Abstract
It is usually posited that the most important factors contributing to sex chromosome evolution in diploids are the suppression of meiotic recombination and the asymmetry that results from one chromosome (the Y) being permanently heterozygous and the other (the X) being homozygous in half of the individuals involved in mating. To distinguish between the roles of these two factors, it would be valuable to compare sex chromosomes in diploid-mating organisms and organisms where mating compatibility is determined in the haploid stage. In this latter group, no such asymmetry occurs because the sex chromosomes are equally heterozygous. Here we show in the fungus Microbotryum violaceum that the chromosomes carrying the mating-type locus, and thus determining haploid-mating compatibility, are rich in transposable elements, dimorphic in size, and carry unequal densities of functional genes. Through analysis of available complete genomes, we also show that M. violaceum is, remarkably, more similar to humans and mice than to yeast, nematodes, or fruit flies with regard to the differential accumulation of transposable elements in the chromosomes determining mating compatibility vs. the autosomes. We conclude that restricted recombination, rather than asymmetrical sheltering, hemizygosity, or dosage compensation, is sufficient to account for the common sex chromosome characteristics.
SEX chromosomes are widely accepted to have evolved from a normal autosome pair by a series of steps, beginning with suppression of recombination in the region that determines mating compatibility and followed by the acquisition of other traits that distinguish sex chromosomes cytologically from the autosomes and from each other (Lahn and Page 1999; Liu et al. 2004). In typical diploid-mating organisms, asymmetry in the degree to which the two sex chromosomes are heterozygous (only the Y being permanently heterozygous) leads to biases in the sheltering of mutations and effective population size that apparently permit functional genes to be lost from the Y chromosome and its deterioration relative to the X (Muller 1914; Rice 1987; Charlesworth 1991). It is therefore surprising that there can be similar size dimorphism of the sex chromosomes in organisms where mating compatibility is determined strictly by the genetic composition of the haploid stage, such as mosses, liverworts, and some fungi (Bull 1978; Hood 2002). In these organisms, asymmetries in the degree of heterozygosity or the sheltering of mutations do not occur because the two sex (or “mating-type”) chromosomes are always heterozygous in the diploid stage. Indeed, genetic analysis of fungi has revealed several characteristics of the chromosome containing the mating-type locus that are similar to sex chromosomes in plants and animals (reviewed by Fraser and Heitman 2004). These include suppressed recombination over most of the chromosome pair (Merino et al. 1996; Gallegos et al. 2000; Hood and Antonovics 2004), sequence divergence (Randall and Metzenberg 1998; Lee et al. 1999), and overall size dimorphism (Hood 2002). The potential of fungi to serve as models of sex chromosome evolution has been strongly promoted (Merino et al. 1996; Lee et al. 1999; Lengeler et al. 2002; Fraser and Heitman 2004). The goals of this study are to compare the nature of DNA sequences on the chromosomes that determine mating compatibility in haploid-mating and diploid-mating organisms and to evaluate which shared evolutionary forces are sufficient to account for their common characteristics.
In addition to asymmetrical sheltering of Y chromosomes, there are additional ways that sex chromosomes are distinct from autosomes in typical diploid-mating organisms. The foremost is restricted recombination, but the degree of hemizygous exposure and dosage compensation may also be important evolutionary forces. For example, it is well established that repetitive elements, particularly retrotransposons, tend to accumulate in regions of suppressed recombination because they cannot be excised by ectopic crossing over and may become fixed via mechanisms such as genetic hitchhiking (Rice 1987) or Muller's ratchet (Charlesworth and Langley 1989). The high density of repetitive DNA in Y chromosomes has been attributed to these processes (Erlandsson et al. 2000; Steinemann and Steinemann 2000). Hemizygosity, specifically in regard to the status of the single X chromosomes in males, is thought to limit retrotransposon accumulation there by exposing such deleterious mutations to selection (Charlesworth and Langley 1989; Duret et al. 2000; Rizzon et al. 2002). However, where X chromosomes have been found to be rich in retrotransposons, such accumulation has been suggested to play a functional role in the gene-silencing mechanisms that compensate for the unequal dosage of X genes in males vs. females, although the precise mechanism is yet unknown (Bailey et al. 2000). Like asymmetrical sheltering, this latter effect on dosage compensation does not occur when mating compatibility is determined in the haploid.
