Abstract
We identified many variable-number tandem repeat (VNTR) loci in the genomes of Neisseria meningitidis serogroups A, B, and C and utilized a number of these loci to develop a multiple-locus variable-number tandem repeat analysis (MLVA). Eighty-five N. meningitidis serogroup B and C isolates obtained from Dutch patients with invasive meningococcal disease and seven reference strains were analyzed using MLVA and multilocus sequence typing (MLST). MLVA, based on eight VNTR loci with limited variability in the number of repeats, yielded clustering of the strains similar to that obtained by MLST, with congruence between both methods amounting to 69%. The ability to recognize clonal complexes makes MLVA a valuable high-throughput method to serve as a tool complementary to MLST. Four highly variable VNTR loci were used in a second assay to analyze N. meningitidis serogroup C strains collected during an outbreak of meningococcal disease in The Netherlands. Typing based on the latter VNTR loci enabled differentiation of isolates with identical MLST sequence types and grouped epidemiologically related strains.
Neisseria meningitidis remains a major cause of meningitis and septicemia worldwide (4, 24). On the basis of the structure of its capsule polysaccharide, 13 serogroups are recognized. Polysaccharide vaccines against serogroups A, C, Y, and W135 are available. Due to poor immunogenicity and cross-reactivity with neural tissue, a vaccine based on the serogroup B polysaccharide is not available. A licensed vaccine against serogroup B meningococci based on other components of the pathogen will not become available for some time. While disease due to serogroup A, W135, and C meningococci is prevalent in Africa and Asia, in Europe and the Americas serogroup B meningococci are causing most of the cases of meningococcal disease. Study of the epidemiology of N. meningitidis increases knowledge about the spread of the bacterium and has identified particular clones with apparent increased virulence (15, 20). Many different typing techniques have been employed to characterize meningococci. This is particularly true for the molecular techniques, which range from multi-locus enzyme electrophoresis to PorA variable region typing and multilocus sequence typing (MLST) (1, 3, 5, 20, 27, 33). MLST can now be considered the gold standard for genotyping N. meningitidis, and a large database is accessible via the Internet (http://pubmlst.org/neisseria/). MLST of N. meningitidis is a method using sequence data obtained from seven housekeeping genes. The alleles from these housekeeping genes are assigned allele numbers, and the combination of these allele numbers makes up a sequence type. MLST is a portable technique yielding unambiguous results and has been shown to be very suited for global epidemiology of meningococci (9, 20, 21, 31). However, despite these obvious advantages, MLST is a costly and labor-intensive typing technique. To type a single strain, seven PCRs and 14 sequence reactions are required. Recently, multiple-locus variable-number tandem repeat analysis (MLVA) has been introduced as a typing method for a large number of bacterial pathogens (6-8, 10, 16, 17, 19, 23, 26, 28, 29, 32, 34, 35) and meningococci (37, 39). In MLVA, the variability in the numbers of short tandem repeated sequences is utilized to create DNA fingerprints for epidemiological studies. Particularly with organisms that have a low rate of horizontal DNA transfer, MLVA often outperforms MLST in discriminatory power. However, MLVA has also proven to be useful for typing of microorganisms that exchange DNA at a high frequency.
Here we describe the use of MLVA to assess molecular epidemiology of N. meningitidis. We show that, given the right choice of repeat loci, MLVA yields a clustering that is similar to that of MLST but at a fraction of the costs associated with MLST. Although it will not replace MLST, MLVA may be a suitable method for the assessment of large collections of meningococcal isolates, allowing the rapid identification of genetically related groups, referred to as clonal complexes.
MATERIALS AND METHODS
Bacterial strains.
A total of 92 N. meningitidis isolates were used in this study. The collection comprised 43 serogroup B strains, 41 serogroup C strains, and a single W135 strain isolated from the spinal fluid samples of Dutch patients with invasive meningococcal disease during 2001 and 2002. Among these clinical isolates were seven serogroup C strains that were isolated during a meningococcal outbreak in the southern part of The Netherlands (36). Furthermore, we included reference strains for the major serotypes (for serogroup A, M1027; for serogroup B, M2092; for serogroup C, NCTC8554; for serogroup Y, Slaterus A72; and for serogroup W135, Artenstein 6308) and two other frequently used reference strains, serogroup B strain H44/76 (13) and serogroup C strain C11 (11).
