Abstract
Interspecies hybrids between distinct species of the genus Xiphophorus are often used in varied research investigations to identify genomic regions associated with the inheritance of complex traits. There are 24 described Xiphophorus species and a greater number of pedigreed strains; thus, the number of potential interspecies hybrid cross combinations is quite large. Previously, select Xiphophorus experimental crosses have been shown to exhibit differing characteristics between parental species and among the hybrid fishes derived from crossing them, such as widely differing susceptibilities to chemical or physical agents. For instance, genomic regions harboring tumor suppressor and oncogenes have been identified via linkage association of these loci with a small set of established genetic markers. The power of this experimental strategy is related to the number of genetic markers available in the Xiphophorus interspecies cross of interest. Thus, we have undertaken the task of expanding the suite of easily scored markers by characterization of Xiphophorus microsatellite sequences. Using a cross between Xiphophorus maculatus and X. andersi, we report a linkage map predominantly composed of microsatellite markers. All 24 acrocentric chromosome sets of Xiphophorus are represented in the assembled linkage map with an average intergenomic distance of 7.5 cM. Since both male and female F1 hybrids were used to produce backcross progeny, these recombination rates were compared between “male” and “female” maps. Although several genomic regions exhibit differences in map length, male- and female-derived maps are similar. Thus Xiphophorus, in contrast to zebrafish, Danio rerio, and several other vertebrate species, does not show sex-specific differences in recombination. The microsatellite markers we report can be easily adapted to any Xiphophorus interspecies and some intraspecies crosses, and thus provide a means to directly compare results derived from independent experiments.
XIPHOPHORUS is a fish genus comprising small fish derived from freshwater habitats of Mexico, Guatemala, Belize, and Honduras. The genus is currently composed of 24 distinct taxa, some of which are sympatric. It was realized several decades ago (Kosswig 1927; Gordon 1931), that Xiphophorus fishes could be readily hybridized in the aquarium setting by simple cohabitation of divergent species or by employing artificial insemination techniques (Clark 1950). Such hybrids are usually fertile, and subsequent generations of hybrid fish are easily produced. However, individual fish can show abnormal phenotypes, and some fishes develop neoplasms spontaneously (Walter and Kazianis 2001). Such neoplasms appear in parental strains only rarely or are not observed at all (Schartl et al. 1995).
Xiphophorus hybrid models have been established to study the genetics underlying cancer development and some of these animal models require treatment with chemical and physical agents. Hybrid fish show an increased susceptibility to cancer development as a result of exposure to X-ray or ultraviolet light radiation or exposure to various chemical agents (Schwab et al. 1978; Setlow et al. 1989; Anders et al. 1991; Nairn et al. 2001). Inclusion of genetic mapping strategies in backcross hybrid experimental cohorts has the great potential to identify genetic regions and loci implicated in differential susceptibilities and tumor phenotypes. Such studies were initiated in the 1970s (Siciliano and Wright 1976; Ahuja et al. 1977) and the amount of available markers increased with technological advancements in molecular biology (Kazianis et al. 1996; Morizot et al. 1998). These and other mapping studies helped in the identification of a sex-linked oncogene referred to as Xmrk (Wittbrodt et al. 1989) in linkage group (LG) 24 and a melanoma tumor suppressor candidate, CDKN2X in LG 5 (Kazianis et al. 1998, 1999, 2000).
In addition to cancer genetics, Xiphophorus fishes and hybrids between Xiphophorus species are actively studied in many diverse disciplines including evolution (Meyer et al. 1994a,b; Meyer 1997; Morizot 2000), sexual determination, endocrinology, ethology and behavioral ecology (Rosenthal and Evans 1998; Hoefler and Morris 1999; Beaugrand and Goulet 2000; Trainor and Basolo 2000), toxicology (De Wolf et al. 1993), parasitology (Schmahl et al. 1996; Dove 2000), and immunology (McConnell et al. 1998).
Development of sequence-tagged site (STS) markers that could be utilized in virtually all Xiphophorus hybrid crosses and may be easily applied to many areas of inquiry would enhance the genetic power of this model system. In addition, once one determines the map position of STS markers, one may optimize resources by performing genome-wide scans, with specific marker panels. For these reasons we initiated microsatellite sequence marker development using Xiphophorus interspecies crosses. Herein, we detail the production of a Xiphophorus microsatellite map in a new interspecies hybrid cross (Xiphophorus maculatus Jp 163 B backcrossed with X. andersi) that has been developed with markers mainly scored using simple agarose-gel technology. We feel these markers establish a genetic tool allowing most laboratories to use the powerful interspecies hybrid genetics afforded by the Xiphophorus model system. For additional information on the Xiphophorus genetic system see http://www.xiphophorus.org.
MATERIALS AND METHODS
Experimental animals and sample processing:
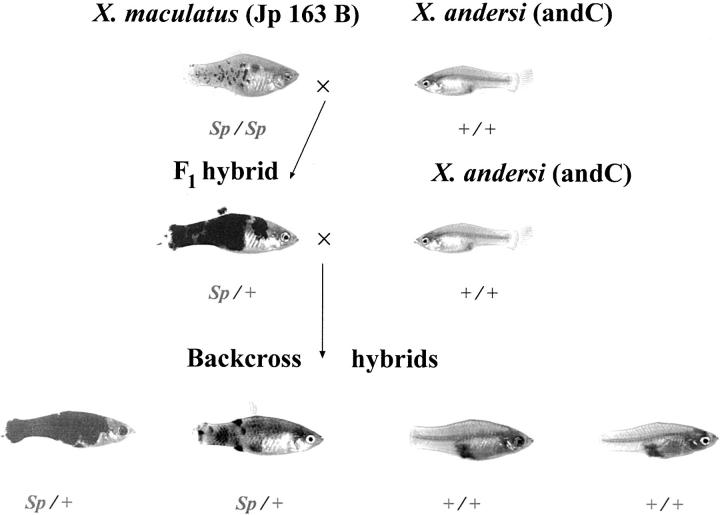
Parental stocks and hybrids used in this project were derived from the Xiphophorus Genetic Stock Center (Texas State University, San Marcos, TX). First-generation backcross hybrids were created between the southern platyfish, X. maculatus (Rio Jamapa, strain Jp 163 B) and X. andersi (Rio Atoyac, strain “andC”) as the recurrent parent. Backcross hybrids (n = 364) were generated using both F1 hybrid female parents (n = 160 BC1 hybrids derived from 14 pedigrees) and male F1 progenitors (n = 204 BC1 hybrids from 16 pedigrees). A total of 364 individual backcross hybrid animals were used in the analysis, although not all individuals were typed for all markers. The majority of the recombination data used to produce the linkage map is derived using DNAs from 160 backcross hybrids, represented in two 96-well microtiter plates (with appropriate parental samples and negative controls). Backcross hybrids were phenotypically scored for macromelanophore [Spot sided (Sp) or +] and micromelanophore (Dot or +) patterning, sex (M or F), and standard length. Fishes were also scored for pigmentation phenotypes derived from phenotypic enhancement of their macromelanophore patterns and placed into one of five categories: heavy, intermediate/heavy, intermediate, intermediate/light, and light. Examples of individual phenotypes corresponding to heavily pigmented animals or lightly pigmented animals are provided in Figure 1.