Here we report on an experimental study characterizing random DNA sequences isolated from the dimorphic mating-type chromosomes and autosomes of the fungus Microbotryum violaceum (the alternative mating types are called “A1” and “A2”). We then compare these results with an analogous comparison of sex chromosomes and autosomes in other organisms whose complete genome sequences are available. Sex chromosomes in haploid- and diploid-mating organisms can be defined as those that determine mating compatibility and that often show inheritance and cytological features distinct from autosomes. We therefore refer to the fungal mating-type chromosomes as sex chromosomes to follow this definition, to be consistent with the recent review on the subject by Fraser and Heitman (2004)(see also Lindegren 1936a,b), and to simplify the use of terminology.
MATERIALS AND METHODS
The basidiomycete fungus M. violaceum causes anther-smut disease in many members of the Caryophyllaceae (the carnation family) and has been widely used as a model system for genetic and population studies of plant pathogens. This fungus has two haploid mating types, A1 and A2, which are inherited as alleles at a single locus found on a highly size-dimorphic pair of chromosomes (Hood 2002). The haploid genome has ∼12 autosomes, ranging in size from 1 to 4 Mbp; the number of autosomes appears to vary among populations yet there is the potential to not differentiate all chromosome bands of similar size that comigrate in pulsed-field gels. Mating between haploid cells of opposite mating type is a prerequisite for infection of new host plants. In different isolates of M. violaceum from the host plant Silene latifolia, the dimorphic sex chromosomes range in size from 2.8 to 3.1 Mbp for the A1 and from 3.4 to 4.2 Mbp for the A2, but within the genome the A2 is always larger (Hood 2002). It appears that recombination is suppressed and DNA sequences are highly divergent throughout much or all the sex chromosome pair in M. violaceum sampled from S. latifolia. For example, across a broad range of samples there is no evidence of recombination between the arms of the A1 and A2 chromosomes that are unequal in length (Hood 2002); i.e., the A2 chromosome is always larger. Also, a recent study involving AFLP mapping and the analysis of meiotic segregation patterns of the AFLP markers indicates that ∼12% of all loci in the genome are heterozygous and linked to mating type, and this percentage is approximately equal to the entire contribution of the sex chromosomes to the total genome size (∼13%; Hood and Antonovics 2004). In addition, a mapping study by Garber and Day (1985) found no markers that recombined with mating type, even though the sex chromosomes are among the largest chromosomes in the genome (Hood 2002). While the systematics of the genus Microbotryum are currently unresolved, we know that strains of the fungus from different hosts or different regions can have variable karyotypes (Hood 2002). The current study used a sample from the host S. latifolia from Lamole, Italy, and the karyotype resembles that of other samples from this host throughout its natural range in having autosomes that are largely homozygous and a size-dimorphic pair of chromosomes carrying the mating-type locus (Hood 2002; Hood and Antonovics 2004).
Since complete sequencing of the genome was not feasible, we isolated random DNA fragments from the sex chromosomes and autosomes of M. violaceum using an electrophoretic method. This method of isolating chromosome bands of pulsed-field gels was previously shown in studies on M. violaceum (Hood 2002) and on other fungi (Brody et al. 1991; Zolan et al. 1992; Geiser et al. 1996) to result in minimal cross-contamination of DNA samples between chromosomes. Chromosomes were separated by pulsed-field gel electrophoresis (as in Hood 2002) and DNA from bands representing the two sex chromosomes and a pooled sample of the bands representing the autosomes were extracted from the gel. With these three samples of DNA from different genomic regions, triple digestions with 4- to 6-bp-cutting restriction enzymes (RsaI, MscI, and DraI) were allowed to run until most fragments were between 200 and 1000 bp. The resulting A1, A2, and autosomal digests were size fractionated by electrophoresis, and fragments in a readily sequenced size range were extracted from the gel, cloned, and sequenced. These DNA sequences (n = 684, mean length = 470 bp, GenBank accession numbers BZ781929, BZ782612) were compared to the global databases available through the National Center for Biotechnology Information (NCBI) using the BLASTx algorithm (http://www.ncbi.nlm.nih.gov/BLAST/). DNA sequences were assigned to categories on the basis of BLASTx hits of e < 0.001. For example, the category of “retrotransposon-related” was assigned when the BLASTx hit matched sequences in the NCBI databases that were retrotransposons, reverse transcriptases, gag or pol genes, polyproteins, or retrovirus sequences. Other categories to which DNA sequences were assigned included “helicases” and other “functional genes,” the latter meaning a BLASTx hit to a known coding region that was other than retrotransposon or helicase. Helicase-related sequences were included as a category of BLAST hits because we had previously found such a sequence on the sex chromosomes of M. violaceum (Hood 2002), which has since been identified as a member of a newly discovered Helitron class of DNA-based transposable element (Poulter et al. 2003). Statistical comparisons of the frequency of sequences in the separate categories were performed between sex chromosomes and autosomes using the PROC LOGISTIC procedures in SAS (SAS Institute, Cary, NC), taking into account the lengths of the DNA sequences being compared.
