Abstract
Cln1p and Cln2p are considered as equivalent cyclins on the basis of sequence homology, regulation, and functional studies. Here we describe a functional distinction between the Cln1p and Cln2p cyclins in the control of the G1/S transition. Inactivation of CLN2, but not of CLN1, leads to a larger-than-normal cell size, whereas overexpression of CLN2, but not of CLN1, results in smaller-than-normal cells. Furthermore, mild ectopic expression of CLN2, but not of CLN1, suppresses the lethality of swi4swi6 and cdc28 mutant strains. In the absence of Cln1p, the kinetics of budding, initiation of DNA replication, and activation of the Start-transcription program are not affected; by contrast, loss of Cln2p causes a delay in bud emergence. A primary role for Cln2p but not for Cln1p in budding is reinforced by the observation that only the cln2 mutation is synthetic lethal with a cdc42 mutation, and only the cln2 mutant strain is hypersensitive to latrunculin B. In addition, we found that Cln1p showed a more prominent nuclear staining than Cln2p. Finally, chimeric proteins composed of Cln1p and Cln2p revealed that Cln2p integrity is required for its functional specificity.
FUNCTIONAL redundancy is a recurrent strategy in cell cycle progression. Over the last few years, the characterization of the molecular mechanism of many cell cycle processes has often revealed the existence of related proteins involved in the same molecular function. This is clearly manifested in the case of the primary regulators of the cell cycle: the cyclins and the cyclin-dependent kinases (CDK). Progression through the cell cycle is governed by the sequential activation of the CDKs, which depends on their physical association with a cyclin regulatory subunit. Eukaryotic cells contain multiple cyclin-CDK complexes (Morgan 1997; Roberts 1999). In the case of the yeast Saccharomyces cerevisiae, nine different cyclins (Cln1p–3p and Clb1p–6p) activate the Cdc28p protein, the only yeast CDK with an essential function in cell cycle progression (reviewed in Nasmyth 1996; Andrews and Measday 1998; Mendenhall and Hodge 1998; Miller and Cross 2001a). Although different complexes are required for distinct cell cycle events, an extensive overlap between the functions of diverse cyclin-CDK kinase activities has often been described. A good example of this is the case of the S. cerevisiae G1 cyclins in the control of the initiation of the cell cycle (Start) at the G1/S transition (Cross 1995). There are three G1 cyclins in S. cerevisiae: Cln1p, Cln2p, and Cln3p (Cross 1988; Nash et al. 1988; Hadwiger et al. 1989). They constitute an essential gene family: only one of the CLN genes is necessary to pass through Start since loss of any two CLNs is tolerated by the cell (Richardson et al. 1989; Cross 1990). However, this genetic redundancy must not be interpreted as functional redundancy in vivo, since Cln3p is clearly functionally distinct from the other two G1 cyclins (Tyers et al. 1993). It shares a low sequence similarity with Cln1p and Cln2p and unlike the CLN1 and CLN2 genes, which are periodically transcribed at the G1/S transition (Wittenberg et al. 1990), the expression of CLN3 increases only slightly at the G1/M border (McInerny et al. 1997). Furthermore, Cln3p is a very-low-abundance protein with a much weaker associated kinase activity relative to Cln1p and Cln2p, and both its protein and kinase activity levels are roughly constant throughout the cell cycle, as opposed to the sharp periodic accumulation of both Cln1p and Cln2p at the G1/S transition (Tyers et al. 1993). The characterization of the molecular mechanism constituting Start revealed that the function of Cln3p is to activate a transcription program mediated by the transcriptional factors SBF and MBF (Dirick et al. 1995; Stuart and Wittenberg 1995). In contrast, Cln1p and Cln2p, which are synthesized as a consequence of this transcriptional activation wave, control the trigger of post-Start processes such as budding, spindle pole body duplication, and the initiation of DNA replication. More recently, intrinsic differences in the specificity of action and subcellular localization between Cln3p-Cdc28p and Cln1,2p-Cdc28p have been reported (Levine et al. 1996; Miller and Cross 2000; Edgington and Futcher 2001), reinforcing the distinction between Cln3p and Cln1,2p.
Cln1p and Cln2p are very similar proteins with an identity of 57% at sequence level, which increases to 74% in the N-terminal half containing the cyclin box. As mentioned above, Cln1p and Cln2p abundance and their associated kinase activities fluctuate with the same periodicity, peaking at the G1/S transition (Tyers et al. 1993). Moreover, their protein levels are controlled by the same molecular mechanisms: CLN1 and CLN2 genes are periodically expressed at the G1/S transition by the SBF transcription factor (Nasmyth and Dirick 1991; Ogas et al. 1991), and Cln1p and Cln2p are very unstable proteins degraded by a ubiquitin-dependent pathway involving common components including Grr1p (Barral et al. 1995; Skowyra et al. 1997) and the Cdc53p (Willems et al. 1996) E3 ubiquitin-protein ligase subunits. Numerous studies from different laboratories have shown an extensive functional overlap between these two cyclins. Both Cln1p and Cln2p play a role in the establishment of growth polarization and budding (Benton et al. 1993; Cvrckova and Nasmyth 1993; Lew and Reed 1993); trigger spindle pole body duplication (Haase et al. 2001); phosphorylate the CDK-inhibitors Far1p and Sic1p, inducing their destruction (Peter et al. 1993; Schwob et al. 1994; Schneider et al. 1996; Henchoz et al. 1997); turn off the degradation of the Clb cyclins by inactivating the anaphase-promoting complex (APC)Cdh1 ubiquitin ligase (Amon et al. 1994; Huang et al. 2001); regulate the Ste20p kinase (Oehlen and Cross 1998); and block the induction of the specific gene transcription by pheromone (Oehlen and Cross 1998; Wu et al. 1998). Also, overexpression of Cln1p and Cln2p stimulates pseudohyphal differentiation (Ahn et al. 2001), while mutation of CLN1 and CLN2 causes many common genetic interactions with the checkpoint gene MEC1 (Vallen and Cross 1999) and the RAD27 endonuclease I gene (Vallen and Cross 1995). All these results have led to the idea that Cln1p and Cln2p have very similar functions. In fact, it has been suggested that functions seen so far for only one of Cln1p or Cln2p are likely shared with the other cyclin as well, such as the role of Cln2p in the activation of the Cdc42p small GTPase (Gulli et al. 2000).
We are investigating different aspects of cell cycle control at the G1/S transition. In the course of our experiments we have detected a clear functional difference between Cln1p and Cln2p. On the basis of several observations, we have established that Cln2p plays the primary role in the control of budding, whereas Cln1p is dispensable for the correct progression at this stage of the cycle and is relevant only whenever Cln2p is absent.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions:
The yeast strains used in this study are shown in Table 1. Strains containing the cln2::LEU2 or cln3::URA3 disruption cassettes were obtained by using plasmid pcln2::LEU2 and pcln3::URA3 (gift from S. I. Reed). Strains containing the cln1::kanr disruption cassette were achieved by using a DNA fragment amplified from plasmid pFA6a-kanMX6 (a gift from J. R. Pringle). The JCY357 and JCY404 were derived from W3031-A and JCY220, respectively, by substituting the SWI4 promoter by a DNA fragment amplified from plasmid pFA6a-kanMX6-GAL-HA (gift from J. R. Pringle). Yeast cells were grown on standard yeast extract-peptone-dextrose (YPD) or YPGal rich medium or synthethic dextrose (SD) and SGal minimal media supplemented as required. To fully repress the tetO2 promoter, doxycycline was added to a concentration of 5 μg/ml; partial repression of the tetO2 promoter was achieved by adjusting doxycycline concentration to 0.5 μg/ml (solid medium) or 0.05 μg/ml (liquid medium) as indicated (we have observed that the effect of the doxycycline is apparently more severe in cells growing in liquid than in solid medium; because of that a higher concentration is required when doxycycline is added to solid medium and, in fact, fresh plates must be used). For growth assays, 10-fold serial dilutions in growth medium were prepared from exponentially growing culture (usually 2–5 × 106 cell/ml) of the different strains; 5 μl of each dilution was then spotted onto the appropriate medium and plates were incubated at the indicated temperature.
