Abstract
Melioidosis, a disease caused by the bacterium Burkholderia pseudomallei, is endemic in southeast Asia and northern Australia. We used suppression subtractive hybridization (SSH) to identify sequences that varied between two B. pseudomallei isolates from Australia and determined the distribution of 45 SSH-derived sequences among a panel of B. pseudomallei and B. thailandensis isolates. Sequences exhibiting variable prevalence were included in a variable amplicon typing (VAT) scheme designed to score the presence or absence of 14 PCR amplicons. VAT analysis was carried out with 48 isolates from Thailand, which were typed by multilocus sequence typing (MLST), and 44 isolates from Australia of known MLST type. The VAT scheme could be used to divide the 48 isolates from Thailand into 23 VAT types and the 44 isolates from Australia into 36 VAT types. Some of the sequences included in the VAT scheme were more commonly PCR positive among isolates from Australia than among isolates from Thailand, and vice versa. No isolate from Australia was PCR positive for genomic island 11 or a putative transposase sequence, whereas four SSH-derived sequences were far more prevalent among the Australian isolates. Analysis based on the VAT scheme indicated that the isolates clustered into groups, some of which were mainly or exclusively from one geographical origin. One cluster included Australian isolates that were mostly associated with severe disease, including rare neurological melioidosis, suggesting that the content of the accessory genome may play an important role in determining the clinical manifestation of the disease.
The disease melioidosis, caused by the gram-negative bacterium Burkholderia pseudomallei, is endemic in southeast Asia and northern Australia (4). B. pseudomallei, a normal inhabitant of soil and surface water in regions of endemicity, infects via direct inoculation or inhalation and can cause severe sepsis or pneumonia. Not only can melioidosis affect many different sites in the body, but it also has a wide spectrum of severity, ranging from acute and often fatal sepsis to more chronic disease (8, 16, 28). Several years ago, B. pseudomallei was separated from an avirulent biotype lacking the ability to assimilate arabinose, now known as B. thailandensis (24). However, it is clear that the levels of virulence exhibited by different B. pseudomallei isolates can vary considerably in animal models (26). Such variations can occur between related strains and do not necessarily correlate with clinical outcome or the source of the isolate (26).
Various typing methods have been applied to the study of genetic variation among B. pseudomallei populations (14, 18), including molecular fingerprinting approaches such as ribotyping (13), random amplified polymorphic DNAs (RAPDs) (17, 26), and macrorestriction analysis coupled with pulsed-field gel electrophoresis (PFGE) (5, 15, 26). More recently, a multilocus sequence typing (MLST) scheme has been developed (11). Extensive typing of isolates by MLST has demonstrated that isolates from Australia differ from those isolated elsewhere (6), but there was no correlation between strain type and clinical presentation, a finding supported by analysis by PFGE (4).
Molecular typing methods often suffer from the lack of portability (RAPDs), the requirement for specialized equipment (PFGE), the length of the procedures (PFGE and MLST), or the cost (MLST). MLST has emerged as a preferred typing method for phylogenetic studies because of its portability and unequivocal output data. However, MLST typing specifically targets the conserved regions of bacterial genomes rather than the accessory genome, which may have an important role to play in virulence. The genome sequence of B. pseudomallei K96243 revealed the presence of 16 genomic islands (GIs) with variable distributions among B. pseudomallei isolates, suggesting that horizontal gene transfer has played an important role in the evolution of this pathogen (12). More recently, 16 regions of difference (RDs) in the genome of strain K96243, 13 of which corresponded to the GIs, were reported following a comparison with strain Bp15682 by the use of microarrays (22). Other studies have provided further evidence of considerable variations in the accessory genome of B. pseudomallei (9, 21).
Subtractive hybridization is a powerful technique for the identification of DNA sequences present in one strain (the tester) but absent from another (the driver or reference), and it has widely been applied to the study of bacterial pathogens (29), including B. pseudomallei and its close relative, B. mallei (9, 10, 20). In this study we describe the use of suppression subtractive hybridization (SSH) to identify sequences that vary between two B. pseudomallei isolates from Australia. We further study the distribution of such sequences among a panel of B. pseudomallei isolates. Using this information and data from previous studies, we describe the development of a portable multiplex PCR (M-PCR)-based method to screen for the presence or absence of 14 PCR amplicons. Finally, we describe the use of this variable amplicon typing (VAT) scheme for its ability to discriminate between isolates from Australia and Thailand.
MATERIALS AND METHODS
Bacterial strains.
The isolates used in this study are listed in Table 1. The Australian isolates chosen for SSH were isolate 338 and isolate 520. Isolate 338 was isolated from the sputum of a 50-year-old man with chronic lung disease who had a mild clinical infection and intermittently positive sputum cultures over several years, despite specific therapy for melioidosis. Isolate 520 was isolated from a 62-year-old woman on corticosteroids for chronic lung disease who died of progressive septicemic melioidosis pneumonia. Uniquely among 49 isolates tested, strain 338 has been found in a mouse model to induce a strong immunoprotective capacity against subsequent infection with another strain (27).
TABLE 1.
Strains used in this studya
| Isolate(s) | Country | Source; notesb | MLST groupc | Allele no.
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ace | gltB | gmhD | lepA | lipA | narK | ndh | ||||
| B. pseudomallei isolates for subtraction and distribution analysis | ||||||||||