Meningococcal isolates were characterized in The Netherlands Reference Laboratory for Bacterial Meningitis by serotyping, MLST (20), and sequencing of the variable regions of porA that encode the PorA epitopes on which the serosubtyping system is based (25). MLVA was performed at the Laboratory for Vaccine-Preventable Diseases of the National Institute of Public Health and the Environment. All data available for the strains used in this study are presented in Table 1.
TABLE 1.
Characteristics of the meningococcal strains used for MLST and MLVAa
| Strain | Serogroup | Serotype | ST | ST complex | PorA VR1 variantc | PorA VR2 variantc | PorA serosubtyped | MT | No. of repeats for VNTR locus, as determined by:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MLVA
|
HV-MLVA
|
|||||||||||||||||||
| V9-1e | V7-2 | V7-1 | V21-2 | V13-1 | V3-2 | V6-1 | V4-5 | V4-4 | V9-2 | V4-2 | V4-3 | |||||||||
| Reference strains | ||||||||||||||||||||
| M1027 | A | 4 | 4 | ST-4 | 1.5-2 | 1.1 | 56 | 2 | 1 | 3 | 2 | 2 | 6 | 3 | 4 | 12 | 9 | 5 | 14 | |
| H44/76 | B | 15 | 32 | ST-32 | ND | ND | P1.7,16 | 59 | 2 | 3 | 3 | 2 | 2 | 6 | 5 | 5 | 10 | 11 | 18 | 14 |
| M2092 | B | NT | 2875 | No | Deleted | Deleted | 45 | 3 | 15 | 3 | 2 | 2 | 5 | 2 | 7 | 3 | 8 | 11 | 5 | |
| C11 | C | 16 | 345 | No | ND | ND | P1.7-1.1 | 60 | 2 | 12 | 2 | 3 | 3 | 4 | 3 | 7 | 11 | 11 | 3 | 6 |
| NCTC8554 | C | NT | 344 | No | 1.5-1 | 1.10-4 | 37 | 2 | 1 | 3 | 3 | 2 | 3 | 2 | 3 | 9 | 12 | 11 | 12 | |
| Artenstein 6308 | W135 | 15 | 174 | No | 1.21 | 1.16 | 23 | 3 | 1 | 2 | 3 | 6 | 6 | 2 | 4 | 10 | 23 | 2 | 20 | |
| Slaterus A72 | Y | NT | 167 | No | 1.5-1 | 1.10-4 | 22 | 3 | 3 | 2 | 3 | 4 | 6 | 2 | 7 | 16 | 11 | 26 | 29 | |
| Strains from cases | ||||||||||||||||||||
| 2011782 | B | 15 | 32 | ST-32 | 1.7 | 1.16-33 | 42 | 2 | 3 | 2 | 2 | 2 | 6 | 2 | 5 | 10 | 8 | 19 | 8 | |
| 2012346 | B | 15 | 32 | ST-32 | 1.7 | 1.16 | 46 | 2 | 3 | 3 | 2 | 2 | 6 | 5 | 8 | 12 | 8 | 23 | 17 | |
| 2011745 | B | NT | 33 | ST-32 | 1.7-1 | 1.4-1 | 53 | 2 | 1 | 3 | 2 | 2 | 6 | 5 | 7 | 8 | 15 | 26 | 11 | |
| 2010530 | B | 4 | 34 | ST-32 | 1.5-2 | 1.10-4 | 51 | 2 | 1 | 3 | 2 | 2 | 6 | 4 | 7 | 9 | 13 | 16 | 19 | |
| 2012139 | B | 4 | 34 | ST-32 | 1.5-2 | 1.1 | 52 | 2 | 1 | 3 | 2 | 2 | 6 | 5 | 5 | 7 | 14 | 18 | 16 | |
| 2011149 | B | 4 | 34 | ST-32 | 1.5 | 1.2 | 54 | 2 | 1 | 3 | 2 | 2 | 6 | 5 | 3 | 12 | 11 | 26 | 9 | |
| 2011921 | B | 4 | 34 | ST-32 | 1.5-2 | 1.1 | 55 | 2 | 1 | 3 | 2 | 2 | 6 | 5 | 11 | 8 | 10 | 18 | 12 | |
| 2010045 | B | 15 | 259 | ST-32 | 1.7 | 1.16 | 46 | 2 | 3 | 3 | 2 | 2 | 6 | 5 | 8 | 13 | 15 | 33 | 12 | |
| 2010447 | B | 15 | 259 | ST-32 | 1.7 | 1.16 | 47 | 2 | 3 | 3 | 2 | 2 | 6 | 5 | 10 | 8 | 11 | 26 | 9 | |
| 2011337 | B | 15 | 259 | ST-32 | 1.7 | 1.16 | 48 | 2 | 3 | 3 | 2 | 2 | 6 | 5 | 7 | 8 | 9 | 32 | 23 | |
| 2010002 | B | 15 | 2696 | ST-32 | 1.5-2 | 1.1 | 52 | 2 | 1 | 3 | 2 | 2 | 6 | 5 | 5 | 9 | 8 | 13 | 30 | |
| 2010016 | B | 4 | 2697 | ST-32 | 1.19 | 1.15 | 50 | 2 | 1 | 3 | 2 | 2 | 6 | 6 | 7 | 16 | 11 | 32 | 22 | |
| 2012567 | B | 15 | 2707 | ST-32 | 1.7-2 | 1.16-33 | 49 | 2 | 4 | 3 | 2 | 2 | 6 | 5 | 9 | 12 | 6 | 12 | 14 | |
| 2010958 | B | 4 | 35 | ST-35 | 1.22-1 | 1.14 | 58 | 2 | 1 | 3 | 8 | 6 | 6 | 2 | 8 | 8 | 19 | 17 | 18 | |
| 2012534 | B | NT | 40 | ST-41/44 | 1.7-2 | 1.13-2 | 8 | 2 | 1 | 2 | 3 | 3 | 5 | 2 | 8 | 10 | 7 | 19 | 11 | |
| 2011486 | B | 4 | 40 | ST-41/44 | 1.7-2 | 1.13-2 | 12 | 2 | 1 | 2 | 3 | 2 | 5 | 2 | 7 | 7 | 7 | 20 | 9 | |
| 2010319 | B | 4 | 41 | ST-41/44 | 1.7-2 | 1.4 | 1 | 2 | 2 | 2 | 2 | 3 | 5 | 2 | 7 | 5 | 8 | 17 | 15 | |
| 2011800 | B | 4 | 41 | ST-41/44 | 1.7-2 | 1.4 | 9 | 2 | 1 | 2 | 2 | 3 | 5 | 2 | 7 | 9 | 6 | 23 | 22 | |
| 2010643 | B | 4 | 41 | ST-41/44 | 1.7-2 | 1.4 | 10 | 2 | 1 | 2 | 2 | 3 | 5 | 2 | 5 | 12 | 21 | 18 | 18 | |
| 2010780 | B | 4 | 42 | ST-41/44 | 1.7-2 | 1.4 | 2 | 2 | 1 | 2 | 2 | 3 | 5 | 8 | 7 | 12 | 6 | 16 | 14 | |
| 2012565 | B | 4 | 42 | ST-41/44 | 1.7-2 | 1.4 | 9 | 2 | 1 | 2 | 2 | 3 | 5 | 2 | 7 | 11 | 6 | 13 | 31 | |
| 2011184 | B | 4 | 42 | ST-41/44 | 1.7-2 | 1.4 | 11 | 2 | 1 | 2 | 2 | 3 | 5 | 2 | 10 | 7 | 4 | 14 | 23 | |
| 2010559 | B | 15 | 571 | ST-41/44 | 1.7-1 | 1.15 | 20 | 2 | 2 | 2 | 3 | 2 | 6 | 5 | 4 | 9 | 12 | 23 | 16 | |
| 2011648 | B | 4 | 839 | ST-41/44 | 1.21 | 1.16 | 21 | 2 | 2 | 2 | 3 | 3 | 3 | 4 | 7 | 17 | 28 | 14 | 17 | |
| 2010044 | B | 4 | 1127 | ST-41/44 | 1.5-2 | 1.10-2 | 3 | 2 | 1 | 2 | 3 | 3 | 5 | 2 | 7 | 10 | 6 | 19 | 17 | |
| 2012251 | B | 4 | 1194 | ST-41/44 | 1.18-1 | 1.3 | 17 | 2 | 1 | 3 | 2 | 3 | 6 | 2 | 5 | 12 | 9 | 3 | 26 | |
| 2012113 | B | 4 | 1255 | ST-41/44 | 1.7-2 | 1.4 | 4 | 2 | 1 | 2 | 3 | 3 | 5 | 2 | 11 | 11 | 9 | 14 | 12 | |
| 2010264 | B | NT | 1374 | ST-41/44 | 1.7-2 | 1.13-2 | 5 | 2 | 1 | 2 | 3 | 3 | 5 | 2 | 4 | 11 | 9 | 11 | 18 | |
| 2011026 | B | 4 | 1374 | ST-41/44 | 1.7-2 | 1.13-2 | 6 | 2 | 1 | 2 | 3 | 3 | 5 | 2 | 3 | 7 | 9 | 13 | 17 | |
| 2010833 | B | 4 | 1960 | ST-41/44 | 1.7-2 | 1.13-2 | 3 | 2 | 1 | 2 | 3 | 3 | 5 | 2 | 7 | 10 | 7 | 17 | 13 | |