To compare the accumulation of retrotransposon-related sequences on the sex chromosomes in M. violaceum with other organisms, an analogous method of randomly sampling DNA fragments was employed using the completed genome sequences available for Homo sapiens, Mus musculus, Saccharomyces cerevisiae, Caenorhabditis elegans, and Drosophila melanogaster (Internet-based resources: Entrez at http://www.ncbi.nlm.nih.gov, Ensembl at htpp://www.ensembl.org, and Wormbase at http://www.wormbase.org). Thus, fragments matching the mean and variance for length in base pairs of cloned sequences from M. violaceum were identified by choosing random start positions throughout the autosomes and X chromosomes. Y chromosome sequences were too incomplete to permit adequate sampling. The random DNA sequences were then compared to the databases available through the NCBI using the BLASTx algorithm in a manner identical to that for the experimentally obtained M. violaceum sequences. The significance criteria, categorization, and statistical analyses were also identical. The initial objective was to obtain ∼200 sequences from both the X chromosome and the pooled autosomes of each organism. For Saccharomyces, Caenorhabditis, and Drosophila the effort was doubled because of the small number of retrotransposon-related sequences. Note that chromosome 3 of Saccharomyces, which contains the mating compatibility locus (MAT), was compared with other chromosomes similarly to the comparison between the sex chromosomes and autosomes of the other organisms. However, because this fungus undergoes mating-type switching, the pattern of inheritance for chromosome 3 of Saccharomyces is not distinct from the other chromosomes (Haber 1998). The Saccharomyces system was therefore a negative control for the effects of restricted recombination and asymmetrical sheltering.
RESULTS
Sex chromosome characteristics in M. violaceum:
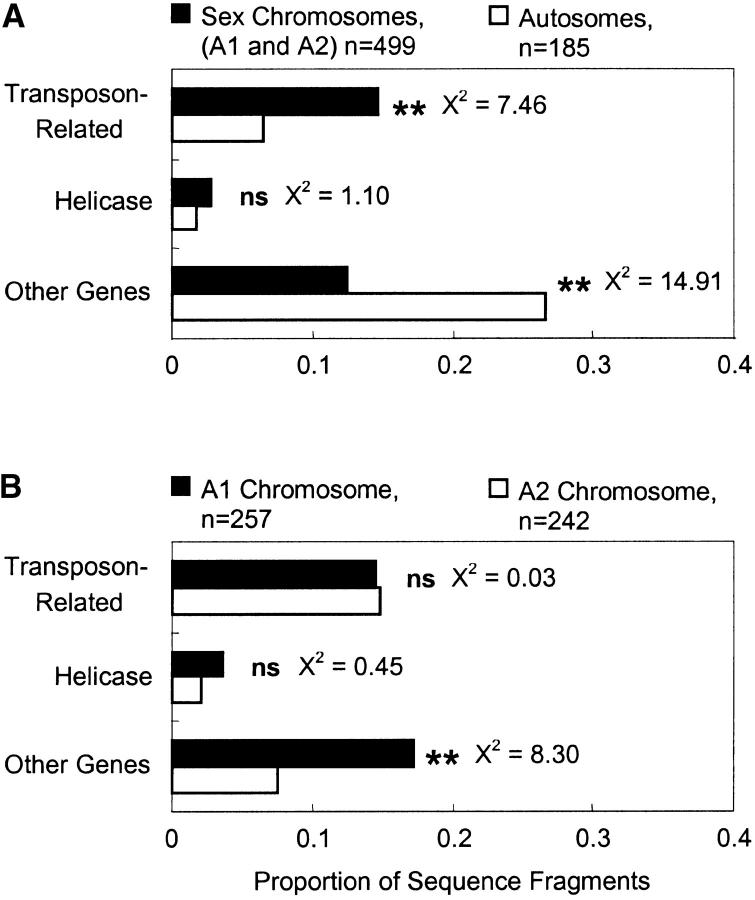
Our results show that the random fragments isolated from the fungal sex chromosomes of M. violaceum were more than twice as likely to be related to transposable elements (specifically retrotransposons) as were random fragments from the autosomes (Figure 1A). Furthermore, sex chromosome sequences were less than half as likely to be related to other functional genes as were random fragments from the autosomes (Figure 1A). There was no significant difference in the distribution of helicase-related sequences between the sex chromosomes and autosomes. The A1 and A2 sex chromosomes were not different from each other with regard to retrotransposons, but the smaller A1 was twice as dense per unit length in sequences related to functional genes as the A2 (Figure 1B). Nevertheless, the A1 was still significantly less dense than the autosomes in functional genes (χ2 = 4.63, d.f. = 1, P = 0.031). We questioned whether the numerous