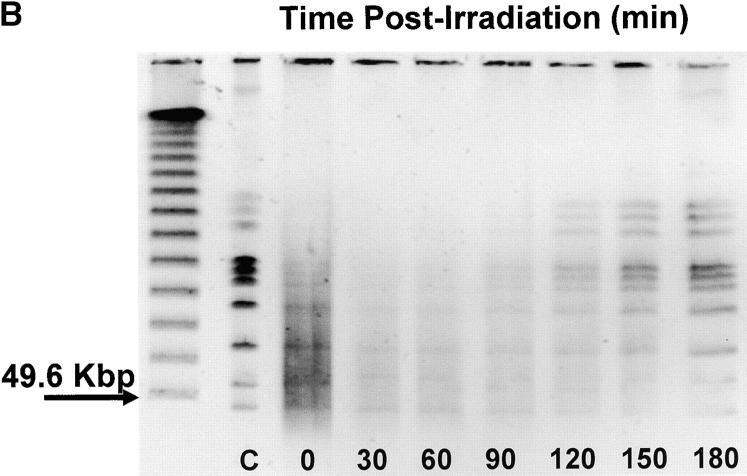
We compared unirradiated R1 cultures in exponential phase growth with age-matched R1 cultures during the first hour after exposure to a nonlethal 3 kGy dose of γ radiation. This time course was deemed appropriate on the basis of data shown in Figure 1, which documents restitution of the R1 genome following 3 kGy γ-radiation. In agreement with previous studies (Udupa et al. 1994; Mattimore et al. 1995; Earl et al. 2002), there is no loss of viability in R1 cultures receiving a 3-kGy dose (Figure 1A). The titer of the cultures prior to irradiation was 1.7 ± 0.5 × 107 cfu/ml, and that determined immediately after irradiation (zero time) was 1.66 ± 0.6 × 107 cfu/ml. After irradiation there is evidence of DNA double-strand breaks, indicated by the reduction in high-molecular-weight DNA and an accompanying increase in low-molecular-weight fragments (Figure 1B). Within 90 min, the high-molecular-weight fragments become more intense, signaling that genome reassembly is well underway. After 1 hr delay, there is a significant increase in cell numbers in the irradiated population (t = −2.52, P = 0.014, d.f. = 58) relative to the number of cells present immediately before irradiation, and the titer of the irradiated population doubles within 3 hr postirradiation, suggesting that all repairs have been completed. On the basis of this time course, it was decided that isolating total RNA immediately after and at 30 and 60 min postirradiation provided the best opportunity to characterize the cell's response to ionizing radiation.
Figure 1.—
(A) Growth of R1 cultures as a function of time following exposure to 3000 Gy γ-radiation. Titers were determined immediately after irradiation (zero time) and at the times indicated. (B) Pulsed-field gel illustrating the restitution of the R1 genome following 3000 Gy γ-radiation. The unlabeled lane corresponds to a lambda ladder standard. Lane C is a Not 1 digest of genomic DNA obtained from the culture prior to irradiation. The remaining lanes contain a Not 1 digest of genomic DNA isolated from irradiated cultures at the times indicated.
To determine how global transcription in D. radiodurans R1 changes in response to ionizing radiation, competitive hybridizations were repeated for six independent experimental trials. Those studies are summarized in supplemental Table 3 at http://www.genetics.org/supplemental/, which identifies the mean Cy5/Cy3 ratios obtained for those genes in the irradiated population that exhibited a threefold or higher increase in expression relative to the unirradiated population. Seventy-two genes (2.2% of the genome) respond with increased expression within the first hour after exposure. A detectable protein product, an accurate mass tag (Lipton et al. 2002), has been reported for 65% of these loci, verifying that these open reading frames encode protein products.
To help establish the validity of the ratios recorded in supplemental Table 3, gene expression was also measured by quantitative RT-PCR for seven loci. Total RNA was isolated from exponential phase cultures of R1 before irradiation and at the three time points following exposure to 3 kGy irradiation (IR). Changes in transcript abundance for recA (DR2340), gap (DR1343), and five genes encoding hypothetical proteins (DR0003, DR0070, DR0326, DR0423, and DRA0346) were determined. An increase in recA gene expression served as a positive control as this deinococcal gene's response to IR had already been established (Earl et al. 2002), and gap, which encodes glyceraldehyde 3-phosphate dehydrogenase, was included among these genes because it was not, on the basis of our microarray data, expected to be a radiation-responsive gene. The relative change in expression for each gene is provided as the mean ratio in supplemental Table 4 at http://www.genetics.org/supplemental/. With one exception, there was remarkable concordance between induction ratios obtained by these methods. The relative change observed for the DR0070 transcript measured by quantitative (Q)-RT-PCR was two to six times higher compared to relative abundance measurements derived from microarray hybridization. Overall the uniformity of measurements from the Q-RT-PCR and microarray analyses suggests that the changes in transcription reported by this microarray are reliable.
The genes induced in response to ionizing radiation were grouped into nine categories on the basis of their similarity to known proteins (supplemental Table 3) with 54 of the 72 loci falling into one of three categories. The largest group (44%) encodes proteins of unknown function. These loci are among the most highly induced, 20 being induced greater than fivefold in the irradiated population. Six genes (recA, ruvB, uvrA, uvrB, gyrA, and gyrB) make up a second group that encodes proteins associated with DNA metabolism. The third group consists of five loci (katA, terB, terZ, mrsA, and dps) that encode proteins that are associated with alleviating the effects of oxidative stress in other species. The katA gene product (DR1998p) is similar to the KatE catalase of E. coli. TerB (DR2220p) and TerZ (DR2224p) are homologs of E. coli proteins that appear to play a role in maintaining the intracellular reducing environment possibly by directly reversing disulfide bonds (Turner et al. 1999). MsrA (DR1849p) shares similarity with E. coli's methionine sulfoxide reductase. Loss of MsrA sensitizes E. coli to hydrogen peroxide (Moskovitz et al. 1995). Dps (DRB0092p) is related to an E. coli protein that nonspecifically binds to DNA, protecting it from oxidative damage (Almiron et al. 1992; Martinez and Kolter 1997).