Figure 1.—
Hybrid cross between X. maculatus and X. andersi. F1 hybrid shows enhancement of the Sp pigment pattern phenotype. Depicted first-generation backcross hybrids show heavy and lightly pigmented offspring with Sp.
Of the 364 BC1 hybrids utilized for linkage mapping, 187 were exposed to N-methyl-N-nitrosourea (MNU) as previously described (Kazianis et al. 2001). All hybrids were killed at 1 year of age except for individuals that had developed exophytic or obviously anaplastic growths, necessitating expedited killing. Such lesions were excised, fixed in 10% formalin, and paraffin-embedded, and 4- to 5-μm-thick sections were stained with hematoxylin and eosin. Histological classification of melanomas and other neoplasms was performed as previously reported (Gimenez-Conti et al. 2001).
Genomic DNA was purified using a Puregene extraction kit (Gentra Systems, Research Triangle Park, NC), suspended in 1× TE (pH 8.0), and quantitated using a BioTek FL600X fluorometer with picogreen (Molecular Probes, Eugene, OR). Working stocks were made at a concentration of 5.0 ng/μl.
Construction of microsatellite libraries:
Four microsatellite-enriched subgenomic libraries were created with the following repeats: CA, ATG, TACA, and TAGA (Genetic Information Services, Chatsworth, CA). These libraries were constructed by inserting X. maculatus (strain Jp 163 A) HindIII-restricted genomic DNA of between 350 and 700 bp in length into pUC19 plasmid vectors. These plasmids were transformed into Escherichia coli strain DH5α. A total of 2394 independent bacterial clones were purified and isolated, potentially harboring plasmids with inserts containing tandem repeats of microsatellite DNA sequences. Each of these clones underwent plasmid minipreparation and DNA isolation, in duplicate. One of these duplicates and the bacterial strain corresponding to the plasmid were cataloged and frozen at −80°. The duplicate plasmid DNA was further purified and subjected to nucleotide sequencing. A total of 2195 plasmids were sequenced in a bidirectional manner using universal forward and reverse primers. From these, nucleotide sequence data were obtained for 2074 plasmid clones. Analyses of these sequences revealed 728 unique (nonduplicate) sequences that contained microsatellite repeats in regions amenable to primer design (i.e., not too close to one end of the sequence). From the 728 unique sequences, 371 oligonucleotide primer pairs were designed and synthesized. These matched primer sets were all designed to anneal at 55° in the polymerase chain reaction (PCR).
PCR and agarose-gel screening of microsatellites:
All primer sets were tested first on X. maculatus (Jp 163 A and Jp 163 B) and X. andersi (andC and andB) parental DNAs (one individual from each highly inbred strain) to enable robust and discrete amplification using PCR. After this, each primer pair was used to establish polymorphic content with DNAs from 10 Xiphophorus species/strains commonly used for interspecies crossing (see Figure 2 legend). All primers were also tested for their ability to amplify PCR products using DNA obtained from Poecilia (strains PVMSK-D and QUL89-A; Sulfur Spring, Venezuela and Quare River, Trinidad, respectively; collaboratively obtained from Felix Breden, Simon Fraser University), Danio (ABC strain, obtained from the Zebrafish Genetic Stock Center), and Oryzias (Carolina Biological). The exact samples used are indicated in the Figure 2 legend and represent the most commonly used parental strains, as well as the aforementioned non-Xiphophorus fishes. Once amplification conditions were optimized, backcross hybrid panels of DNAs were amplified and electrophoresed; a typical agarose-gel image is provided (Figure 3). Each 20-μl reaction contained 1.0 unit of Taq polymerase (Invitrogen, Carlsbad, CA), 1.6× reaction buffer, 0.1 μm of each oligonucleotide, 0.25 mm dNTP mixture, 25 ng of genomic DNA, and final MgCl2 concentrations varying from 1.5 to 3.0 mm.
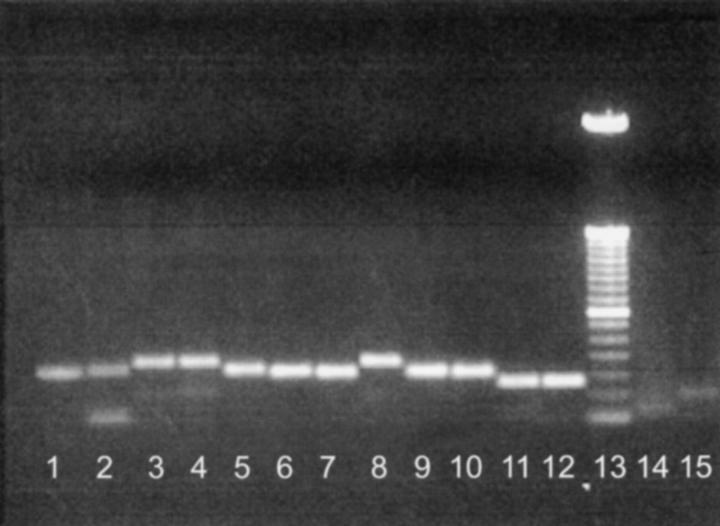
Figure 2.—
Screening of microsatellite locus Msa125 using parental DNAs. Lane 1, X. andersi (andB strain); lane 2, X. couchianus; lane 3, X. maculatus (Jp 163 A); lane 4, X. maculatus (Jp 163 B); lane 5, X. andersi (andC); lane 6, X. helleri (Sara); lane 7, X. variatus (Zarco); lane 8, X. nezahualcoyotl (Ocampo); lane 9, X. helleri (Lance); lane 10, X. variatus (Huich.); lane 11, P. reticulata (PVMSK-D); lane 12, P. reticulata (QUL89-A); lane 13, 50-bp DNA ladder (GIBCO BRL, Gaithersburg, MD); lane 14, O. latipes (strain from Carolina Biological, maintained by the Xiphophorus Genetic Stock Center); and lane 15, D. rerio (AB strain, Zebrafish Genetic Stock Center). Msa125 was determined to be 184 bp for X. maculatus strains Jp 163A and Jp 163 B.