sequences from the sex chromosome, particularly the A2, that had no significant similarity to functional genes were actually sex-related functional genes, which the search may have been less effective at identifying than housekeeping genes. However, we found no difference in the apparent functional degeneracy, as indicated by the frequency of stop codons, of such “no-hit” sequences whether they are isolated from the autosomes or either sex chromosome (χ2, d.f. = 1, P = 0.46). A total of 85 retrotransposon-related sequences were identified (12% of all sequences), with 90% of these having e-values <10−5. By relaxing the matching criterion from e < 0.001 to e < 1, there were no changes to the significances of the observed distributions.
Figure 1.—
Densities of identified sequences from M. violaceum. Indications of statistical significance (**) are at the level of P < 0.01 with 1 d.f.; the smallest P-value among nonsignificant tests was 0.29. (A) Comparison between the fungal sex chromosomes and autosomes. (B) Comparison between the A1 and A2 fungal sex chromosomes.
We also questioned whether the numerous retrotransposon-related sequences were simply replicate copies of an identical DNA sequence. This possibility was tested by aligning the translated BLASTx matches to a model reverse transcriptase from Oryza sativa (NCBI accession AP003217.3). The fragments from M. violaceum were widely distributed across the Oryza reverse transcriptase, with 41 protein alignments from M. violaceum (mean length = 106 amino acids, standard deviation = 37), giving >80% coverage of a 1061-amino-acid region from Oryza. Where the translated fragments overlapped in alignment with the Oryza reverse transcriptase protein, there was substantial dissimilarity among the M. violaceum sequences and they often could not be aligned with one another using their original DNA sequence (using pairwise BLAST available at the NCBI website). When DNA alignment was possible among M. violaceum fragments, there was evidence of purifying selection for functional reverse transcriptase sequences in M. violaceum. Purifying selection was indicated by higher rates of synonymous vs. nonsynonymous nucleotide substitutions; z-tests for the number of substitutions per synonymous site divided by the number of substitutions per nonsynonymous site were >1 at the significance level of P = 0.05 in six of seven alignments for separate regions of M. violaceum retrotransposons and where each alignment contained two or more sequences (Kumar et al. 1994). Thus, the random sampling method was not biased toward one particular DNA sequence and provided a reasonable suggestion of many different transposing elements in the genome of M. violaceum, some of which may have been active recently enough to still show the signature of natural selection in the statistical bias in silent over nonsilent substitutions.
