Abstract
In this study, we developed a one-step, single-tube genogroup-specific reverse transcription-loop-mediated isothermal amplification (RT-LAMP) assay for the detection of norovirus (NoV) genomes targeting from the C terminus of the RNA-dependent RNA polymerase gene to the capsid N-terminal/shell domain region. This is the first report on the development of an RT-LAMP assay for the detection of NoV genomes. Because of the diversity of NoV genotypes, we used 9 and 13 specially designed primers containing mixed bases for genogroup I (GI) and II (GII), respectively. The RT-LAMP assay had the advantages of rapidity, simplicity, specificity, and selectively and could obtain results within 90 min, generally even within 60 min, under isothermal conditions at 62°C. The detection limits for NoV genomes were between 102 and 103 copies/tube for GI and GII with differentiation by genotype, and no cross-reactions among NoV GI and GII and other gastroenteritis viruses, such as sapovirus, human astrovirus, adenovirus type 40 and 41, and group A and C rotavirus, were found. In the evaluation tests with fecal specimens obtained from gastroenteritis outbreaks, the sensitivity and specificity of the RT-LAMP assay with regard to RT-PCR were 100 and 94% for GI and 100 and 100% for GII, respectively. These findings establish that the RT-LAMP assay is potentially useful for the rapid detection of NoV genomes from fecal specimens in outbreaks of food-borne and person-to-person-transmitted gastroenteritis.
Noroviruses (NoVs), which belong to the family Caliciviridae (5), commonly cause human gastroenteritis worldwide (1, 3, 7, 10, 14) and are sometimes detected in oysters (2, 7) and clams (12). In addition, they are also recognized as a cause of person-to-person transmission of gastroenteritis in nursing homes, hospitals, and schools (5, 7, 14). The detection of NoVs is usually carried out by electron microscopy and reverse transcription-PCR (RT-PCR) because they are noncultivable (5).
Recently, the detection of NoV genomes by nucleic acid sequence-based amplification (NASBA) (6, 9, 15) as an isothermal amplification method was reported, and similarly, a transcription-reverse transcription concerted (TRC) assay (8) for the detection of NoV genomes has been commercially developed in Japan. As another isothermal amplification method, the loop-mediated isothermal amplification (LAMP) assay developed by Notomi et al. (18) is a simple method for genome diagnostics and has the advantages of high specificity and selectivity. Six primers containing two loop primers in an RT-LAMP assay are required for rapid and sensitive detection of RNA. Although more primers are needed in the RT-LAMP assay than in other amplification methods, the RT-LAMP assay has recently been applied for the rapid detection of several RNA viruses in humans, such as West Nile virus (23), severe acute respiratory syndrome coronavirus (22, 25), influenza A virus (21), mumps virus (19), measles virus (4), dengue virus (20), and respiratory syncytial virus (26).
Because the NoVs are classified into two genogroups (GI and GII) and further divided into 14 genotypes in GI and 17 genotypes in GII according to the scheme of Kageyama et al. (10), it has been thought likely that the design of consensus primers for many genotypes of NoVs might be difficult.
In this study, we developed a genogroup-specific RT-LAMP assay for detecting NoV genomes using multiple primers containing mixed bases.
MATERIALS AND METHODS
Fecal specimens.
A total of 91 fecal specimens, including 75 samples obtained from 26 gastroenteritis outbreaks and 16 additional NoV-negative samples containing three sapoviruses, three human astroviruses, one adenovirus type 40, three type 41 adenoviruses, three group A rotaviruses, and three group C rotaviruses obtained from sporadic gastroenteritis in children were used for the specificity and evaluation tests of the RT-LAMP assay. Sapoviruses and human astroviruses were detected by RT-PCR with the primers of Vinjé et al. (27) and Sakon et al. (24), respectively. Adenoviruses of types 40 and 41 and group A rotaviruses were detected by enzyme immunoassay (Adenoclone 40/41 and Rotaclone; Meridian Bioscience, Cincinnati, OH), and group C rotaviruses were detected by reversed passive hemagglutination assay (DENKA SEIKEN, Tokyo, Japan).
Viral RNA extraction.
The viral RNA was extracted from 140 μl of the supernatant of approximately 10% fecal specimens with a QIAamp viral RNA mini kit (QIAGEN, Valencia, CA). The extracted RNAs were kept at −80°C until use in tests.
Primer design.