| 338d | Australia | Melioidosis (chronic) | 243 | 1 | 2 | 13 | 4 | 15 | 12 | 1 |
| 520d | Australia | Melioidosis (fulminant) | ND | ND | ND | ND | ND | ND | ND | ND |
| 146 (VE05) | Australia | Goat isolate; same ribotype as 511 (B); LD50 of 9.01 × 102 | ND | ND | ND | ND | ND | ND | ND | ND |
| 511 (VE02) | Australia | Goat isolate, same ribotype as 146 (B); LD50 of 6.32 × 104 | ND | ND | ND | ND | ND | ND | ND | ND |
| 157 (CL26) | Australia | Melioidosis (A); LD50 of 3.00 × 100 | ND | ND | ND | ND | ND | ND | ND | ND |
| 161 (VE06) | Australia | Sheep isolate (A); LD50 of 8.00 × 100 | ND | ND | ND | ND | ND | ND | ND | ND |
| 169 (EN11) | Australia | Soil isolate (A); LD50 of 5.00 × 100 | ND | ND | ND | ND | ND | ND | ND | ND |
| 244 (EN10) | Australia | Soil isolate (B); LD50 of 8.43 × 102 | ND | ND | ND | ND | ND | ND | ND | ND |
| 186 (VE03) | Australia | Soil isolate (B); LD50 of 7.82 × 103 | ND | ND | ND | ND | ND | ND | ND | ND |
| 265 (EN07) | Australia | Soil isolate (B); LD50 of 4.25 × 103 | ND | ND | ND | ND | ND | ND | ND | ND |
| 295 (EN08) | Australia | Soil isolate (B); LD50 of 3.13 × 103 | ND | ND | ND | ND | ND | ND | ND | ND |
| 1655 | Australia | From patient with long-term carriage | ND | ND | ND | ND | ND | ND | ND | ND |
| E503 | Malaysia | Melioidosis | ND | ND | ND | ND | ND | ND | ND | ND |
| E505 | UK/Goa | Clinical isolate | ND | ND | ND | ND | ND | ND | ND | ND |
| E506 | Malaysia | Melioidosis | ND | ND | ND | ND | ND | ND | ND | ND |
| E955 (204), E957 (576) | Thailand | Clinical isolates | ND | ND | ND | ND | ND | ND | ND | ND |
| E958, E8 (E960) | Thailand | Environmental isolates | ND | ND | ND | ND | ND | ND | ND | ND |
| G185 (K96243) | Thailand | Clinical isolate; genome sequence strain | ND | ND | ND | ND | ND | ND | ND | ND |
| B. thailandensis isolates for subtraction and distribution analysis | ||||||||||
| E82 (E959), E32, E111, E125, E132, E135, E216, E251, E253, E254, E255, E260 | Thailand | T. Pitt | ND | ND | ND | ND | ND | ND | ND | ND |
| B. pseudomallei isolates used for VAT analysis (excluding 338) | ||||||||||
| 303 | Australia | Tracheotomy isolate | 36 | 1 | 7 | 14 | 7 | 1 | 12 | 11 |
| 332 | Australia | Human isolate | 106 | 1 | 2 | 3 | 2 | 16 | 21 | 1 |
| 973 | Australia | Human isolate | 107 | 1 | 2 | 3 | 4 | 1 | 8 | 1 |
| 1152 | Australia | Rectal swab isolate | 108 | 1 | 2 | 3 | 2 | 6 | 22 | 1 |
| 64 | Australia | Blood isolate | 109 | 1 | 2 | 13 | 4 | 1 | 19 | 1 |
| 1080 | Australia | Wound swab isolate | 111 | 1 | 2 | 13 | 2 | 1 | 9 | 1 |
| 911 | Australia | Sputum isolate | 112 | 1 | 2 | 13 | 16 | 1 | 22 | 1 |
| 502 | Australia | Soil isolate | 114 | 1 | 3 | 3 | 4 | 1 | 24 | 1 |
| 875 | Australia | Urine isolate | 115 | 1 | 4 | 3 | 2 | 4 | 26 | 1 |
| 1164 | Australia | Blood isolate | 116 | 1 | 4 | 3 | 4 | 1 | 12 | 1 |
| 1153 | Australia | CSF isolate | 117 | 1 | 4 | 13 | 14 | 8 | 22 | 11 |
| 978 | Australia | Blood isolate | 118 | 1 | 4 | 14 | 2 | 1 | 8 | 1 |
| 789 | Australia | Human isolate | 120 | 1 | 4 | 22 | 2 | 5 | 23 | 1 |
| 114 | Australia | Lesion nodule isolate | 121 | 1 | 4 | 23 | 2 | 1 | 8 | 1 |
| 1357 | Australia | Human isolate | 122 | 1 | 6 | 13 | 2 | 1 | 8 | 11 |
| 449 | Australia | Blood isolate | 126 | 1 | 14 | 20 | 1 | 15 | 9 | 15 |
| 944 | Australia | Blood isolate | 127 | 1 | 15 | 3 | 2 | 6 | 27 | 1 |
| 634 | Australia | Human isolate | 128 | 1 | 15 | 13 | 2 | 8 | 12 | 1 |
| 668 | Australia | Blood isolate | 129 | 1 | 15 | 13 | 2 | 1 | 22 | 1 |
| 614 | Australia | Prostate isolate | 132 | 1 | 16 | 13 | 4 | 6 | 21 | 1 |
| 1128 | Australia | Skin lesion isolate | 133 | 1 | 16 | 13 | 4 | 15 | 21 | 1 |
| 130 | Australia | Blood isolate | 134 | 1 | 16 | 14 | 4 | 1 | 19 | 1 |
| 99 | Australia | Human isolate | 135 | 1 | 17 | 13 | 4 | 15 | 22 | 1 |
| 129 | Australia | Right-foot wound isolate | 138 | 4 | 2 | 14 | 4 | 1 | 6 | 1 |
| 239 | Australia | Blood isolate | 140 | 4 | 7 | 3 | 4 | 1 | 19 | 1 |
| 362 | Australia | Urine isolate | 141 | 4 | 16 | 3 | 4 | 1 | 9 | 6 |
| 983 | Australia | Throat isolate | 142 | 8 | 2 | 3 | 4 | 1 | 19 | 1 |
| 1123 | Australia | Blood isolate | 143 | 8 | 2 | 13 | 4 | 1 | 6 | 1 |
| 1168 | Australia | Blood isolate | 144 | 8 | 2 | 13 | 15 | 1 | 27 | 1 |
| 1161 | Australia | Sputum isolate | 146 | 10 | 2 | 3 | 4 | 3 | 2 | 1 |
| 1174 | Australia | Blood isolate | 147 | 10 | 2 | 3 | 4 | 15 | 2 | 1 |
| 62 | Australia | Human isolate | 148 | 10 | 15 | 3 | 4 | 3 | 22 | 1 |
| 356 | Australia | Blood isolate | 149 | 11 | 2 | 14 | 2 | 1 | 6 | 1 |
| 272 | Australia | Sputum isolate | 236 | 1 | 1 | 13 | 1 | 18 | 23 | 11 |
| 210 | Australia | Blood isolate | 238 | 1 | 2 | 3 | 21 | 15 | 9 | 1 |
| 112C | Australia | Human isolate | 241 | 1 | 2 | 13 | 2 | 8 | 6 | 1 |
| 253 | Australia | Right-foot ulcer | 242 | 1 | 2 | 13 | 4 | 1 | 22 | 1 |
| 271 | Australia | Blood isolate | 247 | 1 | 2 | 23 | 2 | 1 | 31 | 1 |
| 222 | Australia | Knee swab isolate | 259 | 1 | 6 | 13 | 2 | 15 | 8 | 11 |
| 504 | Australia | Soil isolate | 268 | 1 | 15 | 31 | 4 | 6 | 19 | 1 |
| 527 | Australia | Blood isolate | 269 | 1 | 21 | 3 | 2 | 5 | 22 | 11 |
| 506 | Australia | Soil isolate | 281 | 12 | 4 | 6 | 2 | 3 | 33 | 1 |
| 157 | Australia | Human isolate | 284 | 13 | 15 | 13 | 4 | 6 | 6 | 11 |
| 139 | PNG | Abscess isolate | 246 | 1 | 2 | 22 | 18 | 1 | 22 | 11 |
| 141 | PNG | Blood isolate | 274 | 4 | 20 | 13 | 2 | 3 | 6 | 11 |
| 140 | Fiji | Abscess isolate | 280 | 12 | 1 | 3 | 1 | 1 | 22 | 1 |
| 314 | Malaysia | Blood isolate | 58 | 3 | 1 | 5 | 1 | 1 | 4 | 1 |
| P1 | Thailand | Blood isolate; northeastern Thailand | u | 1 | 1 | 12 | 2 | 6 | 4 | 1 |
| P2 | Thailand | Pus isolate; northeastern Thailand | 16 | 1 | 2 | 2 | 1 | 1 | 10 | 1 |
| P3 | Thailand | Pus isolate; south Thailand | u | 1 | 1 | 11 | 1 | 6 | 22 | 1 |
| P4 | Thailand | Blood isolate; eastern Thailand | 211 | 3 | 1 | 3 | 1 | 1 | 4 | 1 |
| P5 | Thailand | Pus isolate; northeastern Thailand | u | 3 | 1 | 2 | 1 | 8 | 4 | 3 |
| P6 | Thailand | Blood isolate; south Thailand | 93 | 1 | 1 | 2 | 1 | 1 | 4 | 1 |
| P7 | Thailand | Blood isolate; Bangkok, Thailand | 307 | 1 | 2 | 3 | 1 | 1 | 3 | 1 |
| P8 | Thailand | Sinus isolate; Bangkok, Thailand | 51 | 3 | 1 | 2 | 3 | 1 | 4 | 3 |