TABLE 1.
Yeast strains
| W303-1a | MATa ade2 trp1 leu2 his3 ura3 can1 |
|---|---|
| cdc28-13 | MATa ade1 trp1 leu2 his2 ura3 bar1 cdc28-13 |
| JCY296 | cln2::LEU2 in W303-1a |
| MT244a | cln3::URA3 in W303-1a |
| JCY275 | cln1::kanr in W303-1a |
| JCY306 | cln1::kanr cln2::LEU2 in W303-1a |
| JCY223 | cln3::URA3 cln1::kanr in W303-1a |
| JCY317 | cln3::URA3 cln2::LEU2 in W303-1a |
| CG379a | MATα ade5 trp1 leu2 his7-2 ura3 |
| JCY343 | cln1::kanr in CG379 |
| CGcln2 | cln2::LEU2 in CG379 |
| JCY341 | cln3::URA3 in CG379 |
| JCY357 | kanMX6-GAL-HA-SWI4 in W303-1a |
| JCY404 | kanMX6-GAL-HA-SWI4 swi6::TRP1 in W303-1a |
| DJTD2-16Db | MATα trp1 leu2 his4 ura3 gal2 cdc42-1 |
| JCY396 | cln1::kanr in DJTD2-16D |
| JCY453 | cln2::LEU2 ptetO:CLN2 in DJTD2-16D |
Gift from L. H. Johnston.
Gift from J. R. Pringle.
Centromeric plasmids pCM137 and pCM239, containing HA-tagged versions of Cln1p and Cln2p, respectively, and plasmids pCM258 (ptetO:CLN1 henceforth) and pCM250 (ptetO:CLN2 henceforth) overexpressing the CLN1 and CLN2 genes under the control of the tetO2 promoter, were a gift from E. Herrero and M. Aldea. The plasmids containing the chimeric cyclin genes CLN2N-CLN1C and CLN1N-CLN2C were constructed in a two-step process by cloning the appropriate DNA fragments of CLN2 and CLN1 genes obtained by PCR amplification using pCM239 and pCM137 as templates. In the case of CLN2N-CLN1C, an EcoRI-XbaI fragment expanding from −604 to +600 of the CLN2 gene was first cloned in EcoRI-XbaI-cleaved YCplac33; this plasmid was then digested with XbaI-HindIII and a XbaI-HindIII DNA fragment containing a region from +601 to +1635 of the CLN1 gene fused to a sequence coding for three copies of the HA epitope was subsequently cloned. In the case of CLN1N-CLN2C, an EcoRI-XbaI fragment expanding from −682 to +600 of the CLN1 gene was first cloned into EcoRI-XbaI-cleaved YCplac33; this plasmid was then digested with XbaI-SphI and a XbaI-SphI fragment containing a region from +601 to +1638 of the CLN2 gene fused to a sequence coding for three copies of the HA epitope was subsequently cloned. Note that an insertion of two amino acids occurs at the join point as a consequence of the introduction of a XbaI restriction site.
Immunoprecipitation and kinase activity assay:
Approximately 5 × 108 cells were collected, resuspended in lysis buffer [250 mm NaCl, 5 mm EDTA, 0.1% Triton X-100, 50 mm Tris-HCl pH 8, 1 mm PMSF, complete protease inhibitors from Roche Molecular Biochemicals (Indianapolis)] and broken with glass beads. Protein concentration was determined and adjusted if necessary to use the same amount of total protein in each sample. HA-tagged proteins were immunoprecipitated by the addition of 3F10 monoclonal antibody (Roche) and incubation at 4° overnight with agitation and incubation at 4° for 5 hr with protein G-agarose. Samples were successively washed four times with lysis buffer and twice with buffer K (50 mm Tris-HCl pH 8, 10 mm MgCl2, 1 mm DTT). Immunoprecipitated fractions were split in two aliquots. One half was used to determinate the level of Cln-HA and Cdc28p present in the immunoprecipitated fraction by Western analysis using 12C5A (Roche) and anti-Cdc28p (gift from M. Aldea) antibodies. The other half was used in the kinase activity assay. For this assay, samples were supplemented with 5 μl of buffer K, 2 μl of 5 mm ATP, 2 μl of histone H1, 0.5 mg/ml and 1 μl of [γ-32P] ATP (10 μCi) and incubated at 30° for 30 min. Proteins were resolved by SDS-polyacrylamide gel electrophoresis and phosphorylated H1 was revealed by autoradiography. Specific kinase activity was referred to as the intensity of the phosphorylated H1 band in the kinase assay relative to the intensity of the Cdc28p band detected in the Western analysis of the immunoprecipitated samples.
Cell size analysis:
Cell size was analyzed in exponentially growing cells after brief sonication in a particle count and size Analyzer Z2 (Coulter, Hialeah, FL). Graphs are the mobile average of histograms derived from values from at least six independent cultures.
Miscellaneous:
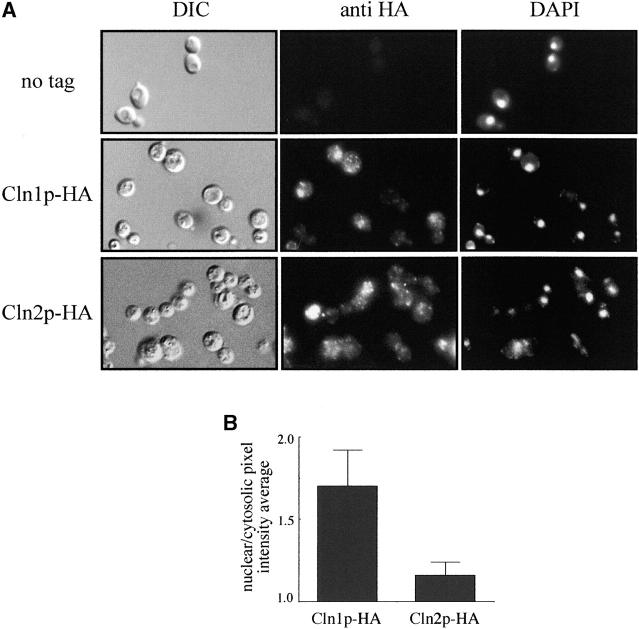
Western blot analysis, Northern analysis, fluorescence-activated cell sorter (FACS) analysis, and indirect immunofluorescence were carried out as described previously (Queralt and Igual 2003). An estimation of the intensity of the cytosolic and nuclear signal in the immunofluorescence assays was achieved by determining the ratio between the average pixel intensity in the cytosolic and that in the nuclear region of the cell (as deduced from DAPI signal); >100 cells from at least three independent assays were analyzed for each strain.