Molecular dissection of D. radiodurans's response to ionizing radiation through an examination of transcription in cells recovering from desiccation:
There is never a perfect correlation between genes that are essential under a given condition and genes that are induced under the same condition (Badarinarayana et al. 2001; Winzeler et al. 1999). Many prokaryotic genes are organized into operons and stress-induced gene expression may generate a polycistronic mRNA, which will be detected as multiple signals on an array, even if only one of the encoded proteins is needed to combat the stress. Therefore, supplemental Table 3 undoubtedly identifies gene products whose expression is incidental and that do not directly participate in the repair of ionizing-radiation-induced intracellular damage. To better characterize D. radiodurans's response to ionizing radiation, we subjected R1 cultures to desiccation and compared the expression pattern to that obtained after ionizing radiation. For a vegetative species, D. radiodurans is quite resistant to desiccation, exhibiting ∼90% viability after being stored dry for 6 weeks (Mattimore and Battista 1996). Since the process of desiccation and rehydration introduces DNA damage (Dose et al. 1992), we assumed that some of the proteins needed to repair ionizing radiation-induced damage, including double-strand breaks, would be identical to proteins used to mend DNA damage introduced following desiccation. The overlap in the cell's response to each stress should specify gene products that directly participate in repair of common DNA damage, potentially identifying novel proteins critical to this process.
The transcriptome of R1 in liquid culture was compared with a culture recovering from 2 weeks desiccation at 5% relative humidity. Samples were obtained from six independent cultures over a three-point time course (0, 0.5, and 1 hr) following rehydration. Under these nonlethal conditions, 73 genes were induced during this time course (supplemental Table 5 at http://www.genetics.org/supplemental/). Although D. radiodurans maintains distinct inducible responses to each stress, there is substantial overlap (Table 2) in the gene expression profiles observed. Thirty-two (45%) of the 73 loci responding to desiccation were also observed during R1's recovery from ionizing radiation.
Deletion of the locus designated pprA (DRA0346) sensitizes D. radiodurans R1 to ionizing radiation:
The overlap in D. radiodurans's response to ionizing radiation and desiccation described by Table 2 is significant because it suggests something long suspected: D. radiodurans radioresistance is the consequence of unprecedented mechanisms of DNA damage tolerance (Battista 1997, 2000; Battista et al. 1999). The preponderance of hypothetical proteins on this list, coupled with only a limited number of well-defined DNA repair proteins, argues that novel processes are facilitating this species' capacity to survive genetic insult. To test the validity of this inference, the five hypothetical genes that were induced to highest level in response to each treatment, ddrA (DR0423), ddrB (DR0070), ddrC (DR0003), ddrD (DR0326), and pprA (DRA0346), were deleted and replaced by an antibiotic resistance marker. The ionizing radiation resistance of each strain was then compared to that of the R1 parent.
With the exception of TNK103 (ΔddrD), the resulting strains were identical in size and appearance to D. radiodurans R1. Like its parent, TNK103 cultures consist of spherical cells that form pairs and tetrads in liquid media, but these cells are approximately one-half the diameter of the parent strain. Deletion of ddrA, ddrB, ddrC, and ddrD does not alter the growth rate of the resulting strains (∼1.5-hr doubling time) in rich liquid media relative to the parent strain, but deletion of pprA results in a strain with a 2.4-hr doubling time (mean of 10 independent measurements). None of the mutant strains exhibits a decrease in the efficiency of natural transformation (∼5 × 10−5 rifampicin-resistant transformants per colony-forming unit) relative to R1, indicating that none of the gene products encoded by these loci is essential for RecA-dependent homologous recombination, which is required for natural transformation in this species.
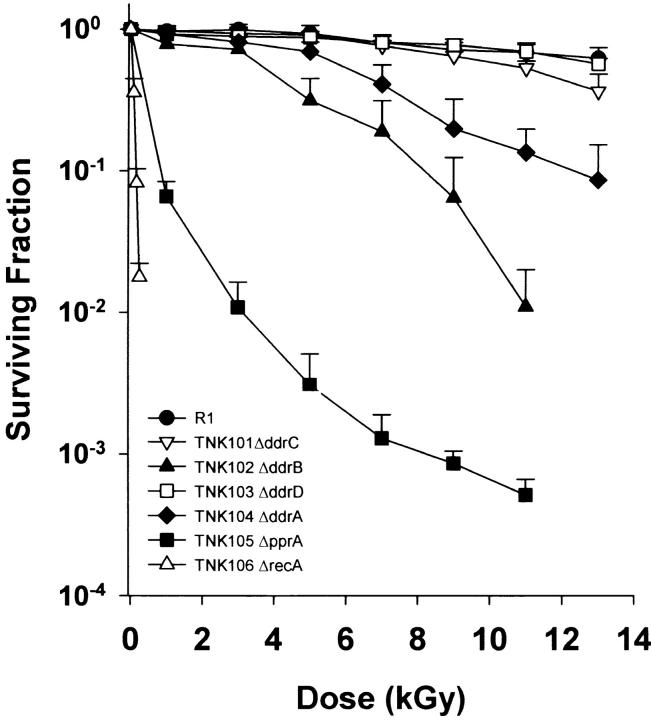
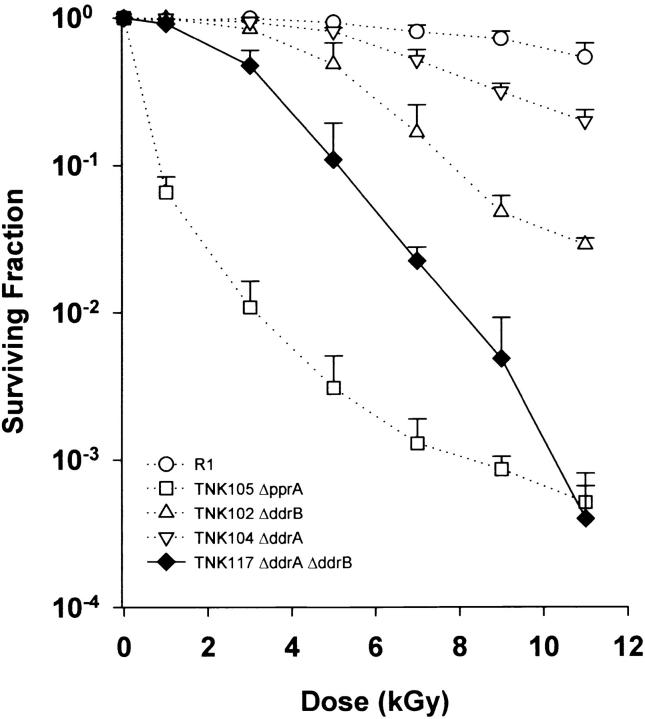
Strains TNK101 (ΔddrC) and TNK103 (ΔddrD) were as resistant to ionizing radiation as their R1 parent, but the other three mutants displayed varying degrees of sensitivity to ionizing radiation (Figure 2). As shown in earlier work, TNK102 (ΔddrB) and TNK104 (ΔddrA) exhibit a reduced capacity to survive these exposures (Liu et al. 2003; Harris et al. 2004), but this is apparent only at doses >2.5 and 5 kGy, respectively. Cultures of TNK105 (ΔpprA) are much more sensitive to ionizing radiation than are TNK102 and TNK104. The shape of the survival curve for TNK105 is atypical. The reduction in viability observed at the lowest doses is not sustained as higher doses of ionizing radiation are applied. At doses above 5 kGy, the slope changes notably, and the rate at which viability declines is reduced. The shoulder of resistance that characterizes R1 cultures is not apparent in this strain; the viability of the irradiated population drops off rapidly at 1 kGy, the lowest dose examined. Very few single mutations have been characterized that result in similar reductions in R1 resistance. To our knowledge only deletions of recA and polA have similar effect (Gutman et al. 1993, 1994). Even so, TNK105 is not as sensitive to γ-radiation (Figure 2) as TNK106, an isogenic ΔrecA strain created for this study.