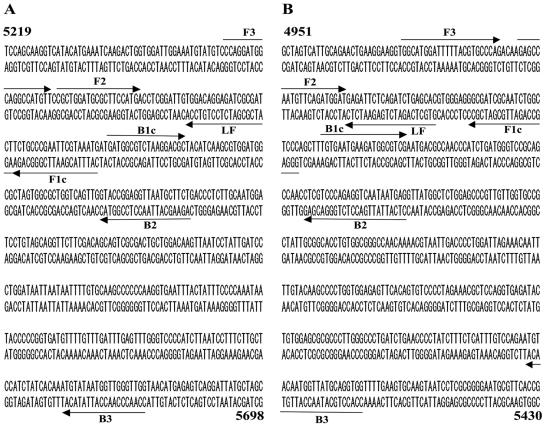
The NoV-specific primers were designed on the basis of the sequence of the region from the C terminus of the RNA-dependent RNA polymerase gene to the capsid N-terminal/shell domain region, and two sets of genogroup-specific primers for GI and GII were designed. For GI, the nine primers consisted of two outer primers (F3 and B3), four inner primers (FIP1, FIP2, BIP1, and BIP2), and three forward loop primers (LF1, LF2, and LF3). For GII, the 13 primers consisted of two outer primers (F3 and B3), seven inner primers (FIP1, FIP2, FIP3, BIP1, BIP2, BIP3, and BIP4), and four forward loop primers (LF1, LF2, LF3, and LF4). FIP contain F1C, a TTTT spacer, and F2, and BIP contain B1C, a TTTT spacer, and B2. The B3 reverse outer primers for GI and GII were the G1SKR and G2SKR primers, respectively, as described elsewhere (13). The FIP and BIP primers were high-performance liquid chromatography-purified primers, and the others were oligonucleotide purification cartridge-purified primers. The sequences and locations of the oligonucleotide primers are shown in Table 1 and Fig. 1.
TABLE 1.
Primer sets for genogroups I and II for detection of noroviruses
| Genogroup | Primer namea | Typeb | Targetc | Sequence(s) (5′-3′)d |
|---|---|---|---|---|
| I | F3 | OF | 5270-5671 | CCRGGNTGGCARGCNATGTT |
| B3 (G1SKR) | OR | CCAACCCARCCATTRTACA | ||
| FIP1 | IF | F1C, CATTTACGAATTCGGGCAGG; F2, CGCTGGATGCGNTTCCATGA | ||
| FIP2 | IF | F1C, CATTTACAAAATCGGGCAGG; F2, CGCTGGATGCGNTTCCATGA | ||
| BIP1 | IR | B1C, GATGGCGTCTAAGGACGC; B2, AGCTGTRTTTGCCTCTGGWAC | ||
| BIP2 | IR | B1C, GATGGCGTCTAAGGACGC; B2, AGCWGTATTAACCTCCGGYAC | ||
| LF1 | LF | AGATYGCGATCYCCTGTCCA | ||
| LF2 | LF | AGATTGCGATCTCCTGCCCA | ||
| LF3 | LF | AGCTCGCGGTCTCCTGTCCA | ||
| II | F3 | OF | 4979-5389 | GGNMTGGANTTTTAYGTGCCMAG |
| B3 (G2SKR) | OR | CCRCCNGCATRHCCRTTRTACAT | ||
| FIP1 | IF | F1C, GGGAGCMAGATTGCGATCGC; F2, GAGBCNATGTTYAGRTGGAT | ||
| FIP2 | IF | F1C, GGGAGCMAGATTGCGATCGC; F2, GAGCCCATGTTCAGRTGGAT | ||
| FIP3 | IF | F1C, GGGAGCGAGATTGCGATCGC; F2, GAGTCAATGTTYAGGTGGAT | ||
| BIP1 | IR | B1C, TGTGAATGAAGATGGCGTCG; B2, CTCATTRTTRVTCTCTGGBACGAG | ||
| BIP2 | IR | B1C, TGTGAATGAAGATGGCGTCG; B2, CTCATTRTTGCYCTCTGGYACGAG | ||
| BIP3 | IR | B1C, TGTGAATGAAGATGGCGTCG; B2, CTCATTGTTGAYCTCTGGKACGAG | ||
| BIP4 | IR | B1C, TGTGAATGAAGATGGCGTCG; B2, CTCATTRTTACTTTCTGGCACGAG | ||
| LF1 | LF | GTGCTCARATCWGARAACCTC | ||
| LF2 | LF | GTGCTGAGGTCWGARAATCTC | ||
| LF3 | LF | GTGCTCAAATCTGAGAATCTC | ||
| LF4 | LF | GTGCTCAAGTCTGAGAAYCTC |
B3 primers used for genogroups I and II were G1SKR and G2SKR, respectively, as described by Kojima et al. (13). FIP and BIP primers consist of F1C plus TTTT plus F2 and B1C plus TTTT plus B2, respectively, where TTTT is a spacer.
OF, outer forward; OR, outer reverse; IF, inner forward; IR, inner reverse; LF, loop forward.
Nucleotide positions according to those of Norwalk/68/US (GenBank accession number M87661) for genogroup I and Lordsdale/93/UK (X86557) for genogroup II.
Mixed bases in degenerate primers are as follows: K, T or G; M, A or C; R, A or G; W, A or T; Y, C or T; B, not A; H, not G; V, not T; N, any.
FIG. 1.
