Abstract
The aim of this study is to determine oncogenic human papillomavirus (HPV) types and HPV type 16 (HPV16) variant distribution in two Spanish population groups, commercial sex workers and imprisoned women (CSW/IPW) and the general population. A multicenter cross-sectional study of 1,889 women from five clinical settings in two Spanish cities was conducted from May to November 2004. Oncogenic HPV infection was tested by an Hybrid Capture II (HC2) test, and positive samples were genotyped by direct sequencing using three different primer sets in L1 (MY09/11 and GP5+/GP6+) and E6/E7. HPV16 variants were identified by sequencing the E6, E2, and L1 regions. Four hundred twenty-five samples were positive for the HC2 test, 31.5% from CSW/IPW and 10.7% from the general population. HPV16 was the most frequent type. Distinct profiles of oncogenic HPV type prevalence were observed across the two populations. In order of decreasing frequency, HPV types 16, 31, 58, 66, 56, and 18 were most frequent in CSW/IPW women, and types 16, 31, 52, 68, 51, and 53 were most frequent in the general population. We analyzed HPV16 intratype variants, and a large majority (78.7%) belonged to the European lineage. AA variants were detected in 16.0% of cases. African variants belonging to classes Af1 (4.0%) and Af2 (1.3%) were detected. Different HPV types and HPV16 intratype variants are involved in oncogenic HPV infections in our population. These results suggest that HPV type distribution differs in CSW/IPW women and in the general population, although further analysis is necessary.
Different epidemiological studies reveal that human papillomavirus (HPV) prevalence depends largely on age and sexual practices and that it shows major differences across geographic areas. HPV prevalence has been described to be higher in Latin America and sub-Saharan Africa and lower in Asia and Europe (12, 15, 17, 30, 32, 35, 40, 43). More than 100 types of HPV have been identified to date, of which at least 42 are associated with infections of the genital tract (42). Different genotype prevalence profiles have also been observed in different geographic areas (8). Vaccines directed against HPV type 16 (HPV16) and HPV18, present in approximately 70% of cervical cancers worldwide, are currently under evaluation and have shown impressive reductions of 90% in the incidence of HPV infection in the case of types 16 and 18 (16, 18, 22, 27, 45). The preliminary data suggest that bivalent HPV16 and HPV18 vaccine may also protect against HPV16-related types (31, 33, 35, 52, and 58) and HPV18-related types (39, 45, 59, 68, and 85) (G. Dubin, 22nd International Papillomavirus Conference and Clinical Workshop 2005, Oral Communication F-03, 2005). In order to design and evaluate the health impact of HPV infections and the potential benefits of the implementation of future vaccines, it is necessary to know the distribution of oncogenic HPV types in different geographic regions and in different populations (29).
In Spain, which has one of the lowest rates of cervical cancer worldwide, there are few studies on HPV epidemiology and even fewer on molecular epidemiology. In a population-based study with a 3% age-adjusted HPV prevalence, performed among 973 women from the general population in Barcelona, de Sanjosé et al. established that HPV16 was the most common type (21%), followed by HPV35 and HPV31, present in 13.8% (12). HPV16 and HPV31 were also the most common in 157 imprisoned women in Barcelona (13). HPV intratype variants have been analyzed, and molecular and epidemiological studies suggest that their oncogenic potential is altered (2, 23, 26, 39). HPV16 intratype variants may be divided on the basis of sequence heterogeneity in L1, L2, LCR, E6, and E2 genes into five phylogenetic branches: European (E), Asian (As), Asian-American (AA), African-1 (Af1), and African-2 (Af2). HPV16 variants also show different geographical distributions (38, 47).
Few data are available in Spain on the distribution of oncogenic HPV types, and none are available on HPV16 intratypic variant distribution. The aim of this study is to describe the distribution of oncogenic HPV types and HPV16 variants in a multicenter cross-sectional study of two Spanish populations, groups with different practices and levels of risk for HPV infection: the general population and imprisoned women (IPW) and commercial sex workers (CSW).
MATERIALS AND METHODS
Patients and samples.
A multicenter cross-sectional study of 1,889 women from five clinical settings in two Spanish cities was conducted from May to November 2004. The study population consisted of women from the general population, recruited at two family planning centers (Centro Municipal de Salud Ciudad Lineal, Madrid, and Centro de Planificación Familiar No II, Alicante), commercial sex workers (CSW) from two sexually transmitted diseases clinics (Centro Sanitario Sandoval, Madrid, and Centro de Información y Prevención del SIDA, Alicante), and imprisoned women (IPW) enrolled within the framework of a gynecological program carried out in a prison. For these analyses, CSW and IPW will be analyzed as one group (CSW/IPW) and the women from the general population will be analyzed in a different group. Sexually transmitted disease and family planning clinics are free of charge to all women, irrespective of their nationality or legal status. Women attending these centers were informed and invited to participate in the study, and none refused.
Among the 1,889 women studied, 1,071 (56.7%) were CSW/IPW (21% of whom were born in Spain) and 818 (43.3%) were women from the general population (83% of whom were born in Spain). The CSW/IPW group was made up of considerably younger women (median age, 28; range, 16 to 63) than those from the general population (median age, 34; range, 14 to 67; P < 0.000). Latin American women represented 61% of the CSW/IPW population, the most common places of birth being Colombia (25%), Ecuador (19%), and the Dominican Republic (7%).
Cervical samples were collected by trained clinicians with a cervical brush (Cervical Sampler; Digene Corporation Gaithersburg, Md.), placed in 1 ml of Specimen Transport Medium (Digene Corporation), sent to the Retroviruses and Papillomavirus Laboratory of the National Centre for Microbiology, and stored at 4°C until required for testing.