RESULTS
The deletion of CLN2, but not CLN1, causes an increase in cell size:
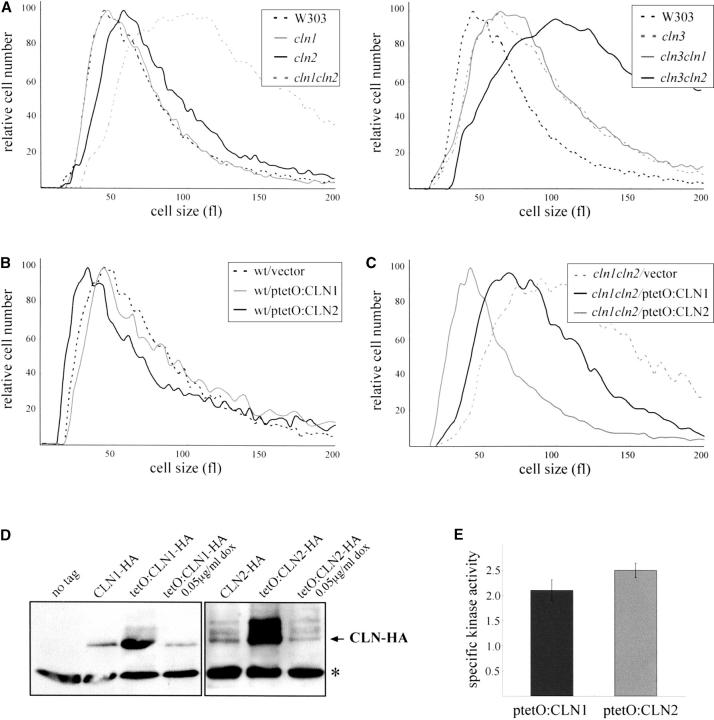
It is well established that the inactivation or overexpression of regulators of Start produces an altered cell size (Cross 1988; Nash et al. 1988, 2001; Hadwiger et al. 1989; Wijnen et al. 2002; Queralt and Igual 2003). We examined the size of the cln1 and cln2 cells in exponentially growing cultures using a Coulter chanellizer. As shown in Figure 1A, deletion of CLN2 caused a dramatic increase in cell size, consistent with previous reports (Tyers et al. 1993). The increase in cell size is similar to that observed in a mutant defective in CLN3. This result was expected considering that Cln2p is a positive regulator of Start downstream processes. Surprisingly, however, the loss of CLN1 had no detectable effect on the cell size. This was an unexpected result, since a previous study reported that cln1 mutant cells were larger than wild type but smaller than cln2 mutant cells (Tyers et al. 1993). We note that a direct comparison with this earlier data is not possible since only the value of the modal cell size and not a complete histogram of cell size distribution is reported by Tyers et al. We also found that the well-established large cell size phenotype of a cln3 mutant strain (Cross 1988; Nash et al. 1988) was unaffected by deletion of CLN1 but deletion of CLN2 caused the double-mutant cells to become even larger (Figure 1A). It was only in the absence of CLN2 that deletion of CLN1 caused an increased cell size. Next, plasmids containing the CLN1 or CLN2 genes expressed under the control of the doxycycline-regulated tetO2 promoter, which leads to a drastic increase in the level of Cln1p or Cln2p present in the cell (see Figure 1D), were introduced in the wild-type strain and the effect of the overexpression of CLN1 and CLN2 on the cell size was investigated. It was known that hyperstable alleles of Cln2p accelerate progression through the G1/S transition, resulting in a smaller-than-normal cell size (Hadwiger et al. 1989). Consistent with this, high dosage of Cln2p induces a reduction in cell size (Figure 1B); however, the overexpression of CLN1 did not produce any detectable alteration in the cell size. These observations suggest that Cln2p, but not Cln1p, is primarily involved in the proper execution of the G1/S transition, whereas Cln1p is required only whenever Cln2p is absent.
Figure 1.—
Analysis of the regulation of the cell size by the CLN1 and CLN2 genes. (A) The size of the cells in exponentially growing cultures on YPD medium of isogenic wild type (W303-1A), cln1 (JCY275), cln2 (JCY296), cln3 (MT244), cln1cln2 (JCY306), cln1cln3 (JCY223), and cln2cln3 (JCY317) was determined in a Coulter cell counter. (B) Wild type (W303-1A) transformed with a control vector (YCp50), or plasmids overexpressing either the CLN1 (ptetO:CLN1) or the CLN2 (ptetO:CLN2) genes under the control of the doxycycline-regulated tetO2 promoter, were grown on synthetic medium, and cell sizes were determined. (C) Size of cln1cln2 cells (JCY306) transformed with the control vector (YCp50), the ptetO:CLN1, or the ptetO:CLN2 plasmid, in exponentially growing cultures in the presence of 0.05 μg/ml doxycycline. (D) Western analysis of Cln1p and Cln2p protein levels in extracts from the cln1cln2 strain (JCY306) bearing a centromeric plasmid containing a HA-tagged version of the CLN1 or CLN2 genes or the ptetO:CLN1 or ptetO:CLN2 centromeric plasmid, which expresses the HA-tagged version of CLN1 or CLN2 under the control of the tetO2 promoter. The cells transformed with either the ptetO:CLN1 or the ptetOCLN2 plasmid were grown in the absence or in the presence of 0.05 μg/ml doxycycline, as indicated. Control extract from cells transformed with an empty vector was included (lane no tag). A nonspecific band that cross-reacts with the 12CA5 antibody is shown as the loading control. (E) Specific kinase activity in HA-immunoprecipitates from extracts of the cln1cln2 strain (JCY306) containing either the ptetO:CLN1 or the ptetO:CLN2 plasmid and grown in the presence of 0.05 μg/ml doxycycline.
The experiments described so far were carried out with yeast strains derived from W303-1a. To ask if the different cell size phenotypes that we observed for the cln1 and cln2 mutants were specific to the strain background, we deleted or overexpressed CLN1 or CLN2 in another unrelated wild-type background (CG379). As for the W303 background, loss or overexpression of CLN1 did not affect the cell-size distribution in an exponentially growing population. By contrast, the loss of CLN2 caused a dramatic increase in cell size, whereas the overexpression of CLN2 caused a reduction in cell size, a result very similar to that observed in the W303-1A background (data not shown). This suggests that the different effect on the cell size of the closely related Cln1p and Cln2p proteins, which must reflect a differential requirement for these two cyclins in the G1/S transition, is a general property of S. cerevisiae.
The observed phenotypic differences could reflect intrinsic functional differences between Cln2p and Cln1p or differences in the levels of the proteins and/or their associated kinase activities. Both Cln1p and Cln2p are unstable proteins, with half-lives in the range of 5–10 min (Wittenberg et al. 1990; Salama et al. 1994; Barral et al. 1995; Lanker et al. 1996; Willems et al. 1996; Schneider et al. 1998), and the specific activity of their associated kinase activity is indeed very similar (Tyers et al. 1993). Therefore it is expected that the expression of both cyclins under the control of the same promoter would provide the cells with a similar amount of protein and associated kinase activity for each cyclin. The different effects of the tetO2:CLN2 and tetO2:CLN1 genes on the cell size described above point in fact to the existence of intrinsic differences between the two cyclins, although the experiment relies on nonnatural dosage conditions, and the genomic CLN1 and CLN2 genes are also present. To obtain a more definitive proof, the plasmids containing the tetO2:CLN2 or tetO2:CLN1 genes were introduced in a cln1cln2 mutant strain, and the doxycycline concentration was adjusted to 0.05 μg/ml. Under this condition, cells contain either Cln1p or Cln2p at a similar amount to that present in wild-type cells. Western analysis and kinase activity assays confirmed that the Cln1p and Cln2p protein levels, along with their associated kinase activity, are similar in these cells (Figure 1, D and E). When cell-size distribution was analyzed in exponentially growing cultures, it was observed that cells expressing CLN2 and lacking CLN1 showed a cell size characteristic of wild-type cells. By contrast, however, cells expressing CLN1, but lacking CLN2, manifested a larger-than-normal cell size (Figure 1C). This result strongly suggests that the observed differences in cell size phenotypes reflect intrinsic functional differences between the Cln1p and Cln2p cyclins and are not due to differences in the protein or kinase activity levels present in the cell.