Comparison of sex chromosomes among model genomes:
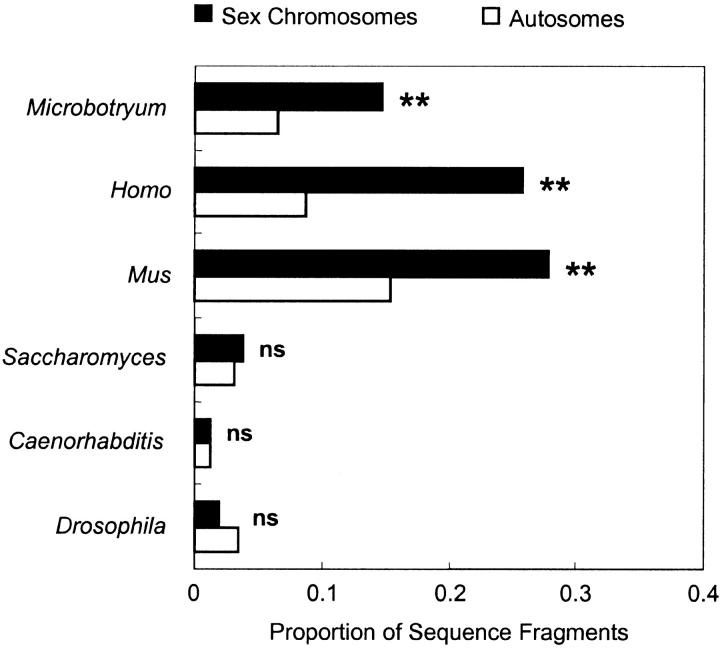
The genomes of M. violaceum, Mus, and Homo have high densities of retrotransposon-related sequences relative to the other organisms (Figure 2; Table 1). Moreover, X chromosomes of Mus and Homo, like the A1 and A2 sex chromosomes of M. violaceum, have two to three times the density of retrotransposon-related sequences as compared to the autosomes (Figure 2). These results are counter to the expectation that hemizygous exposure in X chromosomes would limit retrotransposon accumulation (Erlandsson et al. 2000). However, they confirm the recent analysis of the complete human and mouse genome sequences, which also show a much higher density of LINE elements on the X chromosomes relative to the autosomes (Waterston et al. 2002).
Figure 2.—
Densities of retrotransposon-related sequences on autosomes and sex chromosomes from M. violaceum and X chromosomes of other organisms for which the genome sequences are complete. Sample sizes and other statistics are provided in Table 1. Chromosome 3 of Saccharomyces contains the MAT locus determining mating compatibility, but is otherwise similar to an autosome.
TABLE 1.
Difference in number of retrotransposon-related sequences between sex chromosomes and autosomes
| Source of fragments | n | TE | χ2 | P-value |
|---|---|---|---|---|
| M. violaceum | ||||
| A1 and A2 sex chromosomes | 499 | 73 | 7.46 | <0.01 |
| Autosomes | 185 | 12 | ||
| H. sapiens | ||||
| X chromosome | 214 | 55 | 25.37 | <0.01 |
| Autosomes | 208 | 18 | ||
| M. musculus | ||||
| X chromosome | 180 | 50 | 8.11 | <0.01 |
| Autosomes | 177 | 27 | ||
| S. cerevisiae | ||||
| Chromosome 3 | 396 | 15 | 0.32 | 0.57 |
| Chromosomes 1, 2, 4–16 | 391 | 12 | ||
| C. elegans | ||||
| X chromosome | 399 | 5 | 0.00 | 0.98 |
| Autosome | 401 | 5 | ||
| D. melanogaster | ||||
| X chromosome | 370 | 7 | 1.64 | 0.20 |
| Autosomes | 379 | 13 |
Sequence similarity to retrotransposons was determined by a BLASTx search of the nr databases of the NCBI at the National Institutes of Health. n, number of random DNA fragments per source; TE, number of retrotransposon-related sequences. χ2 tests had 1 d.f. Chromosome 3 of Saccharomyces contains the MAT locus determining mating compatibility, but is otherwise similar to an autosome.
The other organisms (Saccharomyces, Caenorhabditis, and Drosophila) had few retrotransposon-related sequences relative to the other organisms, and the retrotransposon distributions were not significantly different across sex chromosomes and autosomes. This result was expected for Saccharomyces as a negative control (Haber 1998). This result was also consistent with a previous study on Caenorhabditis by Duret et al. (2000), which showed no overall bias in retrotransposons on the X and attributed the result to hemizygous selection against their insertion. Recent work with Drosophila (Rizzon et al. 2002) indicates fewer LTR retrotransposons on the X than on the autosomes, again citing hemizygous exposure as the selective force. Our results with Drosophila were in this direction, although not significantly so.