HPV DNA detection.
Oncogenic HPV infection was determined through the HPV DNA test Hybrid Capture II (HC2; Digene Corporation) using the oncogenic or probe B cocktail (HPV types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68), in accordance with the manufacturer's instructions. The samples with an relative light unit/cutoff ratio greater than 1.0 were considered positive for one or more HPV genotypes included in the probe B cocktail. The specimens with an relative light unit/cutoff ratio of between 1.0 and 2.5 were retested to confirm positive results. For positive calibrator b (a high-risk cocktail), 1 pg/ml of HPV16 DNA was used.
HPV typing.
All HC2-positive samples were typed by PCR and direct sequencing. DNA was extracted from cells recovered from a 500-μl aliquot of the original samples taken before an alkali-denatured solution of HC2 was added. Briefly, the samples were washed with phosphate-buffered saline twice by centrifugation at maximum speed for 2 min. After carefully removing the supernatant, the samples were incubated for 1 h at 58°C in 100 μl of digestion buffer containing 0.05 M KCl, 0.01 M Tris, pH 8.3, 2.5 mM MgCl2, 0.1 mg/ml gelatin, 0.45% NP-40, 0.45% Tween 20, and 200 μg of proteinase K per ml and were then heated to 94°C for 10 min to inactivate the residual protease. The samples were centrifuged briefly, and the 5-μl supernatant was use for PCR amplification.
The quality of the DNA sample was tested with the amplification of the β-actin gene using the primer set 5′-TCACCCACACACTGTGCCCATCTACGA-3′ and 5′-CAGCGGAACCGCTCATTGCCAATGG-3′ (41). The samples successfully amplified were considered optimal for HPV typing.
HPV detection was carried out using MY09/MY11 general consensus primers designed to amplify a 450-bp fragment of the HPV L1 gene, as previously described (28). All PCRs were carried out in a final volume of 50 μl. Negative samples were retested in two different PCR systems in order to increase the positive rate of HPV type detection, the first using the general consensus primer GP5+/GP6+ as nested primers (11, 24) and the second using an E6/E7 consensus primer set (36). Positive samples for these PCR systems were directly sequenced. PCR-amplified products were purified using a QIAquick PCR Purification kit (QIAGEN, Hilden, Germany). PCR products were directly sequenced using a Big Dye Terminator Cycle Sequencing kit (PE Applied Biosystems, Foster City, CA) with inner PCR primers and were resolved using an automated ABI PRISM 3700 analyzer (PE Applied Biosystems, Foster City, CA).
HPV type assignation was accomplished by phylogenetic analysis. The nucleotide sequences obtained were aligned with the reference sequences of different HPV types from the Los Alamos sequence and GenBank database using the Clustal X program. The aligned sequences were used for phylogenetic analysis, which was carried out using programs contained in PHYLIP v3.5.
Identification of HPV16 intratypic variants.
Samples typed as HPV16 were subjected to single-round PCR amplification with a specific set of primers designed to amplify the E6 region (nucleotides [nt] 52 to 575) and E2 gene (nt 3355 to 3872) fragment. The specific primer set for E6 was 16ORF5′ (5′-CGAAACCGGTTAGTATAA-3′) and 16ORF3′ (5′-GTATCTCCATGCATGATT-3′); the primer set for the E2 region was A2 (5′-GGATGCAGTATCAAGATTTGT-3′) and B1 (5′-TAGCAGCAACGAAGTATCCTCTCC-3′) (19). HPV16 variants were identified by direct sequencing. Multiple sequence alignments were performed using the MegAlign program of the Lasergene software (DNASTAR, Madison, Wis.). The sequence as published by Seedorf et al. (HPV16R), which belongs to the European lineage, was used as a reference (34).
Statistical analysis.
Demographic differences were compared through chi-squared tests for normally distributed data and the Mann-Whitney test for abnormally distributed data. HPV type distributions were compared using the chi-squared test, and Fisher's test was used when numbers were low for frequencies. Univariate odds ratios and 95% confidence intervals were used to estimate associations. Data analysis and statistics were produced by using Microsoft Excel and STATA 8.0 statistical software (College Station, Tex.).
RESULTS
Prevalence of oncogenic HPV infection.
As is shown in Table 1, among the 1,889 women tested, 425 (337 from the CSW/IPW group and 88 from the general population) were infected by oncogenic HPV types included in the HC2 test. The oncogenic HPV prevalence was significantly higher in CSW/IPW (31.5%) women than among the women from the general population (10.7%) (odds ratio, 3.81; 95% confidence interval, 2.93 to 4.96). HPV prevalence was higher among CSW/IPW women of all geographical origins except in the rest-of-Europe group, although statistically significant differences were detected only for Spaniards (P < 0.000) and (borderline) for Latin Americans (P = 0.085).
TABLE 1.
Prevalence of oncogenic HPV types determined by HC2 tests in CSW, IPW, and general population women living in Spain
| Place of birth | n | CSW/IPW
|
General population
|
P value | ||
|---|---|---|---|---|---|---|
| n | No. oncogenic (%) | n | No. oncogenic (%) | |||
| Spain | 900 | 224 | 67 (29.9) | 676 | 56 (8.3) | 0.000 |
| Africa | 60 | 52 | 12 (23.1) | 8 | 1 (12.5) | 0.829 |
| Rest of Europe | 130 | 119 | 37 (31.1) | 11 | 4 (36.4) | 0.983 |
| Latin America | 761 | 656 | 215 (32.8) | 105 | 25 (23.8) | 0.085 |
| Unknown/other | 38 | 20 | 6 (30.0) | 18 | 2 (11.1) | 0.304 |
| Total | 1,889 | 1,071 | 337 (31.5) | 818 | 88 (10.7) | 0.000 |
HPV type-specific prevalence.