Figure 2.—
Representative survival curves for D. radiodurans strains TNK101 ΔddrC, TNK102 ΔddrB, TNK103 ΔddrD, TNK104 ΔddrA, TNK105 ΔpprA, TNK106 ΔrecA, and D. radiodurans R1 following exposure to γ-radiation. Values are the means ± standard deviations of three independent experiments. n = 9.
Deletions of the loci designated ddrA (DR0423) or ddrB (DR0070) decrease the ionizing radiation resistance of the ΔpprA strain TNK105:
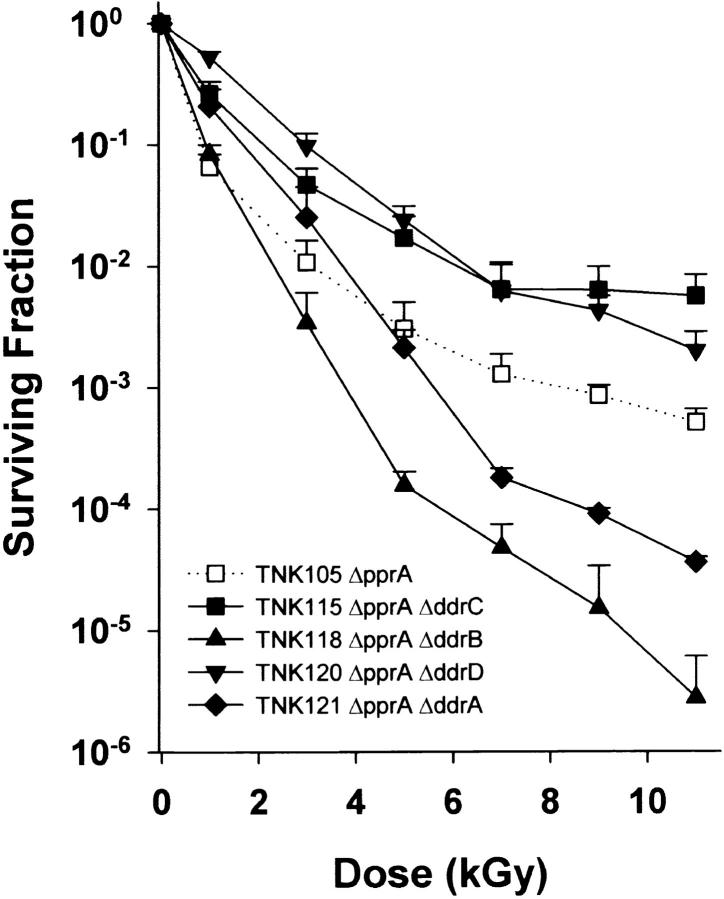
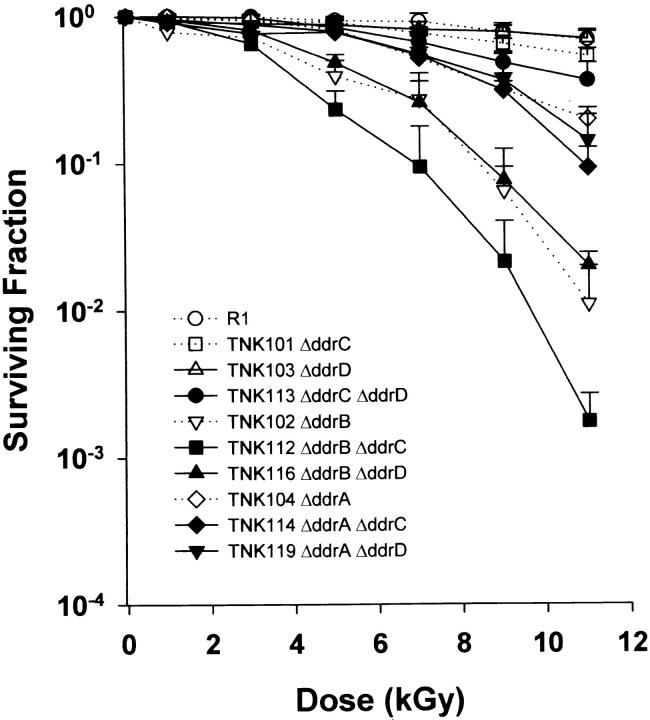
Figure 3 illustrates the survival curves for all combinations of double mutants possible in a ΔpprA background using the genes examined in this study. TNK118 ΔddrB ΔpprA and TNK121 ΔddrA ΔpprA exhibit increased sensitivity to ionizing radiation relative to TNK105 ΔpprA. The loss of DdrA and DdrB is most apparent at doses >3 kGy, largely eliminating the biphasic appearance of the TNK105 survival curve (dotted line in Figure 3). TNK115 ΔddrC ΔpprA and TNK119 ΔddrD ΔpprA appear to decrease the sensitivity of the ΔpprA single mutant to ionizing radiation.