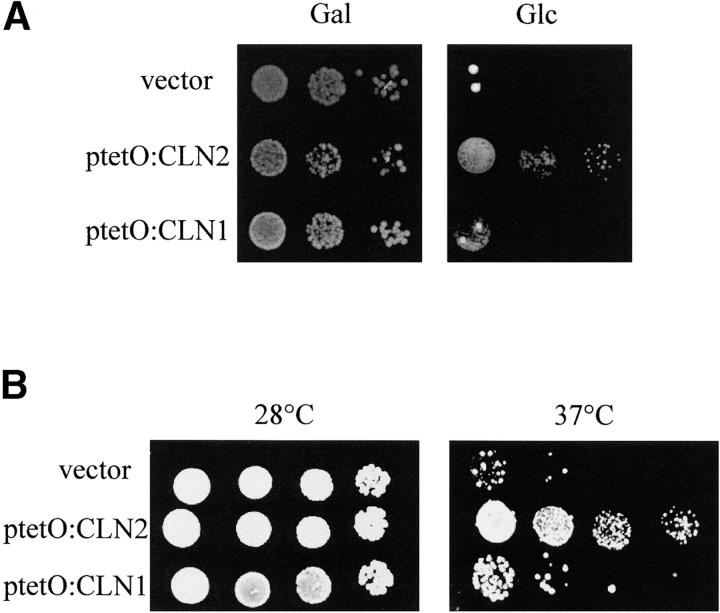
The mild ectopic expression of CLN2, but not CLN1, efficiently suppresses the lethality of Start-mutant strains:
To further explore the functional distinction between Cln1p and Cln2p, we next analyzed suppression of the Start-defective phenotype of a swi4swi6 double-mutant strain by ectopic expression of CLN1 or CLN2. A strain lacking both SWI4 and SWI6 is unable to activate the G1-S-phase transcriptional program; the essential function of these transcription factors is to turn on G1 cyclin gene expression since ectopic expression of CLN2 from the Schizosaccharomyces pombe adh1 promoter restores growth of the double-mutant strain (Nasmyth and Dirick 1991). Consistent with this result, we found that mild ectopic expression of CLN2 from the tetO2 promoter in the presence of 0.5 μg/ml doxycycline also suppresses the growth defect of a swi4swi6 mutant strain (Figure 2A). By contrast, the mild overexpression of CLN1 is unable to efficiently rescue the lethality of the swi4swi6 mutant strain. This result is consistent with previous observations pointing to a better suppression of the swi4mbp1 and swi4swi6ssd1-d mutant strains by CLN2 than by CLN1 (see Wijnen and Futcher 1999, Table 3). Only under conditions of overexpression (i.e., cells grown in the absence of doxycycline) was CLN1 able to suppress the growth defect of the swi4swi6 strain (data not shown). The same result was obtained with another strain defective in Start, the cdc28-13 mutant strain: only the mild ectopic expression of CLN2 was able to allow growth at the restrictive temperature (Figure 2B). These results indicate that moderate levels of Cln2p, but not of Cln1p, can compensate for a defect in the execution of Start, and they reinforce the conclusion that Cln2p, rather than Cln1p, is the primary regulator of the G1/S transition.
Figure 2.—
Suppression analysis of the growth defect of Start-defective mutants by the mild ectopic expression of CLN1 or CLN2. (A) Tenfold serial dilutions from exponentially growing cultures of the GAL:SWI4swi6 strain (JCY404), transformed with a control vector, ptetO:CLN1 or ptetO:CLN2, were spotted onto YPGal or YPD plates supplemented with 0.5 μg/ml doxycycline. Plates were incubated at 28° for 3 days. (B) Tenfold serial dilutions from exponentially growing cultures of the cdc28-13 mutant strain, transformed with a control vector, ptetO:CLN1 or ptetO:CLN2, were spotted onto YPD plates containing 0.5 μg/ml doxycycline and incubated at 28° and at 37° for 3 days.
Bud emergence is primarily controlled by Cln2p, but not by Cln1p:
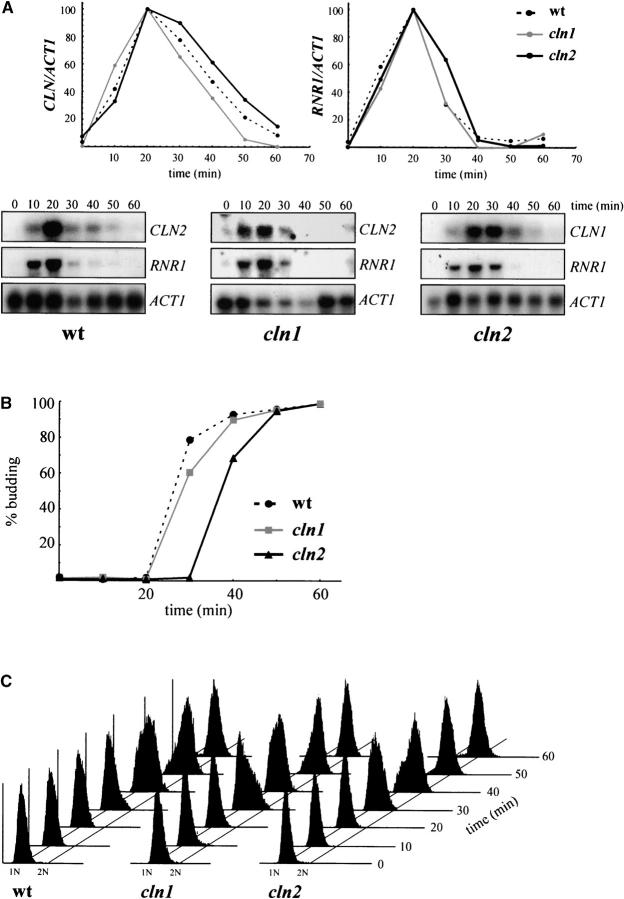
At Start, different processes are switched on depending on the CLN function. To test whether the different requirements for Cln1p and Cln2p affect all Start events or simply one specific process, wild type, cln2, and cln1 cells were arrested with α-factor, and the timing of budding, initiation of DNA replication, and activation of the Start transcription program was analyzed after the release from the arrest. As shown in Figure 3, cln1 mutant cells activate all three processes with similar kinetics as the wild-type strain, confirming that Cln1p plays a secondary role in Start control. With regard to the cln2 mutant cells, no significant differences in the kinetics of the activation of transcription or the initiation of DNA replication were detected when compared to wild-type and cln1 cells, yet, remarkably, budding was delayed by ∼10 min. This is an interesting result, which indicates that Cln2p is the rate-limiting factor for the proper execution of bud emergence, but not for the other Start events. The delay in budding could certainly account for the increased cell size observed in the cln2 mutant strains.
Figure 3.—
Analysis of the G1/S transition in cln1 and cln2 mutant strains. Northern analysis of the expression of CLN and RNR1 genes (A), budding index (B), and FACS analysis of the DNA content (C) in α-factor synchronized cultures of the wild-type (W303-1A), cln1 (JCY275), and cln2 (JCY296) strains are shown. Samples were collected at 0, 10, 20, 30, 40, 50, and 60 min after the release from arrest. In the Northern analysis in A, ACT1 mRNA is shown as the loading control, and plots represent the amounts of CLN and RNR1 mRNA relative to that of ACT1 mRNA.
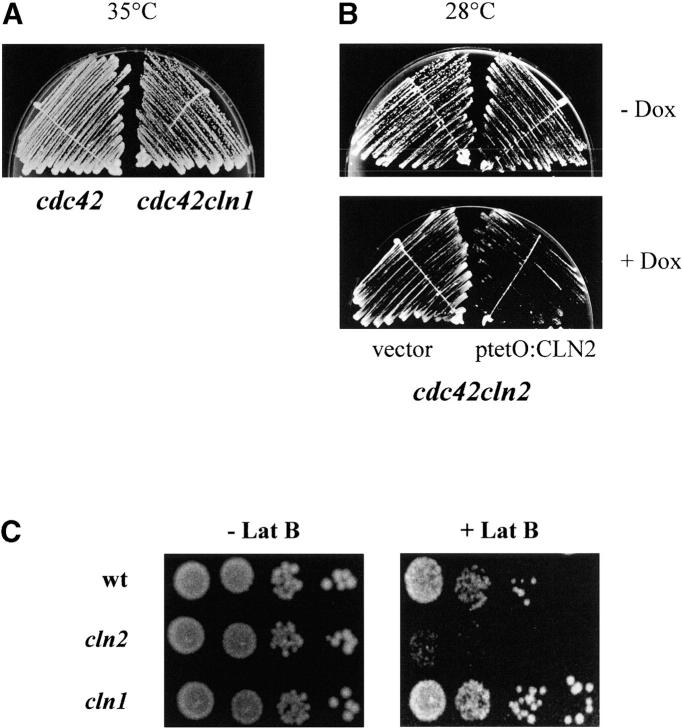
Bud emergence requires polarization of actin cytoskeleton to a particular site in the cell surface. To get a more definitive proof of the functional distinction between Cln2p and Cln1p in the control of bud emergence, we investigated the requirement of each cyclin for viability in cells that are defective in polarization and budding because of a mutation in the CDC42 gene (Adams et al. 1990) or in cells whose actin cytoskeleton is perturbed by the addition of the actin-polymerizing inhibitor latrunculin B to the medium. First, we attempted to delete CLN1 or CLN2 in a strain carrying a thermosensitive allele of CDC42. We readily obtained a CLN1 disruption strain, and the growth analysis at different temperatures of the cln1cdc42 and the parental cdc42 strain revealed that the loss of CLN1 had no effect at all on growth (Figure 4A). In the case of CLN2, however, a direct disruption of the gene in the cdc42 strain could not be obtained after several attempts, which suggested that the inactivation of cln2 could be deleterious in the cdc42 strain. To obtain a cln2cdc42 double mutant, we disrupted the CLN2 gene in a cdc42 strain carrying a plasmid expressing the wild-type CLN2 gene from the tetO2 promoter. In this strain, the viability of the cln2cdc42 double mutant can be assessed by assaying growth in the presence of doxycycline, which repressed the tetO2:CLN2 expression. As observed in Figure 4B, when the CLN2 gene was repressed by the addition of doxycycline to the medium, cell growth was severely impaired even at 28°. These results demonstrate a synthetic interaction between the cln2 and cdc42 mutations and indicate that in the presence of an impaired Cdc42p function, Cln2p, but not Cln1p, becomes essential.