Location of primers used for genogroup I (A) and II (B) for genogroup-specific RT-LAMP assay. (A) Sequence of Norwalk/68/US (GenBank accession number M87661); (B) sequence of Lordsdale/93/UK (X86557). Numbers at the beginning and end of the sequences denote nucleotide positions. Primers FIP and BIP in Table 1 consist of F1C plus TTTT plus F2 and B1C plus TTTT plus B2, respectively, where TTTT is a spacer. The B3 primers for genogroups I and II were primers G1SKR and G2SKR, respectively, as described by Kojima et al. (13).
Genogroup-specific RT-LAMP assay.
The RT-LAMP assay was performed in 25 μl of the reaction mixture with an RNA amplification kit (Eiken Chemical, Tochigi, Japan). The reaction mixture contained 12.5 μl of 2× reaction mix, 1.0 μl of enzyme mix, 2.5 μl of RTmate (Nippon gene, Toyama, Japan), 5 pmol (each) of outer primers F3 and B3, 25 pmol (each) of inner primers FIP1, FIP2, BIP1, and BIP2 for GI or 40 pmol (each) of inner primers FIP1 and BIP1 and 20 pmol (each) of inner primers FIP2, FIP3, BIP2, BIP3, and BIP4 for GII, 20 pmol each of loop primers LF1, LF2, and LF3 for GI or 20 pmol each of loop primers LF1, LF2, LF3, and LF4 for GII, and 2 μl of RNA extract. The reaction mixture was incubated at 62°C for 90 min and heated at 80°C for 5 min in a Loopamp real-time turbidimeter (LA-320C; Teramecs, Kyoto, Japan). For performing the RT-LAMP assay, standard RNAs described below, as positive controls, and distilled water, as a negative control, were used in each test, and a turbidity value of ≥0.1 was considered positive. For measuring the detection limit of the RT-LAMP assay, the samples were tested in triplicate and the lowest concentration of genome copies was taken as the limit when all of the triplicate samples were positive. In addition, the fecal specimens that gave positive results at over 60 min were retested another time. Then, 2 μl of the amplified product from the RT-LAMP assay was electrophoresed on a 3% agarose gel in Tris-acetate-EDTA buffer, stained with ethidium bromide, and visualized under UV light, when necessary.
RT-PCR and real-time quantitative PCR.
A conventional RT-PCR was performed to compare with the sensitivity of the RT-LAMP assay. The RT was first performed in 20 μl of a mixture containing 50 pmol of random nonamer [pd(N)9; Takara Bio, Shiga, Japan], 20 U of RNase inhibitor (TOYOBO, Osaka, Japan), 100 U of reverse transcriptase (ReverTra Ace; TOYOBO), 2.5 mM concentrations of each deoxynucleoside triphosphate, and 9.5 μl of RNA extract. The RT conditions were 30°C for 10 min and then 42°C for 60 min, followed by 99°C for 5 min. After RT, PCR was performed in 50 μl of a reaction mixture containing 0.25 U of Ex Taq DNA polymerase (Takara), 2.5 mM concentrations of each deoxynucleoside triphosphate, 10 pmol (each) of primers G1SKF and G1SKR for GI or G2SKF and G2SKR for GII, as previously described (13), and 3 μl of cDNA.
For the measurement of the number of genome copies in the RT-LAMP-positive samples, real-time quantitative PCR was performed by using primers COG1F and COG1R and fluorogenic probe RING1-TP(a) for GI and primers COG2F and COG2R and fluorogenic probe RING2-TP for GII (11) with a LightCycler (Roche Diagnostics, Penzberg, Germany). The real-time PCR was performed in 20 μl of a reaction mixture containing 2 μl of 10× PCR master mix (LC FastStart DNA hybridization probes; Roche), 10 pmol of each of the primers, 8 pmol fluorogenic probe, 3 mM MgCl2, and 5 μl of cDNA. The following real-time PCR conditions were used: 95°C for 10 min, 50 cycles of 95°C for 15 s, 56°C for 20 s, and 72°C for 4 s, and a final cooling at 40°C for 30 s.
Standard RNA.
The plasmid DNAs containing the target sequences amplified with primer sets F3 and B3 (G1SKR) for GI or F3 and B3 (G2SKR) for GII were first prepared with the pDrive cloning vector (QIAGEN). The standard RNAs were transcribed from the plasmid DNAs by using SP6 polymerase (Ambion, Austin, TX) and were used to determine the sensitivity of the RT-LAMP assay. The standard RNAs for GI and GII belonged to genotypes GI/2 and GII/12, respectively, according to the scheme of Kageyama et al. (10).
Genotyping.
After purification of the RT-PCR-positive products with a QIAquick PCR purification kit (QIAGEN), the sequencing of 260 bp was carried out with a SequiTherm EXCEL II DNA sequencing kit LC for 25- to 41-cm gels (EPCENTRE Technologies, Madison, WI) and a LI-COR 4200 series sequencer (LI-COR, Lincoln, NE). The sequences were compared with those of the reference strains of NoVs obtained from GenBank and classified into 31 genotypes as described by Kageyama et al. (10).