Figure 3.—
Representative survival curves for D. radiodurans strains TNK105 ΔpprA, TNK115 ΔddrC ΔpprA, TNK118 ΔddrB ΔpprA, TNK120 ΔddrD ΔpprA, and TNK121 ΔddrA ΔpprA following exposure to γ-radiation. Values are the means ± standard deviations of three independent experiments. n = 9.
Deletion of the locus designated ddrA (DR0423) decreases the ionizing radiation resistance of the ΔddrB strain TNK102:
At doses >3 kGy, TNK117 ΔddrA ΔddrB is more sensitive to ionizing radiation than TNK102 ΔddrB or TNK104 ΔddrA (Figure 4). TNK117 displays a small shoulder of resistance, noted at 1 kGy, but above this dose survival is substantially reduced relative to each single mutant. At 9 kGy, for example, TNK117 is 10-fold more sensitive than TNK102 and 60-fold more sensitive than TNK104.
Figure 4.—
Representative survival curve for D. radiodurans strain TNK117 ΔddrA ΔddrB. For comparison, the survival curves of TNK102 ΔddrB, TNK104 ΔddrA, TNK105 ΔpprA, and D. radiodurans R1 from Figure 1 are included as dotted lines. Values are the means ± standard deviations of three independent experiments. n = 9.
Deletion of the locus designated ddrC (DR0003) decreases the ionizing radiation resistance of the ΔddrB strain TNK102:
A family of double mutants carrying ΔddrC and ΔddrD was also evaluated for its ability to survive ionizing radiation (Figure 5). In general, the deletion of ddrC and ddrD had little effect on the ionizing radiation resistance of the resulting strain. TNK113, a double mutant lacking ddrC and ddrD, did exhibit a slight (twofold) increase in sensitivity to ionizing radiation, but this phenotype is apparent only at the highest applied doses. In addition, an increase in ionizing radiation sensitivity was noted when TNK112 ΔddrC ΔddrB was compared to TNK102 ΔddrB.
Figure 5.—
Representative survival curves for D. radiodurans strains TNK112 ΔddrB ΔddrC, TNK113 ΔddrC ΔddrD, TNK114 ΔddrA ΔddrC, TNK116 ΔddrB ΔddrD, and TNK119 ΔddrA ΔddrD following exposure to γ-radiation. For comparison, the survival curves of TNK101 ΔddrC, TNK102 ΔddrB, TNK103 ΔddrD, TNK104 ΔddrA, and D. radiodurans R1 from Figure 1 are included as dotted lines. Values are the means ± standard deviations of three independent experiments. n = 9.
Deletions of loci designated ddrA (DR0423), ddrB (DR0070), ddrC (DR0003), or ddrD (DR0326) decrease the resistance of the recA strain TNK106 to ionizing radiation:
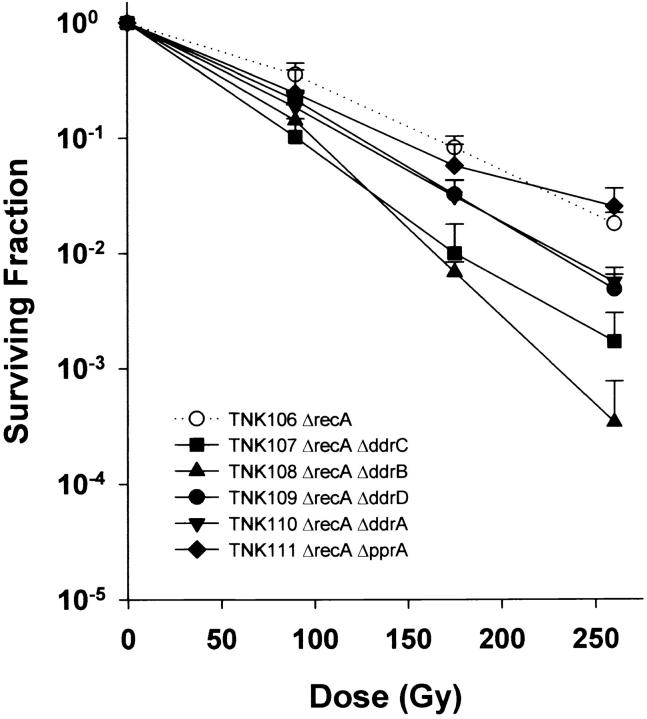
In addition to creating double mutants of all pairwise combinations of deletions of ddrA, ddrB, ddrC, ddrD, and pprA, we also moved a ΔrecA allele into the five strains carrying each deletion. As depicted in Figure 6 inactivation of recA dramatically sensitizes D. radiodurans to ionizing radiation, but the recA strain is made even more sensitive when ddrA, ddrB, ddrC, or ddrD is also inactivated. This increase is statistically significant for each of these double mutants. For example, the slope of the line describing TNK106 (ΔrecA) survival is significantly different (t = 5.5475, P < 0.01, d.f. = 16) from that obtained for TNK109 (ΔrecA ΔddrD), the strain that exhibits the smallest increase (fourfold at 260 Gy) in sensitivity relative to TNK106. TNK108 (ΔrecA ΔddrB) is 50 times more sensitive than TNK106 at 260 Gy. The ΔrecA ΔpprA double mutant (TNK111) is no more sensitive to ionizing radiation than the TNK106 ΔrecA strain (Figure 6).
Figure 6.—
Representative survival curves for D. radiodurans strains TNK106 ΔrecA, TNK107 ΔddrC ΔrecA, TNK108 ΔddrB ΔrecA, TNK109 ΔddrD ΔrecA, TNK110 ΔddrA ΔrecA, and TNK111 ΔpprA ΔrecA following exposure to γ-radiation. Values are the means ± standard deviations of three independent experiments. n = 9.