| P9 | Thailand | Pus isolate; south Thailand | u | 3 | 1 | 2 | 3 | 5 | 2 | 1 |
| P11 | Thailand | Blood isolate; south Thailand | u | 3 | 2 | 3 | 1 | 1 | 4 | 1 |
| P12 | Thailand | Urine isolate; Bangkok, Thailand | 46 | 3 | 1 | 2 | 1 | 1 | 3 | 3 |
| P14 | Thailand | Pleural fluid; Chiang Mai, Thailand | u | 1 | 4 | 3 | 4 | n | n | 3 |
| P15 | Thailand | Pleural fluid; Chiang Mai | 56 | 3 | 1 | 4 | 1 | 1 | 4 | 1 |
| P16, P22 | Thailand | Blood isolate; Chiang Mai | 290 | 3 | 4 | 11 | 3 | 5 | 4 | 1 |
| P17, P21 | Thailand | Blood isolate; Chiang Mai | u | 1 | 1 | 4 | 17 | 1 | 4 | 1 |
| P18, P19 | Thailand | Blood isolate; Chiang Mai | 167 | 1 | 1 | 4 | 1 | 1 | 3 | 1 |
| P20 | Thailand | Blood isolate; Chiang Mai | 70 | 3 | 4 | 11 | 3 | 5 | 4 | 6 |
| P23 | Thailand | Blood isolate; Chiang Mai | u | 1 | 4 | 3 | 3 | 5 | 3 | 3 |
| P24 | Thailand | Blood isolate; Chiang Mai | u | 1 | 1 | 4 | 17 | 1 | 3 | 1 |
| P25 | Thailand | Pus isolate; Chiang Mai | 17 | 1 | 2 | 3 | 1 | 1 | 1 | 1 |
| P26 | Thailand | Pus isolate; Chiang Mai | 10 | 1 | 1 | 13 | 1 | 1 | 1 | 1 |
| P27 | Thailand | Pus isolate; Chiang Mai | u | 1 | 2 | 4 | 1 | 1 | 22 | 1 |
| P28, P31 | Thailand | Pus isolate; Chiang Mai | u | 1 | 1 | 4 | 17 | 1 | 4 | 1 |
| P29 | Thailand | Pus isolate; Chiang Mai | u | 4 | 12 | 13 | 2 | 1 | 2 | 1 |
| P30, P45 | Thailand | Pus isolate; Chiang Mai | u | 3 | 2 | 3 | 1 | 1 | 4 | 1 |
| P32 | Thailand | Pus isolate; Chiang Mai | u | 1 | 2 | 4 | 3 | 6 | 3 | 1 |
| P33, P35 | Thailand | Pus isolate; Chiang Mai | u | 1 | 4 | 6 | 1 | 6 | 3 | 1 |
| P34, P44 | Thailand | Pus isolate; Chiang Mai | 70 | 3 | 4 | 11 | 3 | 5 | 4 | 6 |
| P36 | Thailand | Pus isolate; Chiang Mai | u | 3 | 2 | 4 | 1 | 1 | 4 | 1 |
| P37 | Thailand | Pus isolate; Chiang Mai | u | 4 | 1 | 2 | 2 | 6 | 4 | 1 |
| P38 | Thailand | Sputum isolate; Chiang Mai | 56 | 3 | 1 | 4 | 1 | 1 | 4 | 1 |
| P39 | Thailand | Urine isolate; Chiang Mai | 10 | 1 | 1 | 13 | 1 | 1 | 1 | 1 |
| P40 | Thailand | Urine isolate; Chiang Mai | u | 4 | 1 | 2 | 2 | 6 | 4 | 1 |
| P41 | Thailand | Urine isolate; Chiang Mai | u | 1 | 1 | 4 | 17 | 1 | 4 | 1 |
| P42 | Thailand | Pleural fluid; Chiang Mai | 70 | 3 | 4 | 11 | 3 | 5 | 4 | 6 |
| P43 | Thailand | Blood isolate; Chiang Mai | 10 | 1 | 1 | 13 | 1 | 1 | 1 | 1 |
| P46 | Thailand | Sputum isolate; Chiang Mai | 10 | 1 | 1 | 13 | 1 | 1 | 1 | 1 |
| P47 | Thailand | Sputum isolate; Chiang Mai | u | 3 | 1 | 2 | 3 | 5 | 2 | 1 |
| P48 | Thailand | Sputum isolate; Chiang Mai | 46 | 3 | 1 | 2 | 1 | 1 | 3 | 3 |
| P49 | Thailand | Sputum isolate; Chiang Mai | 70 | 3 | 4 | 11 | 3 | 5 | 4 | 6 |
| P50 | Thailand | Urine isolate; Chiang Mai | 23 | 1 | 2 | 13 | 1 | 1 | 1 | 1 |
| B. thailandensis isolate used in VAT analysis | ||||||||||
| E52 | Thailand | Water isolate; Chiang Mai | ND | ND | ND | ND | ND | ND | ND | ND |
Abbreviations: u, MLST type not present in the database; ND, not determined; n, novel sequence for this locus; UK, United Kingdom; PNG, Papua New Guinea; CSF, cerebrospinal fluid.
The 50% lethal doses (LD50) were determined by using a BALB/c mouse model (26). RAPD subtypes are denoted by letter designations in parentheses.
Where known, the MLST group is indicated.
Isolates in SSH.
The strain panel used to analyze the distribution of subtracted sequences included B. pseudomallei isolates from Australia (26) and B. pseudomallei and B. thailandensis isolates described previously (30). Further collections of 48 uncharacterized isolates from Thailand and 47 isolates mainly from Australia, each of which represented a different MLST group, were used to test sequence distributions by use of the VAT scheme (Table 1). Isolate 338 was included in this analysis. All isolates used in the VAT analysis were isolated from different patients.
Extraction of DNA.
DNA was isolated from strains 338 and 520 for use in SSH by the guanidium thiocyanate method, as described previously (25). Small-scale isolation of DNA from the collection of Thai isolates was carried out with the Wizard Genomic DNA Purification kit (Promega). DNA from the larger collection of mainly Australian isolates was extracted by using the QIAamp DNA mini kit (QIAGEN).
MLST typing.
MLST typing of 48 B. pseudomallei isolates from Thailand was carried out by PCR amplification and DNA sequencing of the seven loci (ace, gltB, gmhD, lepA, lipA, narK, and ndh) used in the published MLST typing scheme (11). The loci were amplified by using the oligonucleotide primers and conditions recommended at the website http://bpseudomallei.mlst.net/ and were sequenced with the same primers. The search facility at http://bpseudomallei.mlst.net/ was used to assign the sequences obtained to allele types and to screen for previously reported MLST types. The MLST types for the 47 isolates mainly from Australia were determined previously (6).
Construction and screening of subtraction libraries.
SSH was carried out with the CLONTECH PCR-Select bacterial genome subtraction kit (Clontech) as recommended by the supplier, but with a hybridization temperature of 73°C to take account of the high G+C content of the organism. PCR products obtained following SSH were cloned into pGEM-T (Invitrogen) to produce a subtracted DNA library of RsaI fragments. Plasmid DNA from individual clones was extracted and sequenced with vector primers by Lark Technologies. Nucleotide sequences were analyzed for their presence in genome sequence strains by using the BLASTN facilities at the websites http://www.sanger.ac.uk/Projects/B_pseudomallei (B. pseudomallei K96243) and http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi (B. mallei ATCC 23344 and B. thailandensis E264). BLASTX searches of the general database were carried out by using the website http://www.ncbi.nlm.nih.gov.
PCR and M-PCR amplification.
The oligonucleotide primers (Sigma-Genosys) used in PCR assays and for the labeling of probes are listed in Table 2 and Table S1 in the supplemental material, along with the annealing temperatures used. Amplifications were carried out in an Eppendorf MasterCycler thermal cycler for 30 cycles consisting of 95°C (1 min), the annealing temperature (1 min), and 72°C (2 min), with an additional extension time at 72°C (10 min) following completion of the 30 cycles.
TABLE 2.