| 2011452 | B | 4 | 1960 | ST-41/44 | 1.22-1 | 1.14 | 3 | 2 | 1 | 2 | 3 | 3 | 5 | 2 | 7 | 7 | 7 | 19 | 24 | |
| 2012176 | B | 4 | 2016 | ST-41/44 | 1.5-1 | 1.10-4 | 7 | 2 | 1 | 2 | 3 | 3 | 5 | 2 | 5 | 7 | 9 | 19 | 8 | |
| 2010915 | B | 4 | 2702 | ST-41/44 | 1.7-2 | 1.4 | 10 | 2 | 1 | 2 | 2 | 3 | 5 | 2 | 5 | 8 | 11 | 3 | 12 | |
| 2011472 | B | 4 | 2703 | ST-41/44 | 1.7-2 | 1.4 | 9 | 2 | 1 | 2 | 2 | 3 | 5 | 2 | 7 | 11 | 7 | 19 | 17 | |
| 2012640 | B | 1 | 2708 | ST-41/44 | 1.7-2 | 1.4 | 19 | 2 | 1 | 2 | 3 | 3 | 6 | 2 | 8 | 7 | 6 | 7 | 9 | |
| 2011319 | B | 1 | 213 | ST-213 | 1.22 | 1.14 | 14 | 2 | 1 | 2 | 2 | 3 | 6 | 4 | 5 | 8 | 9 | 20 | 9 | |
| 2012072 | B | 1 | 213 | ST-213 | 1.22 | 1.14 | 14 | 2 | 1 | 2 | 2 | 3 | 6 | 4 | 5 | 7 | 10 | 19 | 22 | |
| 2011615 | B | 1 | 2705 | ST-213 | 1.22 | 1.14 | 15 | 2 | 1 | 2 | 2 | 3 | 6 | 5 | 5 | 8 | 11 | 14 | 29 | |
| 2011529 | B | NT | 254 | ST-254 | 1.19 | 1.13-2 | 16 | 2 | 1 | 2 | 2 | 3 | 7 | 2 | 5 | 9 | 25 | 9 | 18 | |
| 2010237 | B | 4 | 2701 | ST-254 | 1.19 | 1.15 | 18 | 2 | 1 | 3 | 2 | 3 | 6 | 2 | 3 | 17 | 21 | 19 | 18 | |
| 2010806 | B | 4 | 474 | ST-376 | 1.19-1 | 1.15 | 38 | 2 | 1 | 2 | 3 | 2 | NP | 3 | 3 | 13 | 16 | 2 | 20 | |
| 2012490 | B | 1 | 461 | ST-461 | 1.22 | 1.9 | 13 | 2 | 1 | 2 | 3 | 3 | 5 | 3 | 2 | 11 | 13 | 3 | 18 | |
| 2010147 | B | 16 | 2700 | No | 1.22-1 | 1.14 | 41 | 2 | 1 | 2 | 2 | 2 | 6 | 2 | 6 | 14 | 16 | 19 | 14 | |
| 2010178 | C | 2a | 11 | ST-11 | 1.5 | 1.2 | 24 | 4 | 1 | 2 | 4 | 5 | 4 | 3 | 3 | 10 | 26 | 26 | 14 | |
| 2010443 | C | 2a | 11 | ST-11 | 1.5 | 1.2 | 24 | 4 | 1 | 2 | 4 | 5 | 4 | 3 | 3 | 11 | 27 | 27 | 9 | |
| 2010445 | C | 2a | 11 | ST-11 | 1.5 | 1.2 | 24 | 4 | 1 | 2 | 4 | 5 | 4 | 3 | 3 | 13 | 27 | 30 | 10 | |
| 2010980 | C | 2a | 11 | ST-11 | 1.5 | 1.2 | 25 | 4 | 1 | 2 | 4 | 5 | 4 | 3 | 4 | 7 | 25 | 10 | 10 | |
| 2011215 | C | 2a | 11 | ST-11 | 1.5 | 1.2 | 26 | 4 | 1 | 2 | 4 | 5 | 4 | 3 | 5 | 16 | 16 | 8 | 27 | |
| 2011929 | C | 2a | 11 | ST-11 | 1.5 | 1.2 | 26 | 4 | 1 | 2 | 4 | 5 | 4 | 3 | 5 | 14 | 16 | 9 | 30 | |
| 2012280 | C | 2a | 11 | ST-11 | 1.5 | 1.2 | 26 | 4 | 1 | 2 | 4 | 5 | 4 | 3 | 5 | 10 | 17 | 8 | 29 | |
| 2010563 | C | 2a | 11 | ST-11 | 1.5-1 | 1.10-4 | 27 | 4 | 1 | 2 | 4 | 5 | 6 | 3 | 5 | 14 | 25 | 18 | 22 | |
| 2010938 | C | 2a | 11 | ST-11 | 1.5-1 | 1.10-8 | 27 | 4 | 1 | 2 | 4 | 5 | 6 | 3 | 5 | 14 | 25 | 14 | 9 | |
| 2011205 | C | 2a | 11 | ST-11 | 1.5-1 | 1.10-8 | 27 | 4 | 1 | 2 | 4 | 5 | 6 | 3 | 5 | 12 | 25 | 13 | 9 | |
| 2011784 (Pt 1)b | C | 2a | 11 | ST-11 | 1.5-1 | 1.10-8 | Neg. | 27 | 4 | 1 | 2 | 4 | 5 | 6 | 3 | 5 | 14 | 25 | 8 | 18 |
| 2011785 (Pt 2) | C | 2a | 11 | ST-11 | 1.5-1 | 1.10-8 | Neg. | 27 | 4 | 1 | 2 | 4 | 5 | 6 | 3 | 5 | 10 | 25 | 8 | 18 |
| 2011791 (Pt 3) | C | 2a | 11 | ST-11 | 1.5-1 | 1.10-8 | Neg. | 27 | 4 | 1 | 2 | 4 | 5 | 6 | 3 | 5 | 14 | 25 | 8 | 18 |
| 2011792 (Pt 4) | C | 2a | 11 | ST-11 | 1.5-1 | 1.10-8 | Neg. | 27 | 4 | 1 | 2 | 4 | 5 | 6 | 3 | 5 | 10 | 25 | 8 | 18 |
| 2011801 (Pt 5) | C | 2a | 11 | ST-11 | 1.5-1 | 1.10-8 | Neg. | 27 | 4 | 1 | 2 | 4 | 5 | 6 | 3 | 5 | 14 | 25 | 7 | 18 |
| 2011793 (Pt 6) | C | 2a | 11 | ST-11 | 1.5-1 | 1.10-8 | 27 | 4 | 1 | 2 | 4 | 5 | 6 | 3 | 5 | 14 | 25 | 10 | 17 | |
| 2011814 (Pt 7) | C | 2a | 11 | ST-11 | 1.5-1 | 1.10-8 | 27 | 4 | 1 | 2 | 4 | 5 | 6 | 3 | 5 | 10 | 25 | 20 | 10 | |
| 2011980 | C | 2a | 11 | ST-11 | 1.5-1 | 1.10-8 | 27 | 4 | 1 | 2 | 4 | 5 | 6 | 3 | 5 | 14 | 26 | 19 | 12 | |
| 2012140 | C | 2a | 11 | ST-11 | 1.5-1 | 1.10-8 | 27 | 4 | 1 | 2 | 4 | 5 | 6 | 3 | 5 | 14 | 25 | 25 | 13 | |
| 2012588 | C | 2a | 11 | ST-11 | 1.5-1 | 1.10-8 | 27 | 4 | 1 | 2 | 4 | 5 | 6 | 3 | 5 | 11 | 25 | 12 | 7 | |
| 2012199 | C | 2a | 11 | ST-11 | 1.5-1 | 1.10-1 | 30 | 6 | 1 | 2 | 4 | 2 | 4 | 4 | 5 | 11 | 24 | 13 | 11 | |
| 2011328 | C | 2a | 11 | ST-11 | 1.7-4 | 1.1 | 31 | 2 | 1 | 2 | 4 | 4 | 5 | 3 | 4 | 6 | 24 | 19 | 22 | |
| 2011371 | C | 2a | 11 | ST-11 | 1.5 | 1.2 | 32 | 2 | 1 | 2 | 4 | 4 | 5 | 3 | 7 | 19 | 27 | 19 | 21 | |
| 2011852 | C | 2a | 11 | ST-11 | 1.5 | 1.2 | 43 | 3 | 1 | 2 | 2 | 2 | 5 | 2 | 5 | 13 | 27 | 24 | 24 | |
| 2011978 | C | 2a | 247 | ST-11 | 1.5 | 2 | 28 | 1 | 1 | 2 | 4 | 5 | 4 | 3 | 5 | 15 | 16 | 9 | 18 | |
| 2011975 | C | 2a | 247 | ST-11 | 1.5-1 | 1.10-8 | 29 | 4 | 1 | 2 | 4 | 5 | 6 | 2 | 5 | 14 | 27 | 8 | 18 | |
| 2011872 | C | 2a | 247 | ST-11 | 1.5 | 2-1 | 34 | 7 | 1 | 2 | 4 | 4 | 5 | 3 | 5 | 16 | 24 | 16 | 24 | |
| 2021708 | C | 2a | 247 | ST-11 | 1.5 | 2 | 36 | 5 | 1 | 2 | 3 | 4 | 4 | 2 | 5 | 13 | 28 | 23 | 25 | |
| 2011528 | C | 2a | 2704 | ST-11 | 1.5 | 1.2 | 35 | 5 | 1 | 2 | 3 | 4 | 4 | 3 | 5 | 14 | 26 | 20 | 22 | |
| 2011576 | C | 2a | 2704 | ST-11 | 1.5 | 1.2 | 35 | 5 | 1 | 2 | 3 | 4 | 4 | 3 | 5 | 16 | 26 | 21 | 22 | |
| 2012402 | C | 2a | 2706 | ST-11 | 1.5-1 | 1.10-8 | 27 | 4 | 1 | 2 | 4 | 5 | 6 | 3 | 5 | 13 | 26 | 19 | 9 | |
| 2012673 | C | 2a | 2709 | ST-11 | Deleted | Deleted | 27 | 4 | 1 | 2 | 4 | 5 | 6 | 3 | 5 | 19 | 24 | 21 | 18 | |
| 2020435 | C | 2a | 3298 | ST-11 | 1.5-1 | 1.10-8 | 27 | 4 | 1 | 2 | 4 | 5 | 6 | 3 | 5 | 9 | 27 | 6 | 20 | |