RESULTS
We developed a one-step, single-tube RT-LAMP assay for the detection of NoV genomes with genogroup-specific primers for GI and GII. The RT-LAMP assay was carried out with a set of 9 and 13 primers for GI and GII, respectively. These multiple primers used for the detection of many genotypes contained several mixed bases, which were not used in many other RT-LAMP assays (4, 19, 22, 23, 25). Taking the use of mixed bases into consideration, to achieve high sensitivity, the RT-LAMP reaction in this study was extended to 90 min at 62°C, followed by heat inactivation at 80°C for 5 min. The RT-LAMP assay amplified 402-bp (nucleotides 5270 to 5671 of Norwalk/68/US, GenBank accession number M87661) and 411-bp (nucleotides 4979 to 5389 of Lordsdale/93/UK, X86557) target sequences from the C terminus of the RNA-dependent RNA polymerase gene to the capsid N-terminal/shell domain region for GI and GII, respectively.
Sensitivity and specificity of the RT-LAMP assay.
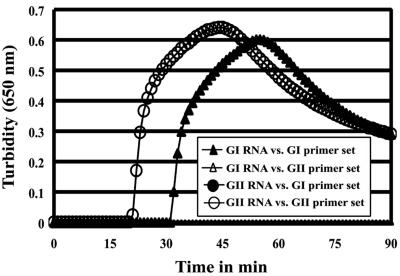
The sensitivity of the RT-LAMP assay for the detection of NoV genomes was tested by using serial 10-fold dilutions of standard RNAs of GI and GII with Easy Dilution solution (Takara) and compared with that of conventional RT-PCR. The detection limits of the RT-LAMP assay for the standard RNAs of GI and GII were found to be approximately 102 and 103 copies/tube, with a time of positive change of 57 to 65 min (average, 61 min) and 57 to 60 min (average, 59 min), respectively, which were the same as for the turbidity assay, but 10-fold less reactivity than that of RT-PCR was observed (Fig. 2). A positive reaction in the RT-LAMP assay was seen as a ladder-like pattern in 3% agarose gel electrophoresis analysis. No cross-reaction was found between GI and GII with the standard RNAs of genotypes GI/2 and GII/12 at 108 copies/tube, respectively. Graphs of the data obtained for real-time amplification in the RT-LAMP assay are shown in Fig. 3. The times until positive change were found to be 31 min for genotype GI/2 and 20 min for GII/12. Furthermore, serial 10-fold dilutions of RNA extracts prepared from the fecal specimens were used to determine the sensitivity of the RT-LAMP assay according to genotype. For GI, the sensitivities of genotypes GI/1, GI/2, GI/4, GI/8, GI/11, and GI/14 tested were 102, 102, 102, 102, 102, and 103 copies/tube, respectively, in the turbidity assay. For GII, the sensitivities of genotypes GII/1, GII/2, GII/3, GII/4, GII/5, GII/6, GII/8, GII/12, and GII/14 tested were 103, 102, 103, 102, 103, 103, 103, 103, and 103 copies/tube, respectively. Thus, the sensitivity of the RT-LAMP assay was different depending on the genotype. Although the fecal specimens containing the genotypes GI/2, GI/11, and GII/5 used in this test also included other genotypes (GII/12, GII/3, and GI/14, respectively), these sensitivities were similar to the others. Furthermore, it was observed that none of the genogroup-specific primers cross-reacted between genogroups with any of the fecal specimens obtained from NoV outbreaks (Table 2).
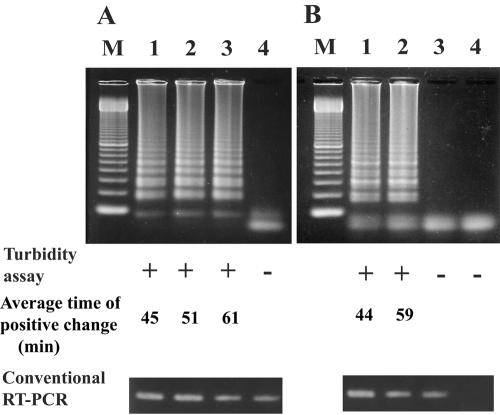
FIG. 2.
Sensitivity of genogroup-specific RT-LAMP assay for standard RNAs of genotype GI/2 for genogroup I (A) and GII/12 for genogroup II (B). Lanes: M, 100-bp DNA ladder; 1, 104 copies/tube; 2, 103 copies/tube; 3, 102 copies/tube; 4, 101 copies/tube. Positive reaction shows a ladder-like pattern in 3% agarose gel electrophoresis. The turbidity was measured using an LA-320C (Teramecs), and RT-PCR was carried out with primers G1SKF and G2SKR for genogroup I and G2SKF and G2SKR for genogroup II (13). +, positive; −, negative.
FIG. 3.
Evaluation of cross-reactivity of genogroup-specific RT-LAMP assay between genogroups I and II. Standard RNAs of genotypes GI/2 and GII/12 at 108 copies/tube were used in genogroups I and II, respectively. The real-time turbidity was determined using an LA-320C (Teramecs).
TABLE 2.