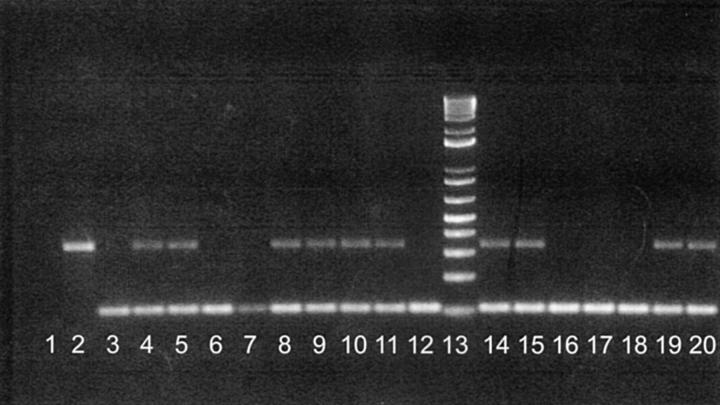
Figure 3.—
Screening of microsat locus Msc018 using backcross hybrid DNAs. Lane 1, negative DNA template (control); lane 2, X. maculatus (Jp 163 A); lane 3, X.andersi (andC); lane 4, F1 hybrid between X. maculatus and X. andersi; lanes 5–12, series of BC1 hybrids; lane 13, 50-bp ladder (GIBCO BRL); and lanes 14–20, series of BC1 hybrids. Msc018 was determined to be 320 bp for X. maculatus strains Jp 163A and Jp 163 B.
In general, the following standard thermal reaction conditions were used in our PCR amplifications: 94° for 5 min, denaturation at 94° for 15 sec, 55° for 30 sec, 72° for 1 min (40 cycles), followed by 72° for 7 min and a 4° hold. Thermal cyclers from Hybaid (PCR Express) and Applied Biosystems (ABI, Foster City, CA; models 2700 and 9700) were utilized and yielded highly similar results. Amplified products were electrophoresed on 2.0% agarose gels (1× TAE buffer; 0.025 μg/ml ethidium bromide). The conditions employed for each microsatellite can be obtained from http://www.xiphophorus.org/microsats/microsat.htm.
Screening using fluorescent primers:
In certain cases, agarose gels did not provide adequate resolution to distinguish between X. andersi and X. maculatus amplification products, although slight size shifts were observed in amplicons from the two taxa. In these instances, new primers were synthesized incorporating fluorescent dyes at their 5′ ends, specifically using either 5-Hex and 5,6-Fam labels. In such cases, the parental screens were repeated with all studied taxa using conditions optimized for downstream analyses using an ABI 377 automated sequencing apparatus. Each 10-μl reaction contained 0.25 units of Taq polymerase (Invitrogen), 1.0× reaction buffer, 0.2 μm of each oligonucleotide, 0.1 mm dNTP mixture, 5 ng of genomic DNA, and final MgCl2 concentration at 2.5 mm. Thermal reaction conditions were as for the PCR amplifications indicated above except that the final 72° elongation step was extended from 7 to 30 min (to reduce heterogeneity in amplicon sizes). Amplification products derived from parental DNAs were diluted 1:20 (with H2O), and 1.0 μl was used along with 3.0 μl of loading dye, which included a GeneScan 500XL (ABI) size standard (0.05 μl/reaction) and deionized formamide (64% final concentration). Once amplimer sizes had been determined, those with appropriately different sizes were labeled with distinct fluorescent dyes and run together (“pool-plexing”) to increase genotyping throughput. Denaturing acrylamide gels (0.2 mm thickness) were prepared with 5.0% Long Ranger (Cabrex, East Rutherford, NJ), 36.0% urea, 0.05% ammonium persulfate, 35 μl TEMED, and 1× TBE final concentration. Samples were denatured at 96° for 5 min, quenched on ice, and then loaded and electrophoresed for 2.5 hr (3000 V, 60 mA) on an ABI377 (96-well conversion; Applied Biosystems, Foster City, CA). Data were collected with GeneScan software and extracted with Gel Processor, and genotypes were tallied with Genotyper software packages (ABI).
Isozyme/allozyme genotyping:
Tissue preparation, vertical starch-gel electrophoresis, histochemical staining, locus abbreviations, and genotypic assignment of allozyme phenotypes followed previously published conventions and protocols (Morizot and Schmidt 1990). In all, 29 total isozymic polymorphisms were informative and were used as known anchor loci for microsatellite genotyping studies detailed herein.
Other PCR-based polymorphisms:
Eleven cloned genes from Xiphophorus were also genotyped in the backcross hybrid cohort and used in the construction of the reported linkage map. These are as follows: UNG (Uracil DNA-N-Glycosylase), TP53 (Tumor Protein p53), LIG1 (DNA Ligase 1), ERCC2/XPD (Excision-Repair Complementation group C-2), FoxO5 (member of the FoxO subfamily of Forkhead transcription factors), CDKN2X (Cyclin-Dependent Kinase Inhibitor-2 from Xiphophorus), RAB27 (RAS-Associated Protein RAB27 homolog), POLB (DNA Polymerase Beta), NF1 (Neurofibromatosis Type I), FEN1 (Flap-structure-specific Endonuclease-1), and APE (Apurinic-Endonuclease). Generally, these loci were mapped by either establishing amplicon size differences that were discernible after agarose-gel electrophoresis or adding a restriction endonuclease step to generate polymorphisms between the species as viewed after agarose-gel electrophoresis. A general list of these conditions and primer sequences is provided in Table 1.
TABLE 1.