| 2011728 | C | 2a | 4283 | ST-11 | 1.5-1 | 1.10-8 | 27 | 4 | 1 | 2 | 4 | 5 | 6 | 3 | 5 | 19 | 17 | 35 | 7 | |
| 2011763 | C | 4 | 34 | ST-32 | 1.5-2 | 1.1 | 52 | 2 | 1 | 3 | 2 | 2 | 6 | 5 | 5 | 8 | 12 | 24 | 17 | |
| 2010118 | C | NT | 60 | ST-60 | 1.5 | 1.2 | 44 | 2 | 1 | 2 | 2 | 2 | 5 | 2 | 3 | 8 | 16 | 14 | 15 | |
| 2012403 | C | 2b | 2680 | ST-8 | 1.5 | 1.2 | 39 | 5 | 1 | 3 | 3 | 2 | 5 | 3 | 7 | 11 | 10 | 25 | 28 | |
| 2010937 | C | 2b | 2680 | ST-8 | 1.5 | 1.2 | 40 | 5 | 1 | 3 | 3 | 2 | 5 | 3 | 5 | 13 | 10 | 27 | 36 | |
| 2010115 | C | 2b | 2699 | ST-8 | 1.5 | 1.2 | 39 | 5 | 1 | 3 | 3 | 2 | 5 | 3 | 7 | 10 | 9 | 14 | 33 | |
| 2010059 | C | NT | 2698 | ST-334 | 1.5-1 | 1.10-1 | 57 | 2 | 1 | 3 | 4 | 2 | 4 | 5 | 2 | 9 | 7 | 15 | 36 | |
| 2020982 | C | 2a | 2732 | ST-35 | 1.5 | 2 | 9 | 2 | 1 | 2 | 2 | 3 | 5 | 2 | 7 | 11 | 19 | 24 | 18 | |
| 2010351 | W135 | 2a | 247 | ST-11 | 1.5 | 2 | 33 | 8 | 1 | 2 | 4 | 4 | 5 | 3 | 10 | 8 | 30 | 26 | 34 | |
Abbreviations: NT, nontypeable; No, not belonging to ST complex or lineage; ND, not determined; Deleted, porA gene deleted; Neg., no PorA expression.
Pt numbers indicate the seven patients from the meningococcal outbreak.
VR1, variable region 1; VR2, variable region 2.
V9-1, VNTR9-1; V7-2, VNTR7-2; and so on for all VNTR loci.
In silico tandem repeat searches.
The genome sequences of N. meningitidis serogroup A isolates (strain Z2491, GenBank accession no. NC_003116), serogroup B isolates (strain MC58, GenBank accession no. NC_003112), and serogroup C isolates (strain FAM18, preliminary genome sequence available at http://www.sanger.ac.uk) were screened for the presence of tandem repeat sequences by using the Tandem Repeats Finder program, version 2.02 (2), and a custom-made script for the Kodon 2.5 beta software (Applied Maths, Sint-Martens-Latem, Belgium).
MLVA.
Variable-number tandem repeat (VNTR) PCRs were performed in 25-μl volumes with Applied Biosystems 9700 PCR machines (Applied Biosystems, Foster City, Calif.). One microliter of purified N. meningitidis genomic DNA (10 ng/μl) or 1 μl of 1:10-diluted, heat-treated N. meningitidis lysate was added to a mixture containing 10 pmol of 5′ 6-carboxyfluorescein-labeled forward primer, 10 pmol unlabeled reverse primer, and 12.5 μl of HotStarTaq mastermix (QIAGEN, Hilden, Germany). All primer sequences are shown in Table 2. VNTR loci were amplified in separate PCRs using the following program: 15 min at 95°C, followed by 20 cycles of amplification that consisted of 20 s at 95°C, 30 s at 55°C, and 30 s at 72°C, and a final step of 30 min at 68°C to ensure complete terminal transferase activity of the Taq DNA polymerase. After PCR, samples were diluted 1:200 in water and 2 μl of the diluted samples was mixed with 10 μl of GeneScan ROX 500 size standard diluted 1:200 in water (Applied Biosystems). After heat denaturation for 5 min at 95°C, fragments were separated with an ABI 3700 DNA sequencer by using the standard GeneScan module with filter set D. The GeneScan data were analyzed with GeneMapper software (Applied Biosystems) to perform sizing and to calculate the number of repeats in the PCR fragments.
TABLE 2.
Primers used for the MLVA
| VNTR locus | Forward primer
|
Reverse primer
|
||||
|---|---|---|---|---|---|---|
| Primer name | Sequence | Genome coordinatesa | Primer name | Sequence | Genome coordinatesa | |
| MLVA | ||||||
| VNTR9-1 | NmVNTR09-01FF | ATTAAAGAAACACACGGCG | 391994-392012 | NmVNTR09-01R | GAAAATTCCTGCACAATCC | 392136-392154 |
| VNTR7-2 | NmVNTR07-02FF | AGCAGCATGGTAGCTCGT | 2161864-2161881 | NmVNTR07-02R | GAAGGAAAATCCGGCAA | 2162011-2162027 |
| VNTR7-1 | NmVNTR07-01FF | GGAATGACGGAATCTTAAGTT | 1907337-1907357 | NmVNTR07-01R | ATACCCCGAACTGAAAATG | 1907423-1907441 |
| VNTR21-2 | NmVNTR21-02FF | GCTTCCTGAGTCGCGTC | 1518264-1518280 | NmVNTR21-02R | GCGAAAGACCAAGTCAAAGAT | 1518377-1518397 |
| VNTR13-1 | NmVNTR13-01FF | TTATCGGTATCCCTTCTTTTATG | 369330-369352 | NmVNTR13-01R | TWTAAGTTGACATATTGTGCGAT | 369418-369440 |
| VNTR3-2 | NmVNTR03-02FF | TAGCTGTTGCAACAACACTT | 2061499-2061518 | NmVNTR03-02R | TCTTGATACCGGCGTAAG | 2061631-2061648 |
| VNTR6-1 | NmVNTR06-01FH | GAAGAATAGTTGCCATCGTCT | 863490-863510 | NmVNTR06-01R | TTTTGCGCTGTATTGCC | 863552-863568 |
| VNTR4-5 | NmVNTR04-05FF | CCAAAAGMGGAGAGGTATAAA | 1576196-1576216 | NmVNTR04-05R | TCAAAGGTTTGAGCAATTTGAT | 1576528-1576549 |
| HV-MLVA | ||||||
| VNTR4-4 | NmVNTR04-03FH | ATACACCGTATCCGCCG | 1556715-1556731 | NmVNTR04-03R | CAGGAAGGGGGTTATAATTTTTT | 1556807-1556829 |
| VNTR9-2 | NmVNTR09-02FN | AAAGGCAAAAACCGCAT | 285887-285903 | NmVNTR09-02R | GATACCGCCCTGCTGTT | 286003-286019 |
| VNTR4-2 | NmVNTR04-02FF | TGTGTTGGATTTTCCGTTT | 1400191-1400209 | NmVNTR04-02R | TGTCTTTTTCGGGTTAATTTAGT | 1400367-1400389 |
| VNTR4-3 | NmVNTR04-03FF | GCAAAACAGCGATAAAACAGATA | 321164-321186 | NmVNTR04-03R | ACAAAGGAAAATTTCTCATGACA | 321079-321101 |
Nucleotide coordinates in N. meningitidis serogroup B genome sequence NC_003112.