As is shown in Table 2, of the 425 oncogenic HC2 test-positive samples, negative results for constitutive β-actin gene PCR amplification were obtained in seven samples (1.6%), while in 39 samples (9.2%) the nucleotide sequence obtained was unclear because multiple sequence traces were superimposed, probably due to multiple HPV infections. HPV types classified as oncogenic or probably oncogenic, according to the Muñoz et al. epidemiologic classification (31), were detected in 346 (81.4%) samples. In 33 samples (7.8%) we detected HPV types epidemiologically classified as nononcogenic or unclassified. These samples contained HPV types 6, 11, 42, 43, 61, 67, 70, 74, 81, 83, and 85. In these samples, we confirmed the presence of multiple HPV infections (data not shown) with oncogenic HPV types which justify HC2 reactivity.
TABLE 2.
HPV type frequency determined by direct sequencing in oncogenic HC2 test-positive samples
| HPV risk | HPV type | Total (%) no. of women | No. (%) of CSW/IPW women | No. (%) from general population | P value |
|---|---|---|---|---|---|
| Oncogenic | 16 | 78 (18.4) | 63 (18.7) | 15 (17.0) | 0.722 |
| 18 | 22 (5.2) | 18 (5.3) | 4 (4.5) | 0.507 | |
| 26 | 1 (0.2) | 1 (0.3) | 0 (0.0) | ||
| 31 | 31 (7.3) | 24 (7.1) | 7 (8.0) | 0.789 | |
| 33 | 16 (3.8) | 12 (3.6) | 4 (4.5) | 0.430 | |
| 35 | 11 (2.6) | 10 (3.0) | 1 (1.1) | 0.298 | |
| 39 | 12 (2.8) | 9 (2.7) | 3 (3.4) | 0.467 | |
| 45 | 16 (3.8) | 12 (3.6) | 4 (4.5) | 0.430 | |
| 51 | 11 (2.6) | 6 (1.8) | 5 (5.7) | 0.040 | |
| 52 | 23 (5.4) | 16 (4.7) | 7 (8.0) | 0.176 | |
| 53 | 17 (4.0) | 12 (3.6) | 5 (5.7) | 0.263 | |
| 56 | 23 (5.4) | 20 (5.9) | 3 (3.4) | 0.261 | |
| 58 | 28 (6.6) | 24 (7.1) | 4 (4.5) | 0.386 | |
| 59 | 9 (2.1) | 7 (2.1) | 2 (2.3) | 0.587 | |
| 66 | 26 (6.1) | 22 (6.5) | 4 (4.5) | 0.490 | |
| 68 | 15 (3.5) | 9 (2.7) | 6 (6.8) | 0.060 | |
| 73 | 2 (0.5) | 2 (0.6) | 0 (0.0) | 0.628 | |
| 82 | 5 (1.2) | 5 (1.5) | 0 (0.0) | ||
| Total | 346 (81.4) | 272 (80.7) | 74 (84.1) | ||
| Nononcogenic | 33 (7.8) | 30 (8.9) | 3 (3.4) | ||
| Undetermined | 39 (9.2) | 29 (8.6) | 10 (11.4) | ||
| β-Actin negative | 7 (1.6) | 6 (1.8) | 1 (1.1) | ||
| Total | 425 (100.0) | 337 (100.0) | 88 (100.0) |
Eighteen different oncogenic HPV types are involved in HPV infections in our population, the individual frequencies being presented in Table 2. The most frequently found was HPV16, present in 18.4% of all positive samples, followed by HPV31 (7.3%), HPV58 (6.6%), HPV66 (6.1%), HPV52 (5.4%), HPV56 (5.4%), and HPV18 (5.2%). HPV types 33, 45, 68, 39, 51, 35, 59, 82, 73, and 26 follow this group, with frequencies lower than 4%. HPV26 was detected in only one sample. When we analyzed individual frequencies stratified by population groups, no significant differences were observed, except for HPV51, which was found to be more frequent in the general population (P = 0.040).
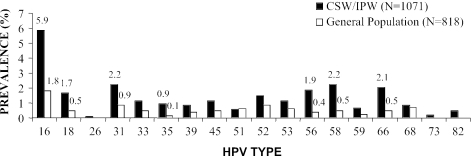
Oncogenic HPV type prevalence among CSW/IPW women and women from the general population is shown in Fig. 1. HPV16 was the most prevalent in both populations (5.9% and 1.8%, respectively). Distinct profiles of oncogenic HPV type prevalence were observed across the two populations. In order of decreasing frequency, HPV types 16, 31, 58, 66, 56, 18, and 52 were most frequent in CSW/IPW women, and types 16, 31, 52, 68, 51, 53, 18, 33, 45, 58, and 66 were most common in women from the general population. HPV26, HPV73, and HPV82 were not detected in women from the general population. Statistically significant differences in HPV type prevalence stratified by population were observed only for HPV types 16, 18, 31, 35, 56, 58, and 66, which were most prevalent in CSW/IPW women (P < 0.05).
FIG. 1.
Prevalence of oncogenic HPV types stratified by population group. Prevalence values are shown only for HPV types with statistically significant prevalence differences in both population groups.
Identification of HPV16 intratypic variants.
Overall, 78 samples were identified as HPV type 16, and 75 were analyzed by direct sequencing of the region coding for the E6 oncoprotein, E2 gene, and MY09/11-amplified highly conserved L1 region to establish variant HPV16 class and subclasses. In the E2 region, no sequence information was obtained in 11 samples. In the remaining three samples, intratypic variant characterization was not possible, since they were only amplified by the GP5+/GP6+ PCR system.