Oligonucleotide primers used for M-PCR amplification
| M-PCR no. and primer | Sequence (5′ to 3′) | Amplicon size (bp) | Target | A.T. (°C)a | Reference or source |
|---|---|---|---|---|---|
| M-PCR1 | 58 | ||||
| 146-5F | ATCTGATCAGGACGCTTG | 666 | gmhA (BPSL2795b) (capsule) | This study | |
| 146-5R | CACTGCTTCCCGAAAATG | ||||
| 338-B7F | ACTGGAATCGGGAAAAAC | 482 | 338-B7 (near GI5) | This study | |
| 338-B7R | ACGATATTTTTCCGCTGC | ||||
| 520-E42F2 | ATGCCGGCAGCGTCATAGA | 257 | 520-E42 (GI7) | This study | |
| 520-E42R2 | ACAACGCATGCTTACAGTA | ||||
| 520-E33F | GATCCATGACCACGGCCA | 135 | 520-E33 | This study | |
| 520-E33R | AGGCCGAGAGTCTGATTG | ||||
| M-PCR2 | 58 | ||||
| TRANSF | TTTACCGAAGTCATGAGC | 657 | Transposase | This study | |
| TRANSR | TTTGAAGTGCTGGTCGAC | ||||
| 338-2C5F | AGCAATAAGCGGGCAAAA | 403 | 338-2C5 | This study | |
| 338-2C5R | ATCACAAGCTATCCGCAG | ||||
| 520-E35F | CTACTAGCCACTGATTCC | 290 | 520-E35 | This study | |
| 520-E35R | ATAGATCATTCGTCCGAG | ||||
| 520-2G9F | ACCTCGATTTTGCGTCTG | 145 | 520-2G9 | This study | |
| 520-2G9R | AGAATGGCGTGGAGATTG | ||||
| M-PCR3 | 58 | ||||
| 338-2D10F2 | ATGTCGTGCCTCCGTTCA | 320 | 338-2D10 | This study | |
| 338-2D10R2 | ATGAGTCGGATCGGATCA | ||||
| GI11BF | TGTCGTGGCCCGGGGATTTGTA | 238 | BPSL3260b (GI11) | 12 | |
| GI11BR | TATTCGTTGCTTTCGCGTGTGGTC | ||||
| 520-E36F | GTAATGACGCAAGACGCCG | 132 | 520-E36 | This study | |
| 520-E36R | ACGGCCGAACACAAGAAC | ||||
| M-PCR4 | 50 | ||||
| GI12F | GCAATGGAATCGACGCAACATTG | 788 | BPSL3349b (GI12) | 12 | |
| GI12R | GACGCTGGCGGGTATGGGTAAG | ||||
| 338-B3F | AATCAGACACTCGAGGAC | 605 | 338-B3 | This study | |
| 338-B3R | ATAACCTGCTCGATTTTCC | ||||
| 520-2E10F | CTCCACCGTGACGCTAAG | 392 | 520-2E10 | This study | |
| 520-2E10R | GAGCACTTCACGCGTCTG |
A.T., annealing temperature.
The GenBank accession number is given.
Dot blot hybridization.
Dot blot hybridization of genomic DNA was carried out as described previously (30) with a digoxigenin labeling and detection system (Roche). Posthybridization washes were carried out by using stringent conditions.
RESULTS
SSH between two Australian isolates.
Two rounds of SSH were carried out between strains 338 and 520, using each strain in turn as the tester strain. In excess of 50 clones were sequenced for each of the subtractions. The presence or absence of each of the nonduplicated subtracted sequences in the tester and the driver strains was assessed by PCR assay. The results of the SSH experiments are summarized in Table 3. We identified 20 sequences that were PCR positive for strain 338 but PCR negative for strain 520 and 19 sequences that were PCR positive for strain 520 but PCR negative for strain 338.
TABLE 3.
Summary of subtractive hybridization
| SSHa sequence and sequence type | Length (bp) | G+C content (%)b | Presence of sequenced genomec
|
Best BLASTX match; comments (GenBank accession no.) | % Identity | Length (no. of amino acids) | E value | ||
|---|---|---|---|---|---|---|---|---|---|
| Bpm | Bt | Bm | |||||||
| Sequences present in strain 338 but not 520 | |||||||||
| Recombination related | |||||||||
| 338-B3 (DQ351720) | 1,097 | 55.7 | − | − | − | DNA helicase-related protein (Xanthomonas campestris) (NP_637459) | 32 | 364 | 2e−45 |
| 338-2D1 | 760 | 57.8 | − | − | − | DNA helicase-related (Xanthomonas campestris) (NP637459); different region of same protein as 338-B3 | 72 | 248 | 7e−83 |
| 338-B20 | 333 | 54.1 | − | − | − | Uncharacterized protein (Rubrivivax gelatinosus) (ZP_00241526); Membrane proteins, DNA recombination protein RmuC (Salmonella and others) (NP_457782) | 80 | 110 | 4e−43 |
| 54 | 110 | 5e−26 | |||||||
| Bacteriophage related | |||||||||
| 338-2C9 | 509 | 51.3 | − | p | − | Putative transmembrane protein (Ralstonia solanacearum) (NP_520413); DNA methylase of bacteriophage Φ E125 (B. thailandensis) (AAL47559) | 74 | 50 | 1e−15 |
| 96 | 28 | 3e−8 | |||||||
| Transcriptional regulators | |||||||||
| 338-B7 | 482 | 54.8 | +1 | − | − | DeoR family transcriptional regulator in RD6/GI5 (BPSL0939) | 99 | 160 | 4.5e−82 |
| 338-2C3 | 305 | 57.4 | − | − | − | Transcriptional regulator (Ralstonia eutropha) (ZP_00169018) | 47 | 59 | 2e−13 |
| Enzymes | |||||||||
| 338-2A12 | 283 | 57.6 | − | − | − | Maleylacetate reductase (Ralstonia sp.) (AAS87585) | 55 | 68 | 4e−16 |
| 338-2D9 | 480 | 59.2 | − | − | − | Alcohol dehydrogenase (Polaromonas sp.) (ZP_00364129) | 72 | 159 | 5e−60 |
| Hypothetical proteins | |||||||||
| 338-B8 | 425 | 50.6 | − | − | − | Hypothetical protein (Escherichia coli O157:H7) (NP_313283) | 32 | 145 | 3e−13 |
| 338-2C4 | 616 | 52.6 | − | − | − | Hypothetical protein (Rhodopseudomonas palustris) (NP_949350) | 31 | 217 | 5e−16 |
| 338-B4 | 282 | 53.9 | +1 | − | − | Hypothetical protein in RD6/GI5 (BPSL0942) | 100 | 88 | 2e−42 |
| 338-2D7 | >624 | 48.9 | − | − | − | Hypothetical protein (Chromobacterium violaceum) (AAQ61798) | 39 | 94 | 3e−10 |
| No significant BLASTX matches | |||||||||
| 338-B1 | 190 | 50.0 | − | − | − | ||||
| 338-B16 | 374 | 48.1 | − | − | − | ||||
| 338-2A7 | 292 | 44.5 | − | − | − | ||||
| 338-2B2 | 429 | 50.4 | − | − | − | ||||
| 338-2B4 | 333 | 51.1 | +2 | + | + | ||||
| 338-2B7 | 426 | 57.5 | − | − | − | ||||
| 338-2B10 | 282 | 43.3 | − | − | − | ||||
| 338-2D3 | 331 | 52.3 | − | − | − | ||||
| Sequences present in strain 520 but not 338 | |||||||||
| Mobile elements | |||||||||
| 520-E15 | 337 | 54.0 | − | − | − | Putative transposase (Burkholderia fungoram) (ZP_00283626) | 75 | 29 | 1e−7 |
| 520-E18 | 335 | 59.4 | +2p | − | p | Putative transposase (BPSS2148) | 97 | 49 | 8e−21 |
| 520-E33 | 158 | 58.2 | +2 | − | − | Putative transposase (BPSS2148); different region but same protein as 520-E18 | 100 | 52 | 8.7e−23 |