To ensure calculation of the number of repeats was correct, PCR products representing at least two variants of each VNTR locus were sequenced. DNA sequencing revealed that calculation of the number of repeats in VNTR3-2 and VNTR6-1 based on fragment sizing was inaccurate. For these VNTR loci, the addition of an extra repeat to the calculated number was required to obtain the true number of repeats. The data with the calculated number of repeats were imported into the Bionumerics version 4 software package (Applied Maths) for further cluster analysis.
Data analysis.
The MLST and MLVA profiles were clustered with the Bionumerics software by using a categorical coefficient and a graphing method called the minimum spanning tree as described before (29). In the minimum spanning tree, the priority rule to first link types that have the highest number of single-locus variants was chosen. In the tree, types are represented by circles and the size of a circle indicates the number of strains with this particular type. Thick, short lines connecting two types denote types differing in a single locus; thin, longer lines connect double-locus variants; and dotted lines indicate the most likely connection between two types differing in more than two loci. For MLST, a maximum neighbor difference of 2 was used to create complexes. For MLVA, a maximum neighbor difference of 1 was used for the creation of groups.
For calculation of the genetic diversity and discrimination index, Simpson's index of diversity (DI) was used (14, 30). 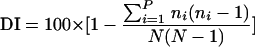 , where ni is the number of strains belonging to the ith type and N is the total number of strains in the sample population.
, where ni is the number of strains belonging to the ith type and N is the total number of strains in the sample population.
DNA sequencing.
For DNA sequencing reactions, fluorescence-labeled dideoxynucleotide technology was used (Applied Biosystems). Sequence reaction mixtures were analyzed with an ABI 3700 automated DNA sequencer.
RESULTS
Identification of VNTR loci in N. meningitidis.
Using the Tandem Repeats Finder program and Kodon software, we identified many tandem repeated sequences in silico in the available genome sequences of N. meningitidis serogroups A, B, and C. We selected and tested 29 different tandem repeat loci for their suitability for typing by using a set of 12 genetically and phenotypically diverse N. meningitidis strains. These strains were a subset of the clinical isolates used for the final analysis and strains H44/76 and C11. Some repeat regions hardly varied in composition, and others could not be amplified from a number of strains tested. In some cases, the PCR yielded multiple bands, either due to the fact that some of the VNTR regions were present in several copies in the genome or due to possible nonspecific PCR. Although more of the 29 tandem repeat loci may be usable if PCR conditions are adapted, 12 different tandem repeat loci that consistently yielded a single band in the PCR and with which at least some polymorphism was observed were selected for further analysis (Tables 3 and 4). Some of the repeat loci carried the same repeat unit but were located at different positions in the genome, with different flanking sequences, and as such are considered to be different VNTR loci. Examples of loci with identical repeats but with different locations and different flanking sequences are VNTR4-3 and VNTR4-5 (Table 3). Using the annotated genome sequences of serogroups A and B, we found that about half of the 12 VNTRs were located in noncoding regions, while the other half were positioned in coding regions, although these were sometimes annotated as open reading frames encoding hypothetical proteins (Table 3).
TABLE 3.
Sequences and positions of VNTR loci used for MLVA of N. meningitidis
| VNTR locus | Size (bp) | Repeat sequence | Detail for N. meningitidis serogroup (GenBank accession no. or strain)
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Serogroup A (NC_003116)
|
Serogroup B (NC_003112)
|
Serogroup C (FAM18)a
|
||||||||
| Genome coordinates | No. of repeats | Annotated putative function of ORFb | Genome coordinates | No. of repeats | Annotated putative function of ORFb | Genome coordinates | No. of repeats | |||
| MLVA | ||||||||||
| VNTR9-1 | 9 | CCGCTACCC | 2052950-2052967 | 2 | Aldose 1-epimerase (mutarotase) | 392028-392045 | 2 | Aldose 1-epimerase | 1819003-1819047 | 5 |
| VNTR7-2 | 7 | TTTCCTG | 365308-365314 | 1 | Hypothetical protein | 2161973-2161993 | 3 | Noncoding | 2074902-2074908 | 1 |
| VNTR7-1 | 7 | TGTTTTC | 632208-632228 | 3 | Noncoding | 1907366-1907386 | 3 | Noncoding | 423366-423379 | 2 |
| VNTR21-2 | 21 | GCTTCAGTTACAGCTTCTTTG | 1603619-1603660 | 2 | Membrane protein | 1518309-1518350 | 2 | Hypothetical protein | 1407985-1408068 | 4 |
| VNTR13-1 | 13 | ACGGGAAAATACG | 2075417-2075442 | 2 | Noncoding | 369378-369403 | 2 | Noncoding | 1844470-1844534 | 5 |
| VNTR3-2 | 3 | GGC | 466815-466832 | 6 | Outer membrane peptidase | 2061531-2061548 | 6 | Serotype 1-specific antigen | 1967113-1967127 | 5 |
| VNTR6-1 | 6 | TTCATC | 1007258-107263 | 1 | ATP-dependent ClpA protease | 863513-863536 | 5 | ATP-dependent ClpA protease | 793093-793104 | 3 |
| VNTR4-5 | 4 | GCTT | 1658340-1658355 | 4 | Noncoding | 1576262-1576281 | 5 | Noncoding | 1463466-1463493 | 7 |
| HV-MLVA | ||||||||||
| VNTR4-4 | 4 | AAGC | 1638926-1638973 | 12 | Noncoding | 1556763-1556806 | 11 | Hypothetical protein | 1444060-1444091 | 8 |
| VNTR9-2 | 9 | AAAGTTGCC | 2158511-2158555 | 5 | Rotamase | 285909-285971 | 7 | Peptidyl-prolyl cis-trans isomerase | 277436-277669 | 26 |
| VNTR4-2 | 4 | AGCC | 1494788-1494799 | 3 | Hypothetical protein | 1400279-1400358 | 20 | Noncoding | 1297888-1297987 | 25 |
| VNTR4-3 | 4 | GCTT | 2123413-2123444 | 8 | Noncoding | 321106-321141 | 9 | Noncoding | 1892701-1892836 | 34 |
Preliminary genome sequence from http://www.sanger.ac.uk; no annotation known yet.
ORF, open reading frame.
TABLE 4.
Sequences and positions of tandem repeat loci not used for MLVA of N. meningitidis
| VNTR locus | Size (bp) | Sequence | Reason for not using VNTR locus in MLVA | Genome coordinatea | Alternate VNTR locus designationb |
|---|---|---|---|---|---|
| VNTR3-1 | 3 | TGC | Low degree of polymorphism, tested on all 92 strains | 1910615 | |
| VNTR4-1 | 4 | GGCA | Low degree of polymorphism, tested on all 92 strains | 1882462 | |
| VNTR4-6 | 4 | AGCA | Several copies of VNTR locus present in genome | 321107 | |
| VNTR4-7 | 4 | AAGG | VNTR locus not present in all test strains | 1994623 | |
| VNTR4-8 | 4 | AAAT | VNTR locus not present in all test strains | 2100218 | |
| VNTR5-1 | 5 | GAAGA | Several copies of VNTR locus present in N. meningitidis serotype B | 455614 | |
| VNTR5-2 | 5 | TTGGG | VNTR locus not present in all test strains | 1273601 | VNTR10 |
| VNTR7-1 | 7 | AAACAAC | Very weak PCR band in N. meningitidis serotype C test strains | 657239 | VNTR01 |
| VNTR7-2 | 7 | ATTTCTC | Not present in all test strains | 773266 | |
| VNTR8-1 | 8 | ATAACAAA | Not present in all test strains | 1437002 | |
| VNTR10-1 | 3 | TAATCCACTA | Polymorphism (deletions) in region flanking locus | 1303221 | VNTR11 |
| VNTR13-1 | 13 | CTGTAGAGATGGG | Several copies of downstream region present in genome | 1131295 | VNTR02 |
| VNTR19-1 | 18 | GGTAATTCCTGACGATTCA | Several copies of VNTR locus present in genome | 1952085 | VNTR04 |
| VNTR19-2 | 19 | CGTCATTCCCACCACTTTT | Several copies of VNTR locus present in genome | 1690479 | |
| VNTR19-3 | 19 | CCGTCATTCCCGCCACTTT | Several copies of VNTR locus present in genome | 1690136 | |
| VNTR21-1 | 21 | GCGTACGGACGTATTCGGCCT | Low degree of polymorphism, tested on all 92 strains | 1425081 | |
| VNTR21-3 | 21 | TTTTTAGATGATGTAAATGTT | Multiple PCR bands, possible PCR artifact | 2228625 |
Variability of VNTRs in N. meningitidis strains.