Figure 4.—
Genetic analysis of morphogenetic defects in the cln1 and cln2 mutant strains. (A) The cdc42 (DJTD2-16D) and the cdc42cln1 (JCY396) strains were streaked onto YPD agar and incubated at 35° for 3 days. (B) The cdc42 (DJTD2-16D) and the cdc42cln2 (JCY453) strains, transformed with the ptetO:CLN2 plasmid, were streaked onto YPD agar and YPD agar supplemented with 5 μg/ml doxycycline and incubated at 28° for 3 days. (C) Tenfold serial dilutions from exponentially growing cultures of the wild-type (W303-1A), cln1 (JCY275), and cln2 (JCY296) strains were spotted onto YPD agar and YPD agar supplemented with 50 μm latrunculin B and then incubated at 28° for 3 days.
Next, the growth of the cln2 and cln1 mutant strains in the presence of different concentrations of the actin-depolymerizing drug latrunculin B was investigated. The loss of CLN2 caused cells to be hypersensitive to the drug: cln2 cells failed to grow in the medium supplemented with 50 μm of latrunculin B, a concentration at which cln1 and wild-type cells grow properly (Figure 4C). This result indicates that when the actin cytoskeleton is perturbed, Cln2p, but not Cln1p, becomes essential, and it reinforces the conclusion that Cln2p is playing a crucial morphogenetic function for a proper G1/S transition and that this function is specific to Cln2p and not shared with Cln1p.
Difference in the subcelluar localization of Cln1p and Cln2p:
It is possible that the functional difference between Cln1p and Cln2p was due to differences in their localization. It has been previously reported that both cyclins are located in the cytoplasm and in the nucleus (Blondel et al. 2000; Miller and Cross 2000, 2001b; Edgington and Futcher 2001) and, in the case of Cln2p, that the nuclear and the cytosolic protein pools have specific functions. However, it is difficult from these studies to rule out the existence of subtle differences in the localization of Cln1p and Cln2p, and, moreover, Cln1p localization was assayed only in cells overexpressing the cyclin. To address this point, we analyzed the subcellular distribution of both cyclins in parallel indirect immunofluorescence assays (Figure 5). When Cln2p subcellular localization was analyzed, the signal was detected through the whole cell, with a punctuate pattern in the cytoplasm, in concordance with published results. Cln1p staining was also observed throughout whole cells, but significant differences were detected when compared to the Cln2p localization: the cytosolic punctuate pattern characteristic of Cln2p was barely detected for Cln1p, and the intensity of nuclear spots was notably higher in the case of Cln1p. In fact, the estimation of the signal intensity in the nucleus and the cytoplasm indicated that the nuclear signal was 1.70 times more intense than the cytosolic signal in the case of Cln1p, in contrast to the value of 1.16 obtained for Cln2p. Thus, small differences in the relative distribution of Cln1p and Cln2p in the cell were observed, which reinforce the conclusion of a functional distinction between these two cyclins.
Figure 5.—
Subcellular localization of Cln1p and Cln2p. (A) Cells from exponentially growing cultures of the cln1 (JCY275) or the cln2 mutant (JCY296) strains transformed with a centromeric plasmid expressing an HA-tagged version of either Cln1p or Cln2p, respectively, were assayed by indirect immunofluorescence as described in materials and methods. DIC images, the Cln1p-HA and Cln2p-HA indirect-fluorescence signals (anti-HA), and the DAPI staining of DNA are shown. The first row corresponds to a control of the untagged wild-type strain. (B) A comparison of the relative distribution of Cln1p and Cln2p between the nucleus and the cytoplasm was achieved by determining the ratio of the average pixel intensity in the nuclear and cytosolic regions of the cell.
Cln2p integrity is required for sustaining its specific function:
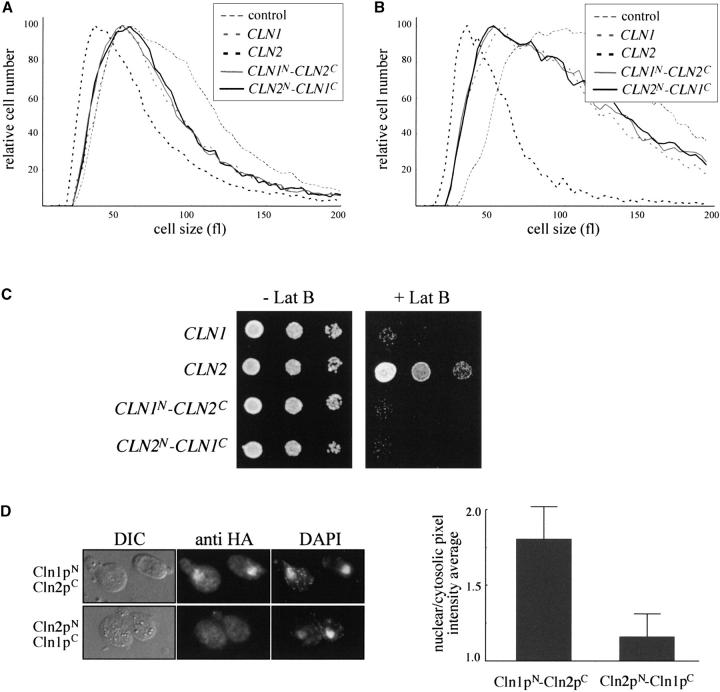
In a first attempt to characterize the determinants of the Cln2p-specific functionality, a swapping experiment was carried out by interchanging the C-terminal two-thirds of the CLN1 and CLN2 genes. Thus, two chimeric cyclins were constructed composed of amino acids 1–200 from Cln1p, amino acids 201–546 from Cln2p, and three copies of the HA epitope (Cln1pN-Cln2pC) or, reciprocally, amino acids 1–200 from Cln2p, amino acids 201–545 from Cln1p, and three copies of the HA epitope (Cln2pN-Cln1pC). Expression of the chimeric cyclins was confirmed by Western analysis, which also revealed that both chimeric cyclins are present at similar protein levels (data not shown). The chimeric cyclins were then introduced into the cln2 and cln1cln2 mutant strains, and their ability to behave like Cln1p or Cln2p was investigated by cell-size determination assays. As is shown in Figure 6, A and B, whereas the presence of a plasmid containing the CLN2 gene restores a wild-type cell size in both strains, the two chimeric cyclins failed to suppress the cell-size defect caused by the cln2 mutation. In fact, the effect of both chimeras on cell size was very similar to that observed with a plasmid containing the CLN1 gene. Next, the ability of the chimeric cyclins to suppress the hypersensitivity of the cln2 mutant strain to latrunculin B was investigated. As can be seen in Figure 6C, whereas a plasmid containing the CLN2 gene restored growth of the cln2 mutant strain in the presence of latrunculin B, plasmids expressing any of the two chimeric cyclin genes or an extra copy of the CLN1 gene failed to support growth. All these results indicated that both chimeric cyclins behave like a genuine Cln1p and none of them is able to carry out the specific function of Cln2p, suggesting that regions located along the whole protein are important to confer functional specificity to Cln2p in relation to Cln1p. In other words, the fact that the substitution of parts of Cln2p for those of Cln1p render chimeric proteins that are active as a cyclin but have lost the specific function of Cln2p confirms the existence of a functional distinction between Cln1p and Cln2p in the control of the mitotic cycle.