Selective gene mapping strategy in Xiphophorus
| Gene | Forward primer | Reverse primer | Polymorphism type |
Restriction endonuclease |
|---|---|---|---|---|
| APE | aggcaagaagacagatgatgggtc | tagaagtttgggaactcggctgtg | PCR/restriction | HaeIII |
| CDKN2X | taaaccagaacaactaagtgg | ctgtattgctcttcgtcca | PCR/size | None |
| ERCC2 | agaacaacgtagacacgctgc | ggcagcatccgtctcctt | PCR/restriction | TaqI |
| FEN1 | tagcaagtgggtaaaccaggctaac | tagatgcacatggaggcatcaatagcaat | PCR/restriction | CfoI |
| FoxO5 | ctctcaccgatgatctactccagtc | gatggaggtgtcaatttgcatggaggatgttc | PCR/restriction | AluI |
| LIG1 | gggcctgatggtgaagactct | tgtgtcgcccacaccgt | PCR/restriction | CfoI |
| NF1 | gcttgctgtggaacaaccagg | tcgcctgccaactgcctt | PCR/restriction | HinfI |
| POLB | gcatccaccattgccaagtatcct | ttccagtttgtaagaactcatcgatcttttctg | PCR/restriction | AluI |
| RAB27 | gcactaaggcagacatcagag | ggttcagagccaggggcccca | PCR/restriction | TaqI |
| TP53 | cttcaggtcaggatttcagtcgc | cccacctccagtcctcttaactct | PCR/restriction | BclI |
| UNG | taggtcaagatccatatcatggtc | gatgctgaaagccgtcga | PCR/size | None |
Map construction:
Genotypic data were entered in Excel (Microsoft, Redmond, WA) in an expanded MAPMAKER (Lander and Green 1987) format. Exported data were analyzed with the aid of MAPMAKER (version 3.0b, DOS version, used in Windows XP), which determined maximum-likelihood map orders. “Framework” loci and map orders were established for each linkage group with alternative orders between them considered >1000 times less likely. Other markers were positioned on the map if alternative positions were 100 times less likely. In cases of tight linkage or insufficient data, loci were placed on the maps using Map Manager QT (Manly and Olson 1999), which was predominantly used to order markers within maps by minimization of double crossovers and map length. This program was also used to create custom filtered and sorted data sets. Graphic map files were generated using MAPMAKER for Macintosh version 2.0 (obtained from S. Tingey, DuPont, Wilmington, DE). Integration of the microsatellite map presented herein with previously published Xiphophorus reference allozyme and gene markers (Morizot et al. 1998) is provided in a supplement to this article at http://www.genetics.org/supplemental/ and is also located at http://www.xiphophorus.org/newmap/map.htm.
RESULTS
Isolation of microsatellite loci:
In total, we isolated and determined the nucleotide sequence of 2074 plasmids (Table 2) from four distinct microsatellite-enriched subgenomic libraries of X. maculatus strain Jp 163 A. These libraries were enriched for sequences having dinucleotide (CA), trinucleotide (ATG), or tetranucleotide (TACA or TAGA) sequences. Analysis of plasmid sequences flanking the microsatellite repeats revealed 728 unique loci that met our criteria for the design of specific oligonucleotide primers (Table 2). A total of 371 oligonucleotide primer pairs were synthesized, and PCR amplification was assessed on parental DNAs that included several species of Xiphophorus and representative DNAs from other aquaria fish species (Poecilia, Danio, and Oryzias).
TABLE 2.
Summation of subgenomic microsatellite-enriched libraries from Xiphophorus
| CA library (%) |
ATG library (%) |
TACA library (%) |
TAGA library (%) |
Total (%) | |
|---|---|---|---|---|---|
| Total clonesa | 850 | 459 | 280 | 485 | 2074 |
| Uniqueb | 418 (49) | 138 (30) | 50 (18) | 122 (25) | 728 (35) |
| Bad sequencec | 49 (6) | 24 (5) | 12 (4) | 40 (8) | 125 (6) |
| No repeatd | 42 (5) | 111 (24) | 88 (31) | 58 (12) | 299 (14) |
| Duplicatee | 204 (24) | 143 (32) | 105 (38) | 222 (46) | 674 (33) |
| Too close to vectorf | 137 (16) | 43 (9) | 25 (9) | 43 (9) | 248 (12) |
The total number of clones that were isolated from the libraries.
The number of clones with unique sequences when compared to all other clones.
Plasmid clones that had been sequenced, but yielded too many ambiguities.
Plasmids that did not show obvious repeats of the desired type (CA, ATG, etc.)
Nonunique clones (see footnote a, above).
Insufficient plasmid-insert nucleotide sequence information available for primer design.
Analysis of the 371 sequences generated in this project was performed using both the Blastn and the Blastx algorithmic approaches (Altschul et al. 1997) against the GenBank, EMBL, DDBJ, and PDB databases. Microsatellite locus Msa094 mapped to a terminal region of LG 13 and showed homology to an independently isolated Triosephosphate Isomerase A from Xiphophorus (Merritt and Quattro 2001). This same linkage group harbors a triosephosphate isomerase polymorphism that has been detected by starch-gel electrophoresis and histochemical staining (locus TPI1; Morizot et al. 1998). This isozyme locus is likely to be represented by the published sequences and the Msa094 marker, although further study is required to be definitive. Similarly, locus Msd049 showed highly significant sequence similarity to both DNA and amino acid sequences derived from the Fundulus Lactate Dehydrogenase-C (LDH-C) locus. Within Xiphophorus, Msd049 as well as isozymic locus LDH1 have been mapped to LG 2, close to the MPI locus (Morizot et al. 1998). It is highly likely that Msd049 is the cloned equivalent of the classically identified LDH1 locus as well.
Several other highly significant matches between sequences within our microsatellite clones and known genes were noted when publicly available nucleic acid and protein databases were searched. Results from these Blastx searches along with the putative homologies are included in the data depository at http://www.xiphophorus.org/Datadep_Supplemental.htm. Although the results are not included in this article, it is noteworthy that numerous Xiphophorus DNA sequences derived from the microsatellite-containing plasmids were extremely similar to nucleotide sequences derived from whole-genome, expressed sequence tag, or other large-scale sequencing efforts from Tetraodon, Danio, Fugu, and Oryzias fish taxa. These sequences have the potential to provide further data regarding syntenic relationships between diverse fish taxa, and thus the microsatellite subgenomic library approach may be informative for genomic comparisons across diverse taxa.