Analysis of the composition of the VNTR loci of the N. meningitidis strain collection revealed that the diversity indices varied among the various VNTR loci. Some loci carried large numbers of repeats, e.g., up to 36 repeats in the VNTR4-3 locus, while other loci carried only a limited number of repeats, e.g., VNTR7-1, with a maximum of 3 repeats.
The diversity indices of the VNTR loci differed between the N. meningitidis serogroup B and C strains. VNTR7-2 varied between one and four repeats among N. meningitidis serogroup B strains, yielding a diversity index of 36.4%. In contrast, the same locus did not vary at all among N. meningitidis serogroup C strains and all strains carried only a single repeat unit in VNTR7-2. Conversely, VNTR9-1 was invariable among N. meningitidis serogroup B strains whereas N. meningitidis serogroup C strains varied considerably (DI = 59.8%). The overall diversities of the VNTR loci were somewhat lower in the N. meningitidis serogroup C strains than in the serogroup B strains.
Initially, all VNTR loci were used in a single assay, but this yielded a different profile for each strain included in the study. Thereafter, several combinations of VNTR loci were used to determine which VNTR loci should be included in the MLVA to obtain sufficiently large groups. Finally, we selected eight VNTR loci (VNTR7-2, VNTR7-1, VNTR9-1, VNTR3-2, VNTR21-2, VNTR6-1, VNTR4-5, and VNTR13-1) for use in an analysis designated the MLVA.
The remaining four repeat loci (VNTR4-4, VNTR9-2, VNTR4-2, and VNTR4-3) on average carried high numbers of repeats and also had a high diversity index among the N. meningitidis strains studied. The use of these highly variable VNTR loci resulted in a high degree of differentiation that is unsuitable for population studies and global epidemiology but may be suitable for outbreak analysis. Therefore, these four loci were combined into a second MLVA, designated the highly variable MLVA (HV-MLVA).
Stability of VNTRs.
The two reference strains H44/76 and C11 were subcultured for 30 consecutive days by streaking single colonies from each strain on chocolate agar plates and incubated overnight at 35°C and 5% CO2. Suspensions of each subculture were made, and the composition of each of the 12 selected VNTRs was determined. With a single exception, all VNTRs remained unchanged during subculture. Only VNTR4-3 in serogroup B strain H44/76 had gained a single repeat on day 27 and remained unaltered during the remaining 3 days of subculturing.
Comparison of MLST and MLVA of N. meningitidis.
To determine the value of MLVA of N. meningitidis for molecular epidemiology, we performed MLST and MLVA of the clinical N. meningitidis isolates obtained from Dutch patients and of the seven reference strains. MLST showed that the collection contained 47 different sequence types (STs), belonging to 12 different previously identified complexes. A number of strains had STs that are not assigned to a known MLST complex as described on the MLST website (http://pubmlst.org/neisseria).
MLVA resulted in a slightly higher differentiation of the N. meningitidis strains than MLST, yielding 60 MLVA types (MTs) as opposed to 47 MLST STs (Fig. 1). The increase was most pronounced with N. meningitidis serogroup C strains, where the diversity index increased from 68.6% for MLST (15 STs) to 84.1% for MLVA (21 MTs). With N. meningitidis serogroup B strains, the diversity index was 97.8% for MLST (30 STs) and 99.0% for MLVA (37 MTs). Similar to the profiles obtained by MLST, the profiles obtained by MLVA were clustered in a minimum spanning tree, creating groups if seven of the eight VNTR loci were identical. The number of strains tested in this study is relatively small, and not all major ST complexes were represented (e.g., ST-269 and ST-5). Therefore, the difference between the diversity indices of MLST and MLVA may change if a larger, more complete set of strains is analyzed.
FIG. 1.
Minimum spanning tree of MLST and MLVA of N. meningitidis isolates obtained from 85 Dutch patients with invasive meningococcal disease and seven reference strains. Each circle denotes a particular MLST or MLVA type, and the size of the circle indicates the number of isolates of that particular type. Thick, solid lines denote connections between single-locus variants; thin, solid lines denote double-locus variants; dotted lines indicate types that differ in more than two loci. The letter within the circle represents the serogroup of the strains with this type. The colors of the circles in the MLST and MLVA trees indicate the MLST clonal complexes as designated on the MLST website (http://pubmlst.org/neisseria). The clonal complexes are also indicated outside the circles in the minimum spanning tree of the MLST. The white circles denote STs for which no clonal complex has been assigned on the MLST website. The halos surrounding the various types denote the groupings obtained by Bionumerics analysis. In Bionumerics MLST, groups (complexes) were created if neighbors differed in no more than two of the seven alleles. MLVA grouping was achieved if neighbors differed in no more than one of the eight VNTR loci. The red halos in the MLVA tree denote three MLVA groups that were not present as separate MLST groups. Two of these groups are made up of isolates belonging to the ST-11 clonal complex. The other MLVA group with a red halo is composed of an MLST ST-254 isolate and an ST-41/44 isolate.
The MLST and MLVA profiles were used in a categorical clustering in Bionumerics, and a minimum spanning tree was constructed. For MLST, groups were created if five of the seven MLST loci were identical, and for MLVA, groups were created if seven of the eight VNTR loci were identical (Fig. 1). The grouping obtained using the MLVA profiles had considerable resemblance to the grouping obtained with MLST (Fig. 1). However, some strains, particularly serogroup C meningococci belonging to the ST-11 complex, did not cluster in a single MLVA group. The ST-254 complex strains and the ST-35 complex strains did not group in separate MLVA groups but had MLVA profiles that were identical or related to those of ST-41/44 strains. Despite obvious differences, there was considerable congruence (69%) between the clustering results obtained by the two typing methods. Although the grouping obtained by MLVA was similar to that obtained by MLST, there were clear differences in the relationships between the strains within such groups. This is exemplified by the distribution of the strains belonging to the ST-32 MLST complex. Of the five strains with ST-34, only two had identical MLVA profiles. Similarly, the three strains with ST-259 differed in their MLVA profiles, although the difference was restricted to VNTR4-5.
High level of discrimination using HV-MLVA.
In order to be able to discriminate strains with identical MLST or MLVA profiles, HV-MLVA with the four highly variable VNTR loci was utilized. For this purpose, we used seven serogroup C strains that were isolated during an outbreak of meningococcal disease in the year 2001 (36). The seven strains were all typed as ST-11, and two strains expressed the PorA protein, while the remaining five were PorA deficient. These 7 strains and 17 PorA-positive serogroup C strains with ST-11 were characterized using HV-MLVA. Figure 2 shows the small minimum spanning trees obtained with the HV-MLVA and with the MLVA. The five PorA-deficient strains from the outbreak were grouped in the HV-MLVA, whereas the two PorA-expressing strains were somewhat more distantly related and were no more related to the PorA-negative strains than several of the other PorA-positive ST-11 strains used in the analysis. The MLVA yielded identical profiles for all seven outbreak strains, and this profile was also found among six other epidemiologically unrelated ST-11 strains.
FIG. 2.
Minimum spanning trees of HV-MLVA and MLVA of N. meningitidis serogroup C outbreak strains and other isolates with ST-11. The dark-gray circles denote five epidemiologically related strains isolated during a meningococcal outbreak in The Netherlands. The light-gray circles denote two other strains isolated during the same outbreak but not epidemiologically linked. The white circles are MLVA types of other unrelated ST-11 strains. Pt 1 through Pt 7 designate the isolates from the patients from the outbreak (also see Table 1). The gray halos surrounding the circles denote the groupings obtained by the Bionumerics software. Further details on grouping and symbols can be found in the legend for Fig. 1.