Figure 6.—
Ability of chimeric cyclins to sustain the Cln2p-specific function. (A) The cln2 mutant strain (JCY296) transformed with a control vector or a centromeric plasmid containing the CLN1, CLN2, CLN1N-CLN2C, or CLN2N-CLN1C gene was grown on synthetic medium and cell sizes were determined. (B) Size of cln1cln2 cells (JCY306) transformed with the control vector or a centromeric plasmid containing the CLN1, CLN2, CLN1N-CLN2C, or CLN2N-CLN1C gene. (C) Tenfold serial dilutions from exponentially growing cultures of the cln2 mutant strain (JCY296) transformed with a control vector or a centromeric plasmid containing the CLN1, CLN2, CLN1N-CLN2C, or CLN2N-CLN1C gene were spotted onto YPD agar and YPD agar supplemented with 50 μm latrunculin B and then incubated at 28° for 3 days. (D) cln1cln2 (JCY306) cells transformed with a centromeric plasmid expressing an HA-tagged version of the chimeric cyclins Cln1pN-Cln2pC or Cln2pN-Cln1pC were assayed by indirect immunofluorescence as described in Figure 5.
Finally, the subcellular localization of the chimeric cyclins was analyzed. Both proteins are distributed in the cytoplasm and in the nucleus. However, the Cln1pN-Cln2pC cyclin is relatively more abundant in the nucleus than the Cln2pN-Cln1pC cyclin (Figure 6D). This result is similar to that observed for Cln1p and suggests that sequences in the N-terminal half of the cyclins are responsible for the more efficient accumulation inside the nucleus of Cln1p in comparison to Cln2p.
DISCUSSION
The main function of the closely related Cln1p and Cln2p cyclins is to switch on the events of bud emergence, spindle pole body (SPB) duplication, and, indirectly, the initiation of DNA replication. To date both cyclins are considered highly related on the basis of their regulation and function. Both Cln1p and Cln2p protein levels are controlled by the same transcriptional (Nasmyth and Dirick 1991; Ogas et al. 1991) and posttranscriptional mechanisms (Barral et al. 1995; Willems et al. 1996; Skowyra et al. 1997) and both cyclins take part in many common functions, such as induction of growth polarization and budding (Benton et al. 1993; Cvrckova and Nasmyth 1993; Lew and Reed 1993), duplication of the SPB (Haase et al. 2001), stimulation of Sic1p and Far1p degradation (Peter et al. 1993; Schwob et al. 1994; Schneider et al. 1996; Henchoz et al. 1997), inactivation of the APC complex (Amon et al. 1994; Huang et al. 2001), repression of pheromone-induced transcription (Oehlen and Cross 1998; Wu et al. 1998), or stimulation of pseudohyphal growth (Ahn et al. 2001). Thus, the extensive work carried out on these two cyclins has revealed an extraordinary similarity between them. However, in this work, we show an important difference in the cellular requirement for Cln1p and Cln2p in executing the G1/S transition. We find that (1) inactivation of CLN2, but not of CLN1, led to an increase in cell size, and reciprocally, overexpression of CLN2, but not of CLN1, caused a reduction in cell size; (2) mild ectopic expression of CLN2, but not of CLN1, suppresses the cdc28 and swi4swi6 Start-defective mutants; (3) the cln2 mutation, but not the cln1 mutation, originates a delay in the timing of bud emergence; and (4) the cln2 mutation, but not the cln1 mutation, is synthetically lethal with a cdc42 mutation and causes hypersensitivity to perturbations in the actin cytoskeleton. On the basis of genetic interactions (Benton et al. 1993; Cvrckova and Nasmyth 1993) and the kinetics of budding (Dirick et al. 1995) in a cln1cln2 mutant strain, it was previously suggested that Cln1p and Cln2p are involved in bud emergence. Our results, however, enable us to conclude that Cln2p is the primary regulator of budding at the G1/S transition during the mitotic cycle, with the Cln1p protein playing a very minor role.
In addition to the numerous reports outlining the similarity between Cln1p and Cln2p, some differences have been previously described. The transient delay in cell cycle progression and the concomitant increase in cell size observed when cells grown on nonfermentable carbon sources are shifted to glucose-containing medium are dependent upon Cln1p, but not on Cln2p (Tokiwa et al. 1994; Flick et al. 1998). However, this difference is due to the specific repression of CLN1 gene transcription after the addition of glucose and does not reflect intrinsic functional differences between Cln1p and Cln2p. In fact, Flick et al. found that Cln2p, when it is expressed under the control of the CLN1 promoter, is as efficient as Cln1p in adapting cell size to a new carbon source, so the mechanism relies on a reduction in CLN expression independently of the nature of the cyclin. In other words, the fact that adaptation of cell size to new conditions is accomplished by a reduction in CLN1 expression suggests that Cln1p could be playing a more prominent role in slowly growing cells on poor carbon sources than in rapidly growing cells on glucose. On the other hand, it has also been reported that Cln1p is required for pseudohyphal development, whereas Cln2p is dispensable for this process (Loeb et al. 1999; Madhani et al. 1999). However, as discussed in Loeb et al., CLN transcripts are not sufficiently abundant for an accurate detection under the experimental conditions used, so it is possible that this difference between Cln1p and Cln2p could also reflect variations in gene expression rather than intrinsic functional differences between the cyclins. In fact, Madhani et al. report that the induction of pseudohyphal development (at least under some conditions) is associated with an increase in CLN1 gene expression (CLN2 gene expression was not analyzed in this study). Finally, a cln2 mutant strain initiates meiosis more rapidly than the wild type, in contrast to the much more modest effect observed in the cln1 mutant strain (Purnapatre et al. 2002). This has led to the conclusion that CLN2 is more active than CLN1 in repressing the transition from cell division to meiotic differentiation. Cells must reach a critical size before they can initiate meiosis and it has been proposed that the effect of CLN in the initiation of meiosis could result from its effect on cell size (Calvert and Dawes 1984; Purnapatre et al. 2002). Thus, cln3 or cln1cln2 mutants are larger than normal and accelerate entry into meiosis, whereas cells overexpressing CLN3 are smaller than normal and delay meiosis initiation. In this context, the different roles of CLN1 and CLN2 in the control of cell size that we have described here could explain the differences observed in the timing of meiosis initiation between the cln1 and cln2 mutant strains: cln2 mutation, but not cln1 mutation, will produce larger cells and, consequently, an accelerated entry in meiosis.
In S. cerevisiae, a single CDK, Cdc28p, associates with nine different cyclins to govern progression through the cell cycle. The mechanism by which the cyclin partner confers functional specificity on the different cyclin-Cdc28p kinases is still a matter of some controversy (reviewed in Roberts 1999; Miller and Cross 2001a). Some experimental data argue against an intrinsic specialization of cyclins and support a scenario in which the apparent functional specificity of the cyclin-CDK complexes reflects simply differences in the timing of expression of the particular cyclin. However, other experimental data support the opposite viewpoint, in which the cyclin identity might be essential for the specialization of the different cyclin-CDK complexes by mediating interactions with specific substrates or regulators and/or localizing the kinases in different subcellular compartments. It is most likely that a combination of these mechanisms, and/or additional factors not yet characterized, contributes to cyclin-CDK specificity.