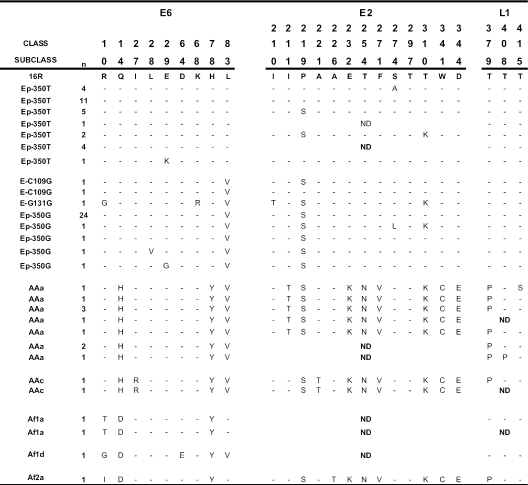
Among the 75 analyzed HPV16 samples, the majority, 59 (78.7%), belonged to European lineage: 28 (37.3%) were lineage Ep-350T, 1 (1.3%) was E-C131G, 2 (2.7%) were E-C109G, and 28 (37.3%) were Ep-350G. AA variants were detected in 12 samples (16.0%), 2 of them being AAc variants (2.7%). African variants were detected in four samples (5.3%), with three belonging to the Af1 class (4.0%) and one to Af2 (1.3%). Asian HPV16 variants (As) were not detected in the present study.
No frame shifts or premature stop codons were observed, and sequence differences between variants are represented in Fig. 2 and 3. Fifteen sequence patterns belong to European classes Ep-350T (Ep-350T and Ep-350T/A188) and Ep-350G (Ep305G, E-C109G, E-G131G/G306, Ep-350G/G185, and Ep-350G/G189); nine sequence patterns belong to AA classes (AAa, AAc, and AAc/C271); three belong to class Af1 (Af1a and Af1d); and one belongs to class Af2. Only 11 HPV16 samples are identical to the HPV16 prototype sequence (Ep-350T) in the three analyzed regions. Seven samples characterized as Ep-350T class present a nucleotide change at nt 3410 (C-T, Pro to Ser at amino acid [aa] 219), and two of them present an additional change at nt 3684 (C-A, Thr to Lys at aa 310) within the E2 region. The majority of the Ep-350G variants (24 out of 31) present the same previously described pattern in the E6, E2, and L1 MY09/11 regions, a nucleotide change (T-G) at nt 350 with replacement of Leu with Val at amino acid position 83 and a single nucleotide change at nt 3684 (C-A, Thr to Lys at aa 310) within the E2 region. Seven of the 10 samples belonging to AAa class present characteristic multiple amino acid changes of this branch in region E6 (Gln to His at aa 14; His to Tyr at aa 78; and Leu to Val at aa 83) and in the E2 region (Ile to Thr at aa 211; Pro to Ser at aa 219; Glu to Lys at aa 232; Thr to Asn at aa 254; Phe to Val at aa 271; Thr to Lys at aa 310; Trp to Cys at aa 341; and Asp to Glu at aa 344). Amino acid changes Ile to Arg at aa 27 in E6 and Ala to Thr at aa 221 in E2 were detected in two samples classified as belonging to the AAc subclass. The Af1 and Af2 HPV16 variants contained multiple nucleotide changes typical of this class in the three analyzed genomic regions. No evidence of genomic recombination was present in any of the analyzed samples.
FIG. 2.
E6, E2, and L1 nucleotide sequence variations among HPV16 clinical samples from women living in Spain. ND, not determined.
FIG. 3.
E6, E2, and L1 amino acid sequence variations among HPV16 clinical samples from women living in Spain. ND, not determined.
The analysis of HPV16 E6 variants by the place of birth of the women is shown in Table 3. In women from Spain, Latin America, and Europe, variants belonging to the European branches are predominant, being present in more than 70%. Frequency differences observed for Ep-350T, Ep-350G, and AA were not statistically significant (P = 0.54, P = 0.29, and P = 0.65, respectively). Af1 variants were detected in women from Spain and Ecuador, and Af2 variants were detected in one woman born in Nigeria.
TABLE 3.
HPV16 variant distribution in women living in Spain, stratified by place of birth
| Variant | No. (%) of tested women from:
|
Total | |||
|---|---|---|---|---|---|
| Spain | Latin America | Rest of Europe | Africa | ||
| Ep-350T | 8 (30.8) | 15 (42.9) | 4 (33.3) | 1 (50.0) | 28 |
| Ep-35OG | 12 (46.1) | 11 (31.4) | 5 (41.7) | 28 | |
| E-G131G | 1 (3.8) | 1 | |||
| E-C109G | 2 (7.7) | 2 | |||
| Af2 | 1 (50.0) | 1 | |||
| Af1 | 1 (3.8) | 2 (5.7) | 3 | ||
| AA | 2 (7.7) | 7 (20.0) | 3 (25.0) | 12 | |
| Total | 26 (100) | 35 (100) | 12 (100) | 2 (100) | 75 |
DISCUSSION
As expected, the prevalence of oncogenic HPV infection was higher in CSW/IPW women than in women from the general population. This was very marked for Spanish women (29.9% versus 8.3%) and African women (23.1% versus 12.5%) but was not that noticeable for women from other European countries (31.1% versus 36.4%) and for Latin Americans (32.8% versus 23.8%). High prevalence in CSW in Spain had been previously described, both in Spanish (44) and in migrant CSW individuals (9). The rate of oncogenic HPV prevalence in the general population for Spanish women agrees with data previously published in Spain (12, 20) and are similar to those of others conducted in European countries, such as Italy, with rates of prevalence of between 6.6 and 7.1% (10, 33).