Assessment of polymorphism in Xiphophorus crosses:
To assess the polymorphic content of microsatellite loci, several species/strains of Xiphophorus were examined for PCR amplification characteristics under identical conditions. Figure 2 depicts a typical agarose gel that includes six species (nine strains). Although oligonucleotide primers were designed on the basis of X. maculatus sequences, the vast majority of primer sets also amplified the other Xiphophorus species, under uniform conditions. All four subgenomic libraries, including di- (CA), tri- (ATG), and tetranucleotide (TACA and TAGA), yielded a high degree of polymorphism within the Xiphophorus taxa. As an example, of the 256 microsatellite loci that were mapped in the X. maculatus (Jp 163 B) × X. andersi (andC) cross described in this report, 203 amplify discrete amplicons and would also be informative in the so-called “Gordon-Kosswig” cross (see Walter and Kazianis 2001) between X. maculatus (Jp 163 A) and X. helleri (Sara). To assess the applicability of microsatellite use in different crosses, we mapped 104 of these microsatellite markers in this later cross. In all cases, these tested markers mapped to the same Xiphophorus linkage groups (data not shown); therefore, linkage group designations in this manuscript are in accord with other Xiphophorus crosses and previous publications.
The microsatellite markers also can be utilized in intraspecific crossing schemes as well. For example, within X. helleri, the Sarabia and Lancetilla strains show 65 polymorphisms in 168 (38.7%) agarose-gel scorable amplicons. These 65 markers are at least 20 bp different in size from the nearest marker and could be readily employed for mapping within intraspecific crosses. If one considers polymorphisms while using the ABI377, 35 of 72 (48.6%) assessed markers between these same strains are polymorphic (as defined by a conservative 5-bp resolution). Thus, these microsatellite markers can prove useful in intraspecific in addition to the more commonly studied interspecific crosses.
Within the parental panels, we also included the non-Xiphophorus genera, Poecilia, Oryzias and Danio. Poecilia is closely related to Xiphophorus in phylogenetic studies (Meyer et al. 1994a,b; Breden et al. 1999) and single amplification products were observed for ∼20% of the Xiphophorus microsatellite markers tested (data not shown). Thus, these markers may be of some use in Poecilia crosses and in comparing Poecilia and Xiphophorus gene maps. In contrast, most of the primer sets based on our Xiphophorus sequence failed to amplify discrete amplicons from distantly related fishes, such as Oryzias and Danio.
Map statistics:
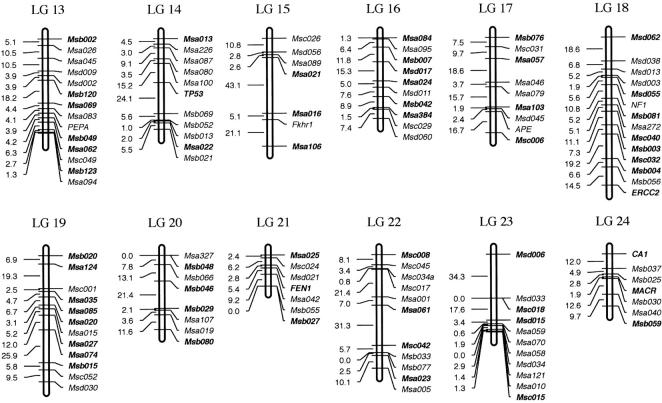
In total, 290 markers were mapped in a backcross between X. maculatus and X. andersi. These included 22 isozyme polymorphisms as detected by starch-gel electrophoretic techniques, 256 microsatellite loci, and 11 distinct gene loci. The macromelanophore locus derived from X. maculatus and coding for the Sp pigment pattern phenotype was additionally mapped as well. In all, 24 distinct linkage groups were established from the cumulative data set corresponding to the 24 sets of acrocentric chromosomes in Xiphophorus (Figure 4; Foerster and Anders 1977). Only 7 of 263 established microsatellite polymorphisms with >50 genotyped animals failed to map to the established linkage groups. Thus, it is believed that the genome is well represented by the 290 total mapped markers within this cross. The defined linkage groups have overlapping markers within each linkage group of previous Xiphophorus gene maps (Morizot et al. 1998), which predominantly utilized a cross between X. maculatus (strains Jp 163 A and Jp 163 B) and X. helleri (Rio Sarabia strain). Thus, a standardized linkage group designation (as a given LG number) corresponds between these crosses and others utilizing Xiphophorus fishes. The LG number designations for microsatellite markers in relation to previously published allozyme and gene markers (Morizot et al. 1998) are provided at http://www.xiphophorus.org/newmap/map.htm, and in supplementary material at http://www.genetics.org/supplemental/.
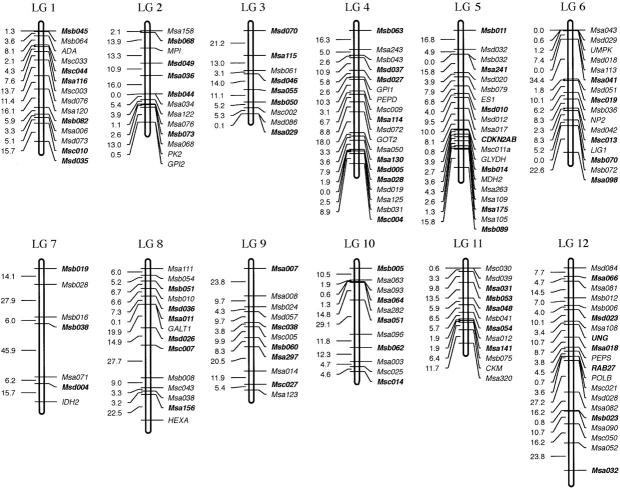
Figure 4.—
(A) Linkage map derived from an interspecific hybrid cross (LGs 1–12). Assigned names of loci and linkage groups are consistent with a previous publication (Morizot et al. 1998). Loci indicated in boldface type within each linkage group represent framework loci (see materials and methods ). (B) Linkage map derived from an interspecific hybrid cross (LGs 13–24).
The overall map depicted in Figure 4 is derived from pedigrees with either F1 male or females and is therefore considered to be “sex averaged.” It corresponds to a map size of 2178.0 cM overall size, using the Kosambi mapping function. The average marker coverage is about one marker per 7.5 cM and the average linkage group size is 90.7 cM, ranging from 26.1 (LG 21) to 178.2 (LG 4).