Which mechanisms account for the functional distinction between Cln1p-Cdc28p and Cln2p-Cdc28p that we have characterized? First, Cln1p/Cln2p functional specificity may reflect differences in the period of the cycle in which the cyclin is expressed or in their protein levels. However, this explanation can probably be ruled out because Cln1p and Cln2p are both present in the cell during the same period of the cycle, and the difference between both cyclins is still observed when they are ectopically expressed under the control of the same promoter. Moreover, the Cln2pN-Cln1pC chimeric cyclin, which is expressed from the CLN2 promoter, has lost the specific functionality of Cln2p. Second, Cln1p and Cln2p may have differences in their association with or the activation of the Cdc28p kinase. However, published results (Tyers et al. 1993), as well as our work, do not support this model because the specific activities of the Cln1p-Cdc28p and Cln2p-Cdc28p kinases in the cell are very similar. A third possibility that could explain the functional specificity of Clnp-Cdc28p complexes is the control of the subcellular localization of the complex by the cyclin partner. Strong evidence for this mechanism comes from studies on the specific functions of Cln2p- and Cln3p-associated kinases (Edgington and Futcher 2001; Miller and Cross 2000, 2001b). In the case of Cln1p and Cln2p, both cyclins are distributed in the nucleus and in the cytoplasm, but there are significant differences since Cln1p shows a higher proportion of nuclear accumulation than Cln2p does. Budding is a process that involves reorganization of the cytoskeleton and the cell surface, so the relative higher abundance of Cln2p in the cytosol could help to explain the specific role that Cln2p plays in this process. Finally, an additional mechanism that could contribute to the functional distinction between cyclin-CDK complexes is the ability of cyclins to mediate critical protein interactions, which target the kinases to specific substrates. It is interesting to note that the N-terminal regions of Cln1p and Cln2p, which contain the cyclin box, are highly similar (74% identity). However, conservation is relaxed in their C-terminal regions (45% identity). This would suggest that the elements responsible for the distinction between both cyclins (for instance, specific protein-interacting domains) could be located in the C-terminal portions of the proteins. In support of this idea, we found that substitution of the C-terminal part of Cln2p for that of Cln1p in the Cln2pN-Cln1pC chimera causes the loss of the Cln2p-specific functionality, in spite of the fact that the chimera is expressed under the control of the CLN2 promoter and shows a similar pattern of localization to Cln2p. The C-terminal two-thirds of Cln2p, however, are not sufficient to confer Cln2p-specific functionality, as deduced from the failure of the Cln1pN-Cln2pC chimeric cyclin to suppress cln2 deletion-specific defects like increased cell size or hypersensitivity to latrunculin B. This result might point to the presence in the N-terminal region of Cln2p of additional sequences required for its specific functionality; nevertheless, the fact that the Cln1pN-Cln2pC chimeric cyclin shows a pattern of subcellular localization characteristic of Cln1p and different from Cln2p might be masking its potential ability to function as a genuine Cln2p cyclin.
In summary, we have provided evidence in favor of intrinsic functional specialization between the Cln1p and Cln2p cyclins during the mitotic cycle of S. cerevisiae. Considering that these are closely related cyclins, our results suggest that the cyclin protein family might be more specialized than previously suspected.
Acknowledgments
We are very grateful to E. Herrero, M. Aldea, J. R. Pringle, and L. H. Johnston for kindly supplying plasmids and strains. This work was supported by grant BMC2001-0446-CO2-02 from the Ministerio de Ciencia y Tecnología of the Spanish Government. E.Q. is a recipient of a fellowship from the Ministerio de Ciencia y Tecnología of the Spanish Government.
References
- Adams, A. E. M., D. I. Johnson, R. M. Longnecker, B. F. Sloat and J. R. Pringle, 1990. CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J. Cell Biol. 111: 131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn, S. H., B. T. Tobe, J. N. Fitz Gerald, S. L. Anderson, A. Acurio et al., 2001. Enhanced cell polarity in mutants of the budding yeast cyclin-dependent kinase Cdc28p. Mol. Biol. Cell 12: 3589–3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amon, A., S. Irniger and K. Nasmyth, 1994. Closing the cell cycle circle in yeast: G2 cyclin proteolysis initiated at mitosis persists until the activation of G1 cyclins in the next cycle. Cell 77: 1037–1050. [DOI] [PubMed] [Google Scholar]
- Andrews, B., and V. Measday, 1998. The cyclin family of budding yeast: abundant use of a good idea. Trends Genet. 14: 66–72. [DOI] [PubMed] [Google Scholar]
- Barral, Y., S. Jentsch and C. Mann, 1995. G1 cyclin turnover and nutrient uptake are controlled by a common pathway in yeast. Genes Dev. 9: 399–409. [DOI] [PubMed] [Google Scholar]
- Benton, B. K., A. H. Tinkelenberg, D. Jean, S. D. Plump and F. R. Cross, 1993. Genetic analysis of Cln/Cdc28 regulation of cell morphogenesis in budding yeast. EMBO J. 12: 5267–5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel, M., J. M. Galan, Y. Chi, C. Lafourcade, C. Longaretti et al., 2000. Nuclear-specific degradation of Far1 is controlled by the localization of the F-box protein Cdc4. EMBO J. 19: 6085–6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvert, G. R., and I. W. Dawes, 1984. Cell size control of development in Saccharomyces cerevisiae. Nature 312: 61–63. [DOI] [PubMed] [Google Scholar]
- Cross, F. R., 1988. DAF1, a mutant gene affecting size control, pheromone arrest, and cell cycle kinetics of Saccharomyces cerevisiae. Mol. Cell. Biol. 8: 4675–4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross, F. R., 1990. Cell cycle arrest caused by CLN gene deficiency in Saccharomyces cerevisiae resembles START-I arrest and is independent of the mating-pheromone signalling pathway. Mol. Cell. Biol. 10: 6482–6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross, F. R., 1995. Starting the cell cycle: What's the point? Curr. Opin. Cell Biol. 7: 790–797. [DOI] [PubMed] [Google Scholar]
- Cvrckova, F., and K. Nasmyth, 1993. Yeast G1 cyclins CLN1 and CLN2 and a GAP-like protein have a role in bud formation. EMBO J. 12: 5277–5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirick, L., T. Bohm and K. Nasmyth, 1995. Roles and regulation of Cln-Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. EMBO J. 14: 4803–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgington, N. P., and B. Futcher, 2001. Relationship between the function and the location of G1 cyclins in S. cerevisiae. J. Cell Sci. 114: 4599–4611. [DOI] [PubMed] [Google Scholar]
- Flick, K., D. Chapman-Shimshoni, D. Stuart, M. Guaderrama and C. Wittenberg, 1998. Regulation of cell size by glucose is exerted via repression of the CLN1 promoter. Mol. Cell. Biol. 18: 2492–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulli, M. P., M. Jaquenoud, Y. Shimada, G. Niederhauser, P. Wiget et al., 2000. Phosphorylation of the Cdc42 exchange factor Cdc24 by the PAK-like kinase Cla4 may regulate polarized growth in yeast. Mol. Cell 6: 1155–1167. [DOI] [PubMed] [Google Scholar]
- Haase, S. B., M. Winey and S. I. Reed, 2001. Multi-step control of spindle pole body duplication by cyclin-dependent kinase. Nat. Cell Biol. 3: 38–42. [DOI] [PubMed] [Google Scholar]