HPV16 is the most prevalent oncogenic HPV genotype, and its prevalence is similar in different areas. However, the prevalence of HPV16 relative to other HPV types varies by region, being highest in Europe and lowest in Africa (8). The prevalence of other HPV types varies across geographical locations: HPV35, HPV45, HPV52, HPV56, and HPV58 are more common in HPV-positive women in sub-Saharan Africa than in Europe, HPV52 and HPV58 are more prevalent in Central Asia, and HPV39 and HPV59 were almost entirely confined to central and South America (3, 4, 7, 14, 21, 46).
Several molecular methods based on PCR amplification have been used to determine specific HPV types. Analysis of PCR products was carried out mainly by type-specific probe hybridizations and direct sequencing. In order to compare type-specific HPV prevalence in different geographical areas, it is necessary to bear in mind the molecular methods used, because some HPV genotypes are not successfully amplified with some primer sets. Thus, the PGMY09/11 system is more sensitive than MY09/11 for the detection of HPV types 26, 35, 42, 45, 52, 54, 55, 59, and 83 (4), and primer set GP5+/GP6+ is able to detect HPV types 30, 42, 43, 51, 59, 67, 74, 90, and 91, which are not successfully amplified by MY09/11, but it is less sensitive for some HPV types, such as HPV53 and HPV61 (6). For this reason, in the present study we used three different primer sets in order to improve the sensitivity of PCR detection. Although the type-specific HPV test based on PCR and direct sequencing has the disadvantage of not being able to detect multiple infections, it is a helpful and reliable tool in HPV screening and allows for the estimation of the predominant circulating HPV types in clinical samples.
This is the largest study conducted in Spain on the distribution of HPV types. HPV16 was the most common type found in our study, present in 18.4% of positive HC2 samples, followed by HPV types 31, 58, 66, 52, 56, and 18.
Several studies conducted in European countries, such as Germany, Denmark, and Belgium, on the distribution of HPV types describe type-specific frequencies that agree with our results, with HPV31 being the second most common type, with frequencies of 10.1%, 8.0%, and 7.3%, respectively (1, 25, 37). The next most common type in our population is HPV58, with a frequency of 6.6%, probably due to the high proportion of women from South America, in which this type is the most frequent after HPV16 (8). HPV66, which is not included in the oncogenic HC2 cocktail probe, was detected in 6.1% of cases. This is remarkable, since this type has been classified as probably high risk. HPV52 is detected in 5.4% and is the fifth most frequently detected HPV type. In other studies, the prevalence of this genotype might be underestimated because the GP5+/GP6+ primer set, widely used for HPV typing, amplifies HPV52 inefficiently (5).
HPV16 and HPV31 were the most common types, both in CSW/IPW women and in women from the general population, although the relative distribution of the rest of the HPV types varied among these populations. As expected, HPV type-specific prevalences were higher in CSW/IPW women for most HPV types, except for HPV51. In this group of women, types 58, 66, 56, 18, and 52 followed 16 and 31 in frequency, and 52, 68, 51, 53, 18, 33, 45, 58, and 66 accounted for the most common types after HPV16 and HPV31 in women from the general population.
The different profiles of oncogenic HPV type prevalence observed across CSW/IPW women and women in the general population need to be further validated in larger studies with prospective follow-up, and it is necessary to adjust the type-specific prevalence by age and country of birth of the women because of the high proportion of migrant women in the CSW/IPW population and of Spaniards in the general population.
Taking into account that HPV genotyping was performed by direct sequencing, the observed HPV distribution reflects the majority of HPV types present in the studied population, and coinfecting HPV genotypes with a low number of copies would not be detected.
Detection of HPV16 natural variants by PCR and direct sequencing analysis allows for the identification of subclasses and their geographical distribution pattern. Classically, HPV16 variants were characterized in samples with cervical cancer in order to establish the relationship between these variants and their potential oncogenicity. In the present study, we characterized HPV16 variants in cervical samples from routine screening in order to evaluate the intratype variants circulating in our populations. All HPV16 variants detected have been previously described. The large majority of HPV16 variants circulating in our population belong to European branches (78.8%) that are found in all geographical regions except for Africa. In our study, we detected one prototype variant in a woman born in Morocco. AA variants were found to the same extent in women born in Spain as in women from other geographical areas. There are no significant differences in HPV16 intratype variant distributions by the place of birth of the women, probably because of the limited number of samples analyzed.
The study of HPV type distribution and intratypic variants is important in order to establish whether the increase of women in our population from countries with high prevalence of HPV infection might modify this pattern.
Acknowledgments
This work was supported by a grant from Proyecto Intramural ISCIII MPY-1117/03 and from funds provided by Generalitat Valenciana, Conselleria de Sanidad, and RED Centros de Estudio de Salud Pública RCESP C03/09.