DISCUSSION
D. radiodurans is one of the most DNA damage tolerant organisms known. The species' ability to withstand what is a sterilizing dose of ionizing radiation for almost all other microbes has led to a great deal of superficial speculation about the origins of the Deinococcaceae and the mechanisms they employ to repair DNA damage. Despite the fact that the genome has been sequenced and annotated (White et al. 1999; Makarova et al. 2001), and analyses of the transcriptome (Liu et al. 2003) and proteome published (Lipton et al. 2002), there is very little specific information available that characterizes the unique properties of deinococcal species (Edwards and Battista 2003; Narumi 2003). It has been suggested that the extreme radioresistance of this family is a consequence of unprecedented DNA repair mechanisms, and that the absence of well-defined explanations for the radioresistance of the Deinococcaceae reflects our lack of knowledge about DNA repair in this species (Battista et al. 1999; Battista 2000).
As discussed by Eisen and Hanawalt (1999), our perception of what qualifies as a DNA repair protein in a prokaryote is determined by our knowledge of DNA repair in E. coli. Since prokaryotes are remarkably diverse, occupying every conceivable environmental niche, it is all but certain that the E. coli paradigm for DNA repair does not apply to all species. Among characterized prokaryotes, D. radiodurans's repair functions seem very likely to deviate from that of E. coli. D. radiodurans tolerates levels of DNA damage (with little or no loss of viability) that are capable of completely eradicating an E. coli population. Genome analysis (White et al. 1999; Makarova et al. 2001) revealed that D. radiodurans encodes the same complement of DNA repair proteins found in E. coli, suggesting that there are unidentified features of D. radiodurans physiology that facilitate its resistance to DNA damage. Since almost half of the open reading frames described for D. radiodurans encode proteins of unknown function, it is reasonable to assume that some of these uncharacterized proteins are critical for DNA damage tolerance. In this study, we have described phenotypes associated with the inactivation of five “hypothetical” proteins encoded by D. radiodurans R1 that clearly indicate that the action of novel proteins contributes to this species' capacity to tolerate exposure to ionizing radiation.
D. radiodurans's transcriptional responses to ionizing radiation and desiccation overlap:
We have defined the genomic expression profile of D. radiodurans R1 cultures as they recover from a sublethal dose of ionizing radiation, comparing that profile with R1 cultures recovering from desiccation (Mattimore and Battista 1996). Table 2 lists 32 loci that are induced as cultures recover from ionizing radiation and desiccation. This group includes only four genes (recA, ruvB, uvrA, and uvrB) that encode proteins that have been previously associated with DNA repair in other species, indicating that increased transcription from most genes encoding identifiable DNA repair proteins is not necessary for D. radiodurans's radioresistance. The reasons for this are not apparent from this study, but two possible explanations seem likely. Either constitutive expression of these proteins is sufficient for the repair of DNA damage or the cell does not depend on these proteins to repair ionizing radiation-induced DNA damage. At present, neither possibility can be excluded. These data argue that D. radiodurans's ability to repair the damage caused by ionizing radiation and desiccation is not related to dramatic shifts in gene expression or large-magnitude changes in transcript abundance.
This result differs from a previous study that monitored ionizing-radiation-induced global gene expression in D. radiodurans. Liu et al. (2003) reported that 832 genes were induced during a 24-hr recovery period in D. radiodurans cultures exposed to 15 kGy γ-radiation. These numbers are an order of magnitude larger than those we are reporting, and we believe there are two principle reasons for the difference. First, in the earlier study stationary-phase cultures of D. radiodurans were irradiated and this culture was transferred to fresh media to recover (Liu et al. 2003). These authors compared the RNA isolated from the recovering cultures to RNA isolated from the unirradiated stationary-phase control culture. The ratios obtained were, therefore, detecting transcription occurring in response to ionizing radiation as well as the transcription needed to transition the cell from stationary phase to exponential-phase growth. The cell's need to deal with both conditions simultaneously may help explain the large number of differentially expressed transcripts reported relative to this study. Second, we have chosen to stringently exclude any transcript from this analysis that does not exhibit at least a threefold differential relative to the unirradiated control. The previous study included all changes in transcripts that were twofold or higher (Liu et al. 2003).
The RecA and RuvB proteins function in homologous recombination, and UvrA and UvrB are subunits of the endonuclease responsible for nucleotide excision repair in prokaryotes. Of these proteins only RecA is necessary for ionizing radiation resistance in D. radiodurans. Genetic inactivation of RuvB (Kitayama et al. 1997) and UvrA (Agostini et al. 1996) does not substantively alter the radioresistance of R1, suggesting that if these proteins participate in protecting D. radiodurans from ionizing radiation-induced DNA damage, their activities are redundant. In contrast, deletion of recA from R1 results in a strain that is extremely sensitive to ionizing radiation (Narumi et al. 1999), a result reiterated in Figure 2. Experimental evidence has identified two physiological functions that are RecA dependent, homologous recombination (Daly and Minton 1995, 1996) and cleavage of LexA repressor (Satoh et al. 2002), and arguments have been made that both activities are necessary for the ionizing radiation resistance of this species.
Genes encoding the two subunits of the DNA gyrase, gyrA (DR1913) and gyrB (DR0906), are also induced in response to ionizing radiation. DNA gyrase forms negative supercoils in the DNA helix to alleviate excess positive supercoiling caused by transcription, DNA replication, and repair processes. In E. coli, treatments that reduce negative supercoiling increase expression of gyrA and gyrB (Menzel and Gellert 1983), and the introduction of DNA double-strand breaks will trigger their expression.
Sixty percent of the loci that are part of D. radiodurans's common response to ionizing radiation and desiccation encode proteins of unknown function (Table 2). As indicated in Table 2, some of the predicted proteins share motifs with characterized proteins, but the degree of similarity is not sufficiently high to assign a putative function to the gene product. Five of these hypothetical genes (designated ddrA, ddrB, ddrC, ddrD, and pprA) were, on average, induced >10-fold when cells were irradiated.