DISCUSSION
Molecular typing has become an important tool for the study of the epidemiology and population structure of N. meningitidis. When MLST was introduced in 1998, N. meningitidis was the first bacterial pathogen used with this robust and portable molecular typing technique (20). Since then, many molecular epidemiology studies of N. meningitidis have been performed using MLST, corroborating its usefulness for typing (15, 18, 22, 32). This has resulted in a large N. meningitidis MLST database, which currently contains roughly 5,300 different sequence types. In MLST, related STs, differing in only one or two alleles, are grouped into clonal complexes. Some of these clonal complexes have been shown to be related to disease, while others are related to carriage (38). The major advantages of MLST are its unambiguous results and suitability to the construction of international databases that can be electronically exchanged. The major drawbacks of this typing technique are the relatively labor-intensive nature and the high costs associated with DNA sequencing. In the study presented here, we evaluated the applicability of MLVA for molecular typing of N. meningitidis. We propose the use of eight different VNTR loci with limited polymorphism for MLVA. Using a set of 85 N. meningitidis strains isolated from Dutch patients with invasive disease and 7 reference strains, we found that clustering of MLVA profiles yielded groupings similar to those obtained by MLST. MLVA had a slightly higher discriminatory power than MLST, particularly for group C meningococci. Although there were clear differences in the distributions of the various strains over the minimum spanning trees created with both typing methods, the congruence between the two methods is high (69%). The major advantages of MLVA over MLST are its speed, relatively simple processing and interpretation of the data, and considerably lower costs. Similar to MLST, MLVA yields unambiguous numeric profiles that can easily be electronically exchanged. Currently we are adapting MLVA to be able to analyze the composition of the eight VNTR loci in two multiplex PCRs, each amplifying four loci. Using this multiplex approach, 48 strains can be analyzed by MLVA in a single run with an ABI 3700 sequencer. In contrast, a single run would not even suffice to determine the MLST profiles of seven strains. In addition, the reagents used for MLVA are less costly than the dye terminator mixtures required for DNA sequencing. However, there are two important advantages of MLST over MLVA. First, MLST is considered the gold standard for molecular typing and is already used by many research groups worldwide for epidemiological studies, and a large public MLST database is readily available over the Internet. Second, MLST yields DNA sequences and if the concatenated MLST sequences are used for comparison of strains, the resolution of the assay increases and probably outperforms MLVA (12). Since MLVA yields groupings similar to those obtained by MLST, MLVA may be used as a tool complementary to MLST to screen large collections of meningococcal isolates at relatively low cost. Subsequently, MLST can be used to assign clonal complexes to the MLVA groupings.
The MLVA described in this study was composed of eight VNTR loci with limited variation in the number of repeats. In contrast, the HV-MLVA was done using four VNTR loci with highly variable repeat numbers, making these loci unsuitable for population studies. However, the HV-MLVA may be suitable for the analysis of outbreaks, whereas MLST cannot discriminate sufficiently. This was demonstrated by the HV-MLVA of a number of ST-11 N. meningitidis serogroup C strains from a meningococcal outbreak in The Netherlands. The HV-MLVA grouped the PorA-deficient strains that were isolated from five patients living in the same village and that were epidemiologically linked, while two other PorA-expressing strains, isolated during the same period from patients living near the five other patients, were as related to this group as a number of other epidemiologically unrelated Dutch ST-11 N. meningitidis strains. Such subtle relationships could not be visualized by MLVA or MLST.
Recently, Yazdankhah et al. presented two studies on the use of MLVA for molecular typing of N. meningitidis (37, 39). In their reports, they show that their MLVA can be used for fine typing of meningococcal strains and that outbreaks can be delineated by VNTR analysis. The MLVA developed by Yazdankhah and coworkers differs considerably from the MLVA we describe. They initially tested 15 different VNTR loci and eventually selected 4 VNTR loci for MLVA. In our study, we used only Yazdankhah's VNTR07 (our VNTR6-1) for MLVA and VNTR08 (our VNTR9-2) for our HV-MLVA. In their final selection of four loci, Yazdankhah and coworkers included VNTR06 and VNTR08 as two separate loci in their MLVA. However, after close inspection we found that the primer sets designed for VNTR06 and VNTR08 both amplify the VNTR08 locus but not the VNTR06 locus. As a result, only three VNTR loci were used for their MLVA, as opposed to the eight loci of the MLVA presented in our study. We analyzed the VNTR PCR products with an automated DNA sequencer, allowing the discrimination of fragments differing in size by only a single base pair. Hence, we were able to calculate the number of repeats of each VNTR locus and translate these data into a profile similar to the allele profile obtained by MLST. This would be impossible with mixtures of four VNTR PCR products on 2% agarose gels. The resolution of such gels is too low to discriminate PCR fragments differing in size by repeat lengths of only 7 to 13 bp, and comigrating bands would perturb the interpretation of the results. Indeed, Yazdankhah and coworkers did not determine the number of repeats of the VNTR loci for their analyses but used scanned images of the gels and Dice-based band clustering instead. Also, no true comparison between profiles obtained by MLVA and MLST was made.
The MLVA presented in this study is a suitable method for the screening of large collections of meningococcal isolates, after which isolates can be selected for MLST and assigned clonal complexes to MLVA groupings. Thus, MLVA should be considered a tool complementary to MLST. We are developing a multiplex variant of the MLVA that should provide us with a high-throughput method enabling us to analyze a large number of strains in a relatively short time. The results with the HV-MLVA suggest that this method may be suitable for a high degree of discrimination of strains, enabling identification of outbreak strains. However, we have typed only a single outbreak and several collections of N. meningitidis outbreak strains will have to be typed to assess the true value of HV-MLVA.