REFERENCES
- 1.Beerens, E., L. Van Renterghem, M. Praet, Y. Sturtewagen, S. Weyers, M. Temmerman, H. Depypere, P. Claeys, and C. A. Cuvelier. 2005. Human papillomavirus DNA detection in women with primary abnormal cytology of the cervix: prevalence and distribution of HPV genotypes. Cytopathology 16:199-205. [DOI] [PubMed] [Google Scholar]
- 2.Berumén, J., R. M. Ordoñez, E. Lazcano, J. Salmerón, S. C. Galván, R. A. Estrada, E. Yunes, A. García-Carranca, G. Gonzalez-Lira, and A. Madrigal-de la Campa. 2001. Asian-American variants of human papillomavirus 16 and risk for cervical cancer: a case-control study. J. Natl. Cancer Inst. 93:1325-1330. [DOI] [PubMed] [Google Scholar]
- 3.Bosch, F. X., M. M. Manos, N. Muñoz, M. Sherman, A. M. Jansen, J. Peto, M. H. Schiffman, V. Moreno, R. Kurman, K. V. Shah, and International Biological Study on Cervical Cancer Study Group. 1995. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. J. Natl. Cancer Inst. 87:796-802. [DOI] [PubMed] [Google Scholar]
- 4.Castellsagué, X., C. Menéndez, M. P. Loscertales, J. R. Kornegay, F. dos Santos, F. X. Gómez-Olive, B. Lloveras, N. Abarca, N. Vaz, A. Barreto, F. X. Bosch, and P. Alonso. 2001. Human papillomavirus genotypes in rural Mozambique. Lancet 358:1429-1430. [DOI] [PubMed] [Google Scholar]
- 5.Chan, P. K., T. H. Cheung, A. O. Tam, K. W. Lo, S. F. Yim, M. M. Yu, K. F. To, Y. F. Wong, J. L. Cheung, D. P. Chan, M. Hui, and M. Ip. 2006. Biases in human papillomavirus genotype prevalence assessment associated with commonly used consensus primers. Int. J. Cancer 118:243-245. [DOI] [PubMed] [Google Scholar]
- 6.Chaturvedi, A. K., J. Dumestre, A. M. Gaffga, K. M. Mire, R. A. Clark, P. S. Braly, K. Dunlap, T. E. Beckel, A. F. Hammons, P. J. Kissinger, and M. E. Hagensee. 2005. Prevalence of human papillomavirus genotypes in women from three clinical settings. J. Med. Virol. 75:105-113. [DOI] [PubMed] [Google Scholar]
- 7.Clifford, G. M., J. S. Smith, T. Aguado, and S. Franceschi. 2003. Comparison of HPV type distribution in high grade cervical lesions and cervical cancer: a meta-analysis. Br. J. Cancer 89:101-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clifford, G. M., S. Gallus, R. Herrero, N. Muñoz, P. J. Snijders, S. Vaccarella, P. T. Anh, C. Ferreccio, N. T. Hieu, E. Matos, M. Molano, R. Rajkumar, G. Ronco, S. de Sanjosé, H. R. Shin, S. Sukvirach, J. O. Thomas, S. Tunsakul, C. J. Meijer, S. Franceschi, and IARC HPV Prevalence Surveys Study Group. 2005. Worldwide distribution of human papillomavirus types in cytologically normal women in the International Agency for Research on Cancer HPV prevalence surveys: a pooled analysis. Lancet 366:991-998. [DOI] [PubMed] [Google Scholar]
- 9.del Amo, J., C. González, J. Losana, P. Clavo, L. Muñoz, J. Ballesteros, A. García-Sáiz, M. J. Belza, M. Ortiz, B. Menéndez, J. del Romero, and F. Bolumar. 2005. Influence of age and geographical origin in the prevalence of high risk human papillomavirus in migrant female sex workers in Spain. Sex Transm. Infect. 81:79-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Francesco, M. A., F. Gargiulo, C. Schreiber, G. Ciravolo, F. Salinaro, and N. Manca. 2005. Detection and genotyping of human papillomavirus in cervical samples from Italian patients. J. Med. Virol. 75:588-592. [DOI] [PubMed] [Google Scholar]
- 11.De Roda Husman, A. M., J. M. Walboomers, E. Hopman, O. P. Bleker, T. M. Helmerhost, L. Rozendal, F. J. Voorhost, and C. J. Meijer. 1995. HPV prevalence in cytomorphologically normal cervical scrapes of pregnant women as determined by PCR: the age-related pattern. J. Med. Virol. 46:97-102. [DOI] [PubMed] [Google Scholar]
- 12.de Sanjosé, S., R. Almirall, B. Lloveras, R. Font, M. Díaz, N. Muñoz, I. Catalá, C. J. Meijer, P. J. Snijders, R. Herrero, and F. X. Bosch. 2003. Cervical human papillomavirus infection in the female population in Barcelona, Spain. Sex Transm. Dis. 30:788-793. [DOI] [PubMed] [Google Scholar]
- 13.de Sanjosé, S., I. Valls, P. Canadinas, B. Lloveras, M. J. Quintana, K. V. Shah, and F. X. Bosch. 2000. Human papillomavirus and human immunodeficiency virus infections as risk factors for cervix cancer in women prisoners. Med. Clin. (Barcelona) 115:81-84. [DOI] [PubMed] [Google Scholar]
- 14.de Villiers, E. M. 1994. Human pathogenic papillomavirus types: an update. Curr. Top. Microbiol. Immunol. 186:1-12. [DOI] [PubMed] [Google Scholar]
- 15.Ferreccio, C., R. B. Prado, A. V. Luzoro, S. L. Ampuero, P. J. Snijders, C. J. Meijer, S. V. Vaccarella, A. T. Jara, K. I. Puschel, S. C. Robles, R. Herrero, S. F. Franceschi, and J. M. Ojeda. 2004. Population-based prevalence and age distribution of human papillomavirus among women in Santiago, Chile. Cancer Epidemiol. Biomark. Prev. 13:2271-2276. [PubMed] [Google Scholar]