The hypothetical proteins DdrA, DdrB, and PprA confer resistance to killing by ionizing radiation:
The coding sequences of ddrA, ddrB, ddrC, ddrD, pprA, and recA were deleted and replaced with an antibiotic resistance marker. In addition, all possible combinations of double mutants with these six alleles were constructed in an attempt to establish genetic evidence for potential interactions between the encoded gene products. All of these genes appear to contribute to D. radiodurans's capacity to survive exposure to ionizing radiation. Pending further investigation, we are assuming that the increases in ionizing radiation sensitivity are due to the loss of the gene product encoded by the gene that has been deleted. We note that the ddrA, ddrC, ddrD, and pprA genes are located within small clusters of genes, and in each of these situations the downstream gene also encodes a hypothetical protein. Although we did not observe an increase in transcription from these downstream genes when R1 was exposed to ionizing radiation, there is a formal possibility that the gene replacement might affect expression of the downstream coding sequence.
Our analyses of the effect of how combining alleles influences the ionizing radiation resistance of the resulting strain have allowed us to operationally define the founding members of three epistasis groups involved in the D. radiodurans response to ionizing radiation, which we have chosen to name the recA, ddrA, and ddrB groups. The recA epistasis group includes recA and pprA. The effect of the combined loss of recA and pprA on the radioresistance of D. radiodurans is quantitatively the same as the loss observed when only recA has been deleted (Figure 6). Deletion of ddrA and ddrB increases the sensitivity of the recA strain, indicating that their respective gene products contribute to radioresistance by mechanisms that are distinct from that of RecA. Consistent with this grouping, pprA is not epistatic to ddrA or ddrB. The shape of the survival curves for the ddrA pprA and ddrB pprA strains and that of the pprA strain are dissimilar, the double mutants exhibiting substantially greater sensitivity to doses of ionizing radiation in excess of 5 kGy (Figure 3).
The ddrA and ddrB genes are clearly part of separate epistasis groups. The combined effect of inactivation of both loci results in a decrease in ionizing radiation resistance comparable to that observed when the pprA locus is inactivated (Figure 4). Taken together, the survival data in Figures 3, 4, and 6 argue that three genetically separable processes significantly contribute to the ionizing radiation resistance of D. radiodurans and that the level of resistance observed in this species is a consequence the combined effect of all three activities.
At present, it is not possible to definitively place ddrC or ddrD in one of these groups. Since the ddrC and ddrD single mutants do not exhibit any increased susceptibility relative to R1 (Figure 2), the failure of double mutants, generated using these alleles, to express a phenotype different from that of a sensitive single mutant cannot be taken as proof of epistasis. Thus, while there is evidence that DdrC and DdrD participate in ionizing radiation resistance (Figures 3, 5, and 6), the relationships between these proteins and DdrA, DdrB, and PprA cannot be unequivocally defined.
The ddrA and ddrB gene products:
Previous studies established that DdrA (Harris et al. 2004) and DdrB (Liu et al. 2003) contribute to the ionizing radiation resistance of D. radiodurans R1, and those results are confirmed here. DdrA is distantly, but specifically, related to the Rad52 protein of eukaryotes and the Erf protein of some cryptic phage (Iyer et al. 2002), and in vitro studies have established that purified DdrA binds to single-stranded DNA with affinity for 3′ ends, protecting those ends from nuclease degradation (Harris et al. 2004). In contrast, DdrB has no obvious similarity to any characterized protein, and its biochemical function has not been elucidated. Since TNK117 ΔddrA ΔddrB is much more sensitive to ionizing radiation than TNK102 ΔddrB or TNK104 ΔddrA (Figure 4), it appears that the loss of one of these proteins is partially compensated for by the activity of the other, suggesting that DdrA and DdrB have overlapping functions. We have proposed that DdrA is part of a DNA end-protection system that helps to preserve genome integrity following exposure to ionizing radiation (Harris et al. 2004). On the basis of this interpretation, we predict that DdrB may also act to protect the genome of the irradiated cell possibly by preventing degradation of genomic DNA postirradiation by an alternate mechanism.
The pprA gene product:
Narumi et al. described PprA as a DNA repair protein when they deposited the coding sequence in 1997 (NCBI accession no. O32504), but with the exception of anecdotal reference to this peptide in two recent articles (Gao et al. 2003; Hua et al. 2003), there has not been a published description of the protein's activity or effect on the DNA damage tolerance of D. radiodurans. A search for homologs of PprA (DRA0346p) using its predicted amino acid sequence failed to identify any significant matches between this protein and any other listed in current databases or to any described sequence motif. In this study, we demonstrate that the pprA gene product is required for ionizing radiation resistance (Figure 2), and that it is epistatic to RecA (Figure 6). Also, we have determined that PprA is not necessary for natural transformation. The ΔpprA strain TNK105 takes up and integrates genetic markers as efficiently as R1, indicating that this protein functions to facilitate DNA repair and not homologous recombination per se. This observation suggests that PprA modifies the process of homologous recombination in a way that enhances the cell's capacity to deal with ionizing-radiation-induced DNA damage. The nature of this enhancement is unknown, but any mechanism that improves the efficiency of recombinational repair would also improve cell survival. PprA could, for example, protect the substrates for recombination (such as free ends generated by DNA double-strand breaks) from exonuclease digestion. In many species, including D. radiodurans, exposed free ends serve as substrates for intracellular exonucleases that begin degrading DNA from these sites. If it is genomic DNA that is being degraded, the loss of genetic information will ultimately lead to cell death. D. radiodurans appears to have the ability to control DNA degradation postirradiation by synthesizing proteins that prevent extensive digestion of the genome (Dean et al. 1966; Lett et al. 1967; Vukovic-Nagy et al. 1974). Perhaps PprA is one of these proteins.
A RecA-independent contribution to the radioresistance of D. radiodurans:
As illustrated in Figures 2 and 6, a recA strain of D. radiodurans is very sensitive to ionizing radiation when compared with the R1 parent. Clearly, RecA-mediated homologous recombination is necessary for ionizing radiation resistance in this species, but the ΔrecA strain TNK106 can be made more sensitive by also deleting ddrA, ddrB, ddrC, or ddrD. This result provides genetic evidence that D. radiodurans utilizes RecA-independent mechanisms that enhance cellular resistance to ionizing-radiation-induced damage. The significance of these mechanisms is apparent when one compares the sensitivity of the TNK117 ΔddrA ΔddrB with the other strains evaluated in this study. At the highest dose tested this strain was as sensitive as the ΔpprA strain TNK105 (Figure 4). The only strains showing higher sensitivity were the double mutants that combined ΔddrA or ΔddrB with ΔpprA (Figure 3) and those strains that lack recA. Thus, it appears that at least two recA-independent processes contribute to ionizing radiation resistance in D. radiodurans, one dependent on DdrA and the other dependent on DdrB.