Previously published work has established the per cell, haploid or diploid DNA content for Xiphophorus fishes. One study established that a haploid cell contains ∼0.755 pg of DNA, corresponding to ∼8.3 × 108 bp (Tiersch et al. 1989). Using 2178.0 cM as the size of the overall map, it is estimated that 1.0 cM in Xiphophorus roughly corresponds to 381.3 kb of DNA. Since recent expansion of genomic techniques has resulted in the cloning of the entire genome in bacterial artificial chromosomes (Froschauer et al. 2002) as well as yeast artificial chromosomes (Kirchner et al. 2001), this estimate of physical distance derived from recombination data should be directly testable in the near future.
Comparison of male vs. female maps:
There can be large differences in recombination rates in linkage maps depending upon the sex of the informative progenitor meioses. For example, in zebrafish, mapping of alleles derived from male meioses results in distinct suppression of recombination when compared to maps derived from females, and this is particularly evident around centromeric regions (Singer et al. 2002). The practical manifestation of this biological phenomenon is that all tested zebrafish linkage groups show smaller overall length when tallying recombination from male meioses vs. female meioses. A “male-derived” zebrafish map showed an overall map size of 942.5 cM as compared to 2582.7 cM derived from female meioses that were informative (Singer et al. 2002). In Oryzias latipes (medaka), differences in recombination frequencies have also been detected on the sex chromosomes (Kondo et al. 2001).
In Xiphophorus fishes, the data set presented within this report represents the first comprehensive direct assessment of male vs. female recombination. In this case, the informative meioses are derived from the F1 progenitors. BC1 hybrids (n = 364) were derived from both male (n = 204) and female (n = 160) F1 parents. It should be noted that these F1 progenitors are also hybrid fishes between two allopatric species, thus the data yield insight as to genome organization in a hybrid context.
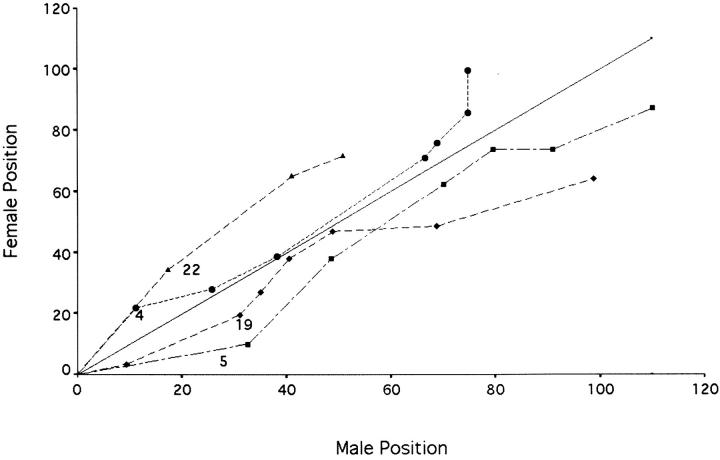
The derived data are partially represented in Table 3. In sum, 11 of the 24 linkage groups show that overall LG length is larger in male maps vs. female maps. When these data are pooled and the entire data set is assessed using framework loci, the male map is just slightly longer than the female map (1567.3 vs. 1506.8 cM, respectively). However, examination of distinct linkage groups reveals that the variability seen in overall size is not uniform over the entire length of the LGs. As an example, Figure 5 reveals four distinct LGs that show either a shorter overall length in the male-derived data set (LGs 5 and 19) vs. female or the opposite effect (LGs 4 and 22). It should be noted that this lack of uniformity between markers was observed most within linkage groups (data not shown). Such a lack of uniformity has been reported in medaka and zebrafish (Kondo et al. 2001; Singer et al. 2002).
TABLE 3.
Comparative recombination rates between male and female meioses
| Linkage group |
No. of framework loci |
Total length (male meioses) |
Total length (female meioses) |
|---|---|---|---|
| 1 | 6 | 55.7 | 57.3 |
| 2 | 5 | 49.4 | 58.8 |
| 3 | 6 | 83.3 | 64.4 |
| 4 | 8 | 74.8 | 99.5 |
| 5 | 7 | 110.1 | 87.2 |
| 6 | 5 | 78.0 | 41.5 |
| 7 | 3 | 68.2 | 75.0 |
| 8 | 6 | 81.6 | 63.7 |
| 9 | 5 | 73.2 | 53.9 |
| 10 | 5 | 67.8 | 56.5 |
| 11 | 6 | 46.8 | 60.3 |
| 12 | 7 | 89.3 | 97.5 |
| 13 | 6 | 66.0 | 54.6 |
| 14 | 3 | 62.0 | 48.6 |
| 15 | 4 | 60.1 | 61.7 |
| 16 | 6 | 60.0 | 47.9 |
| 17 | 4 | 58.9 | 73.1 |
| 18 | 8 | 94.8 | 90.0 |
| 19 | 8 | 98.9 | 64.0 |
| 20 | 4 | 40.9 | 58.6 |
| 21 | 3 | 15.5 | 30.2 |
| 22 | 4 | 50.9 | 71.6 |
| 23 | 4 | 56.2 | 48.2 |
| 24 | 3 | 24.9 | 42.7 |
| Totals | 126 | 1567.3 | 1506.8 |
Figure 5.—
Selective comparison of four male and female LGs. The position of framework loci within the male F1-derived BC1 cohort (x-axis) vs. the position of the identical loci in female F1-derived fish (y-axis). ▴, LG 22; •, LG 4; ♦, LG19; and ▪, LG 5.
DISCUSSION
Interest in using the powerful genetics provided by the Xiphophorus model system to study the hereditary factors associated with multigenetic phenotypes is increasing. The classical genetic power of the Xiphophorus model system for approaching complex scientific problems is attractive due to the extreme species variability and concomitant ability to perform interspecies crosses and backcrosses. Thus, development of genetic tools, such as microsatellite genetic markers that are easily scored among diverse species in the Xiphophorus, is of interest. Also, as the number of DNA and amino acid sequences increases and projects involving the complete genome sequencing continue, sequence-tagged sites from organisms such as Xiphophorus have the potential to yield comparative data regarding putative homology/orthology of interesting loci in other less genetically powerful models. Such endeavors can also potentially yield data with respect to conserved syntenic regions of vertebrate genomes.