DISCUSSION
The selective force that is common to organisms in which retrotransposons have accumulated to a greater extent on the sex chromosomes (specifically the X chromosomes in the case of animal models) was a correspondingly greater restriction of recombination on the sex chromosomes relative to the autosomes. This correlation between retrotransposon accumulation and restricted recombination, in contrast to the other forces affecting sex chromosome evolution, can be revealed in the patterns of sex chromosome inheritance across the taxa, noting that superficially similar genetic systems may belie major differences in the forces that drive retrotransposon accumulation. For example, recombination of X chromosomes is reduced to two-thirds of that for autosomes in Mus and Homo and likely to a greater extent in the sex chromosomes of M. violaceum (Hood 2002; Hood and Antonovics 2004). In these species there is a biased accumulation of retrotransposons on the sex chromosomes. However, in the remaining organisms, opportunities for recombination are more equivalent between the sex chromosomes and autosomes and there is no biased retrotransposon accumulation. In Caenorhabditis, the XO males are extremely rare, and ∼99% of germline passages for the X are in self-fertile XX hermaphrodites (Cutter and Payseur 2003). Therefore, the Caenorhabditis X should be very much like an autosome in recombination frequency (as well as in hemizygous exposure). Accordingly, we see no bias in retrotransposon distribution in Caenorhabditis, which is similar to the negative control Saccharomyces in which the MAT-containing chromosome 3 behaves essentially as an autosome. In Drosophila, neither X nor autosomal recombination occurs in males (Betrán et al. 2002), and again the ability to excise retrotransposons from the X by recombination is equivalent to that of the autosomes.
In contrast, hemizygosity and dosage compensation are not consistent with the observed patterns of retrotransposon accumulation. While these forces may affect sex chromosome evolution in the organisms where they occur, our data show that they are not required to explain a biased accumulation of retrotransposons. Hemizygous exposure occurs in Mus, Homo, and Drosophila, but only in Drosophila have retrotransposons not accumulated to a greater extent on the X than on the autosomes. In fact, a more exhaustive study by Rizzon et al. (2002) showed that retrotransposons were actually less common on the X of Drosophila than on the autosomes, and this result was attributed to hemizygous exposure limiting their accumulation. Furthermore, if we assume that the differences in sex chromosome size and apparent densities of identifiable genes in M. violaceum lead to some hemizygous exposure, it is clear that such exposure also has not limited biased accumulation of retrotransposons on the sex chromosomes in this species. Taken together, these results suggest that hemizygous exposure may be sufficient to reduce retrotransposon accumulation on the X only in the absence of a differential in restricted recombination, as in Drosophila. This suggestion is consistent with the previous statement by Charlesworth and Langley (1989) that “while there may be some fitness effects of insertional mutations, they are not necessarily the chief factor involved in containing the spread of transposable elements.” Similarly, the observed retrotransposon distributions cannot be consistently explained by their involvement in dosage compensation (Bailey et al. 2000) since such a process is not expected to occur in M. violaceum but does occur in Mus and Homo. Any direct role of retrotransposons in the silencing of X-linked genes may have developed through a co-opting of an already biased retrotransposon distribution, but it is difficult to separate direct proximal effects on retrotransposon distributions from selection for derived functions of these elements.
The three species (M. violaceum, Mus, and Homo) that showed biased accumulation of retrotransposons on sex or X chromosomes also have higher overall densities of such elements in their genomes, including the autosomes. It is unclear whether this result represents a causal relationship, but it would be consistent with the idea that a higher density on the sex chromosomes would provide a reservoir for a net movement from the sex chromosomes onto the autosomes (which in turn may be exacerbated by the resulting increase in transposition back to the sex chromosomes). If these relationships are causal, then sex chromosome evolution may have broader implications for the dynamics of eukaryotic genomes through effects on the retrotransposon densities on autosomes and the central role retrotransposons play in karyotypic change as well as in gene duplication and regulation (Wolf-Ekkehard and Saedler 2002).
Our results with M. violaceum also address the issue of how differences between the sex chromosomes themselves may have evolved. Theoretical consideration of Y chromosome degeneration has resulted in very satisfying arguments based upon the fact that it is only the Y and not the X that is permanently heterozygous (Muller 1914; Rice 1987; Charlesworth 1991). The consistency with which Y chromosomes have lost functional genes relative to X chromosomes strongly supports the asymmetrical sheltering models. However, our results show that alternative mechanisms, acting without asymmetrical sheltering, can give rise to similar characteristics. Specifically, the A1 and A2 sex chromosomes of M. violaceum are dimorphic in size and the A1 has a significantly higher density of identifiable genes. Which mechanisms are at work here remains to be determined, but our data indicate that the dimorphism in M. violaceum is not due to unequal rates of retrotransposon accumulation (as we had originally suspected and as has been proposed in the case of sex chromosome dimorphism in S. latifolia; Filatov et al. 2000). Because the A1 chromosome in M. violaceum both is smaller and appears more dense with functional genes, a possible scenario is that the A2 chromosome is larger because it has gained noncoding sequences, rather than the selective loss of noncoding regions of the A1. Mating types in fungi do have particular developmental roles, such as initiating conjugation structures or determining uniparental inheritance of mitochondria, but M. violaceum is the first fungus in which the sex chromosomes have been shown to differ in size or in the overall nature of the functional sequences they contain. Additional studies are needed to establish whether sex chromosome properties as found in M. violaceum might correlate with mating-type specific functions across a broader range of fungi, particularly involving species where suppressed recombination also appears to have played a role in the differentiation of the fungal sex chromosomes (e.g., Neurospora tetrasperma; Merino et al. 1996; Gallegos et al. 2000). Interestingly, in bryophytes, it is often the Y that is smaller (Bull 1978) and may contain more repetitive DNA (Okada et al. 2001).