Comparison of RT-PCR and RT-LAMP assays for detection of noroviruses in gastroenteritis outbreaks
| Outbreak no. | Genogroup I
|
Genogroup II
|
Epidemiology (transmission) | ||||
|---|---|---|---|---|---|---|---|
| RT-PCRa | RT-LAMPa | Genotype(s)b | RT-PCRa | RT-LAMPa | Genotype(s)b | ||
| 40 | ••• | ••• | GI/1, GI/1, GI/1 | ○○○ | ○○○ | Person to person | |
| 38 | ••○ | ••○ | GI/1, GI/11 | ••• | ••• | GII/3, GII/3, GII/3 | Food borne |
| 35 | ○○• | ••• | GI/2 | ••• | ••• | GII/5, GII/5, GII/5 | Food borne |
| 20 | ○•• | ○•• | GI/2, GI/2 | ••• | ••• | GII/12, GII/12, GII/14 | Food borne |
| 32 | ○○• | ○○• | GI/2 | ••• | ••• | GII/14, GII/14, GII/14 | Food borne |
| 61 | ••• | ••• | GI/4, GI/4, GI/4 | ○○○ | ○○○ | Person to person | |
| 43 | •• | •• | GI/4, GI/4 | •• | •• | GII/3, GII/3 | Food borne |
| 25 | ○• | ○• | GI/4 | •• | •• | GII/12, GII/12 | Person to person |
| 16 | ••• | ••• | GI/4, GI/4, GI/8 | ○○○ | ○○○ | Food borne | |
| 48 | ○○• | ○○• | GI/8 | ••○ | ••○ | GII/4, GII/12 | Food borne |
| 42 | •○ | •○ | GI/14 | ○• | ○• | GII/4 | Food borne |
| 27 | ••• | ••• | GI/14, GI/14, GI/14 | ○•• | ○•• | GII/5, GII/5 | Food borne |
| 15 | ○○○ | ○○○ | ••○ | ••○ | GII/1, GII/1 | Food borne | |
| 45 | ○○○ | ○○○ | •○• | •○• | GII/1, GII/1 | Food borne | |
| 60 | ○○○ | ○○○ | ••• | ••• | GII/2, GII/2, GII/2 | Person to person | |
| 39 | ○○○ | ○○○ | ••• | ••• | GII/3, GII/3, GII/3 | Person to person | |
| 49 | ○○○ | ○○○ | •○• | •○• | GII/4, GII/4 | Person to person | |
| 50 | ○○○ | ○○○ | ••• | ••• | GII/4, GII/4, GII/4 | Person to person | |
| 51 | ○○○ | ○○○ | ••○ | ••○ | GII/4, GII/4 | Person to person | |
| 55 | ○○○ | ○○○ | ••• | ••• | GII/4, GII/4, GII/4 | Person to person | |
| 57 | ○○○ | ○○○ | ○•• | ○•• | GII/4, GII/4 | Person to person | |
| 59 | ○○○ | ○•• | ••○ | ••○ | GII/4, GII/6 | Food borne | |
| 28 | ○○○ | ○○○ | ••• | ••• | GII/5, GII/5, GII/5 | Food borne | |
| 37 | ○○○ | ○○○ | ••• | ••• | GII/5, GII/5, GII/5 | Food borne | |
| 54 | ○○○ | ○○○ | ••• | ••• | GII/6, GII/6, GII/6 | Person to person | |
| 33 | ○○○ | ○○○ | ••• | ••• | GII/8, GII/8, GII/8 | Person to person | |
Same samples in order in RT-PCR and RT-LAMP assay. •, positive; ○, negative. Each circle refers to the results from a single fecal specimen in the outbreak. Underlined results denote that positivity was detected at over 60 min in the RT-LAMP assay.
The genotyping of amplicons by RT-PCR was carried out according to the scheme of Kageyama et al. (10). The column represents the genotypes identified in the outbreak and the genotypes of the positives by RT-PCR are shown in order. Eleven outbreaks were due to multiple genotypes, and one or two genotypes in each fecal specimen were identified.
The specificity of the RT-LAMP assay was checked using 16 fecal specimens containing the following: sapovirus (3 specimens), human astrovirus (3 specimens), adenovirus type 40 and 41 (4 specimens), and group A and C rotavirus (6 specimens). The fecal specimens containing these gastroenteritis viruses gave negative results for the RT-LAMP assay within a 90-min run (data not shown). Thus, the genogroup-specific RT-LAMP assay with the multiple primers containing mixed bases had a high degree of specificity for NoV genomes.
Evaluation of RT-LAMP assay with fecal specimens.