- Hadwiger, J. A., C. Wittenberg, H. E. Richardson, M. De Barros Lopes and S. I. Reed, 1989. A family of cyclin homologs that control the G1 phase in yeast. Proc. Natl. Acad. Sci. USA 86: 6255–6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henchoz, S., Y. Chi, B. Catarin, I. Herskowitz, R. J. Deshaies et al., 1997. Phosphorylation- and ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor Far1p in budding yeast. Genes Dev. 11: 3046–3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J. N., I. Park, E. Ellingson, L. E. Littlepage and D. Pellman, 2001. Activity of the APC(Cdh1) form of the anaphase-promoting complex persists until S phase and prevents the premature expression of Cdc20p. J. Cell Biol. 154: 85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanker, S., M. H. Valdivieso and C. Wittenberg, 1996. Rapid degradation of the G1 cyclin Cln2 induced by CDK-dependent phosphorylation. Science 271: 1597–1601. [DOI] [PubMed] [Google Scholar]
- Levine, K., K. Huang and F. R. Cross, 1996. Saccharomyces cerevisiae G1 cyclins differ in their intrinsic functional specificities. Mol. Cell. Biol. 16: 6794–6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew, D. J., and S. I. Reed, 1993. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J. Cell Biol. 120: 1305–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb, J. D. J., T. A. Kerentseva, T. Pan, M. Sepulveda-Becerra and H. Liu, 1999. Saccharomyces cerevisiae G1 cyclins are differentially involved in invasive and pseudohyphal growth independent of the filamentation mitogen-activated protein kinase pathway. Genetics 153: 1535–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani, H. D., T. Galitski, E. S. Lander and G. R. Fink, 1999. Effectors of a developmental mitogen-activated protein kinase cascade revealed by expression signatures of signaling mutants. Proc. Natl. Acad. Sci. USA 96: 12530–12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcinerny, C. J., J. F. Partridge, G. E. Mikesell, D. P. Creemer and L. Breeden, 1997. A novel Mcm1-dependent element in the SWI4, CLN3, CDC6, and CDC47 promoters activates M/G1-specific transcription. Genes Dev. 11: 1277–1288. [DOI] [PubMed] [Google Scholar]
- Mendenhall, M. D., and A. E. Hodge, 1998. Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62: 1191–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, M. E., and F. R. Cross, 2000. Distinct subcellular localization patterns contribute to functional specificity of the Cln2 and Cln3 cyclins of Saccharomyces cerevisiae. Mol. Cell. Biol. 20: 542–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, M. E., and F. R. Cross, 2001. a Cyclin specificity: How many wheels do you need on a unicycle? J. Cell Sci. 114: 1811–1820. [DOI] [PubMed] [Google Scholar]
- Miller, M. E., and F. R. Cross, 2001. b Mechanisms controlling subcellular localization of the G(1) cyclins Cln2p and Cln3p in budding yeast. Mol. Cell. Biol. 21: 6292–6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan, D. O., 1997. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 13: 261–291. [DOI] [PubMed] [Google Scholar]
- Nash, R., G. Tokiwa, S. Anand, K. Erickson and A. B. Futcher, 1988. The WHI1+ gene of Saccharomyces cerevisiae tethers cell division to cell size and is a cyclin homolog. EMBO J. 7: 4335–4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash, R. S., T. Volpe and B. Futcher, 2001. Isolation and characterization of WHI3, a size-control gene of Saccharomyces cerevisiae. Genetics 157: 1469–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth, K., 1996. At the heart of the budding yeast cell cycle. Trends Genet. 12: 405–412. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K., and L. Dirick, 1991. The role of SWI4 and SWI6 in the activity of G1 cyclins in yeast. Cell 66: 995–1013. [DOI] [PubMed] [Google Scholar]
- Oehlen, L. J., and F. R. Cross, 1998. Potential regulation of Ste20 function by the Cln1-Cdc28 and Cln2-Cdc28 cyclin-dependent protein kinases. J. Biol. Chem. 273: 25089–25097. [DOI] [PubMed] [Google Scholar]
- Ogas, J., B. J. Andrews and I. Herskowitz, 1991. Transcriptional activation of CLN1, CLN2, and a putative new G1 cyclin (HCS26) by SWI4, a positive regulator of G1-specific transcription. Cell 66: 1015–1026. [DOI] [PubMed] [Google Scholar]
- Peter, M., A. Gartner, J. Horecka, G. Ammerer and I. Herskowitz, 1993. FAR1 links the signal transduction pathway to the cell cycle machinery in yeast. Cell 73: 747–760. [DOI] [PubMed] [Google Scholar]
- Purnapatre, K., S. Piccirillo, B. L. Schneider and S. M. Honigberg, 2002. The CLN3/SWI6/CLN2 pathway and SNF1 act sequentially to regulate meiotic initiation in Saccharomyces cerevisiae. Genes Cells 7: 675–691. [DOI] [PubMed] [Google Scholar]
- Queralt, E., and J. C. Igual, 2003. Cell cycle activation of the Swi6p transcription factor is linked to nucleocytoplasmic shuttling. Mol. Cell. Biol. 23: 3126–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, H. E., C. Wittenberg, F. Cross and S. I. Reed, 1989. An essential G1 function for cyclin-like proteins in yeast. Cell 59: 1127–1133. [DOI] [PubMed] [Google Scholar]
- Roberts, J. M., 1999. Evolving ideas about cyclins. Cell 98: 129–132. [DOI] [PubMed] [Google Scholar]
- Salama, S. R., K. B. Hendricks and J. Thorner, 1994. G1 cyclin degradation: the PEST motif of yeast Cln2 is necessary, but not sufficient, for rapid protein turnover. Mol. Cell. Biol. 14: 7953–7966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, B. L., Q. H. Yang and A. B. Futcher, 1996. Linkage of replication to start by the Cdk inhibitor Sic1. Science 272: 560–562. [DOI] [PubMed] [Google Scholar]
- Schneider, B. L., E. E. Patton, S. Lanker, M. D. Mendenhall, C. Wittenberg et al., 1998. Yeast G1 cyclins are unstable in G1 phase. Nature 395: 86–89. [DOI] [PubMed] [Google Scholar]
- Schwob, E., T. Bohm, M. D. Mendenhall and K. Nasmyth, 1994. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell 79: 233–244. [DOI] [PubMed] [Google Scholar]
- Skowyra, D., K. L. Craig, M. Tyers, S. J. Elledge and J. W. Harper, 1997. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91: 209–219. [DOI] [PubMed] [Google Scholar]
- Stuart, D., and C. Wittenberg, 1995. CLN3, not positive feedback, determines the timing of CLN2 transcription in cycling cells. Genes Dev. 9: 2780–2794. [DOI] [PubMed] [Google Scholar]
- Tokiwa, G., M. Tyers, T. Volpe and B. Futcher, 1994. Inhibition of G1 cyclin activity by the Ras/cAMP pathway in yeast. Nature 371: 342–344. [DOI] [PubMed] [Google Scholar]
- Tyers, M., G. Tokiwa and B. Futcher, 1993. Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J. 12: 1955–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallen, E. A., and F. R. Cross, 1995. Mutations in RAD27 define a potential link between G1 cyclins and DNA replication. Mol. Cell. Biol. 15: 4291–4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallen, E. A., and F. R. Cross, 1999. Interaction between the MEC1-dependent DNA synthesis checkpoint and G1 cyclin function in Saccharomyces cerevisiae. Genetics 151: 459–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnen, H., and B. Futcher, 1999. Genetic analysis of the shared role of CLN3 and BCK2 at the G1-S transition in Saccharomyces cerevisiae. Genetics 153: 1131–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnen, H., A. Landman and B. Futcher, 2002. The G1 cyclin Cln3 promotes cell cycle entry via the transcription factor Swi6. Mol. Cell. Biol. 22: 4402–4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems, A. R., S. Lanker, E. E. Patton, K. L. Craig, T. F. Nason et al., 1996. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell 86: 453–463. [DOI] [PubMed] [Google Scholar]
- Wittenberg, C., K. Sugimoto and S. I. Reed, 1990. G1-specific cyclins of S. cerevisiae: cell cycle periodicity, regulation by mating pheromone, and association with the p34CDC28 protein kinase. Cell 62: 225–237. [DOI] [PubMed] [Google Scholar]
- Wu, C., T. Leeuw, E. Leberer, D. Y. Thomas and M. Whiteway, 1998. Cell cycle- and Cln2p-Cdc28p-dependent phosphorylation of the yeast Ste20p protein kinase. J. Biol. Chem. 273: 28107–28115. [DOI] [PubMed] [Google Scholar]