- 16.Franceschi, S. 2002. Human papillomavirus: a vaccine against cervical carcinoma uterine. Epidemiol. Prev. 26:140-144. [PubMed] [Google Scholar]
- 17.Franco, E. L., L. L. Villa, J. P. Sobrinho, J. M. Prado, M. C. Rousseau, M. Desy, and T. E. Rohan. 1999. Epidemiology of acquisition and clearance of cervical human papillomavirus infection in women from a high-risk area for cervical cancer. J. Infect. Dis. 180:1415-1423. [DOI] [PubMed] [Google Scholar]
- 18.Frazer, I. 2002. Vaccines for papillomavirus infection. Virus Res. 89:271-274. [DOI] [PubMed] [Google Scholar]
- 19.Giannoudis, A., M. Duin, P. J. Snijders, and C. S. Herrington. 2001. Variation in the E2-binding domain of HPV 16 is associated with high-grade squamous intraepithelial lesions of the cervix. Br. J. Cancer 84:1058-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonzalez, C., M. Ortiz, J. Canals, L. Muñoz, I. Jarrin, M. Garcia de la Hera, A. García-Sáiz, and J. del Amo. Higher prevalence of human papillomavirus infection in migrant women from Latin America in Spain. Sex Transm. Infect., in press. [DOI] [PMC free article] [PubMed]
- 21.Gravitt, P. E., A. M. Kamath, L. Gaffikin, Z. M. Chirenje, S. Womack, and K. V. Shah. 2002. Human papillomavirus genotype prevalence in high-grade squamous intraepithelial lesions and colposcopically normal women from Zimbabwe. Int. J. Cancer 100:729-732. [DOI] [PubMed] [Google Scholar]
- 22.Harper, D. M., E. L. Franco, C. Wheeler, D. G. Ferris, D. Jenkins, A. Schuind, T. Zahaf, B. Innis, P. Naud, N. S. De Carvalho, C. M. Roteli-Martins, J. Teixeira, M. M. Blatter, A. P. Korn, W. Quint, G. Dubin, and GlaxoSmithKline HPV Vaccine Study Group. 2004. Efficacy of a bivalent L1 virus-like particle vaccine in prevention of infection with human papillomavirus types 16 and 18 in young women: a randomised controlled trial. Lancet 364:1731-1732. [DOI] [PubMed] [Google Scholar]
- 23.Hildesheim, A., M. Schiffman, C. Bromley, S. Wacholder, R. Herrero, A. Rodriguez, M. C. Bratti, M. E. Sherman, U. Scarpidis, Q. Q. Lin, M. Terai, R. L. Bromley, K. Buetow, R. J. Apple, and R. D. Burk. 2001. Human papillomavirus type 16 variants and risk of cervical cancer. J. Natl. Cancer Inst. 93:315-318. [DOI] [PubMed] [Google Scholar]
- 24.Jacobs, M. V., P. J. Snijders, A. J van den Brule, T. J. Helmerhorst, C. J. Meijer, and J. M. Walboomers. 1997. A general primer GP5+/GP6+-mediated PCR-enzyme immunoassay method for rapid detection of 14 high-risk and 6 low-risk human papillomavirus genotypes in cervical scrapings. J. Clin. Microbiol. 35:791-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson, T., K. Bryder, S. Corbet, and A. Fomsgaard. 2003. Routine genotyping of human papillomavirus samples in Denmark. APMIS 111:398-404. [DOI] [PubMed] [Google Scholar]
- 26.Kammer, C., M. Tommasino, S. Syrjanen, H. Delius, U. Hebling, U. Warthorst, H. Pfister, and I. Zehbe. 2002. Variants on long control region and E6 oncogene in European human papillomavirus type 16 isolates: implications on cervical disease. Br. J. Cancer 86:269-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koutsky, L. A., K. A. Ault, C. M. Wheeler, D. R. Brown, E. Barr, F. B. Alvarez, L. M. Chiacchierini, and K. U. Jansen. 2002. A controlled trial of a human papillomavirus type 16 vaccine. N. Engl. J. Med. 347:1645-1651. [DOI] [PubMed] [Google Scholar]
- 28.Manos, M. M., Y. Ting, D. K. Wright, A. J. Lewis, T. R. Broker, and S. M. Wolinsky. 1989. Use of polymerase chain reaction amplification for the detection of genital human papillomavirus. Cancer Cell 7:209-214. [Google Scholar]
- 29.Manos, M. M., W. K. Kinney, L. B. Hurley, M. E. Sherman, J. Shieh-Ngai, R. J. Kurman, J. E. Ransley, J. Fetterman, J. S. Hartinger, K. M. Mclntosh, G. F. Pawlick, and R. A. Hiatt. 1999. Identifying women with cervical neoplasia: using human papillomavirus DNA testing for equivocal Papanicolau results. JAMA 281:1605-1610. [DOI] [PubMed] [Google Scholar]
- 30.Molano, M., H. Posso, E. Weiderpass, A. J. van den Brule, M. Ronderos, S. Franceschi, C. J. Meijer, A. Arslan, N. Muñoz, and H. P. V. Study Group. 2002. Prevalence and determinants of HPV infection among Colombian women with normal cytology. Br. J. Cancer 87:324-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muñoz, N., F. X. Bosch, S. de Sanjosé, R. Herrero, X. Castellsagué, K. V. Shah, P. J. Snijders, C. J. Meijer, and International Agency for Research on Cancer Multicenter Cervical Cancer Study Group. 2003. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 348:518-527. [DOI] [PubMed] [Google Scholar]
- 32.Pham, T. H., T. H. Nguyen, R. Herrero, S. Vaccarella, J. S. Smith, T. T. Nguyen Thuy, H. N. Nguyen, B. D. Nguyen, R. Ashley, P. J. Snijders, C. J. Meijer, N. Muñoz, D. M. Parkin, and S. Franceschi. 2003. Human papillomavirus infection among women in South and North Vietnam. Int. J. Cancer 104:213-220. [DOI] [PubMed] [Google Scholar]