| 520-2F1 | 420 | 59.5 | +2 | − | − | Putative transposase (BPSS2148); different region but same protein as 520-E18 and 520-E33 | 99 | 128 | 2e−66 |
| Secretion related | |||||||||
| 520-E12 | 765 | 53.9 | − | − | − | Hypothetical SecA-related protein (Photobacterium profundum) (YP_133346) | 48 | 218 | 2e−53 |
| Lipoprotein | |||||||||
| 520-E44 | 202 | 56.4 | +1 | + | − | Putative lipoprotein (BPSL2045) | 97 | 45 | 5e−19 |
| Enzymes | |||||||||
| 520-E19 | 759 | 53.9 | − | − | − | Appr-1-p processing enzyme family (Nitrosomonas europaea) (NP_841411); conserved hypothetical protein (Synechocystis spp.) (NP_942395) | 77 | 127 | 5e−53 |
| 65 | 119 | 3e−43 | |||||||
| 520-E1 | 374 | 50.0 | +1p | − | p | Conserved hypothetical protein (Synechocystis spp.) (NP_942395); Appr-1-p processing enzyme family (Nitrosomonas europia) (NP841411.1) | 68 | 29 | 4e−5 |
| 56 | 30 | 0.008 | |||||||
| 520-2F8 | 314 | 56.7 | +1 | p | + | Molybdopterin oxidoreductase (BPSL2207) | 100 | 54 | 3e−25 |
| Hypothetical or uncharacterized proteins | |||||||||
| 520-2E7 | 314 | 60.2 | − | − | − | Uncharacterized protein (Microbulbifer degradans) (ZP_00318360) | 55 | 103 | 7e−25 |
| 520-2F2 | 773 | 54.1 | +1 | − | − | Hypothetical protein in RD7/GI6 (BPSL1146); variation in the C terminus | 80 | 111 | 7e−40 |
| 520-2E10 (DQ351721) | 519 | 54.0 | +1p | + | − | Hypothetical protein (BPSL2048) | 49 | 123 | 6e−26 |
| 520-E35 (DQ351716) | 308 | 52.9 | − | + | − | Hypothetical protein (BPSL2048A) | 59 | 101 | 3e−24 |
| 520-2G6 | 373 | 53.9 | − | − | − | Hypothetical protein (B. mallei) (YP_105718) | 47 | 113 | 1e−23 |
| No significant BLASTX matches | |||||||||
| 520-E16 | 134 | 57.0 | − | − | − | ||||
| 520-E10 | 529 | 54.6 | − | − | − | ||||
| 520-2E1 | 233 | 52.8 | − | − | − | ||||
| 520-2F11 | 602 | 54.5 | − | − | − | ||||
| 520-2F6 | 814 | 50.7 | − | − | − | ||||
| Sequences present in strains 338 and 520 | |||||||||
| 338-2D10 (DQ351718) | 370 | 56.2 | − | + | − | Bacteriophage protein from Φ1026b (B. pseudomallei 1026b) (NP_945078); bacteriophage protein from ΦE125 (B. thailandensis) (NP_536399) | 87 | 100 | 3e−47 |
| 84 | 100 | 6e−45 | |||||||
| 338-2B9 | 663 | 47.1 | +2 | − | − | Putative exported protein (BPSS0658) | 100 | 162 | 4e−78 |
| 520-E36 (DQ351719) | 150 | 58.0 | − | − | − | Putative transposase (Streptomyces avermitilis) (NP_821845) | 48 | 45 | 0.023 |
| 520-2G9 (DQ351717) | 159 | 57.2 | − | − | − | ISRSO16 transposase ORFB (R. solanacearum) (NP_523187) | 80 | 51 | 4e−18 |
| 338-B14 | 181 | 54.1 | − | − | − | Hypothetical protein (Methylococcus capsulatus) (YP_115042) | 90 | 31 | 6e−9 |
| 338-B18 | 206 | 60.2 | +2 | − | − | Hypothetical protein in GI14 (BPSS0655) | 98 | 68 | 4e−34 |
| 338-2A1 | >630 | 47.0 | +2 | p | + | Hypothetical proteins (BPSS1753) | 100 | 68 | 2e−33 |
| 520-E42 | 303 | 54.1 | +1 | − | − | Hypothetical protein in GI7 (BPSL1385) | 100 | 71 | 7e−34 |
| No significant BLASTX matches | |||||||||
| 338-B12 | 692 | 55.6 | − | − | − | ||||
| 338-2C5 | 429 | 51.3 | +1 | − | + | Overlaps BPSL2558 by 10 bp but lies mainly in the gap between BPSL2558 and BPSL2559 | |||
GenBank accession numbers are indicated in parentheses for those novel sequences used in the VAT analysis.
G+C content for the subtracted sequence.
The presence (+) or absence (−) of the subtracted sequence, based on >90% sequence identity by using BLASTN, is indicated for the genome-sequenced strains of B. pseudomallei (Bpm), B. thailandensis (Bt), and B. mallei (Bm). For B. pseudomallei the number of the matching chromosomes is indicated. p, a partial match, where the match does not extend over the entire length of the subtracted sequence.
Distribution of subtracted sequences among a panel of strains.
Using PCR amplification assays, we studied the distribution of 45 sequences from the subtractions between strain 338 and strain 520 among a panel of 19 B. pseudomallei isolates and 14 B. thailandensis isolates (Table 1). The sequences screened included 10 sequences that were PCR positive for both strain 338 and strain 520 (Table 3), 5 of which were absent from the genome sequence strain (K96243). Among B. pseudomallei isolates, only strain 338 was PCR positive for sequences 338-B1, 338-B3, 338-B8, 338-B20, 338-2A7, 338-2B2, 338-2B4, 338-2B10, 338-2C3, and 338-2D9, whereas only strain 520 was PCR positive for sequences 520-E12, 520-2E7, 520-2F2, and 520-E18. The distribution of the remaining sequences among the B. pseudomallei isolates, based on PCR assays, is shown in Fig. 1. Interestingly, a single strain of B. thailandensis was PCR positive for the sequences 338-B3 and 338-B8, both of which had been detected only in a single strain of B. pseudomallei. B. thailandensis isolates were also PCR positive for sequence 520-E35 (all 14 isolates), sequence 520-2H3 (12 isolates), sequence 520-2E10 (11 isolates), sequence 338-2C5 (10 isolates), sequence 338-2D10 (8 isolates), sequence 520-2F6 (3 isolates), sequence 520-E16 (1 isolate), and sequence 520-E19 (1 isolate). All other PCR amplification tests conducted with the B. thailandensis isolates were negative.
FIG. 1.
PCR-based distribution analysis of SSH sequences. Filled boxes indicate PCR-positive results.
Sequences 338-B4 and 338-B7 shared common distribution profiles and matched putative genes located in close proximity to each other, upstream from the previously reported genomic island GI5 (12), and within RD6 (22) in strain K96243 (Table 3; Fig. 1). Sequence 338-B18, included in the distribution analysis, matched a sequence located in the previously reported genomic island GI14 (12). All but three of the isolates tested were PCR positive for this sequence (Fig. 1). All but one of the isolates tested were PCR positive for sequence 520-E42, located within GI7 (12).
Development of a VAT scheme.