A total of 75 fecal specimens obtained from 26 NoV outbreaks were tested by the genogroup-specific RT-LAMP assay, and the results were compared to those of conventional RT-PCR. A concordance of 95% (71 of 75) between the RT-LAMP assay and RT-PCR was observed. Of the 75 samples, 23 and 57 were positive for GI and GII, respectively, and 48 and 18 were negative for GI and GII, respectively, in both assays. The RT-LAMP assay, however, gave positive results for four RT-PCR-negative specimens of two outbreaks and detected NoV GI in these specimens. These fecal specimens were also positive by the RT-LAMP assay in retests. The sensitivity and specificity of the RT-LAMP assay with regard to RT-PCR were 100 and 94% for GI and 100 and 100% for GII, respectively. Of 27 NoV-positive specimens in GI, 22 (81%) reacted within 60 min in the RT-LAMP assay and 50 (88%) of 57 positive specimens in GII reacted within 60 min (Table 2). The genotypes detected from the 26 NoV outbreaks were divided into six types (GI/1, GI/2, GI/4, GI/8, GI/11, and GI/14) for GI and nine types (GII/1, GII/2, GII/3, GII/4, GII/5, GII/6, GII/8, GII/12, and GII/14) for GII. The RT-LAMP assay was able to detect all of these genotypes of NoV with a high degree of specificity. The number of genome copies in the RT-LAMP-positive specimens was estimated to range from 1.7 × 102 to 3.7 × 108 copies/tube in GI and from 1.3 × 103 to 3.2 ×106 copies/tube in GII (Table 3). The numbers of copies of NoV genomes in the fecal specimens obtained from gastroenteritis outbreaks in this study were higher than the levels of sensitivity of the RT-LAMP assay.
TABLE 3.
Numbers of genome copies in RT-LAMP-positive fecal specimens, as determined by RT-PCR
| Genogroup | Genotypea | No. of copies/tubeb
|
|
|---|---|---|---|
| Minimum value | Maximum value | ||
| I | GI/1 | 4.2 × 105 | 6.8 × 106 |
| GI/2 | 7.2 × 102 | 1.4 × 105 | |
| GI/4 | 1.7 × 102 | 3.7 × 108 | |
| GI/8 | 2.0 × 104 | 4.5 × 105 | |
| GI/11c | 8.0 × 105 | ||
| GI/14 | 1.6 × 103 | 3.0 × 106 | |
| II | GII/1 | 2.0 × 104 | 1.6 × 106 |
| GII/2 | 1.5 × 105 | 1.1 × 106 | |
| GII/3 | 1.4 × 104 | 3.2 × 106 | |
| GII/4 | 1.3 × 103 | 3.7 × 105 | |
| GII/5 | 3.2 × 103 | 1.1 × 106 | |
| GII/6 | 1.9 × 104 | 3.7 × 105 | |
| GII/8 | 7.2 × 105 | 8.2 × 105 | |
| GII/12 | 2.8 × 104 | 7.0 × 105 | |
| GII/14 | 5.0 × 105 | 7.0 × 105 | |
Genotypes were determined according to the scheme of Kageyama et al. (10).
Genome copy numbers were determined by real-time PCR with primers COG1F and COG1R and fluorogenic probe RING1-TP(a) for genogroup I and primers COG2F and COG2R and probe RING2-TP for genogroup II, as described by Kageyama et al. (11).
One fecal specimen was tested in genotype GI/11.
DISCUSSION
NoVs are major causative agents of nonbacterial gastroenteritis in many countries in the cold season (16). Therefore, the rapid detection of NoV genomes is needed when acute gastroenteritis outbreaks occur. The detection of NoV genomes is usually performed by RT-PCR (5), which requires 3 to 4 h. Isothermal amplification methods such as LAMP, NASBA, and TRC are available for the detection of NoV genomes within a shorter time (6, 8, 9, 15, 18), but the NASBA assay requires a further test for the confirmation of NoV genomes and the TRC assay requires a precision instrument such as a real-time fluorometer. On the other hand, the RT-LAMP assay has extremely high specificity and selectivity because of its use (generally) of six primers, with two loop primers recognizing eight distinct regions on the target sequence. The specificity and selectively of the RT-LAMP assay have been documented in previous studies (4, 19-23, 25, 26). Furthermore, the RT-LAMP reaction yields a white precipitate of magnesium pyrophosphate in the reaction mixture, and the positive reaction can be identified at the end point of the assay by the appearance of this white precipitate without a turbidimeter.
We could achieve rapid detection of NoV genomes by using a one-step, single-tube genogroup-specific RT-LAMP assay. This is the first report of the development of an RT-LAMP assay for the detection of NoV genomes. The specificity of the RT-LAMP assay for NoV genomes was established by examining the cross-reaction with standard RNAs of GI and GII, RNA templates of 15 genotypes of NoVs, and RNAs of other viruses causing gastroenteritis. The RT-LAMP assay for the detection of NoV genomes was genogroup and genus specific and can be used for the detection of NoVs from outbreaks of food-borne and person-to-person-transmitted gastroenteritis and sporadic gastroenteritis cases.