One great advantage of the Xiphophorus model system stems from its diversity of hybrid crossing schemes that each have distinct characteristics. For example, within some crosses, melanomas occur in BC1 hybrids at appreciable incidences in the absence of inductive chemical or physical agents (Nairn et al. 2001). In others the background incidence is low and reaches a high percentage only after treatment with MNU (Kazianis et al. 2001). These diverse hybrid models, however, have not been fully exploited since the genomic tools of Xiphophorus have not been fully developed, particularly when compared to tools established in zebrafish, medaka, and the Tetraodontiform fishes including Fugu.
The establishment of the large number of highly polymorphic mapped microsatellites presented within this report should prove invaluable to future linkage mapping/genomic studies in any hybrid crossing schemes employing Xiphophorus. Since these microsatellite loci are highly polymorphic between Xiphophorus taxa, they are particularly useful in such studies. The genetic distance between numerous Xiphophorus taxa results in a high percentage of informative microsatellite primer sets per given cross. In most cases, this variation is readily detectable using agarose-gel/ethidium bromide staining. Other, more efficient modes of genotyping, such as inclusion of mixed fluorescent dyes in “automated” sequence detectors, can greatly expedite such genotyping studies. The availability of highly inbred genetic lines such as those housed at the Xiphophorus Genetic Stock Center, greatly reduces potential problems derived from allelic variation, therefore increasing the applicability of the microsatellite approach. The mapped microsatellite markers that were simultaneously typed in several distinct species and strains will enable researchers to carry targeted genotyping studies or genome-wide scans with much greater efficiency.
Acknowledgments
We sincerely thank the employees of the Xiphophorus Genetic Stock Center for their hard work and dedication in maintaining the inbred parental lines and raising the hybrid fishes used in this study. We appreciate the assistance of Michael D. Rudd, Andrew P. Butler, and Donald C. Morizot (UTMD Anderson Cancer Center) in the mapping of certain gene loci. We thank Felix Breden (Simon Fraser University) for the Poecilia samples and the Zebrafish Genetic Stock Center for the Danio samples. This work was supported by National Institutes of Health grants CA75137, CA76674, RR017072 and the Mitte Foundation.
References
- Ahuja, M. R., M. Schwab and F. Anders, 1977. Tissue-specific esterases in the Xiphophorine fish Platypoecilus maculatus, Xiphophorus helleri, and their hybrid. Biochem. Genet. 15: 601–610. [DOI] [PubMed] [Google Scholar]
- Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang et al., 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders, A., C. Zechel, B. Schlatterer, H. Groger, D. Schmidt et al., 1991. Genetic and molecular approach for breeding and use of laboratory fish for the detection of agents with carcinogenic and/or promoting activity. Bull. Cancer 78: 415–433. [PubMed] [Google Scholar]
- Beaugrand, J. P., and C. Goulet, 2000. Distinguishing kinds of prior dominance and subordination experiences in males of green swordtail fish (Xiphophorus helleri). Behav. Processes 50: 131–142. [DOI] [PubMed] [Google Scholar]
- Breden, F., M. B. Ptacek, M. Rashed, D. Taphorn and C. A. Figueiredo, 1999. Molecular phylogeny of the live-bearing fish genus Poecilia (Cyprinodontiformes: Poeciliidae). Mol. Phylogenet. Evol. 12: 95–104. [DOI] [PubMed] [Google Scholar]
- Clark, E., 1950. A method for artificial insemination in viviparous fishes. Science 112: 722–723. [DOI] [PubMed] [Google Scholar]
- De Wolf, W., W. Seinen and J. L. M. Hermens, 1993. N-acetyltransferase activity in rainbow trout liver and in vitro biotransformation of chlorinated anilines and benzenes in fish. Xenobiotica 23: 1045–1056. [DOI] [PubMed] [Google Scholar]
- Dove, A. D., 2000. Richness patterns in the parasite communities of exotic poeciliid fishes. Parasitology 120: 609–623. [DOI] [PubMed] [Google Scholar]
- Foerster, W., and F. Anders, 1977. Chromosome complements from different subspecies and species of Xiphophorus. Zool. Anz. 198: 167–177. [Google Scholar]
- Froschauer, A., C. Korting, T. Katagiri, T. Aoki, S. Asakawa et al., 2002. Construction and initial analysis of bacterial artificial chromosome (BAC) contigs from the sex-determining region of the platyfish Xiphophorus maculatus. Gene 295: 247–254. [DOI] [PubMed] [Google Scholar]
- Gimenez-Conti, I. B., A. D. Woodhead, J. C. Harshbarger, S. Kazianis, R. B. Setlow et al., 2001. A proposed classification scheme for Xiphophorus melanomas based on histopathologic analyses. Mar. Biotechnol. 3: S100–S106. [DOI] [PubMed] [Google Scholar]
- Gordon, M., 1931. Hereditary basis of melanosis in hybrid fishes. Am. J. Cancer 15: 1495–1523. [Google Scholar]
- Hoefler, C. D., and M. R. Morris, 1999. A technique for the temporary application and augmentation of pigment patterns in fish. Ethology 105: 431–437. [Google Scholar]
- Kazianis, S., D. C. Morizot, B. B. McEntire, R. S. Nairn and R. L. Borowsky, 1996. Genetic mapping in Xiphophorus hybrid fish: assignment of 43 AP- PCR/RAPD and isozyme markers to multipoint linkage groups. Genome Res. 6: 280–289. [DOI] [PubMed] [Google Scholar]
- Kazianis, S., H. Gutbrod, R. S. Nairn, B. B. McEntire, L. Della-Coletta et al., 1998. Localization of a CDKN2 gene in linkage group V of Xiphophorus fishes defines it as a candidate for the DIFF tumor suppressor. Genes Chromosomes Cancer 22: 210–220. [PubMed] [Google Scholar]