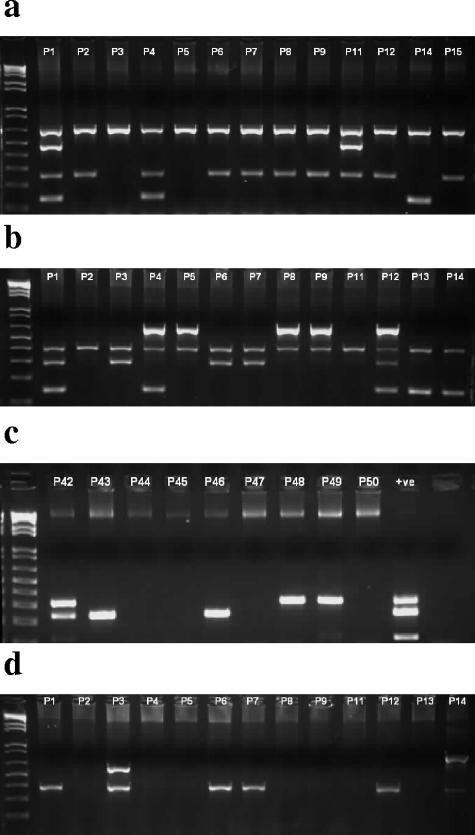
Using the information derived from the distribution analysis we chose several sequences that exhibited variation between strains of B. pseudomallei and tested various primer set combinations with a view to developing M-PCR assays designed to give variable amplicon profiles. As a positive control we included a PCR assay for a capsule gene (gmhA; Table 2). In addition, we included a PCR assay for a putative transposase gene originally identified in strain E503 (23) and known to have a variable prevalence among B. pseudomallei strains (unpublished data) and PCR assays for some of the genomic islands identified previously (12). After testing numerous combinations of primers, we developed a strategy involving four separate M-PCRs (Fig. 2). M-PCR1 is designed to assay for the gmhA-positive control and the variable sequences 338-B7, 520-E42, and 520-E33. M-PCR2 assays for the putative transposase from strain E503 and the variable sequences 338-2C5, 520-E35, and 520-2G9. M-PCR3 assays for GI11 and the variable sequences 338-2D10 and 520-E36. Finally, M-PCR4 assays for GI12 and the variable sequences 338-B3 and 520-2E10. Amplicon sizes are given in Table 2. Overall, four sequences matched within or near genomic islands, four sequences matched transposases, one sequence was bacteriophage related, one sequence was DNA helicase related, two sequences matched hypothetical proteins of unknown function, and one sequence had no significant match (Table 3).
FIG. 2.
M-PCR analysis of B. pseudomallei isolates. The figure shows agarose gels of example PCR amplicons derived from Thai isolates (indicated above individual lanes) by M-PCR1 (a), M-PCR2 (b), M-PCR3 (c), and M-PCR4 (d). The first lane on each gel contains a 1 kb-plus DNA ladder (Invitrogen). +ve, positive control.
Application of VAT to isolates from Australia and Thailand.
The 48 isolates of B. pseudomallei from Thailand were assigned to an allele type for each of the loci ace, gltB, gmhD, lepA, lipA, narK, and ndh (Table 1). With the exception of the lipA and narK loci of one isolate (P14), all alleles matched a sequence already deposited in the MLST database. The allele profiles were used to assign 24 of the isolates to previously reported MLST groups (Table 1). The 48 isolates could be subdivided into 29 MLST groups, 11 of which contained more than one strain. Eighteen isolates had MLST profiles that were unique among the collection of Thai isolates. The largest MLST groups (sequence type 70 [ST70] and one MLST type previously unreported) comprised five isolates (isolates P20, P34, P44, P42, and P49 and isolates P17, P21, P28, P31, and P41, respectively). Four isolates (isolates P26, P39, P43, and P46) shared MLST type ST10. None of the isolates from Thailand shared an MLST type with any of the isolates from Australia.
The M-PCR assays for VAT were applied to DNA extracted from strains of the two collections. Positive control DNA comprising individual or mixed DNA samples known to contain the relevant sequences were included in each of the PCR amplifications, and each DNA sample was tested on a minimum of two occasions. An amplicon was obtained from all DNA preparations for the positive control capsule gene (M-PCR1) with the exception of the preparation for B. thailandensis E52. The full VAT profiles are available in Table S2 in the supplemental material. The 95 B. pseudomallei isolates could be separated into a total of 57 VAT types. The 48 isolates from Thailand, comprising 29 different MLST groups, could be separated into 23 VAT types. The 44 isolates from Australia, comprising 44 different MLST groups, could be separated into 36 VAT types.
The five isolates from Thailand of ST70 could be subdivided by their VAT profiles into three groups. Only one isolate (isolate P42) was PCR positive for GI11, and only isolates P20 and P44 were PCR negative for sequence 338-2D10. Of isolates P17, P21, P28, P31, and P41, which shared an MLST type, isolates P17, P28, and P31 also shared common VAT profiles. However, isolate P21 differed by being PCR positive for GI11, and isolate P41 differed by being PCR positive for sequence 338-B7, located upstream of GI5. Of the four isolates sharing ST10, one isolate (isolate P39) differed from the others in that it was PCR negative for GI11. Another pair of isolates (isolates P33 and P35) that shared a common MLST type also differed in their VAT profiles. However, four other pairs of isolates (isolates P12 and P48, P9 and P47, P30 and P45, and P37 and P40) shared a common MLST type and identical VAT profiles.
The prevalences of the VAT PCR amplicons are summarized in Table 4. Some of the sequences included in the VAT scheme were more commonly PCR positive among isolates from one of the main geographical locations than among isolates from the other. In particular, no isolate from Australia was PCR positive for the putative transposase sequence (TRANS) or GI11; a far higher proportion of Australian isolates were PCR positive for sequences 520-E33, 520-2G9, and 520-E36 (all transposase related) and sequence 338-B3 (DNA helicase related); a higher proportion of the isolates from Thailand were PCR positive for sequences 338-B7 (near GI5) and 520-2E10. Of the known genomic islands, GI11 was present in only five isolates from Thailand and GI12 was present in five isolates from Thailand and nine isolates from Australia. In contrast, the distribution of GI7, as inferred from the PCR assay results for sequence 520-E42, was more widespread (Table 4).
TABLE 4.
Distribution of sequences among B. pseudomallei isolates
| Isolate | % PCR positive
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| gmhA | 338-B7 | 520-E42 | 520-E33 | TRANS | 338-2C5 | 520-E35 | 520-2G9 | 338-2D10 | GI11 | 520-E36 | GI12 | 338-B3 | 520-2E10 | |
| Original panel of B. pseudomallei | 100 | 48 | 95 | 11 | NDa | 90 | 10 | 67 | 38 | ND | 48 | ND | 5 | 14 |
| B. thailandensis (including E52) | 0 | 0 | 0 | 0 | ND | 73 | 100 | 0 | 50 | ND | 0 | ND | 7 | 80 |
| Australian isolates (n = 44) | 100 | 7 | 68 | 64 | 0 | 95 | 34 | 91 | 27 | 0 | 39 | 20 | 27 | 23 |
| Thai isolates (n = 48) | 100 | 21 | 94 | 27 | 31 | 100 | 31 | 42 | 23 | 10 | 4 | 10 | 4 | 50 |
| Other isolates (n = 4) | 100 | 25 | 75 | 50 | 0 | 100 | 25 | 100 | 0 | 0 | 75 | 0 | 0 | 25 |
| Combined (n = 96) | 100 | 15 | 82 | 45 | 16 | 98 | 41 | 67 | 24 | 5 | 23 | 15 | 15 | 36 |
ND, not determined.
The distribution of the sequences among the isolates from Thailand indicated by PCR assays was confirmed by dot blot hybridizations with digoxigenin-labeled probes for the sequences gmhA, 520-E33, 338-B7, and 520-E42 (from M-PCR1); TRANS, 520-E35, and 520-2G9 (from M-PCR2); and 338-B3 and 520-2E10 (from M-PCR4) (data not shown). The PCR assays for GI11 and GI12 had been validated previously (12). From M-PCR3, the sequence 520-E36 dot blots were less clear due to background hybridization. We tested this sequence with a second primer set and obtained the same distribution as before. Because the PCR assay and dot blot data based on our initial primer set for 338-2D10 did not agree, new primers (primers 338-2D10F2 and 338-2D10R2) were designed and tested with the Thai isolates. The distribution results corresponded to those obtained by using dot blots; therefore, the new primer set was incorporated into M-PCR3.