Our study shows that considerable insight can be gained into the evolution of sex chromosomes of plants and animals through the study of alternative systems in which the chromosomal determination of mating compatibility occurs in the haploid stage. Additionally, our study shows that shotgun sampling of genomic regions can greatly extend the scope of comparative studies that bear on theories of sex chromosome evolution. Such studies do not have to be limited to organisms in which complete genome sequences are already available, particularly in fungi where it is possible to electrophoretically isolate chromosome-specific DNA (Brody et al. 1991; Zolan et al. 1992; Geiser et al. 1996).
Acknowledgments
This work was supported by grant MCB-0129995 from the National Science Foundation. We thank reviewers for their most helpful comments.
References
- Bailey, J. A., L. Carrel, A. Chakravarti and E. E. Eichler, 2000. Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: the Lyon repeat hypothesis. Proc. Natl. Acad. Sci. USA 97: 6634–6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betrán, E., K. Thornton and M. Long, 2002. Selection at linked sites in the partial selfer Caenorhabditis elegans. Genome Res. 12: 1854–1859.12466289 [Google Scholar]
- Brody, H., J. Griffith, A. J. Cuticchia, J. Arnold and W. E. Timberlake, 1991. Chromosome-specific recombinant DNA libraries from the fungus Aspergillus nidulans. Nucleic Acids Res. 19: 3105–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull, J., 1978. Sex chromosomes in haploid dioecy: a unique contrast to Muller's theory for diploid dioecy. Am. Nat. 112: 245–250. [Google Scholar]
- Charlesworth, B., 1991. The evolution of sex chromosomes. Science 251: 1030–1033. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., and C. H. Langley, 1989. The population genetics of Drosophila transposable elements. Annu. Rev. Genet. 23: 251–287. [DOI] [PubMed] [Google Scholar]
- Cutter, A. D., and B. A. Payseur, 2003. Selection at linked sites in the partial selfer Caenorhabditis elegans. Mol. Biol. Evol. 20: 665–673. [DOI] [PubMed] [Google Scholar]
- Duret, L., G. Marais and C. Biémont, 2000. Transposons but no retrotransposons are located preferentially in regions of high recombination rate in Caenorhabditis elegans. Genetics 156: 1661–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlandsson, R., J. F. Wilson and S. Pääbo, 2000. Sex chromosomal transposable element accumulation and male-driven substitutional evolution in humans. Mol. Biol. Evol. 17: 804–812. [DOI] [PubMed] [Google Scholar]
- Filatov, D. A., F. Moneger, I. Negrutiu and D. Charlesworth, 2000. Low variability in a Y-linked plant gene and its implications for Y-chromosome evolution. Nature 404: 388–390. [DOI] [PubMed] [Google Scholar]
- Fraser, J. A., and J. Heitman, 2004. Evolution of fungal sex chromosomes. Mol. Microbiol. 51: 299–306. [DOI] [PubMed] [Google Scholar]
- Gallegos, A., D. J. Jacobson, N. B. Raju, M. P. Skupski and D. O. Natvig, 2000. Suppressed recombination and a pairing anomaly on the mating-type chromosome of Neurospora tetrasperma. Genetics 154: 623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber, E. D., and A. W. Day, 1985. Genetic-mapping of a phytopathogenic basidiomycete, Ustilago-violacea. Botanical Gazette 146: 449–459. [Google Scholar]
- Geiser, D. M., M. L. Arnold and W. E. Timberlake, 1996. Wild chromosome variants in Aspergillus nidulans. Curr. Genet. 29: 293–300. [DOI] [PubMed] [Google Scholar]
- Haber, J. E., 1998. Mating-type switching in Saccharomyces cerevisiae. Annu. Rev. Genet. 32: 561–599. [DOI] [PubMed] [Google Scholar]