At least 14 and 17 genotypes have been found for GI and GII, respectively (10). It is difficult to detect many genotypes using an RT-LAMP assay system with a high degree of sensitivity because of the diversity of NoV genotypes. Our RT-LAMP assay was able to detect NoV genomes with sensitivity levels of approximately 102 to 103 copies/tube in serial 10-fold dilution tests of RNA templates and had good performance with regard to concordance, specificity, and sensitivity in comparison to RT-PCR. Although the sensitivity of the RT-LAMP assay differed by genotype and was not influenced by the presence of multiple genotypes in a fecal specimen, our RT-LAMP assay was sufficient to detect NoV genomes from fecal specimens. However, it may be necessary to further lower the limit of detection because the numbers of copies of the NoV genome in foods such as oysters are low (17).
A total of 15 genotypes (GI/1, GI/2, GI/4, GI/8, GI/11, and GI/14 of GI and GII/1, GII/2, GII/3, GII/4, GII/5, GII/6, GII/8, GII/12, and GII/14 of GII) tested could be detected by our RT-LAMP assay. The sensitivity of the detection of the remaining 16 genotypes (GI/3, GI/5, GI/6, GI/7, GI/9, GI/10, GI/12, and GI/13 of GI and GII/7, GII/9, GII/10, GII/11, GII/13, GII/15, GII/16, and GII/17 of GII) by our RT-LAMP assay must be tested in further studies, although the primers used in the RT-LAMP assay were probably designed as consensus primers for the NoV sequences obtained from GenBank, and it is expected that the RT-LAMP assay will also be able to detect the remaining genotypes not tested in this study.
We expect that the genogroup-specific RT-LAMP assay will be routinely used in most laboratories because of its simplicity, specificity, and selectively. The greatest advantage of the RT-LAMP assay is the substantial reduction in required time compared to that required for RT-PCR. Our simple-to-use genogroup-specific RT-LAMP assay shortens the amplification time to within 90 min or even 60 min in most cases.
REFERENCES
- 1.Bon, F., K. Ambert-Balay, H. Giraudon, J. Kaplon, S. Le Guyader, M. Pommepuy, A. Gallay, V. Vaillant, H. de Valk, R. Chikhi-Brachet, A. Flahaut, P. Pothier, and E. Kohli. 2005. Molecular epidemiology of caliciviruses detected in sporadic and outbreak cases of gastroenteritis in France from December 1998 to February 2004. J. Clin. Microbiol. 43:4659-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng, P. K. C., D. K. K. Wong, T. W. H. Chung, and W. W. L. Lim. 2005. Norovirus contamination found in oysters worldwide. J. Med. Virol. 76:593-597. [DOI] [PubMed] [Google Scholar]
- 3.Fankhauser, R. L., S. S. Monroe, J. S. Noel, C. D. Humphrey, J. S. Bresee, U. D. Parashar, T. Ando, and R. I. Glass. 2002. Epidemiologic and molecular trends of “Norwalk-like viruses” associated with outbreaks of gastroenteritis in the United States. J. Infect. Dis. 186:1-7. [DOI] [PubMed] [Google Scholar]
- 4.Fujino, M., N. Yoshida, S. Yamaguchi, N. Hosaka, Y. Ota, T. Notomi, and T. Nakayama. 2005. A simple method for the detection of measles virus genome by loop-mediated isothermal amplification (LAMP). J. Med. Virol. 76:406-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green, K. Y., R. M. Chanock, and A. Z. Kapikian. 2001. Human caliciviruses, p. 841-874. In D. M. Knipe, P. M. Howley, D. E. Griffin, et al. (ed.), Fields virology, 4th ed. Lippincott-Raven, Philadelphia, Pa.
- 6.Greene, S. R., C. L. Moe, L.-A. Jaykus, M. Cronin, L. Grosso, and P. van Aarle. 2003. Evaluation of the NucliSens® Basic kit assay for detection of Norwalk virus RNA in stool specimens. J. Virol. Methods 108:123-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inouye, S., K. Yamashita, S. Yamadera, M. Yoshikawa, N. Kato, and N. Okabe. 2000. Surveillance of viral gastroenteritis in Japan: pediatric cases and outbreak incidents. J. Infect. Dis. 181(Suppl. 2):S270-S274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishiguro, T., J. Saitoh, R. Horie, T. Hayashi, T. Ishizuka, S. Tsuchiya, K. Yasukawa, T. Kido, Y. Nakaguchi, M. Nishibuchi, and K. Ueda. 2003. Intercalation activating fluorescence DNA probe and its application to homogeneous quantification of a target sequence by isothermal sequence amplification in a closed vessel. Anal. Biochem. 314:77-86. [DOI] [PubMed] [Google Scholar]