- Kazianis, S., D. C. Morizot, L. D. Coletta, D. A. Johnston, B. Woolcock et al., 1999. Comparative structure and characterization of a CDKN2 gene in a Xiphophorus fish melanoma model. Oncogene 18: 5088–5099. [DOI] [PubMed] [Google Scholar]
- Kazianis, S., L. D. Coletta, D. C. Morizot, D. A. Johnston, E. A. Osterndorff et al., 2000. Overexpression of a fish CDKN2 gene in a hereditary melanoma model. Carcinogenesis 21: 599–605. [DOI] [PubMed] [Google Scholar]
- Kazianis, S., I. Gimenez-Conti, R. B. Setlow, A. D. Woodhead, J. C. Harshbarger et al., 2001. MNU induction of neoplasia in a platyfish model. Lab. Invest. 81: 1191–1198. [DOI] [PubMed] [Google Scholar]
- Kirchner, J. M., V. Ivanova, A. Samson, V. N. Noskov, J.-N. Volff et al., 2001. Transformation-associated recombination (TAR) cloning of tumor-inducing Xmrk2 gene from Xiphophorus maculatus. Mar. Biotechnol. 3: S168–S176. [DOI] [PubMed] [Google Scholar]
- Kondo, M., E. Nagao, H. Mitani and A. Shima, 2001. Differences in recombination frequencies during female and male meioses of the sex chromosomes of the medaka, Oryzias latipes. Genet. Res. 78: 23–30. [DOI] [PubMed] [Google Scholar]
- Kosswig, C., 1927. Uber bastarde der teleostier Platypoecilus und Xiphophorus. Z. Indukt. Abstammungs-Vererbungsl. 44: 253. [Google Scholar]
- Lander, E., and P. Green, 1987. Construction of multilocus genetic maps in humans. Proc. Natl. Acad. Sci. USA 84: 2363–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manly, K. F., and J. M. Olson, 1999. Overview of QTL mapping software and introduction to map manager QT. Mamm. Genome 10: 327–334. [DOI] [PubMed] [Google Scholar]
- McConnell, T. J., U. B. Godwin, S. F. Norton, R. S. Nairn, S. Kazianis et al., 1998. Identification and mapping of two divergent, unlinked major histocompatibility complex class II B genes in Xiphophorus fishes. Genetics 149: 1921–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt, T. J., and J. M. Quattro, 2001. Evidence for a period of directional selection following gene duplication in a neurally expressed locus of triosephosphate isomerase. Genetics 159: 689–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, A., 1997. The evolution of sexually selected traits in male swordtail fishes (Xiphophorus: Poeciliidae). Heredity 79: 329–337. [Google Scholar]
- Meyer, A., J. M. Morrissey and M. Schartl, 1994. a Recurrent origin of a sexually selected trait in Xiphophorus fishes inferred from a molecular phylogeny. Nature 368: 539–542. [DOI] [PubMed] [Google Scholar]
- Meyer, A., J. M. Morrissey and M. Schartl, 1994. b Molecular phylogeny of fishes of the genus Xiphophorus suggests repeated evolution of a sexually selected trait. Nature 368: 539–541. [DOI] [PubMed] [Google Scholar]
- Morizot, D., 2000 Reconstructing the genome of the vertebrate ancestor, pp. 43–60 in Genomes, edited by P. Gustafson. Kluwer Academic/Plenum Publishers, New York.
- Morizot, D., and R. Schmidt, 1990 Starch gel electrophoresis and histochemical visualization of proteins, pp. 23–80 in Applications of Electrophoresis and Isoelectric Focusing in Fisheries Management, edited by D. Whitmore. CRC Press, Boca Raton, FL.
- Morizot, D. C., R. S. Nairn, R. B. Walter and S. Kazianis, 1998. The linkage map of Xiphophorus fishes. Inst. Lab. Anim. Res. 39: 237–248. [DOI] [PubMed] [Google Scholar]
- Nairn, R. S., S. Kazianis, L. D. Coletta, D. Trono, A. P. Butler et al., 2001. Genetic analysis of susceptibility to spontaneous and UV-induced carcinogenesis in Xiphophorus hybrid fish. Mar. Biotechnol. 3: S24–S36. [DOI] [PubMed] [Google Scholar]
- Rosenthal, G. G., and C. S. Evans, 1998. Female preference for swords in Xiphophorus helleri reflects a bias for large apparent size. Proc. Natl. Acad. Sci. USA 95: 4431–4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schartl, A., B. Malitschek, S. Kazianis, R. Borowsky and M. Schartl, 1995. Spontaneous melanoma formation in nonhybrid Xiphophorus. Cancer Res. 55: 159–165. [PubMed] [Google Scholar]
- Schmahl, G., H. Schmidt and G. Ritter, 1996. The control of ichthyophthiriasis by a medicated food containing quinine: efficacy tests and ultrastructure investigations. Parasitol. Res. 82: 697–705. [DOI] [PubMed] [Google Scholar]
- Schwab, M., S. Haas, M. R. Ahuja, G. Kollinger, A. Anders et al., 1978. Genetic basis of susceptibility for development of neoplasms following treatment with N-methyl-N-nitrosourea (MNU) or x-rays in the platyfish/swordtail system. Experientia 34: 780–782. [DOI] [PubMed] [Google Scholar]
- Setlow, R. B., A. D. Woodhead and E. Grist, 1989. Animal model for ultraviolet radiation-induced melanoma: platyfish-swordtail hybrid. Proc. Natl. Acad. Sci. USA 86: 8922–8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siciliano, M. J., and D. A. Wright, 1976. Biochemical genetics of the platyfish-swordtail hybrid melanoma system. Prog. Exp. Tumor Res. 20: 398–411. [DOI] [PubMed] [Google Scholar]
- Singer, A., H. Perlman, Y. L. Yan, C. Walker, G. Corley-Smith et al., 2002. Sex-specific recombination rates in zebrafish (Danio rerio). Genetics 160: 649–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiersch, T. R., R. W. Chandler, K. D. Kallman and S. S. Wachtel, 1989. Estimation of nuclear DNA content by flow cytometry in fishes of the genus Xiphophorus. Comp. Biochem. Physiol. [B] 94: 465–468. [DOI] [PubMed] [Google Scholar]
- Trainor, B. C., and A. L. Basolo, 2000. An evaluation of video playback using Xiphophorus helleri. Anim. Behav. 59: 83. [DOI] [PubMed] [Google Scholar]
- Walter, R. B., and S. Kazianis, 2001. Xiphophorus interspecies hybrids as genetic models of induced neoplasia. Inst. Lab. Anim. Res. 42: 299–321. [DOI] [PubMed] [Google Scholar]
- Wittbrodt, J., D. Adam, B. Malitschek, W. Maueler, F. Raulf et al., 1989. Novel putative receptor tyrosine kinase encoded by the melanoma- inducing Tu locus in Xiphophorus. Nature 341: 415–421. [DOI] [PubMed] [Google Scholar]