- 33.Ronco, G., V. Ghisetti, N. Segnan, P. J. Snijders, A. Gillio-Tos, C. J. Meijer, F. Merletti, and S. Franceschi. 2005. Prevalence of human papillomavirus infection in women in Turin, Italy. Eur. J. Cancer 41:297-305. [DOI] [PubMed] [Google Scholar]
- 34.Seedorf, K., G. Krammer, M. Durst, S. Suhai, and W. G. Rowekamp. 1985. Human papillomavirus type 16 DNA sequence 1. Virology 145:181-185. [DOI] [PubMed] [Google Scholar]
- 35.Shin, H. R., S. Franceschi, S. Vaccarella, J. W. Roh, Y. H. Ju, J. K. Oh, H. J. Kong, S. H. Rha, S. I. Jung, J. L. Kim, K. Y. Jung, L. J. van Doorn, and W. Quint. 2004. Prevalence and determinants of genital infection with papillomavirus, in female and male university students in Busan, South Korea. J. Infect. Dis. 190:468-476. [DOI] [PubMed] [Google Scholar]
- 36.Sotlar, K., D. Diemer, A. Dethleffs, Y. Hack, A. Stubner, N. Vollmer, S. Menton, M. Menton, K. Dietz, D. Wallwiener, R. Kandolf, and B. Bultmann. 2004. Detection and typing of human papillomavirus by E6 nested multiplex PCR. J. Clin. Microbiol. 42:3176-3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Speich, N., C. Schmitt, R. Bollmann, and M. Bollmann. 2004. Human papillomavirus (HPV) study of 2916 cytological samples by PCR and DNA sequencing: genotype spectrum of patients from the West German area. J. Med. Microbiol. 53:125-128. [DOI] [PubMed] [Google Scholar]
- 38.Stewart, A. C., A. M. Eriksson, M. M. Manos, N. Muñoz, F. X. Bosch, J. Peto, and C. M. Wheeler. 1996. Intratype variation in 12 human papillomavirus types: a worldwide perspective. J. Virol. 70:3127-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stoppler, M. C., K. Ching, H. Stoppler, K. Clancy, R. Schlegel, and J. Icenogle. 1996. Natural variants of the human papillomavirus type 16 E6 protein differ in their abilities to alter keratinocyte differentiation and to induce p53 degradation. J. Virol. 70:6987-6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sukvirach, S., J. S. Smith, S. Tunsakul, N. Muñoz, V. Kesararat, O. Opasatian, S. Chichareon, V. Kaenploy, R. Ashley, C. J. Meijer, P. J. Snijders, P. Coursaget, S. Franceschi, and R. Herrero. 2003. Population-based human papillomavirus prevalence in Lampang and Songkla, Thailand. J. Infect. Dis. 187:1246-1256. [DOI] [PubMed] [Google Scholar]
- 41.Takenouchi, N., Y. Yamano, K. Usuku, M. Osame, and S. Izumu. 2003. Usefulness of proviral load measurement for monitoring of disease activity in individual patients with human T-lymphotropic virus type 1-associated myelopathy/tropical spastic paraparesis. J. Neurol. Virol. 9:29-35. [DOI] [PubMed] [Google Scholar]
- 42.Terai, M., and R. D. Burk. 2001. Characterization of a novel genital human papillomavirus by overlapping PCR: candHPV86 identified in cervicovaginal cells of a woman with cervical neoplasia. J. Gen. Virol. 82:2035-2040. [DOI] [PubMed] [Google Scholar]
- 43.Thomas, J. O., R. Herrero, A. A. Omigbodun, K. Ojemakinde, I. O. Ajayi, A. Fawole, O. Oladepo, J. S. Smith, A. Arslan, N. Muñoz, P. J. Snijders, C. J. Meijer, and S. Franceschi. 2004. Prevalence of papillomavirus infection in women in Ibadan, Nigeria: a population-based study. Br. J. Cancer 90:638-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Touze, A., S. de Sanjosé, P. Coursaget, M. R. Almirall, V. Palacio, C. J. Meijer, J. Kornegay, and F. X. Bosch. 2001. Prevalence of anti-human papillomavirus type 16, 18, 31, and 58 virus-like particles in women in general population and in prostitutes. J. Clin. Microbiol. 39:4344-4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Villa, L. L., R. L. Costa, C. A. Petta, R. P. Andrade, K. A. Ault, A. R. Giuliano, C. M. Wheeler, L. A. Koutsky, C. Malm, M. Lehtinen, F. E. Skjeldestad, S. E. Olsson, M. Steinwall, D. R. Brown, R. J. Kurman, B. M. Ronnett, M. H. Stoler, A. Ferenczy, D. M. Harper, G. M. Tamms, J. Yu, L. Lupinacci, R. Railkar, F. J. Taddeo, K. U. Jansen, M. T. Esser, H. L. Sings, A. J. Saah, and E. Barr. 2005. Prophylactic quadrivalent human papillomavirus (types 6, 11, 16, and 18) L1 virus-like particle vaccine in young women: a randomised double-blind placebo-controlled multicentre phase II efficacy trial. Lancet Oncol. 6:256-257. [DOI] [PubMed] [Google Scholar]
- 46.Xi, L. F., P. Toure, C. W. Critchlow, S. E. Hawes, B. Dembele, P. S. Sow, and N. B. Kiviat. 2004. Prevalence of specific types of human papillomavirus and cervical squamous intraepithelial lesions in consecutive, previously unscreened, West African women over 35 years of age. Int. J. Cancer 103:803-809. [DOI] [PubMed] [Google Scholar]
- 47.Yamada, T., M. M. Manos, J. Peto, C. E. Greer, N. Muñoz, F. X. Bosch, and C. M. Wheeler. 1997. Human papillomavirus type 16 sequence variation in cervical cancer: a worldwide perspective. J. Virol. 71:2463-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]