DISCUSSION
SSH between the two Australian isolates 338 and 520 identified 39 sequences that varied between the two isolates. The subtracted sequences varied between 134 bp and 1,097 bp in length, and all had a G+C content (<60.3%) below the average for the organism (68.1%). Among the 39 sequences were 7 matching sequences in chromosome 1 and 4 matching sequences in chromosome 2 of strain K96243. Analysis of the genome sequence of B. pseudomallei K96243 suggests that chromosome 1 contains a higher proportion of genes involved in core functions, while chromosome 2 contains a higher proportion of genes encoding accessory functions (12). However, we found no bias toward chromosome 2 among those subtracted sequences that matched the sequences of strain K96243. In common with previous SSH analysis between non-Australian isolates of B. pseudomallei, we identified several variable sequences that matched transposases and bacteriophages (9). Prophages make a significant contribution to genetic diversity in pathogenic bacteria (1-3). The temperate bacteriophage ΦE125 was originally identified in B. thailandensis as specific for B. mallei (31). More recently, bacteriophage Φ1026b was identified in B. pseudomallei and carries genes for DNA packaging, tail morphogenesis, host lysis, integration, and DNA replication nearly identical to those of ΦE125, while those genes involved in head morphogenesis differ from those of ΦE125 (9). Two sequences exhibiting variable prevalence among B. pseudomallei and B. thailandensis shared similarity but not 100% identity with sequences from these bacteriophages, suggesting that strain 338 and other isolates of B. pseudomallei may carry related bacteriophages. Both sequences matched a region shared by Φ1026b and ΦE125 at a nucleotide sequence identity of 94% (9). Interestingly, the two subtracted sequences (338-2C9 and 338-2D10) did not share the same distribution among the panel strains (Fig. 1), and 338-2D10 was present in both strain 338 and strain 520 (Table 3).
We identified transposase-related sequences from the subtraction only using strain 520 as the tester (Table 3). Interestingly, because of the approach to the initial screening of the subtracted libraries that we chose, we identified a number of variable sequences, including two putative transposase-related sequences, that were not genuinely subtracted. Often, libraries generated following SSH are prescreened to identify tester-specific sequences. Initially, for convenience, we took an approach whereby subtracted sequences were first sequenced and then used to screen the genome sequence strain (B. pseudomallei K96243) prior to the design of oligonucleotide primers for PCR screening of tester and driver DNA. Our observations suggest that the SSH procedure may enrich for regions with low G+C contents in a G+C-rich genome among those sequences not genuinely subtracted. Hence, we were able to identify some interesting sequences that were absent from the genome sequence strain or that were variable among the panel of isolates but that were not true subtracted sequences.
Our SSH analysis identified some sequences carried by the genomic islands previously identified in strain K96243 (12). Two additional islands were included in the VAT scheme. The prophage-like islands GI7 (sequence 520-E42) and GI12 differed considerably in overall prevalence, with GI12 sharing a similarly low prevalence with GI11, a putative integrated plasmid, or a conjugative element (12). However, our findings and those of others (9) suggest that there may be other genomic islands that are not present in strain K96243 but that exhibit a variable prevalence between isolates. The contribution of such islands and variable sequences to the variations in virulence or clinical manifestations exhibited by different strains remains unclear. However, our observations lend support to the notion that horizontal gene transfer has played an important role in the evolution of this pathogen.
Our VAT scheme is designed to give some indication of the mobilomes of isolates while also providing a cheap, reproducible, and portable method for strain discrimination. We chose to test the scheme with collections of isolates from Australia and Thailand. The isolates from Australia, all of which were different by MLST typing, have previously been used to demonstrate a difference between B. pseudomallei isolates from Australia and isolates from other regions of endemicity (6). The isolates from Thailand were first characterized by MLST typing in this study. Thus, some isolates that share the same MLST types were included among the isolates from Thailand. However, although some of these isolates also shared VAT profiles, some had different VAT profiles, suggesting that identical molecular strain types may vary in their mobilomes. Similarly, in some cases isolates of different strain types shared VAT profiles.
Cluster analysis was used to gain an insight into the relationships between strains based on VAT profiles (Fig. 3). The isolates cluster into groups, some of which are mainly or exclusively from one of the main geographical origins and some of which are mixed. Overall, there was a tendency for isolates to cluster with those isolates from the same geographical location, suggesting a divergence in the mobilomes between isolates from Australia and Thailand that is in agreement with the apparently distinctive nature of isolates from Australia compared to the nature of the isolates from southeast Asia (6). However, the presence of minority isolates within clusters dominated by one geographical origin and the existence of some mixed clusters suggest that the picture is more complex. In order to resolve this, it will be necessary to sequence more B. pseudomallei genomes, especially those of Australian isolates, and conduct comprehensive microarray surveys of collections of isolates from different geographical locations.
FIG. 3.
Cluster analysis of VAT profiles. The VAT profiles were clustered by using a multivariate analysis for clustering of observations in the MINITAB software package. A dendrogram was constructed by using average linkage and Pearson distance. One isolate of B. thailandensis (isolate E52) was included in the analysis as an outlier. B. pseudomallei isolates P1 to P50 were from Thailand. All other isolates with the exception of isolates 139 to 141 and 314 were from Australia. The cluster that includes the neurotropic isolates is bracketed.
Interestingly, one cluster included five isolates (isolates 668, 62, 983, 1153, and 332) from patients with the rare neurological melioidosis presentation, which is a specific entity that can occur in patients without risk factors (neurotropic isolates) (7). Of these, three isolates (isolates 62, 983, 1153) had identical VAT profiles, yet these three isolates and the other two neurological melioidosis-related isolates all had different MLST profiles (MLST groups ST129, ST148, ST142, ST117, and ST106, respectively), with no two isolates sharing more than three of the seven MLST alleles. To date, correlations between isolates associated with melioidosis encephalomyelitis have not been found by MLST or PFGE typing (4, 6). ST129, ST148, ST142, ST117, and ST106 were widely distributed in a dendrogram showing the results of cluster analysis based on MLST allele profiles and, apart from ST117 and ST129, were also widely distributed on a tree constructed from concatenated sequences (6). Furthermore, other isolates within the VAT cluster containing the five neurotropic isolates were mostly associated with more severe disease, including three isolates (isolates 1161, 64, and 944) that were associated with bacteremic pneumonia with septic shock. This suggests that the content of the accessory genome may play an important role in determining the clinical manifestations of some forms of melioidosis and that this can be independent of the conserved genome. Although the data obtained in this study are insufficient to identify specific genes or activities that might contribute to the success of the isolates causing disease of the central nervous system, we can discount the need for genomic islands GI11 and GI12, which were absent from these isolates.
Supplementary Material
Acknowledgments
K.D. acknowledges sponsorship from the Royal Thai Government.
We thank Prasit Tharavichitkul and Nuanchan Chittasobhon (Department of Microbiology, Faculty of Medicine, Chiang Mai University, Thailand) for providing strains. We also thank Gary Lum and the microbiology laboratory staff at Royal Darwin Hospital for B. pseudomallei isolation and identification. We are grateful to Brian Spratt and Daniel Godoy for establishment and curation of the MLST database and determining the STs of the Australian strains.