Evidence for recA-independent repair of DNA double-strand breaks has been provided previously. Daly and Minton (1996) reported that as much as 30% of the genome of D. radiodurans could be reassembled by a mechanism that did not require RecA. These authors argued that single-strand annealing was responsible for the limited restitution observed, in part because, despite the damage caused by high-dose irradiation, D. radiodurans cultures recover without evidence of mutation. As opposed to other known RecA-independent processes, which can repair double-strand DNA breaks, such as nonhomologous end joining (Lees-Miller and Meek 2003), single-strand annealing is a largely error-free process. However, direct evidence for single-strand annealing participating in the repair of DNA damage in D. radiodurans has never been provided.
The radioresistance of D. radiodurans:
The results presented here indicate that part of the explanation for the radioresistance of D. radiodurans is found among the proteins of unknown function encoded by this species. As discussed above, this result was not unexpected. Analysis of the D. radiodurans genome sequence established that most of the DNA repair proteins, found in E. coli, are also present in D. radiodurans (White et al. 1999). Since E. coli is far more sensitive to ionizing radiation than D. radiodurans, it has been considered likely that D. radiodurans expresses unique mechanisms for dealing with ionizing-radiation-induced DNA damage. Here, we have provided evidence that at least five of these hypothetical proteins contribute to the radioresistance of this species, and in the process we have taken the preliminary steps toward defining what appears to be a complex interaction between RecA-dependent and RecA-independent mechanisms of DNA repair in this species.
Acknowledgments
The authors gratefully acknowledge Michael M. Cox for helpful discussions and for critically reading the manuscript and we appreciate the technical assistance of L. Alice Simmons. This work was supported by the U.S. Department of Energy grants DEFG0201ER63151 to J.R.B., S.N.P., and J.A.E. The studies of desiccation were supported by a Multidisciplinary University Research Initiative subcontract award (N000014-01-1-0852) to J.R.B. from the Naval Research Laboratory.
References
- Agostini, H. J., J. D. Carroll and K. W. Minton, 1996. Identification and characterization of uvrA, a DNA repair gene of Deinococcus radiodurans. J. Bacteriol. 178: 6759–6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almiron, M., A. J. Link, D. Furlong and R. Kolter, 1992. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 6: 2646–2654. [DOI] [PubMed] [Google Scholar]
- Badarinarayana, V., P. W. Estep, III, J. Shendure, J. Edwards, S. Tavazoie et al., 2001. Selection analyses of insertional mutants using subgenic-resolution arrays. Nat. Biotechnol. 19: 1060–1065. [DOI] [PubMed] [Google Scholar]
- Battista, J. R., 1997. Against all odds: the survival strategies of Deinococcus radiodurans. Annu. Rev. Microbiol. 51: 203–224. [DOI] [PubMed] [Google Scholar]
- Battista, J. R., 2000. Radiation resistance: the fragments that remain. Curr. Biol. 10: R204–R205. [DOI] [PubMed] [Google Scholar]
- Battista, J. R., A. M. Earl and M. J. Park, 1999. Why is Deinococcus radiodurans so resistant to ionizing radiation? Trends Microbiol. 7: 362–365. [DOI] [PubMed] [Google Scholar]
- Daly, M. J., and K. W. Minton, 1995. Interchromosomal recombination in the extremely radioresistant bacterium Deinococcus radiodurans. J. Bacteriol. 177: 5495–5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly, M. J., and K. W. Minton, 1996. An alternative pathway of recombination of chromosomal fragments precedes recA-dependent recombination in the radioresistant bacterium Deinococcus radiodurans. J. Bacteriol. 178: 4461–4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean, C. J., P. Feldschreiber and J. T. Lett, 1966. Repair of x-ray damage to the deoxyribonucleic acid in Micrococcus radiodurans. Nature 209: 49–52. [DOI] [PubMed] [Google Scholar]
- Dean, C. J., M. G. Ormerod, R. W. Serianni and P. Alexander, 1969. DNA strand breakage in cells irradiated with X-rays. Nature 222: 1042–1044. [DOI] [PubMed] [Google Scholar]
- Dose, K., A. Bieger-Dose, M. Labusch and M. Gill, 1992. Survival in extreme dryness and DNA-single strand breaks. Adv. Space Res. 12: 221–229. [DOI] [PubMed] [Google Scholar]
- Earl, A. M., M. M. Mohundro, I. S. Mian and J. R. Battista, 2002. The IrrE protein of Deinococcus radiodurans R1 is a novel regulator of recA expression. J. Bacteriol. 184: 6216–6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, J. S., and J. R. Battista, 2003. Using DNA microarray data to understand the ionizing radiation resistance of Deinococcus radiodurans. Trends Biotechnol. 21: 381–382. [DOI] [PubMed] [Google Scholar]
- Eisen, J. A., and P. C. Hanawalt, 1999. A phylogenomic study of DNA repair genes, proteins, and processes. Mutat. Res. 435: 171–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funayama, T., I. Narumi, M. Kikuchi, S. Kitayama, H. Watanabe et al., 1999. Identification and disruption analysis of the recN gene in the extremely radioresistant bacterium Deinococcus radiodurans. Mutat. Res. 435: 151–161. [DOI] [PubMed] [Google Scholar]
- Gao, G., B. Tian, L. Liu, D. Sheng, B. Shen et al., 2003. Expression of Deinococcus radiodurans PprI enhances the radioresistance of Escherichia coli. DNA Repair 2: 1419–1427. [DOI] [PubMed] [Google Scholar]
- Gutman, P. D., P. Fuchs, L. Ouyang and K. W. Minton, 1993. Identification, sequencing, and targeted mutagenesis of a DNA polymerase gene required for the extreme radioresistance of Deinococcus radiodurans. J. Bacteriol. 175: 3581–3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutman, P. D., J. D. Carroll, C. I. Masters and K. W. Minton, 1994. Sequencing, targeted mutagenesis and expression of a recA gene required for the extreme radioresistance of Deinococcus radiodurans. Gene 141: 31–37. [DOI] [PubMed] [Google Scholar]