REFERENCES
- 1.Bash, M. C., K. B. Lesiak, S. D. Banks, and C. E. Frasch. 1995. Analysis of Neisseria meningitidis class 3 outer membrane protein gene variable regions and type identification using genetic techniques. Infect. Immun. 63:1484-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benson, G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bygraves, J. A., and M. C. Maiden. 1992. Analysis of the clonal relationships between strains of Neisseria meningitidis by pulsed field gel electrophoresis. J. Gen. Microbiol. 138:523-531. [DOI] [PubMed] [Google Scholar]
- 4.Cartwright, K., N. Noah, and H. Peltola. 2001. Meningococcal disease in Europe: epidemiology, mortality, and prevention with conjugate vaccines. Report of a European advisory board meeting Vienna, Austria, 6-8 October, 2000. Vaccine 19:4347-4356. [DOI] [PubMed] [Google Scholar]
- 5.Caugant, D. A., B. E. Kristiansen, L. O. Froholm, K. Bovre, and R. K. Selander. 1988. Clonal diversity of Neisseria meningitidis from a population of asymptomatic carriers. Infect. Immun. 56:2060-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coletta-Filho, H. D., M. A. Takita, A. A. de Souza, C. I. Aguilar-Vildoso, and M. A. Machado. 2001. Differentiation of strains of Xylella fastidiosa by a variable number of tandem repeat analysis. Appl. Environ. Microbiol. 67:4091-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farlow, J., D. Postic, K. L. Smith, Z. Jay, G. Baranton, and P. Keim. 2002. Strain typing of Borrelia burgdorferi, Borrelia afzelii, and Borrelia garinii by using multiple-locus variable-number tandem repeat analysis. J. Clin. Microbiol. 40:4612-4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farlow, J., K. L. Smith, J. Wong, M. Abrams, M. Lytle, and P. Keim. 2001. Francisella tularensis strain typing using multiple-locus, variable-number tandem repeat analysis. J. Clin. Microbiol. 39:3186-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feavers, I. M., S. J. Gray, R. Urwin, J. E. Russell, J. A. Bygraves, E. B. Kaczmarski, and M. C. Maiden. 1999. Multilocus sequence typing and antigen gene sequencing in the investigation of a meningococcal disease outbreak. J. Clin. Microbiol. 37:3883-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frothingham, R., and W. A. Meeker-O'Connell. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology 144:1189-1196. [DOI] [PubMed] [Google Scholar]
- 11.Goldschneider, I., E. C. Gotschlich, and M. S. Artenstein. 1969. Human immunity to the meningococcus. II. Development of natural immunity. J. Exp. Med. 129:1327-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanage, W. P., T. Kaijalainen, E. Herva, A. Saukkoriipi, R. Syrjanen, and B. G. Spratt. 2005. Using multilocus sequence data to define the pneumococcus. J. Bacteriol. 187:6223-6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holten, E. 1979. Serotypes of Neisseria meningitidis isolated from patients in Norway during the first six months of 1978. J. Clin. Microbiol. 9:186-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jolley, K. A., J. Kalmusova, E. J. Feil, S. Gupta, M. Musilek, P. Kriz, and M. C. Maiden. 2000. Carried meningococci in the Czech Republic: a diverse recombining population. J. Clin. Microbiol. 38:4492-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keim, P., A. M. Klevytska, L. B. Price, J. M. Schupp, G. Zinser, K. L. Smith, M. E. Hugh-Jones, R. Okinaka, K. K. Hill, and P. J. Jackson. 1999. Molecular diversity in Bacillus anthracis. J. Appl. Microbiol. 87:215-217. [DOI] [PubMed] [Google Scholar]
- 17.Klevytska, A. M., L. B. Price, J. M. Schupp, P. L. Worsham, J. Wong, and P. Keim. 2001. Identification and characterization of variable-number tandem repeats in the Yersinia pestis genome. J. Clin. Microbiol. 39:3179-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kriz, P., J. Kalmusova, and J. Felsberg. 2002. Multilocus sequence typing of Neisseria meningitidis directly from cerebrospinal fluid. Epidemiol. Infect. 128:157-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, Y., M.-A. Lee, E.-E. Ooi, Y. Mavis, A.-L. Tan, and H.-H. Quek. 2003. Molecular typing of Salmonella enterica serovar Typhi isolates from various countries in Asia by a multiplex PCR assay on variable-number tandem repeats. J. Clin. Microbiol. 41:4388-4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murphy, K. M., K. A. O'Donnell, A. B. Higgins, C. O'Neill, and M. T. Cafferkey. 2003. Irish strains of Neisseria meningitidis: characterisation using multilocus sequence typing. Br. J. Biomed. Sci. 60:204-209. [DOI] [PubMed] [Google Scholar]
- 22.Nicolas, P., L. Decousset, V. Riglet, P. Castelli, R. Stor, and G. Blanchet. 2001. Clonal expansion of sequence type (ST-)5 and emergence of ST-7 in serogroup A meningococci, Africa. Emerg. Infect. Dis. 7:849-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pourcel, C., Y. Vidgop, F. Ramisse, G. Vergnaud, and C. Tram. 2003. Characterization of a tandem repeat polymorphism in Legionella pneumophila and its use for genotyping. J. Clin. Microbiol. 41:1819-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosenstein, N. E., B. A. Perkins, D. S. Stephens, L. Lefkowitz, M. L. Cartter, R. Danila, P. Cieslak, K. A. Shutt, T. Popovic, A. Schuchat, L. H. Harrison, and A. L. Reingold. 1999. The changing epidemiology of meningococcal disease in the United States, 1992-1996. J. Infect. Dis. 180:1894-1901. [DOI] [PubMed] [Google Scholar]
- 25.Russell, J. E., K. A. Jolley, I. M. Feavers, M. C. Maiden, and J. Suker. 2004. PorA variable regions of Neisseria meningitidis. Emerg. Infect. Dis. 10:674-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sabat, A., J. Krzyszton-Russjan, W. Strzalka, R. Filipek, K. Kosowska, W. Hryniewicz, J. Travis, and J. Potempa. 2003. New method for typing Staphylococcus aureus strains: multiple-locus variable-number tandem repeat analysis of polymorphism and genetic relationships of clinical isolates. J. Clin. Microbiol. 41:1801-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sacchi, C. T., A. M. Whitney, M. W. Reeves, L. W. Mayer, and T. Popovic. 2002. Sequence diversity of Neisseria meningitidis 16S rRNA genes and use of 16S rRNA gene sequencing as a molecular subtyping tool. J. Clin. Microbiol. 40:4520-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schouls, L. M., A. van der Ende, I. van de Pol, C. Schot, L. Spanjaard, P. Vauterin, D. Wilderbeek, and S. Witteveen. 2005. Increase in genetic diversity of Haemophilus influenzae serotype b (Hib) strains after introduction of Hib vaccination in The Netherlands. J. Clin. Microbiol. 43:2741-2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schouls, L. M., H. G. van der Heide, L. Vauterin, P. Vauterin, and F. R. Mooi. 2004. Multiple-locus variable-number tandem repeat analysis of Dutch Bordetella pertussis strains reveals rapid genetic changes with clonal expansion during the late 1990s. J. Bacteriol. 186:5496-5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simpson, E. H. 1949. Measurement of diversity. Nature 163:688. [Google Scholar]
- 31.Sullivan, C. B., M. A. Diggle, and S. C. Clarke. 2005. Multilocus sequence typing: data analysis in clinical microbiology and public health. Mol. Biotechnol. 29:245-254. [DOI] [PubMed] [Google Scholar]
- 32.Takahashi, H., T. Kuroki, Y. Watanabe, H. Tanaka, H. Inouye, S. Yamai, and H. Watanabe. 2004. Characterization of Neisseria meningitidis isolates collected from 1974 to 2003 in Japan by multilocus sequence typing. J. Med. Microbiol. 53:657-662. [DOI] [PubMed] [Google Scholar]
- 33.Tondella, M. L., C. T. Sacchi, and B. C. Neves. 1994. Ribotyping as an additional molecular marker for studying Neisseria meningitidis serogroup B epidemic strains. J. Clin. Microbiol. 32:2745-2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Top, J., L. M. Schouls, M. J. Bonten, and R. J. Willems. 2004. Multiple-locus variable-number tandem repeat analysis, a novel typing scheme to study the genetic relatedness and epidemiology of Enterococcus faecium isolates. J. Clin. Microbiol. 42:4503-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Belkum, A., S. Scherer, W. van Leeuwen, D. Willemse, L. van Alphen, and H. Verbrugh. 1997. Variable number of tandem repeats in clinical strains of Haemophilus influenzae. Infect. Immun. 65:5017-5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Ende, A., C. T. Hopman, W. C. Keijzers, L. Spanjaard, E. B. Lodder, P. H. van Keulen, and J. Dankert. 2003. Outbreak of meningococcal disease caused by PorA-deficient meningococci. J. Infect. Dis. 187:869-871. [DOI] [PubMed] [Google Scholar]
- 37.Yazdankhah, S. P., K. Kesanopoulos, G. Tzanakaki, J. Kremastinou, and D. A. Caugant. 2005. Variable-number tandem repeat analysis of meningococcal isolates belonging to the sequence type 162 complex. J. Clin. Microbiol. 43:4865-4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yazdankhah, S. P., P. Kriz, G. Tzanakaki, J. Kremastinou, J. Kalmusova, M. Musilek, T. Alvestad, K. A. Jolley, D. J. Wilson, N. D. McCarthy, D. A. Caugant, and M. C. Maiden. 2004. Distribution of serogroups and genotypes among disease-associated and carried isolates of Neisseria meningitidis from the Czech Republic, Greece, and Norway. J. Clin. Microbiol. 42:5146-5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yazdankhah, S. P., B. A. Lindstedt, and D. A. Caugant. 2005. Use of variable-number tandem repeats to examine genetic diversity of Neisseria meningitidis. J. Clin. Microbiol. 43:1699-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]