- 9.Jean, J., D. H. D'Souza, and L.-A. Jaykus. 2004. Multiplex nucleic acid sequence-based amplification for simultaneous detection of several enteric viruses in model ready-to-eat foods. Appl. Environ. Microbiol. 70:6603-6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kageyama, T., M. Shinohara, K. Uchida, S. Fukushi, F. B. Hoshino, S. Kojima, R. Takai, T. Oka, N. Takeda, and K. Katayama. 2004. Coexistence of multiple genotypes, including newly identified genotypes, in outbreaks of gastroenteritis due to Norovirus in Japan. J. Clin. Microbiol. 42:2988-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kageyama, T., S. Kojima, M. Shinohara, K. Uchida, S. Fukushi, F. B. Hoshino, N. Takeda, and K. Katayama. 2003. Broadly reactive and high sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 41:1548-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kingsley, D. H., G. K. Meade, and G. P. Richards. 2002. Detection of both hepatitis A virus and Norwalk-like virus in imported clams associated with food-borne illness. Appl. Environ. Microbiol. 68:3914-3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kojima, S., T. Kageyama, S. Fukushi, F. B. Hoshino, M. Shinohara, K. Uchida, K. Natori, N. Takeda, and K. Katayama. 2002. Genogroup-specific PCR primers for detection of Norwalk-like viruses. J. Virol. Methods 100:107-114. [DOI] [PubMed] [Google Scholar]
- 14.Lopman, B. A., G. K. Adak, M. H. Reacher, and D. W. G. Brown. 2003. Two epidemiologic patterns of norovirus outbreaks: surveillance in England and Wales, 1992-2000. Emerg. Infect. Dis. 9:71-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore, C., E. M. Clark, C. I. Gallimore, S. A. Corden, J. J. Gray, and D. Westmoreland. 2004. Evaluation of a broadly reactive nucleic acid sequence based amplification assay for the detection of noroviruses in faecal material. J. Clin. Virol. 29:290-296. [DOI] [PubMed] [Google Scholar]
- 16.Mounts, A. W., T. Ando, M. Koopmans, J. S. Bresee, J. Noel, and R. I. Glass. 2000. Cold weather seasonality of gastroenteritis associated with Norwalk-like viruses. J. Infect. Dis. 181(Suppl. 2):S284-S287. [DOI] [PubMed] [Google Scholar]
- 17.Nishida, T., H. Kimura, M. Saitoh, M. Shinohara, M. Kato, S. Fukuda, T. Munemura, T. Mikami, A. Kawamoto, M. Akiyama, Y. Kato, K. Nishi, K. Kozawa, and O. Nishio. 2003. Detection, quantitation, and phylogenic analysis of noroviruses in Japanese oyster. Appl. Environ. Microbiol. 69:5782-5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Notomi, T., H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino, and T. Hase. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okafuji, T., N. Yoshida, M. Fujino, Y. Motegi, T. Ihara, Y. Ota, T. Notomi, and T. Nakayama. 2005. Rapid diagnostic method for detection of mumps virus genome by loop-mediated isothermal amplification. J. Clin. Microbiol. 43:1625-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parida, M., K. Horioke, H. Ishida, P. K. Dash, P. Saxena, A. M. Jana, M. A. Islam, S. Inoue, N. Hosaka, and K. Morita. 2005. Rapid detection and differentiation of dengue virus serotypes by a real-time reverse transcription-loop-mediated isothermal amplification assay. J. Clin. Microbiol. 43:2895-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poon, L. L. M., C. S. W. Leung, K. H. Chan, J. H. C. Lee, K. Y. Yuen, Y. Guan, and J. S. M. Peiris. 2005. Detection of human influenza A viruses by loop-mediated isothermal amplification. J. Clin. Microbiol. 43:427-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poon, L. L. M., C. S. W. Leung, M. Tashiro, K. H. Chan, B. W. Y. Wong, K. Y. Yuen, Y. Guan, and J. S. M. Peiris. 2004. Rapid detection of the severe acute respiratory syndrome (SARS) coronavirus by a loop-mediated isothermal amplification assay. Clin. Chem. 56:1050-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prida, M., G. Posadas, S. Inoue, F. Hasebe, and K. Morita. 2004. Real-time reverse transcription loop-mediated isothermal amplification for rapid detection of West Nile virus. J. Clin. Microbiol. 42:257-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakon, M., K. Yamazaki, E. Utagawa, Y. Okuno, and I. Oishi. 2000. Genomic characterization of human astrovirus type 6 Katano virus and the establishment of a rapid and effective reverse transcription-polymerase chain reaction to detect all serotypes of human astrovirus. J. Med. Virol. 61:125-131. [DOI] [PubMed] [Google Scholar]
- 25.Thai, H. T. V., M. Q. Le, C. D. Vuong, M. Parida, H. Minekawa, T. Notomi, F. Hasebe, and K. Morita. 2004. Development and evaluation of a novel loop-mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 42:1956-1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ushio, M., I. Yui, N. Yoshida, M. Fujino, T. Yonekawa, Y. Ota, T. Notomi, and T. Nakayama. 2005. Detection of respiratory syncytial virus genome by subgroup-A, B specific reverse transcription loop-mediated isothermal amplification (RT-LAMP). J. Med. Virol. 77:121-127. [DOI] [PubMed] [Google Scholar]
- 27.Vinjé, J., H. Deijl, R. van der Heide, D. Lewis, K.-O. Hedlund, L. Svensson, and M. P. G. Koopmans. 2000. Molecular detection and epidemiology of Sappro-like viruses. J. Clin. Microbiol. 38:530-536. [DOI] [PMC free article] [PubMed] [Google Scholar]