- Hood, M. E., 2002. Dimorphic mating-type chromosomes in the fungus Microbotryum violaceum. Genetics 160: 457–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood, M. E., and J. Antonovics, 2004. Mating within the meiotic tetrad and the maintenance of genomic heterozygosity. Genetics 166: 1751–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S., K. Tamura and M. Nei, 1994. Mega-molecular evolutionary genetics analysis software for microcomputers. Comput. Appl. Biosci. 10: 189–191. [DOI] [PubMed] [Google Scholar]
- Lahn, B. T., and D. C. Page, 1999. Four evolutionary strata on the human X chromosome. Science 251: 964–967. [DOI] [PubMed] [Google Scholar]
- Lee, N., G. Bakkeren, K. Wong, J. E. Sherwood and J. W. Kronstad, 1999. The mating-type and pathogenicity locus of the fungus Ustilago hordei spans a 500-kb region. Proc. Natl. Acad. Sci. USA 96: 15026–15031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengeler, K. B., D. S. Fox, J. A. Fraser, A. Allen, K. Forrester et al., 2002. Mating-type locus of Cryptococcus neoformans: a step in the evolution of sex chromosomes. Eukaryot. Cell 1: 704–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindegren, C. C., 1936. a A six-point map of the sex chromosome of Neurospora crassa. J. Genet. 32: 243–256. [Google Scholar]
- Lindegren, C. C., 1936. b The structure of sex chromosomes of Neurospora crassa. J. Hered. 27: 251–259. [Google Scholar]
- Liu, Z. Y., P. H. Moore, H. Ma, C. M. Ackerman, M. Ragiba et al., 2004. A primitive Y chromosome in papaya marks incipient sex chromosome evolution. Nature 427: 348–352. [DOI] [PubMed] [Google Scholar]
- Merino, S. T., M. A. Nelson, D. J. Jacobson and D. O. Natvig, 1996. Pseudohomothallism and evolution of the mating-type chromosome in Neurospora tetrasperma. Genetics 143: 789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, H. J. J., 1914. A gene for the fourth chromosome of Drosophila. Exp. Zool. 17: 325–336. [Google Scholar]
- Okada, S., T. Sone, M. Fujisawa, S. Nakayama, M. Takenaka et al., 2001. The Y chromosome in the liverwort Marchantia polymorpha has accumulated unique repeat sequences harboring a male-specific gene. Proc. Natl. Acad. Sci. USA 98: 9454–9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulter, R. T. M., T. J. D. Goodwin and M. I. Butler, 2003. Vetrebrate helentrons and other novel Helitrons. Gene 313: 201–213. [DOI] [PubMed] [Google Scholar]
- Randall, T. A., and R. L. Metzenberg, 1998. The mating type locus of Neurospora crassa: identification of an adjacent gene and characterization of transcripts surrounding the idiomorphs. Mol. Gen. Genet. 259: 615–621. [DOI] [PubMed] [Google Scholar]
- Rice, W. R., 1987. Genetic hitchhiking and the evolution of reduced genetic activity of the Y sex chromosome. Genetics 116: 161–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzon, C., G. Marais, M. Gouy and C. Biemont, 2002. Recombination rate and the distribution of transposable elements in the Drosophila melanogaster genome. Genome Res. 12: 400–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinemann, M., and S. Steinemann, 2000. Common mechanisms of Y chromosome evolution. Genetica 109: 105–111. [DOI] [PubMed] [Google Scholar]
- Waterston, R. H., K. Lindblad-Toh, E. Birney, J. Rogers, J. F. Abril et al., 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562. [DOI] [PubMed] [Google Scholar]
- Wolf-Ekkehard, L., and H. Saedler, 2002. Chromosome rearrangements and transposable elements. Annu. Rev. Genet. 36: 389–410. [DOI] [PubMed] [Google Scholar]
- Zolan, M. E., J. R. Crittenden, N. K. Heyler and L. C. Seitz, 1992. Efficient isolation and mapping of RAD genes of the fungus Coprinus cinereus using chromosome-specific libraries. Nucleic Acids Res. 20: 3993–3999. [DOI] [PMC free article] [PubMed] [Google Scholar]