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Brussow, H., and R. W. Hendrix. 2002. Phage genomics: small is beautiful. Cell 108:13-16. [DOI] [PubMed] [Google Scholar]
- 2.Canchaya, C., G. Fournous, S. Chibani-Chennoufi, M. L. Dillmann, and H. Brussow. 2003. Phage as agents of lateral gene transfer. Curr. Opin. Microbiol. 6:417-424. [DOI] [PubMed] [Google Scholar]
- 3.Casjens, S. 2003. Prophages and bacterial genomics: what have we learned so far? Mol. Microbiol. 49:277-300. [DOI] [PubMed] [Google Scholar]
- 4.Cheng, A. C., and B. J. Currie. 2005. Melioidosis: epidemiology, pathophysiology, and management. Clin. Microbiol. Rev. 18:383-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng, A. C., N. P. Day, M. J. Mayo, D. Gal, and B. J. Currie. 2005. Burkholderia pseudomallei strain type, based on pulsed-field gel electrophoresis, does not determine disease presentation in melioidosis. Microbes Infect. 7:104-109. [DOI] [PubMed] [Google Scholar]
- 6.Cheng, A. C., D. Godoy, M. Mayo, D. Gal, B. G. Spratt, and B. J. Currie. 2004. Isolates of Burkholderia pseudomallei from northern Australia are distinct by multilocus sequence typing, but strain types do not correlate with clinical presentation. J. Clin. Microbiol. 42:5477-5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Currie, B. J., D. A. Fisher, D. M. Howard, and J. N. Burrow. 2000. Neurological melioidosis. Acta Trop. 74:145-151. [DOI] [PubMed] [Google Scholar]
- 8.Dance, D. A. 2002. Melioidosis. Curr. Opin. Infect. Dis. 15:127-132. [DOI] [PubMed] [Google Scholar]
- 9.Deshazer, D. 2004. Genomic diversity of Burkholderia pseudomallei clinical isolates: subtractive hybridization reveals a Burkholderia mallei-specific prophage in B. pseudomallei 1026b. J. Bacteriol. 186:3938-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fushan, A., G. Monastyrskaya, I. Abaev, M. Kostina, O. Filyukova, E. Pecherskih, and E. Sverdlov. 2005. Genome-wide identification and mapping of variable sequences in the genomes of Burkholderia mallei and Burkholderia pseudomallei. Res. Microbiol. 156:278-288. [DOI] [PubMed] [Google Scholar]
- 11.Godoy, D., G. Randle, A. J. Simpson, D. M. Aanensen, T. L. Pitt, R. Kinoshita, and B. G. Spratt. 2003. Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei. J. Clin. Microbiol. 41:2068-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holden, M. T., R. W. Titball, S. J. Peacock, A. M. Cerdeno-Tarraga, T. Atkins, L. C. Crossman, T. Pitt, C. Churcher, K. Mungall, S. D. Bentley, M. Sebaihia, N. R. Thomson, N. Bason, I. R. Beacham, K. Brooks, K. A. Brown, N. F. Brown, G. L. Challis, I. Cherevach, T. Chillingworth, A. Cronin, B. Crossett, P. Davis, D. Deshazer, T. Feltwell, A. Fraser, Z. Hance, H. Hauser, S. Holroyd, K. Jagels, K. E. Keith, M. Maddison, S. Moule, C. Price, M. A. Quail, E. Rabbinowitsch, K. Rutherford, M. Sanders, M. Simmonds, S. Songsivilai, K. Stevens, S. Tumapa, M. Vesaratchavest, S. Whitehead, C. Yeats, B. G. Barrell, P. C. Oyston, and J. Parkhill. 2004. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc. Natl. Acad. Sci. USA 101:14240-14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inglis, T. J., L. O'Reilly, N. Foster, A. Clair, and J. Sampson. 2002. Comparison of rapid, automated ribotyping and DNA macrorestriction analysis of Burkholderia pseudomallei. J. Clin. Microbiol. 40:3198-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koh, T. H., L. S. Yong Ng, J. L. Foon Ho, L. H. Sng, G. C. Wang, and R. V. Tzer Pin Lin. 2003. Automated identification systems and Burkholderia pseudomallei. J. Clin. Microbiol. 41:1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koonpaew, S., M. N. Ubol, S. Sirisinha, N. J. White, and S. C. Chaiyaroj. 2000. Genome fingerprinting by pulsed-field gel electrophoresis of isolates of Burkholderia pseudomallei from patients with melioidosis in Thailand. Acta Trop. 74:187-191. [DOI] [PubMed] [Google Scholar]
- 16.Leelarasamee, A. 2004. Recent development in melioidosis. Curr. Opin. Infect. Dis. 17:131-136. [DOI] [PubMed] [Google Scholar]
- 17.Leelayuwat, C., A. Romphruk, A. Lulitanond, S. Trakulsomboon, and V. Thamlikitkul. 2000. Genotype analysis of Burkholderia pseudomallei using randomly amplified polymorphic DNA (RAPD): indicative of genetic differences amongst environmental and clinical isolates. Acta Trop. 77:229-237. [DOI] [PubMed] [Google Scholar]
- 18.Lowe, P., C. Engler, and R. Norton. 2002. Comparison of automated and nonautomated systems for identification of Burkholderia pseudomallei. J. Clin. Microbiol. 40:4625-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reference deleted.
- 20.Monastyrskaya, G., A. Fushan, I. Abaev, O. Filyukova, M. Kostina, E. Pecherskih, and E. Sverdlov. 2004. Genome-wide comparison reveals great inter- and intraspecies variability in B. pseudomallei and B. mallei pathogens. Res. Microbiol. 155:781-793. [DOI] [PubMed] [Google Scholar]
- 21.Ong, C., C. H. Ooi, D. Wang, H. Chong, K. C. Ng, F. Rodrigues, M. A. Lee, and P. Tan. 2004. Patterns of large-scale genomic variation in virulent and avirulent Burkholderia species. Genome Res. 14:2295-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ou, K., C. Ong, S. Y. Koh, F. Rodrigues, S. H. Sim, D. Wong, C. H. Ooi, K. C. Ng, H. Jikuya, C. C. Yau, S. Y. Soon, D. Kesuma, M. A. Lee, and P. Tan. 2005. Integrative genomic, transcriptional, and proteomic diversity in natural isolates of the human pathogen Burkholderia pseudomallei. J. Bacteriol. 187:4276-4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rainbow, L., C. A. Hart, and C. Winstanley. 2002. Distribution of type III secretion gene clusters in Burkholderia pseudomallei, B. thailandensis and B. mallei. J. Med. Microbiol. 51:374-384. [DOI] [PubMed] [Google Scholar]
- 24.Smith, M. D., B. J. Angus, V. Wuthiekanun, and N. J. White. 1997. Arabinose assimilation defines a nonvirulent biotype of Burkholderia pseudomallei. Infect. Immun. 65:4319-4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith-Vaughan, H. C., D. Gal, P. M. Lawrie, C. Winstanley, K. S. Sriprakash, and B. J. Currie. 2003. Ubiquity of putative type III secretion genes among clinical and environmental Burkholderia pseudomallei isolates in northern Australia. J. Clin. Microbiol. 41:883-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ulett, G. C., B. J. Currie, T. W. Clair, M. Mayo, N. Ketheesan, J. Labrooy, D. Gal, R. Norton, C. A. Smith, J. Barnes, J. Warner, and R. G. Hirst. 2001. Burkholderia pseudomallei virulence: definition, stability and association with clonality. Microbes Infect. 3:621-631. [DOI] [PubMed] [Google Scholar]
- 27.Ulett, G. C., J. T. Labrooy, B. J. Currie, J. L. Barnes, and N. Ketheesan. 2005. A model of immunity to Burkholderia pseudomallei: unique responses following immunization and acute lethal infection. Microbes Infect. 7:1263-1275. [DOI] [PubMed] [Google Scholar]
- 28.White, N. J. 2003. Melioidosis. Lancet 361:1715-1722. [DOI] [PubMed] [Google Scholar]
- 29.Winstanley, C. 2002. Spot the difference: applications of subtractive hybridisation to the study of bacterial pathogens. J. Med. Microbiol. 51:459-467. [DOI] [PubMed] [Google Scholar]
- 30.Winstanley, C., and C. A. Hart. 2000. Presence of type III secretion genes in Burkholderia pseudomallei correlates with Ara− phenotypes. J. Clin. Microbiol. 38:883-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woods, D. E., J. A. Jeddeloh, D. L. Fritz, and D. Deshazer. 2002. Burkholderia thailandensis E125 harbors a temperate bacteriophage specific for Burkholderia mallei. J. Bacteriol. 184:4003-4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.