- Harris, D. R., M. Tanaka, S. V. Saveliev, E. Jolivet, A. M. Earl et al., 2004. Preserving genome integrity: the DdrA protein of Deinococcus radiodurans R1. PLOS Biol. 2: (in press). [DOI] [PMC free article] [PubMed]
- Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen and L. R. Pease, 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77: 61–68. [DOI] [PubMed] [Google Scholar]
- Hua, Y., I. Narumi, G. Gao, B. Tian, K. Satoh et al., 2003. PprI: a general switch responsible for extreme radioresistance of Deinococcus radiodurans. Biochem. Biophys. Res. Commun. 306: 354–360. [DOI] [PubMed] [Google Scholar]
- Iyer, L. M., E. V. Koonin and L. Aravind, 2002. Classification and evolutionary history of the single-strand annealing proteins, RecT, Redβ, ERF and RAD52. BMC Genomics 3: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitayama, S., and A. Matsuyama, 1971. Double-strand scissions in DNA of gamma-irradiated Micrococcus radiodurans and their repair during postirradiation incubation. Agric. Biol. Chem. 35: 644–652. [Google Scholar]
- Kitayama, S., M. Kohoroku, A. Takagi and H. Itoh, 1997. Mutation of D. radiodurans in a gene homologous to ruvB of E. coli. Mutat. Res. 385: 151–157. [DOI] [PubMed] [Google Scholar]
- Lees-Miller, S. P., and K. Meek, 2003. Repair of DNA double strand breaks by non-homologous end joining. Biochimie 85: 1161–1173. [DOI] [PubMed] [Google Scholar]
- Lett, J. T., P. Feldschreiber, J. G. Little, K. Steele and C. J. Dean, 1967. The repair of x-ray damage to the deoxyribonucleic acid in Micrococcus radiodurans: a study of the excision process. Proc. R. Soc. Lond. Ser. B Biol. Sci. 167: 184–201. [DOI] [PubMed] [Google Scholar]
- Lipton, M. S., L. Pasa-Tolic, G. A. Anderson, D. J. Anderson, D. L. Auberry et al., 2002. Global analysis of the Deinococcus radiodurans proteome by using accurate mass tags. Proc. Natl. Acad. Sci. USA 99: 11049–11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., J. Zhou, M. V. Omelchenko, A. S. Beliaev, A. Venkateswaran et al., 2003. Transcriptome dynamics of Deinococcus radiodurans recovering from ionizing radiation. Proc. Natl. Acad. Sci. USA 100: 4191–4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova, K. S., L. Aravind, Y. I. Wolf, R. L. Tatusov, K. W. Minton et al., 2001. Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol. Mol. Biol. Rev. 65: 44–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, A., and R. Kolter, 1997. Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J. Bacteriol. 179: 5188–5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattimore, V., and J. R. Battista, 1996. Radioresistance of Deinococcus radiodurans: functions necessary to survive ionizing radiation are also necessary to survive prolonged desiccation. J. Bacteriol. 178: 633–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattimore, V., K. S. Udupa, G. A. Berne and J. R. Battista, 1995. Genetic characterization of forty ionizing radiation-sensitive strains of Deinococcus radiodurans: linkage information from transformation. J. Bacteriol. 177: 5232–5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel, R., and M. Gellert, 1983. Regulation of the genes for E. coli DNA gyrase: homeostatic control of DNA supercoiling. Cell 34: 105–113. [DOI] [PubMed] [Google Scholar]
- Moskovitz, J., M. A. Rahman, J. Strassman, S. O. Yancey, S. R. Kushner et al., 1995. Escherichia coli peptide methionine sulfoxide reductase gene: regulation of expression and role in protecting against oxidative damage. J. Bacteriol. 177: 502–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narumi, I., 2003. Unlocking radiation resistance mechanisms: still a long way to go. Trends Microbiol. 11: 422–425. [DOI] [PubMed] [Google Scholar]
- Narumi, I., K. Satoh, M. Kikuchi, T. Funayama, S. Kitayama et al., 1999. Molecular analysis of the Deinococcus radiodurans recA locus and identification of a mutation site in a DNA repair-deficient mutant, rec30. Mutat. Res. 435: 233–243. [DOI] [PubMed] [Google Scholar]
- Peterson, S., R. T. Cline, H. Tettelin, V. Sharov and D. A. Morrison, 2000. Gene expression analysis of the Streptococcus pneumoniae competence regulons by use of DNA microarrays. J. Bacteriol. 182: 6192–6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan, B., H. Nakano, M. Tanaka, J. A. Mills, J. A. DeVito et al., 2004. Cysteinyl-tRNA(Cys) formation in Methanocaldococcus jannaschii: the mechanism is still unknown. J. Bacteriol. 186: 8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh, K., I. Narumi, M. Kikuchi, S. Kitayama, T. Yanagisawa et al., 2002. Characterization of RecA424 and RecA670 proteins from Deinococcus radiodurans. J. Biochem. 131: 121–129. [DOI] [PubMed] [Google Scholar]
- Turner, R. J., J. H. Weiner and D. E. Taylor, 1999. Tellurite-mediated thiol oxidation in Escherichia coli. Microbiology 145(9): 2549–2557. [DOI] [PubMed] [Google Scholar]
- Udupa, K. S., P. A. O'Cain, V. Mattimore and J. R. Battista, 1994. Novel ionizing radiation-sensitive mutants of Deinococcus radiodurans. J. Bacteriol. 176: 7439–7446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukovic-Nagy, B., B. W. Fox and M. Fox, 1974. The release of a deoxyribonucleic acid fragment after x-irradiation of Micrococcus radiodurans. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 25: 329–337. [DOI] [PubMed] [Google Scholar]
- Ward, J. F., 1975. Molecular mechanisms of radiation-induced damage to nucleic acids. Adv. Radiat. Biol. 5: 181–239. [Google Scholar]
- White, O., J. A. Eisen, J. F. Heidelberg, E. K. Hickey, J. D. Peterson et al., 1999. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286: 1571–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson et al., 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906. [DOI] [PubMed] [Google Scholar]